Abstract
It is known that cancers are caused by accumulated mutations in various genes and consequent functional alterations of proteins that are important for maintenance of normal cellular functions. The changes in nucleotide sequences and expression patterns of cancer‐related genes are being extensively studied to better understand the mechanisms of tumorigenesis and to develop methods for DNA protein diagnosis and drug discovery. At present, a number of computer databases for molecular information on cancer‐related genes are available publicly through the internet. These databases deal with familial cancer and sporadic cancer at the levels of germline mutation or somatic mutation, genomic or chromosomal abnormalities, and changes in the expression levels of relevant genes. Previously, we constructed a human gene mutation database named MutationView (http://mutview.dmb.med.keio.ac.jp/) and have accumulated mutation data for ∼300 genes that are involved mainly in monogenic diseases. Forty‐two genes are cancer‐related and therefore a separate cancer database named KMcancerDB was constructed. MutationView/KMcancerDB utilizes a graphic display function for both queries and search results much more often than other existing databases, making the system quite user friendly. MutationView/KMcancerDB provides a highly sophisticated search function for all genes through a single internet URL. In the present paper, we briefly review various useful databases for cancer‐related genes, and describe MutationView/KMcancerDB in more detail. (Cancer Sci 2007; 98: 259–267)
The majority of cancers occur sporadically in individuals. These are thought to be the result of multistep tumorigenesis, during which somatic mutations occur in a series of genes that play important roles in the regulation of growth, differentiation and apoptosis. However, some cancers occur rarely but repeatedly in different members of the same family, and this is considered to be familial cancer in which germline mutations are known to be involved. To date, numerous genes have been identified as causative genes for both sporadic and familial cancers and in some cases mutation types are correlated to malignancy and prognosis. It is obvious that molecular information on cancer‐related genes is indispensable for further investigating the mechanisms of tumorigenesis and developing methods for DNA diagnosis, gene therapy and drug discovery. At present, a number of computer databases are publicly available publicly throughout the world. However, in spite of their usefulness and high quality there are still many issues to be solved for establishing comprehensive and integrated cancer gene databases which can only be put into reality through international cooperation. Here, we briefly review the current status of gene mutation databases and describe in more detail some cancer‐related gene mutation databases, including our original system called MutationView/KMcancerDB.
Locus‐Specific Mutation Database and Central Mutation Database
Establishing coordinated databases for human disease‐associated gene mutations has been discussed repeatedly at the annual meetings of the Human Genome Variation Society (formerly the HUGO Mutation Database Initiative), which we joined as an active database‐constructing group from its initial phase.( 1 ) Two types of databases have been considered useful: the Locus‐Specific Mutation Database (LSDB) and the Central Mutation DataBase (CMDB). LSDB basically deals with a particular gene (genetic locus) that is known to be involved in the pathogenesis of a certain disease, and usually it is created and maintained by individual investigators (or curators) as a part of research activities. At present, there are 571 LSDBs managed by 390 curators.( 2 ) A partial list of LSDBs and representative portal sites for them are shown in Table 1. There are many merits of LSDB such as appropriateness of collected data items, accuracy of collected information and frequent updating, which are possible only by the efforts of curators. However, because each LSDB was established independently, a variety of software, data formats and user interfaces have been created and utilized. Data items are variable depending on the disease, but coordination is necessary at least for a minimal set of items such as gene symbol, nucleotide position of mutation, consequent change of amino acid sequence of protein, and description of phenotype. It is desired that the data format of such common items and the user interface are standarized. Another drawback of most LSDBs is that data is presented mainly using table format with character‐based information. Although the table format is indispensable for systematic data collection and comparison of mutations, graphical or visual ways to present search results will be useful to intuitively understand the mutation event and its biological meaning.
Table 1.
