Abstract
Head and neck squamous cell carcinoma (HNSCC) is one prevalent human cancer worldwide. No molecular markers are presently used for predicting prognosis in HNSCC. Krüppel‐like factor 4 (KLF4) is a transcription factor with diverse physiological functions, and possesses opposing roles in different human cancers. The expression and roles of KLF4 in HNSCC remain to be elucidated. In this study, immunohistochemical (IHC) analysis of KLF4 in 62 HNSCC was firstly performed. IHC results demonstrated that 42 (67.7%) had decreased KLF4 expression compared with surrounding normal epithelium, while persistent KLF4 expression was demonstrated in 20 (32.3%). The IHC results were further verified by Western blot and real‐time PCR analyses to confirm the robustness of staining and interpretation. Interestingly, persistent KLF4 expression independently correlated with a worse disease‐specific survival (P = 0.005), especially in patients with advanced disease. In consistent with clinical observation, all five HNSCC cell lines tested revealed a low level of baseline KLF4 expression. Moreover, enforced KLF4 expression in cell line SAS significantly increased in vitro migration/invasion abilities, multi‐drug resistance, and in vivo tumorigenicity. These results clearly illustrate that persistent KLF4 expression predicts poor prognosis and confers aggressiveness in HNSCC. Our data therefore provides valuable information that HNSCC with persistent KLF4 expression might require intensified combination treatment in future practice. (Cancer Sci 2011; 102: 895–902)
Head and neck squamous cell carcinoma (HNSCC) represents the sixth most common malignancy worldwide, and has a profound impact on quality of life.( 1 ) Despite advances in diagnosis and treatment, survival rates of HNSCC have only marginally improved in recent decades.( 2 ) TNM staging system is the current standard for prognostication, which relies heavily on clinical, radiologic and histopathologic parameters. However, substantial heterogeneity is frequently observed in patients of the same risk group. Thus, identifying novel biomarkers will be very useful to improve clinical diagnosis and patient stratification.( 3 )
Krüppel‐like factor 4 (KLF4), also known as gut‐enriched Krüppel‐like factor or epithelial zinc finger, is a zinc finger transcription factor that is physiologically expressed in differentiated epithelium of the gastrointestinal (GI) tract, skin and vascular endothelium.( 4 , 5 ) KLF4 is primarily regarded as a negative regulator of cell growth, with its ability to regulate the expression of a number of genes involved in cell cycle progression.( 6 , 7 ) It also plays crucial roles in other physiologic processes, including development, differentiation and tissue homeostasis.( 8 ) Recently, KLF4 has been recognized as one of the genes that can reprogram somatic cells into inducible pluripotent stem cells.( 9 , 10 , 11 ) Overall, these data indicate that the functions of KLF4 are diverse and cell‐type specific.
Malignant transformation and progression of cancer cells require the accumulation of myriads of dysregulated genetic alterations.( 12 , 13 ) Functions of genes can thus be influenced by complex interactions of altered signaling under specific genetic context.( 14 ) KLF4 is one gene which has opposing roles in different cancers, either as a tumor suppressor gene or as an oncogene.( 15 ) It is implicated as a tumor suppressor gene in GI tract epithelium, given the fact that KLF4 is decreased in human colon and gastric cancers( 16 , 17 , 18 ) and loss of KLF4 is associated with poor survival.( 17 ) A similar tumor suppressor role is also observed in cancers originating from other organs, including the esophagus,( 19 ) lung,( 20 ) bladder cancers,( 21 ) and cancers like medulloblastoma( 22 ) and T‐cell leukemia.( 23 ) Conversely, KLF4 can function as a transforming oncogene. KLF4‐transformed rat kidney epithelial cells demonstrate morphologic transformation and increased tumorigenicity in athymic mice.( 24 ) In human HNSCC and breast cancer, increased KLF4 expression has been reported.( 24 , 25 ) Moreover, KLF4 expression has been demonstrated to be a poor prognostic factor for early breast cancer and skin cancer,( 26 , 27 ) corroborating its oncogenic roles.
KLF4 has been speculated to be an oncogene in HNSCC because of increased KLF4 mRNA expression.( 24 ) However, the status and functional roles of KLF4 protein expression has not been elucidated yet. To address this question, systematic analysis of KLF4 expression in HNSCC tissues and cell lines was performed. Our results demonstrate that KLF4 protein expression is decreased in 42 out of 62 HNSCC, while persistent KLF4 expression is observed in the remainder 20 samples. Low level of KLF4 expression is also demonstrated in all five HNSCC cell lines tested. In addition, persistent KLF4 expression in HNSCC is associated with a worse disease‐specific survival and enforced KLF4 expression induces in vitro and in vivo aggressiveness. This study therefore demonstrates for the first time that the role of KLF4 can be switched from tumor suppressive in normal epithelium into oncogenic in HNSCC.
Materials and Methods
Patients and tissues. Sixty‐two patients who underwent curative surgery for HNSCC at Taipei Veterans General Hospital, Taiwan, from October 2004 to November 2006 were enrolled. Their demographic data were summarized in Table 1. Patients were staged according to the criteria of the American Joint Committee on Cancer.( 28 ) The median follow‐up duration was 24.3 months (range, 2–33 months). Primary tumor and adjacent normal epithelium were collected during surgery and confirmed by pathology. Four precancerous lesions with pathological diagnosis of severe dysplasia were also analyzed. Oropharyngeal epithelium from non‐cancerous patients undergoing tonsillectomy was used as normal control. The hospital’s institutional review board approved the study and all patients provided informed consent.