Various cancer‐related databases
| Database name | Gene | Internet address | Ref |
|---|---|---|---|
| Locus‐specific Database | |||
| Familial Adenomatous Polyposis at GeneDis | APC | http://life2.tau.ac.il/GeneDis/Tables/APC/apc.html | |
| UMD Locus Specific Databases | APC, MEN1, VHL, others | http://www.umd.be/ | 4 |
| ATbase – a registry of patients with ataxia‐telangiectasia | ATM | http://www.cnt.ki.se/ATbase | |
| Ataxia‐Telangiectasia Mutation Database | ATM | http://www.benaroyaresearch.org/bri_investigators/atm.htm | |
| Bloom Syndrome‐BLMbase | BLM | http://bioinf.uta.fi/BLMbase/index2.html | 15 |
| Breast Cancer Mutation Database | BRCA1, BRCA2 | http://research.nhgri.nih.gov/bic/ | |
| CDKN2a Database Project (a human p16 database with annotation) | CDKN2A | http://biodesktop.uvm.edu/perl/p16 | |
| EGFR Mutation Database | EGFR | http://www.cityofhope.org/cmdl/egfr_db/index.html | |
| Fanconi Anaemia Mutation Database | FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, others | http://www.rockefeller.edu/fanconi/mutate/ | |
| LYSTbase: Mutation registry for Chediak‐Higashi syndrome | LYST | http://bioinf.uta.fi/LYSTbase/ | |
| MMR Databank | MLH1, MLH3, MSH2, MSH6, PMS1, PMS2 | http://www.insight‐group.org/ | |
| Mismatch Repair Genes Variant Database | MLH1, MSH6, MSH6 | http://www.med.mun.ca/mmrvariants/ | |
| NF1 International Mutation Databas | NF1 | http://www.nfmutation.org/ | |
| NF2 International Mutation Database | NF2 | http://www.nfmutation.org/ | |
| PTCH Mutation Database | PTCH | http://www.cybergene.se/cgi‐bin/w3‐msql/ptchbase/index.html | |
| Retinoblastoma Genetics | RB1 | http://www.verandi.de/joomla/ | 16 |
| SH2D1Abase: Mutation registry for X‐linked lymphopreliferative syndrome (XLP) | SH2D1A | http://bioinf.uta.fi/SH2D1Abase/ | |
| IARC TP53 Mutation Database | TP53 | http://www‐p53.iarc.fr/ | 3 |
| p53 web site | TP53 | http://p53.free.fr | |
| Database of Germline p53 Mutations | TP53 | http://www.lf2.cuni.cz/projects/germline_mut_p53.htm | 17 |
| p53 Mutation Database Analysis & Search | TP53 | http://p53.genome.ad.jp/ | |
| TP53 mutation database (UMD LSDB) | TP53 | http://www.umd.be:2072/ | 5 |
| Human p53 database and software | TP53 | http://sunsite.unc.edu/dnam/des_p53.htm | 18 |
| TSC Mutation Database | TSC1, TSC2 | http://chromium.liacs.nl/lovd/index.php?select_db=TSC1 | 19 |
| The Cardiff‐Rotterdam Tuberous sclerosis mutation database | TSC2 | http://www.uwcm.ac.uk/uwcm/mg/tsc_db/ | |
| WASbase: Mutation registry for Wiskott‐Aldrich syndrome (WAS) | WAS | http://bioinf.uta.fi/WASbase/ | |
| Database of WS‐associated WRN mutations | WRN | http://www.pathology.washington.edu/werner/ws_wrn.html | |
| GeneDis; Human Genetic Disease Database | http://life2.tau.ac.il/GeneDis/ | ||
| MutationView | 1 † | http://mutview.dmb.med.keio.ac.jp/ | 12,13 |
| Databases specialized for cancers | |||
| COSMIC; Catalogue of Somatic Mutations in Cancer | http://www.sanger.ac.uk/genetics/CGP/cosmic/ | 6 | |
| Atlas of Genetics and Cytogenetics in Oncology and Haematology | http://www.infobiogen.fr/services/chromcancer/ | 7 | |
| Cancer Genome Anatomy Project, CGAP | http://cgap.nci.nih.gov/ | 8 | |
| The Tumor Gene Database | http://condor.bcm.tmc.edu/ermb/tgdb/tgdb.html | 9 | |
| Cancer Gene Expression Database, CGED | http://cged.hgc.jp/ | 20 | |
| Central Mutation Databases | |||
| Genome Database, GDB | http://www.gdb.org/ | 21 | |
| Genatlas | http://www.dsi.univ‐paris5.fr/genatlas/ | 22 | |
| GeneCards Database | http://www.genecards.org/index.shtml | 23 | |
| Human Genome Variation database, HGVbase | http://hgvbase.cgb.ki.se/ | 24 | |
| Human Organised Whole Genome Database, HOWDY | http://howdy.jst.go.jp/ | 25 | |
| Online Mendelian Inheritance in Man, OMIM | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM | 10 | |
| Human Gene Mutation Database,HGMD | http://www.hgmd.cf.ac.uk/ac/index.php | 11 | |
| Sequence Tag Alignment and Consensus Knowledgebase, STACK | http://www.sanbi.ac.za/Dbases.html | 26 | |
CMDB is an integrated database for multiple genes, which facilitates searches across genes. In the ideal CMDB, all mutation data for known disease genes would be collected with one common database format, and the user interface must be unified in a visual manner by effective use of graphics. Search and analysis should be possible using the common data fields described above in addition to mutation type (missense, deletion, etc.), effect on splicing, inheritance (dominant/recessive) and mutation origin (germline/somatic). If any CMDB is established by compiling data accumulated in LSDB after getting consensus of worldwide curators, the quantity and quality of the data will be impressive and the database must be quite useful. However, CMDB should not be considered a replacement for LSDBs but rather it must be an organized data pool to be used for genome‐wide integrated analysis across genes. It should function as a highly sophisticated portal site to each LSDB. There are several databases that have some characteristics of CMDB (see section ‘Databases with CMDB‐like characteristics’, which will give many suggestions to establish CMDB). Recently, an international meeting entitled ‘The Human Variome Project’ was held with the support of the World Health Organization and Australian Government to discuss establishing the CMDB as an international cooperation.( 2 )
The genes responsible for familial cancers and cancer‐related genes have been important targets of LSDBs (Table 1) and will be the main components of CMDB. For cancers, there are various molecular genetic databases for information other than mutations, including abnormal karyotypes and changes of gene expression. In the following sections, LSDB specialized for cancer as well as several CMDB‐like databases will be overviewed and discussed.
Databases specialized for cancers
IARC TP53 Mutation Database. IARC TP53 Mutation Database is a typical LSDB that was created by the International Agency for Research on Cancer (IARC) at Lyon, France, since 1994.( 3 ) It has collected all of the TP53 mutations identified in human cancer tissues or cell lines since 1989. Available data sets include TP53 somatic mutations in sporadic cancers, TP53 germline mutations in familial cancers, TP53 polymorphisms identified in human populations, functional properties of TP53 mutant proteins, and TP53 gene status in human cell lines. Data entries in the latest version R10 (released in July 2005) are 21 587 somatic mutations reported in 1876 original publications, 283 germline mutations reported in 129 publications (1989 to December 2004) and functional properties of more than 425 mutant proteins. Moreover, statistical information is presented with graphs such as growth rate of database, distribution of mutations in tumor sites, type of mutations, and codon distribution of mutations. From these statistics, it was found that approximately 75% of TP53 mutations are ‘missense’ type for both somatic and germline cases. This is a prominent nature of TP53 because mutation types of somatic cases in other tumor suppressor genes are more abundant in ‘nonsense’ or ‘frameshift’ mutations (see section ‘Statistical Function’ under ‘MutationView/KMcancerDB’ for the description of the von Hippel–Lindau syndrome gene [VHL]).