Table 1.
Patient characteristics and the status of KLF4 expression by IHC
| Variables | n (%) | KLF4 expression (no.%) | P‐values* | |
|---|---|---|---|---|
| KLF4 (−) | KLF4 (+) | |||
| Patient number | 62 (100) | 42 (67.7) | 20 (32.3) | |
| Age | ||||
| <50 | 25 (40.3) | 14 (56.0) | 11 (44.0) | 0.104 |
| ≥50 | 37 (59.7) | 28 (75.7) | 9 (24.3) | |
| Gender | ||||
| Male | 58 (93.5) | 39 (67.2) | 19 (32.8) | 1.000 |
| Female | 4 (6.5) | 3 (75.0) | 1 (25.0) | |
| T classification | ||||
| T1–2 (4, 25) | 29 (46.8) | 19 (65.5) | 10 (34.5) | 0.725 |
| T3–4 (17, 16) | 33 (53.2) | 23 (69.7) | 10 (30.3) | |
| N classification | ||||
| N0–1 (30, 6) | 36 (58.1) | 25 (69.4) | 11 (30.6) | 0.736 |
| N2–3 (26, 0) | 26 (41.9) | 17 (65.4) | 9 (34.6) | |
| TNM stage | ||||
| Stage I–II (3, 14) | 17 (27.4) | 11 (64.7) | 6 (35.3) | 0.753 |
| Stage III–IV (15, 30) | 45 (72.6) | 31 (68.9) | 14 (31.1) | |
| Histologic differentiation | ||||
| WD | 44 (71.0) | 29 (65.9) | 15 (34.1) | 0.629 |
| MD to PD | 18 (29.0) | 13 (72.2) | 5 (27.8) | |
*Estimated by Pearson chi‐squared or Fisher’s exact tests. IHC, immunohistochemistry; KLF4, Krüppel‐like factor 4; KLF4 (−), decreased KLF4 expression; KLF4 (+), persistent KLF4 expression; MD, moderately‐differentiated; PD, poorly‐differentiated; WD, well‐differentiated.
Immunohistochemistry and immunoscoring. Sections of paraffin‐embedded tissue specimens (5‐μm thick) were deparaffinized, rehydrated and microwaved in citrate buffer (pH 6.0). After proper blocking, primary antibodies were applied 4°C overnight, including anti‐KLF4 (H‐180; Santa Cruz, CA, USA), 9 , 17 anti‐Ki‐67 (MIB‐1; DAKO, Glostrup, Denmark), and anti‐cyclin D1 (SP4; Abcam, Cambridge, UK). After reacting with a biotinylated secondary antibody, horseradish peroxidase conjugated streptavidin (Biogenex, San Ramon, CA, USA) was employed and 3,3′‐diaminobenzidine was used as the chromogen. The staining specificity was confirmed by omitting the primary antibody.
Two investigators (W.Y.L. and S.K.T.) independently scored the slides in a blinded manner. KLF4 staining was scored according to the staining percentage and intensity. The percentage scores were defined as 0–4: (0), <10%; (1), 10–25%; (2), 25–50%; (3), 50–75; and (4), >75%. The intensity scores were defined as 0–3: (0), no staining; (1), light brown; (2), brown; and (3), dark brown. Overall score was determined by multiplying the percentage score and intensity score, and was divided into negative (≤3), weak positive (>3 and ≤6) and strong positive (>6), as reported.( 17 , 29 ) Ki‐67 and cyclin D1 staining were scored according to the percentage of positively stained tumor cell nuclei in 10 high‐powered tumor fields. Tumors with high Ki‐67 index were defined as those with ≥20% Ki‐67 positive tumor cells. For cyclin D1, tumors were scores as positive if ≥10% of tumor cells were positively stained.
Protein extraction and Western blot analysis. Tissues were homogenized in 200 μl of Cell Culture Lysis Reagent (Promega, Madison, WI, USA). Cell lysates containing 30 μg of proteins were separated on a 10% sodium dodecyl sulfate‐polyacrylamide gel and transferred to nitrocellulose membrane. The membrane was probed with the same KLF4 or cyclin D1 antibodies. An anti‐GAPDH antibody was used to probe the same membrane as the loading control.
RNA purification and real‐time PCR analysis. Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA), and DNase I‐treated RNA (2 μg) was used for first‐strand cDNA synthesis. Quantitative real‐time PCR was done in an ABI PRISM 7700 sequence detection system with the preset PCR program, using SYBR green master mix (Applied Biosystems, Foster City, CA, USA). The primer sequences used in real‐time PCR were: KLF4, 5′‐ACCCACACAGGTGAGAAACC‐3′ and 5′‐ATGTGTAAGGCGAGGTGGTC‐3′; GAPDH, 5′‐TTGCCCTCAACGACCACTTT‐3′ and 5′‐TGGTGGTCCAGGGGTCTTAC‐3′. The reaction was done in a total volume of 50 μl, with 40 ng of cDNA and 80 μM of each primer in 1× SYBR green mixture. Quantitative data was analyzed by the ratio between the value of KLF4 and GAPDH.