Universal Mutation Databases. Universal Mutation Databases (UMDs) are LSDBs constructed using a UMD software package.( 4 ) UMDs include mutations of cancer‐related genes such as TP53, APC, BRCA1, BRCA2, MEN1 and VHL. UMDs also include mutations of other genes such as FBN1, LDLR, LMNA, EMD and ATP7B, which are responsible for genetic diseases. Although most of these genes are accommodated in the same website (http://www.umd.be/), each gene is maintained by individual curators. The UMD‐TP53 was created from the p53 website that collected somatic and germline mutations and polymorphisms documented in the literature since 1989 and unpublished data submitted by the curators.( 5 ) The UMD package contains various software tools to dynamically link among LSDBs, with a database structure suitable for collecting and analyzing a wide range of clinical data, which should be instructive to geneticists and biologists. Most UMD‐LSDBs are freely accessible, except some UMD‐LSDBs require user ID and password.
COSMIC. The Catalog of Somatic Mutations in Cancer (COSMIC) was developed by The Sanger Center to provide integrated genetic data for cancer genes.( 6 ) COSMIC contains various information on definite cases of cancer tissues and cell lines, such as tissue/cell line ID, derived organ, tissue or cancer subtype, analyzed gene name, and mutation details. It is possible to display mutation position and frequency, and to list genes with mutations for particular cancer cases. The current data contents (version 21, released 14 September 2006) are: 3611 references; 164 905 cancer tissues and cell lines; 1339 tested genes; 86 cancer genes with mutations; and 344 cancer‐related genes with mutations.
Atlas of Genetics and Cytogenetics in Oncology and Haematology. The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a database for genetic and cytogenetic information on cancer, leukemia and ‘cancer‐prone’ diseases such as xeroderma pigmentosum and Li–Fraumeni syndrome.( 7 ) The contents include clinical characteristics, associated chromosomal abnormalities, and various information on the responsible genes. The numbers of collected genes are approximately 900 for cancer genes and 3600 for possible cancer‐related genes. The user is able to search by disease, gene and chromosome. A unique feature of this database is the collection of educational materials for molecular biology and genetics related to oncology. Link pages covering a wide range of websites are useful. The contents of this database have been collected by many collaborators (477 as of September 2006) throughout the world and are controlled by editors in various fields (39 as of September 2006).
CGAP. The Cancer Genome Anatomy Project (CGAP) is a project run by the National Cancer Institute, USA, and the National Center for Biotechnology Information (NCBI), USA, to determine the gene expression profiles of normal, precancer and cancer cells.( 8 ) The website of this project consists of seven modules: genes, tissues, pathways, RNA interference (RNAi), chromosomes, SAGE Genie and tools. Of these, the data on RNAi constructs and gene expression profiles with SAGE present especially valuable information for pursuing molecular research on cancer.
Tumor Gene Database. The Tumor Gene Database has been maintained at the Baylor College of Medicine (USA).( 9 ) The database collects various information on the proto‐oncogenes and tumor suppressor genes that are targets for cancer‐causing mutations. The information in this database is categorized into 20–30 groups such as ‘clinical’, ‘DNA structure’, ‘function’, ‘oncogenicity’, ‘protein binding’, ‘protein structure’ and ‘tumor type’. Each category has concise descriptions (occasionally more than 80) as an individual ‘fact’ extracted from the literature. Currently, this database has 2600 facts for 300 genes, therefore it is useful as a reference for concentrated knowledge on cancer‐causing genes.