Cell culture, plasmid construction and stable transfection. Human tongue cancer cell line SAS was obtained from the Japanese Collection of Research Bioresources (JCRB, Osaka, Japan), and cultured with DMEM with 10% FBS. Other HNSCC cell lines, including CAL27 and FADU from American Type Culture Collection (Manassas, VA, USA), OECM‐1 and OC3 provided by Dr Chang K.W. (National Yang‐Ming University, Taiwan), were cultured as recommended at 37°C and 5% CO2.
Full‐length human KLF4 coding sequence was amplified by PCR and subcloned into the HindIII and BamHI sites of pcDNA3 vector (Invitrogen). The construct pcDNA3KLF4 was verified by sequencing. To create stable clones, SAS were transfected with pcDNA3KLF4 or pcDNA3 using Lipofectamine 2000 (Invitrogen), and selected in the presence of G418 (400 μg/mL; Sigma, St. Louis, MO, USA). Each stable clone was confirmed for the expression of KLF4 by Western blot analysis.
Cell migration and invasion assays. Eight micrometer pore size 24‐well modified Boyden transwell chamber (Costar, Cambridge, MA, USA) was used for migration and invasion assays. In the upper chamber, 5 × 104 cells suspended in medium containing 0.5% FBS were plated for migration assay. For invasion assay, the upper side of the filter was covered with Matrigel (Collaborative Research Inc., Bedford, MA, USA) (1:3 dilution with routine medium) before plating 5 × 105 cells in the upper chamber. In the lower chamber, medium containing 15% FBS was added as a chemoattractant. After 24 h of migration or 48 h of invasion, cells on the upper side of the filter were removed and cells remained adherent to the underside of membrane were fixed in 4% formaldehyde and stained with DAPI. Ten separate fields under ×40 magnification were counted and averaged. The experiments were performed in triplicate.
In vivo tumorigenesis assay. Subcutaneous injection of 2 × 106 cells into the flanks of 8‐week‐old athymic nude mice was done for testing tumorigenicity. Tumor growth was measured twice weekly after appearance. Tumor volume was calculated using the formula (length × width × height)/2. The mice were sacrificed on day 45 and the tumors were weighed after excision. The hospital’s institutional animal care and usage committee approved the study.
Chemosensitivity assay. Cells were plated in 96‐well plates at a density of 1 × 104 cells per well. After overnight incubation, cells were treated with 0.1 mL serum‐free medium (SFM) containing various drug concentrations. Drug‐containing SFM was maintained for 24 h for cisplatin and docetaxel, and 48 h for 5‐fluorouracil (5‐FU), before measuring cell viability using alamarBlue assay (Serotec, Oxford, UK). Absorbance at 570 and 600 nm was measured 4 h after adding the redox indicator. The IC50 was calculated graphically as the drug concentration causing a 50% decrease in cell viability. All measurements were repeated in triplicate for each stable clone.
Statistical analysis. Dichotomous variables were compared by using Pearson chi‐squared or Fisher’s exact tests, where appropriate. Student’s t‐test was used to compare the continuous variables between groups. The Kaplan–Meier estimate was used for survival analysis, and log‐rank test was used to compare the survival difference. Multivariate Cox’s proportional hazards model was applied to test the independent prognostic factors. All tests were two‐sided and the level of statistical significance was set at 0.05.
Results
KLF4 is decreased in a majority of HNSCC. Previous study showed that KLF4 mRNA is elevated in dysplastic epithelium, and laryngeal and oral SCC.( 24 ) Because mRNA expression level does not always correlate with protein level, we analyzed KLF4 protein expression by immunohistochemistry (IHC). In normal epithelium, KLF4 was expressed in the nuclei with overall score of 6.23 ± 1.45 (mean ± SD), weaker at the proliferative basal layer and strongest at the non‐proliferative, differentiated spinous layer (Fig. 1a). In severe dysplasia, slightly decreased KLF4 expression was observed under similar pattern, with overall score of 5.12 ± 1.03 (Fig. 1b). In HNSCC, 42 (67.7%) showed negative KLF4 staining (overall score 1.33 ± 0.90) and were assigned as decreased KLF4 expression group (KLF4 (−); Fig. 1c). The other 20 (32.3%) showed positive KLF4 staining (overall score 4.65 ± 1.31), including one strong positive and 19 weak positive staining. They were assigned as persistent KLF4 expression group (KLF4 (+); Fig. 1d). To verify the results, Western blot and quantitative real‐time PCR analyses were performed using 24 available fresh frozen tissue pairs from the 62 HNSCC. In accord with IHC, Western blot analysis showed that KLF4 expression was decreased in 17 (70.8%) of the samples tested (Fig. 1e), and consistent expression pattern was shown again by quantitative real‐time PCR analysis (Fig. 1f). All these results indicate that KLF4 expression is decreased in a majority of HNSCC.
Figure 1.
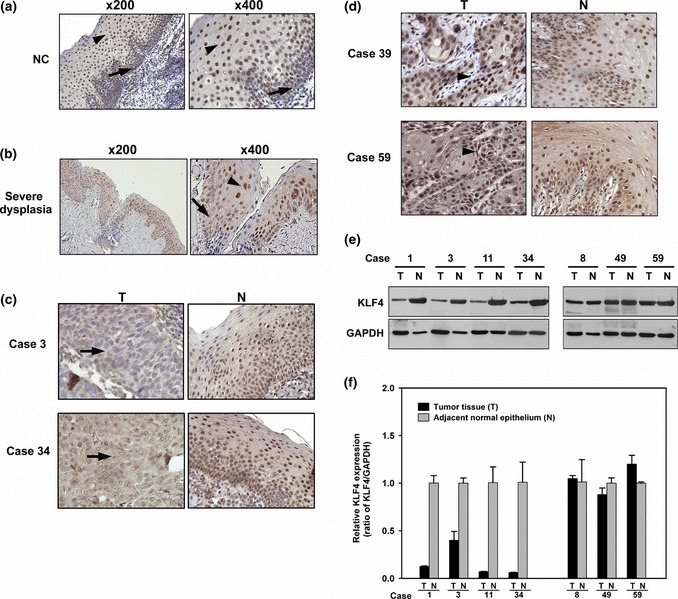
Analysis of Krüppel‐like factor 4 (KLF4) expression by immunohistochemical, Western blot and real‐time PCR. (a) Normal epithelium (NC) with positive KLF4 expression, weaker at basal layer (arrow) and strongest at spinous layer (arrowhead). (b) Severe dysplasia with slightly decreased KLF4 expression under similar pattern. Representative head and neck squamous cell carcinoma (HNSCC) with (c) decreased KLF4 expression (arrow) and (d) persistent KLF4 expression (arrowhead) were shown (magnification, ×400). (e) Western blot analysis and (f) quantitative real‐time PCR analysis in seven representative HNSCC and adjacent normal epithelium. N, adjacent normal epithelium; T, tumor tissue.
Persistent KLF4 expression in HNSCC is associated with poor prognosis. To clarify the impacts of KLF4 expression in HNSCC, clinicopathological features and survival outcomes were analyzed. There was no difference in KLF4 expression when stratified by age, gender, tumor staging and differentiation (Table 1). However, a significantly higher rate of neck recurrence (P = 0.031) and a trend toward higher rate of distant metastasis (P = 0.094) was observed in KLF4 (+) group (Table 2). In survival analysis, KLF4 (+) group demonstrated a significantly worse 3‐year disease‐specific survival (P = 0.018; Fig. 2a). Multivariate analysis showed that KLF4 (+) remained one independent predictor for a worse disease‐specific survival (Table 3; P = 0.016, hazard ratio = 3.72, 95% confidence interval 1.27–10.84). Subgroup analysis in advanced patients with T3–4, N2–3 or stage III–IV disease consistently showed that KLF4 (+) identified another poor prognostic subgroup in such advanced patients, suggesting its role in progression (Fig. 2b,c,d). Together, these results indicate that persistent KLF4 expression in HNSCC predicts worse prognosis, especially in patients with advanced disease.
Table 2.
Relationship between KLF4 expression and disease control variables
| Variable | KLF4 expression (n [%]) | P‐values* | |
|---|---|---|---|
| KLF4 (−) | KLF4 (+) | ||
| Local recurrence | |||
| Yes | 2 (4.8) | 3 (15.0) | 0.317 |
| No | 40 (95.2) | 17 (85.0) | |
| Neck recurrence | |||
| Yes | 2 (4.8) | 5 (25.0) | 0.031 |
| No | 40 (95.2) | 15 (75.0) | |
| Distant metastasis | |||
| Yes | 6 (14.3) | 7 (35.0) | 0.094 |
| No | 36 (85.7) | 13 (65.0) | |
*Estimated by Fisher’s exact test. KLF4, Krüppel‐like factor 4; KLF4 (−), decreased KLF4 expression; KLF4 (+), persistent KLF4 expression.
Figure 2.
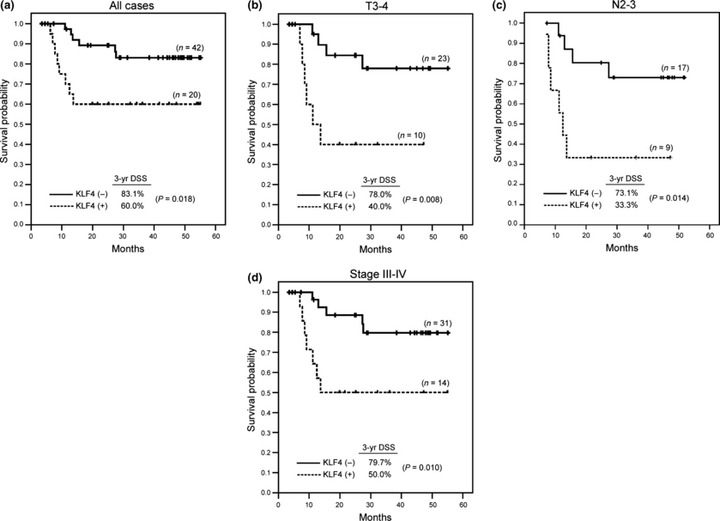
Prognostic significance of persistent Krüppel‐like factor 4 (KLF4) expression in head and neck squamous cell carcinoma (HNSCC). Comparison of Kaplan–Meier survival curves of disease‐specific survival (DSS) were done by log‐rank test for (a) all 62 HNSCC, (b) T3–4, (c) N2–3, and (d) stage III–IV patients. KLF4 (−), decreased KLF4 expression; KLF4 (+), persistent KLF4 expression.