Databases with CMDB‐like characteristics
Online Mendelian Inheritance in Man. Mendelian Inheritance in Man (MIM) is an encyclopedia of human genetic traits written by Professor Victor A. McKusick (Johns Hopkins University) in 1966.( 10 ) It has been edited continuously and is currently in its 12th edition. Genetic traits in this book include hereditary diseases such as hemophilia and familial cancers, non‐disease but inherited traits such as blood groups, and various genes with known function. Descriptions of each trait cover phenotype, biological facts including chromosomal map, molecular cloning and representative mutations, clinical symptoms, family history and population genetics. Many entries have descriptions of the discovery‐research history of the disease‐gene and details of patient cases. Online Mendelian Inheritance in Man (OMIM) is a web‐based computer search system of MIM, which is available through the NCBI, USA. OMIM is updated daily by Professor McKusick. Currently, the number of OMIM entries has reached 17 000 (July 2006), including ∼1800 diseases and ∼1300 disease‐causing genes (these numbers are inconsistent because occasionally multiple diseases with different names are caused by the same gene). The number of entries regarding cancer is more than 250, most of which are familial cancers. Since OMIM contains virtually all known diseases, it is considered as a CMDB. The information in OMIM is quite useful for learning about frontier research on hereditary diseases, gene‐related disorders and other phenotypes. However, use of OMIM as a ‘database’ requires some effort because it does not have a typical database structure. Additionally, mutation data in OMIM is not comprehensive but is limited to representative cases, so that only characteristic mutations have been listed and documented.
Human Gene Mutation Database. The Human Gene Mutation Database (HGMD) is an online database for mutations in disease‐causing and disease‐susceptibility genes created at University of Wales College of Medicine.( 11 ) In HGMD, a description of each mutation is given with the name of the disease, a few commentary words and references. Nucleotide sequence of cDNA for each gene and a simple diagram of mutation distribution can be displayed. Current data has reached 53 208 mutations in 2056 genes (July 2006), which are accessible free of charge with a user ID and password. Data on mutations for more genes are provided but at a cost. The present total information is 64 251 mutations for 2362 genes (October 2006). HGMD is the largest mutation database and is considered to be a useful CMDB‐like database. It will be much nicer if more information such as frequency and ethnic origin are added and graphical display is employed. There is a difference between OMIM and HGMD in the number of collected genes with mutations, but this can be accounted for by the fact that HGMD collected more disease‐susceptibility genes than OMIM.
MutationView/KMcancerDB
As described above, each LSDB is useful, but data format, user interface and software are not unified. An authentic CMDB has not been established yet. There are CMDB‐like databases that are valuable and practical, but they have various drawbacks regarding sufficiency of data items, database format, completeness of mutation collection and coordination with LSDBs. Both LSDB and CMDB‐like databases have deficiencies in the effective use of graphical data representation. To overcome these drawbacks, we have developed a database system named MutationView (http://mutview.dmb.med.keio.ac.jp/) and constructed the cancer‐related gene mutation database KMcancerDB using the Mutation‐View system.( 12 , 13 ) The characteristics of MutationView and current data contents in the KMcancerDB are described below.
Database organization and software of MutationView. Mutation View is basically in the category of LSDB, but all genes and diseases collected in MutationView utilize the same database format and common user interface, hence they are accessible through the single URL. In this regard, MutationView is not an ordinary LSDB but an integrated LSDB system. Also, Mutation‐View operates as a distributed database. Data files for each gene can be kept at the curator's site so that only the curator can open or close the website and add, erase or change the data. We call these sites that are created and maintained by curators as ‘disease servers’. Data of disease servers are coordinated by ‘coordinating servers’. The coordinating server has the right to manage user access, accept users’ requests and retrieve data from appropriate disease servers. The coordinating server creates characteristic graphic data representations (see the following sections). Software for both coordinating server and disease servers is written in JAVA language, and can be implemented to various computer systems including Windows XP, LINUX and Solaris. Currently, the coordinating server is at Keio University School of Medicine and several disease servers are located at different sites.