Table 3.
Prognostic factors for disease‐specific survival
| Variable | 3‐year DSS (%) | P‐values | |
|---|---|---|---|
| Univariate† | Multivariate‡ | ||
| KLF4 expression | |||
| KLF4 (−) | 83.1 | 0.018 | 0.016 |
| KLF4 (+) | 60.0 | ||
| T classification | |||
| T1–2 | 85.5 | 0.069 | 0.150 |
| T3–4 | 64.9 | ||
| N classification | |||
| N0–1 | 92.9 | 0.004 | 0.018 |
| N2–3 | 58.4 | ||
| Perineural invasion | |||
| No | 81.8 | 0.066 | 0.891 |
| Yes | 63.0 | ||
| Lymphovascular invasion | |||
| No | 77.3 | 0.567 | – |
| Yes | 72.1 | ||
| ECS§ | |||
| No | 87.8 | 0.003 | 0.019 |
| Yes | 53.1 | ||
†Log‐rank test; ‡Cox’s proportional hazards regression analysis; §Fifty‐seven patients who underwent neck dissection were available to be analyzed. DSS, disease‐specific survival; ECS, extracapsular spread; KLF4, Krüppel‐like factor 4; KLF4 (−), decreased KLF4 expression; KLF4 (+), persistent KLF4 expression.
KLF4 overexpression increases migration and invasion abilities of HNSCC cells. To investigate the mechanisms how KLF4 contributes to poor prognosis in HNSCC, KLF4 protein expression in human HNSCC cell lines was examined by Western blot analysis. In consistent with clinical observation, all five HNSCC cell lines tested demonstrated a low level of KLF4 expression, compared to the normal control epithelium (Fig. 3a). We therefore performed overexpression experiment in HNSCC cell line SAS. KLF4 expression vector pcDNA3KLF4 was transfected to select cell lines stably overexpressing KLF4, and enforced KLF4 expression was confirmed in two SAS‐KLF4 (SAS‐K) clones, compared to the pcDNA3 vector transfected clones (SAS‐C) (Fig. 3b). Cell proliferation ability was checked and showed that enforced KLF4 expression did not inhibit the proliferation of SAS‐K clones (Fig. 3c). We next evaluated the migration and invasion abilities by transwell migration and invasion assays, and found that enforced KLF4 expression significantly increased the migration and invasiveness of SAS (Fig. 3d). Compared with SAS‐C clones, the SAS‐K clones showed a seven‐ to 12‐fold increase in the migration ability (Fig. 3e), and five to sixfold increase in the invasion ability (Fig. 3f). These data demonstrate that enforced KLF4 expression in SAS substantially increases the migration and invasion abilities without obvious growth inhibition.
Figure 3.
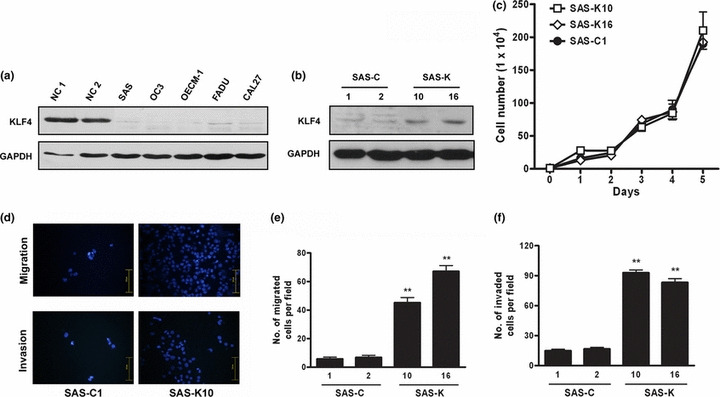
Krüppel‐like factor 4 (KLF4) overexpression increased the migration/invasion abilities of SAS. (a) Western blot analysis of KLF4 in five head and neck squamous cell carcinoma (HNSCC) cell lines. (b) Western blot of KLF4 expression in SAS‐K versus SAS‐C clones. (c) Growth curves indicated no growth inhibition in SAS‐K clones. (d) Representative pictures of cells that migrated/invaded through the transwell membrane. (e) The number of cells migrated across the membrane per field. (f) The number of cells invaded through Matrigel per field. The average and standard deviations were shown for three independent experiments. **P < 0.001. NC, normal epithelium.
KLF4 overexpression enhances tumorigenicity of HNSCC cells in vivo. To determine whether the KLF4‐mediated enhancement of migration and invasion could also be observed in vivo, 2 × 106 of SAS‐K10 and SAS‐C1 cells were implanted on the right and left flanks of 8‐week‐old athymic nude mice, respectively. Compared with SAS‐C1 cells, larger subcutaneous tumor by size and weight were observed in the flanks inoculated with SAS‐K10 cells on day 45 (Fig. 4a,b). The tumor growth curve revealed that tumor volume difference became obvious by day 14 after implantation (Fig. 4c). These results indicate that enforced KLF4 expression in SAS not only increases in vitro migration and invasion abilities, but also substantially enhances in vivo tumorigenicity.
Figure 4.
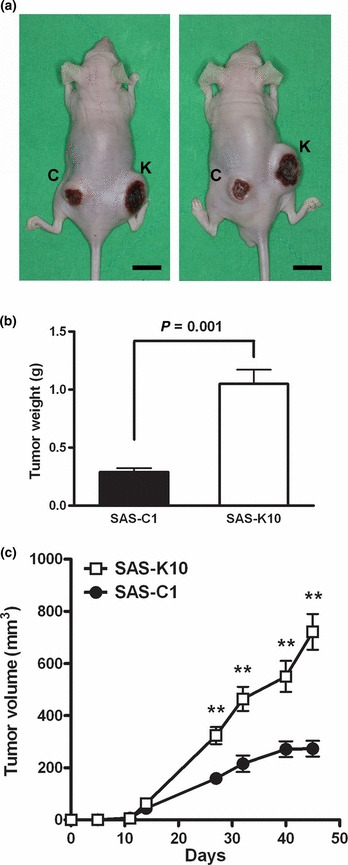
Krüppel‐like factor 4 (KLF4) overexpression enhanced SAS tumorigenicity. (a) Photographs of subcutaneous tumors derived from injection of 2 × 106 SAS‐C1 (C) and SAS‐K10 (K) in two representative athymic nude mice (scale bar: 10 mm). (b) Tumors were dissected and weighed on day 45 after implantation. (c) Tumor growth curve indicated the enhanced tumorigenicity of SAS‐K10, compared to the vector control clone SAS‐C1. n = 6, **P < 0.001.
High KLF4 expression induces multi‐drug resistance in HNSCC cells. Cisplatin, 5‐FU and docetaxel has been the major components of standard chemotherapy regimen for HNSCC. We treated the established stable clones with these chemotherapeutic agents to ask whether enforced KLF4 expression modulates chemosensitivity. After treatment for 24 h, both SAS‐K10 and SAS‐K16 showed higher resistance to cisplatin and docetaxel (Fig. 5a,b). The cisplatin IC50 values of SAS‐K10 and SAS‐K16 were 1.5‐ and three‐folds higher than that of SAS‐C1, respectively. In addition, the docetaxel IC50 values of SAS‐K10 and SAS‐K16 were 2.7‐ and four‐folds higher than that of SAS‐C1, respectively. The difference in 5‐FU sensitivity was not obvious at higher concentration, although a trend toward higher resistance against 5‐FU was observed at lower concentration for SAS‐K clones (Fig. 5c). These results indicated that enforced KLF4 expression in SAS induces multi‐drug resistance, especially for cisplatin and docetaxel.
Figure 5.
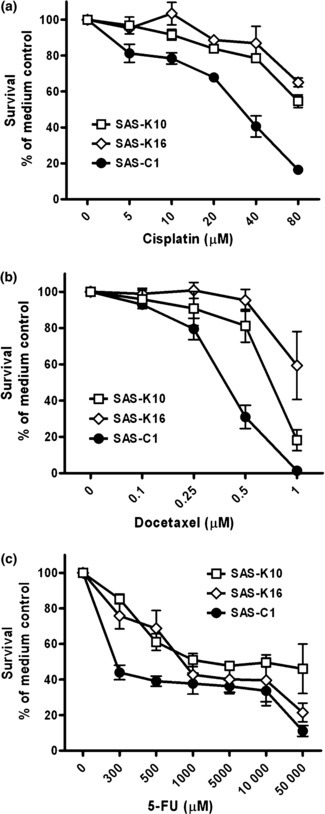
Krüppel‐like factor 4 (KLF4) overexpression induced multi‐drug resistance in SAS. Cells in 96‐well plate (1 x 104 each well) were treated with varying concentrations of (a) cisplatin and (b) docetaxel for 24 h, and (c) 5‐FU for 48 h. Survival was determined by comparing with the medium control wells. Results represent the average of three replicates. Error bars indicate standard deviation for each data point. 5‐FU, 5‐fluorouracil.
Discussion
Given the role of KLF4 as a negative cell cycle regulator,( 5 , 30 ) persistent KLF4 expression could be unfavorable for tumor growth. In consistent with this concept, decreased KLF4 expression was observed in several human cancers, such as those of the GI tract.( 18 ) However, because of its diverse and cell‐type specific functions, increased KLF4 expression has been reported in breast cancer and HNSCC.( 24 , 25 ) In HNSCC, KLF4 mRNA expression was studied previously by northern blot analysis and in situ hybridization.( 24 ) In the present study, KLF4 expression was evaluated at both protein and mRNA levels. Our observation that KLF4 is decreased in the majority of HNSCC is not in accord with previous reported data. This discrepancy may be due to: (i) the larger cohort of samples and different techniques for the evaluation, potentially contributing to more relevant results; and (ii) KLF4 mRNA level may be unable to reflect the protein expression level in tumor samples.