Access to genes. Figure 1 shows various graphical displays to search and select gene(s) of interest. By clicking on any chromosome in Fig. 1a, all of the diseases or disease‐causing genes assigned to that chromosome described in OMIM are displayed (Fig. 1b). Entries with blue color represent cancer‐related genes. The ‘chromosome overview’ menu shows a list of all the genes stored in MutationView (Fig. 1c). Keyword search is also possible in this menu. In Fig. 1c, a search result of ‘cancer’ is shown with red or green color. Forty‐two genes were found including VHL. Link buttons to other databases (OMIM, Genome Data Base [GDB], HGMD, Human Genome Variation Database [HGVbase], and Human full‐length CDNA annotation invitational Database [H‐Inv DB]) appear by clicking each entry (right button) (Fig. 1e). The anatomical chart of the human body is also useful, in which names of organs and tissues are clickable to list diseases and causative genes (Fig. 1d). A menu using a protein model is available for some diseases, showing a diagram of a protein complex or the relationship of related proteins. In Fig. 1f, a protein complex involved in nucleotide excision repair is shown, in which disease‐causative genes are colored blue and are clickable. Clicking a selected gene (Fig. 1b–d,f) opens the ‘gene structure window’.
Figure 1.
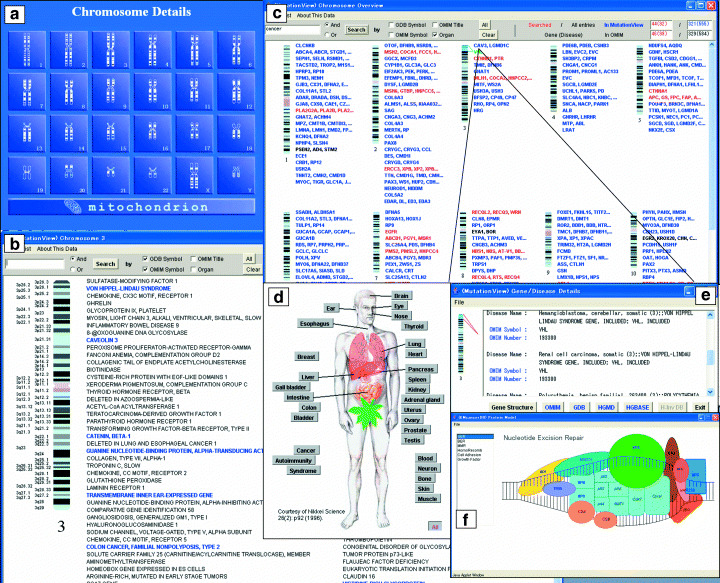
Access to genes. (a) Chromosome detail window. Clicking each chromosome schema opens a disease and gene list. (b) Disease and gene list of chromosome 3 opened by clicking chromosome 3 in (a). (c) Chromosome overview window. Cancer‐related genes are shown in red. The pointer indicates the VHL gene, which is in green color. Clicking each gene symbol shows a gene structure window (Fig. 2). (d) Anatomy window. Clicking on organ‐tissue names shows a list of diseases and causative genes. (e) Gene/disease details window, opened by clicking (right button) the VHL gene in (c). (f) Protein model window for the nucleotide excision repair complex. Clicking each member of the protein complex shows a gene structure window (Fig. 2).
Gene structure window. The gene structure window was designed to graphically display mutations in the primary structure of genes and cDNA sequences with various associated information. In Fig. 2a, the gene structure window of the VHL gene is shown. Each mutation is located at the appropriate cDNA position as a histogram, in which the height is proportional to the number of cases in the literature. The symbol table lists the type of mutation such as nonsense, missense and deletion (inset of Fig. 2a). The displays for cDNA/protein coding region/genomic structure can be alternatively switched from a pull‐down menu. In Fig. 2a, the protein coding region is shown on the x‐axis together with the protein functional domain, which is useful to analyze the relationship between mutation position and possible defect of protein function. In this case, ‘acidic domain’ and ‘Sp1 binding domain’ are seen. The x‐axis can be zoomed in until the nucleotide sequence and amino acid sequence appear. In fact, when zoomed in, other functional domains become visible such as the elongin binding and VMP1 binding domains. The genomic structure mode (Fig. 2b) displays mutations in the introns and flanking regions. The gene structure window also shows the size of the genomic/cDNA/coding region and the number of cases/mutations/polymorphisms (Fig. 2a).