Our results demonstrate a gradual decline of KLF4 from normal epithelium to most HNSCC, suggesting a gradual loss of growth inhibition in the process of carcinogenesis. In colon and gastric cancers, mechanisms for KLF4 downregulation include point mutation, loss of heterozygosity, promoter hypermethylation and loss of adenomatous polyposis coli (APC) or β‐catenin activation.( 16 , 17 ) How KLF4 is downregulated in most HNSCC but is persistently expressed in some cases remains unclear. We speculate that epigenetic control might play a role for the variable KLF4 expression levels in HNSCC, and is worthy of future investigations. In this study, persistent KLF4 expression in HNSCC is associated with poor prognosis, especially in advanced patients who carry higher rates of cancer‐specific death. Thus, persistent KLF4 expression may become a useful prognostic marker to identify a subgroup of advanced patients who require intensified treatment and follow up in the future.
In accord with clinical data, in vitro and in vivo models reveal that enforced KLF4 expression leads to tumor progression in cell line SAS (Figs 3,4). Moreover, enforced KLF4 expression also induces drug resistance (Fig. 5). These clearly imply that persistent KLF4 expression contributes to tumor progression in HNSCC, although KLF4 is expressed in normal epithelium and commonly decreased in most HNSCC. Such ambiguous condition has not been reported in other human malignancies yet. Thus, simple dichotomization between tumor suppressor gene and oncogene may not be appropriate for KLF4 in HNSCC. It is plausible that genetic alterations accumulate during tumorigenesis and switch the role of KLF4 from tumor suppressive in normal epithelium, into transforming and oncogenic in HNSCC.
Mechanisms underlying the oncogenic roles of KLF4 in HNSCC remain unclear. We performed Ki‐67 staining to clarify the relation between KLF4 expression and cell proliferation ability in HNSCC. In normal and dysplastic epithelia, opposite expression pattern of KLF4 and Ki‐67 was observed. Areas high for KLF4 was generally low for Ki‐67, and vice versa (Fig. 6a). This is in consistent with the reported concept that KLF4 dominantly upregulates p21 to induce growth arrest (Fig. 6b),( 14 , 15 ) although it also represses p53 and BAX expression to confer anti‐apoptosis.( 6 , 31 ) However, we did not observe such opposite expression pattern in KLF4 (+) HNSCC (Fig. 6c). High Ki‐67 index was observed in 45% of KLF4 (+) HNSCC, not significantly lower than the KLF4 (−) group (54.8%, P = 0.472). Thus, it is plausible that distinct mitogenic signals contribute to bypassing the growth inhibition by KLF4, and unmasking its transforming, anti‐apoptotic activities, leading to tumor progression. A hypothetical model is proposed in Figure 6d, using cyclin D1 as a presumptive mitogenic signal, because cyclin D1 can counteract G1 arrest, can sequester and inactivate p21,( 32 ) and is overexpressed in 30–50% of HNSCC.( 33 , 34 ) Supporting the hypothesis, cell line SAS, which remains proliferative and becomes more aggressive after enforced KLF4 expression in this study, demonstrates cyclin D1 overexpression (Fig. 6e). However, IHC of cyclin D1 showed positive staining in only six (30%) of the 20 KLF4 (+) HNSCC. This suggests that other mitogenic signals might also participate in switching the role of KLF4 and further investigation is required.
Figure 6.
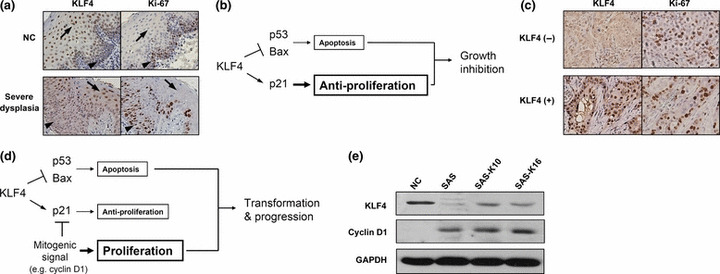
Hypothetic model for pleiotropic functions of Krüppel‐like factor 4 (KLF4). (a) Opposite expression pattern of KLF4 and Ki‐67 in spinous (arrow) and basal (arrowhead) layers of normal and dysplastic epithelia (magnification, ×400). (b) Reported growth inhibitory role of KLF4 through dominant upregulation of p21.( 14 , 15 ) (c) Lost of opposite expression pattern of KLF4 and Ki‐67 in head and neck squamous cell carcinoma (HNSCC) (magnification, ×400). (d) Hypothetic model for the role of KLF4 in KLF4 (+) HNSCC. (e) Cyclin D1 overexpression in cell line SAS by Western blot analysis. KLF4 (−), decreased KLF4 expression; KLF4 (+), persistent KLF4 expression; NC, normal epithelium.