Figure 2.
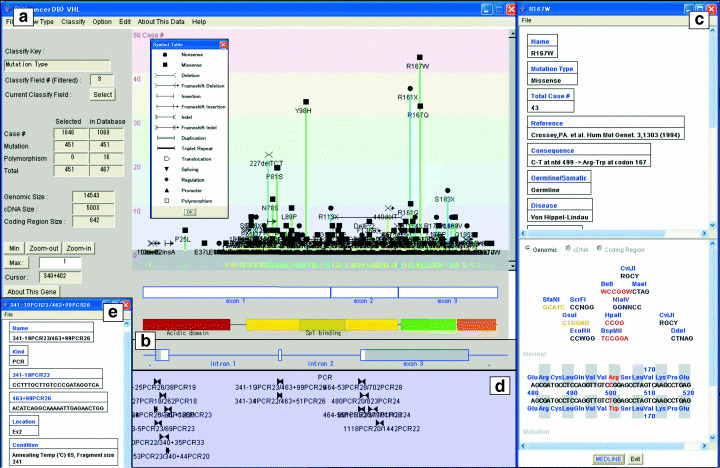
Gene structure window. (a) Default gene structure window of the VHL gene. A histogram of mutation frequency, exon boundaries and protein domain structure is shown. Inset: a mutation symbol table. (b) Exon/intron structure of the VHL gene shown by switching from cDNA/coding region mode to genomic mode. (c) Mutation details window of mutation R167W. (d) Probe window showing polymerase chain reaction (PCR) primer pairs to amplify various parts of the VHL gene. (e) Probe detail window showing the nucleotide sequence of PCR primers and reaction conditions.
The mutation detail window is shown by clicking on the mutation symbol, for example mutation R167W (Fig. 2c). The nucleotide sequence and deduced amino acid sequence of the region around the mutation are shown with restriction sites for comparison between normal and mutated sequences. Changes in the nucleotide sequence, amino acid sequence and restriction sites are highlighted in red. Additional information is shown in this window, such as total case number reported for this mutation, mutation heredity (germline or somatic) and name of associated disease.
Information for polymerase chain reaction (PCR) primers and allele‐specific oligomers (ASOs) is shown in the probe‐displaying field of genomic structure (Fig. 2b,d). Pairs of triangles show the PCR primers to amplify the corresponding region in between. Clicking each primer pair opens another window to show the primer sequences, PCR conditions and reference papers (Fig. 2e). Detection of certain mutations can be carried out easily using the probe information and mutation details in Fig. 2c.
Statistical function. Each mutation data is associated with various other information as shown in Fig. 2c. Figure 3a shows a data table for the VHL gene, which was set in the disease server computer but is invisible to users. In this data table, one line represents one particular case. Columns on the left half represent common fields such as mutation type, position, mutation name and reference, whereas columns on the right half represent extended information such as ethnic origin, disease name and subtype, germline or somatic, and other disease‐specific data. The MutationView system analyzes this table in real time and performs statistical calculations. In the default condition, it sums up a number of cases with the same mutation name and draws histograms in the gene structure window (2, 3). Users can carry out statistical analysis on demand. The statistical function of MutationView has various activities such as filtering, grouping and sorting. The classify window (Fig. 3b) sets the condition for statistical analysis of VHL. In this case, seven columns are shown including germline/somatic, disease/details, ethnic origin and tumor type/organ, each of which corresponds to each column with the same name in Fig. 3a. The result of statistical calculations is shown in the classify window itself and in the gene structure window (Fig. 3b–d). Descriptions in each column of the classify window are field values and the number on the head of each line is frequency.