In conclusion, this study demonstrates that, in HNSCC, persistent KLF4 expression is independently associated with progression and poor prognosis. Enforced KLF4 expression in HNSCC cells consistently confers aggressiveness, with increased migration/invasion abilities, multidrug resistance and tumorigenicity. These results therefore provide valuable information that HNSCC with persistent KLF4 expression might require intensified combination treatment in future practice.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This work was in part supported by Grant NSC 96‐2314‐B‐010‐017, NSC 98‐2314‐B‐010‐013‐MY3, NSC 98‐2320‐B‐010‐022‐MY3 from the National Science Council; Grant V96C1‐066, V97S5‐001, V98C1‐169 and V98S5‐001 from Taipei Veterans General Hospital; Grant VN98‐01, VN99‐04, VN100‐06 from TVGH‐NTUH Joint Research Program; Grant 96A‐D‐D132 from the National Yang‐Ming University, MOE, Taiwan. We are grateful to technical assistance from the Division of Experimental Surgery of Department of Surgery, and the Clinical Research Core Laboratory, Taipei Veterans General Hospital.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 2. Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med 2001; 345: 1890–900. [DOI] [PubMed] [Google Scholar]
- 3. Lothaire P, de Azambuja E, Dequanter D et al. Molecular markers of head and neck squamous cell carcinoma: promising signs in need of prospective evaluation. Head Neck 2006; 28: 256–69. [DOI] [PubMed] [Google Scholar]
- 4. Garrett‐Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc‐finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem 1996; 271: 31384–90. [DOI] [PubMed] [Google Scholar]
- 5. Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut‐enriched Kruppel‐like factor expressed during growth arrest. J Biol Chem 1996; 271: 20009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W, Geiman DE, Shields JM et al. The gut‐enriched Kruppel‐like factor (Kruppel‐like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem 2000; 275: 18391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut‐enriched Kruppel‐like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res 2000; 28: 2969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans PM, Liu C. Roles of Krupel‐like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin (Shanghai) 2008; 40: 554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takahashi K, Tanabe K, Ohnuki M et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–72. [DOI] [PubMed] [Google Scholar]
- 10. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–76. [DOI] [PubMed] [Google Scholar]
- 11. Park IH, Zhao R, West JA et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008; 451: 141–6. [DOI] [PubMed] [Google Scholar]
- 12. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61: 759–67. [DOI] [PubMed] [Google Scholar]
- 13. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: 57–70. [DOI] [PubMed] [Google Scholar]
- 14. Rowland BD, Peeper DS. KLF4, p21 and context‐dependent opposing forces in cancer. Nat Rev Cancer 2006; 6: 11–23. [DOI] [PubMed] [Google Scholar]
- 15. Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context‐dependent oncogene. Nat Cell Biol 2005; 7: 1074–82. [DOI] [PubMed] [Google Scholar]
- 16. Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel‐like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 2004; 23: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei D, Gong W, Kanai M et al. Drastic down‐regulation of Kruppel‐like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res 2005; 65: 2746–54. [DOI] [PubMed] [Google Scholar]
- 18. Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis 2006; 27: 23–31. [DOI] [PubMed] [Google Scholar]
- 19. Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN, Wu M. Down‐regulation of gut‐enriched Kruppel‐like factor expression in esophageal cancer. World J Gastroenterol 2002; 8: 966–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu W, Hofstetter WL, Li H et al. Putative tumor‐suppressive function of Kruppel‐like factor 4 in primary lung carcinoma. Clin Cancer Res 2009; 15: 5688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohnishi S, Ohnami S, Laub F et al. Downregulation and growth inhibitory effect of epithelial‐type Kruppel‐like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun 2003; 308: 251–6. [DOI] [PubMed] [Google Scholar]
- 22. Nakahara Y, Northcott PA, Li M et al. Genetic and epigenetic inactivation of Kruppel‐like factor 4 in medulloblastoma. Neoplasia 2010; 12: 20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yasunaga J, Taniguchi Y, Nosaka K et al. Identification of aberrantly methylated genes in association with adult T‐cell leukemia. Cancer Res 2004; 64: 6002–9. [DOI] [PubMed] [Google Scholar]
- 24. Foster KW, Ren S, Louro ID et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A‐immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c‐MYC and the zinc finger protein GKLF. Cell Growth Differ 1999; 10: 423–34. [PubMed] [Google Scholar]
- 25. Foster KW, Frost AR, McKie‐Bell P et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res 2000; 60: 6488–95. [PubMed] [Google Scholar]
- 26. Pandya AY, Talley LI, Frost AR et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early‐stage breast cancer. Clin Cancer Res 2004; 10: 2709–19. [DOI] [PubMed] [Google Scholar]
- 27. Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC, Chen CM. Nuclear Kruppel‐like factor 4 expression is associated with human skin squamous cell carcinoma progression and metastasis. Cancer Biol Ther 2008; 7: 783–5. [DOI] [PubMed] [Google Scholar]
- 28. Greene FL, Page DL, Fleming ID et al. AJCC Cancer Staging Manual. New York, NY: Springer, 2002. [Google Scholar]
- 29. Wang L, Wei D, Huang S et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res 2003; 9: 6371–80. [PubMed] [Google Scholar]
- 30. Chen X, Johns DC, Geiman DE et al. Kruppel‐like factor 4 (gut‐enriched Kruppel‐like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem 2001; 276: 30423–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Kruppel‐like factor 4 exhibits antiapoptotic activity following gamma‐radiation‐induced DNA damage. Oncogene 2007; 26: 2365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1‐phase progression. Genes Dev 1999; 13: 1501–12. [DOI] [PubMed] [Google Scholar]
- 33. Bova RJ, Quinn DI, Nankervis JS et al. Cyclin D1 and p16INK4A expression predict reduced survival in carcinoma of the anterior tongue. Clin Cancer Res 1999; 5: 2810–9. [PubMed] [Google Scholar]
- 34. Vielba R, Bilbao J, Ispizua A et al. p53 and cyclin D1 as prognostic factors in squamous cell carcinoma of the larynx. Laryngoscope 2003; 113: 167–72. [DOI] [PubMed] [Google Scholar]