Figure 3.
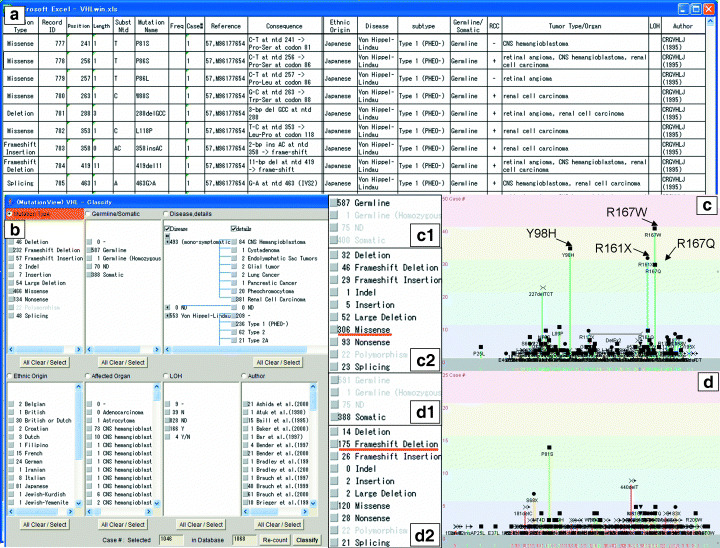
Classify function of MutationView. (a) A raw data file of the mutation description table of the VHL gene in Excel format. (b) Classify window. (c) A histogram with mutation data filtered by only ‘germline’. (c1) A part of the classify window to filter the mutations to only ‘germline’. (c2) The classify window after filtering by only ‘germline’. (d) A histogram with mutation data filtered by only ‘somatic’. (d1) A part of the classify window to filter the mutations to only ‘somatic’. (d2) The classify window after filtering by only ‘somatic’.
Words such as ‘germline’ and ‘somatic’ in the case of the germline/somatic column are field values and the numbers are frequency. ‘Filtering’ eliminates parts of the data. Elimination and inclusion of data can be done by clicking each field value. For example, it is possible to limit the analysis to only Japanese cases with germline mutations. ‘Grouping’ is a function to create new field value from existing ones, for example to create ‘base change’ from missense and nonsense. ‘Sorting’ is a function to classify the mutations by bar colors of a histogram. In the default condition, the sorting key item is ‘mutation type’ as shown with the orange color in the column header (Fig. 3b). It can be changed by clicking any one of the column headers.
It is known that germline mutations of the VHL gene are often missense and nonsense, whereas somatic mutations of the VHL gene are frameshift‐type deletions and insertions.( 14 ) In MutationView, these results can be graphically demonstrated after statistical calculation in real time. In the classify window, only ‘Germline’ can be selected by filtering (Fig. 3c1) and immediately shows that the majority of cases are mutation type ‘missense’ (Fig. 3c2). Also in the histogram, hot spots of missense and nonsense mutations are seen as tall bars such as Y98H, R161X, R167Q and R167W (Fig. 3c). Filtering by ‘somatic’ (Fig. 3d1) shows an increase in ‘frameshift deletion’ and a decrease in ‘missense’ (Fig. 3d2). Thus, the statistical function of MutationView is powerful for immediate analysis and display of various associations of the deposited data.
Current data contents. To date, we have collected 16 567 entries of mutations from 2306 published papers dealing with 294 genes involved in 510 diseases. These data are classified into nine categories: eye, heart, brain, ear, muscle, blood, syndrome, autoimmunity and cancer (2, 3). The categorized subsets of data can be accessed separately through the same URL. The subset database of cancer‐related genes is named KMcancerDB.
Table 2.
Current data amount in MutationView (1 December 2006)
| Data type | No. entries |
|---|---|
| Gene | 294 |
| Disease | 510 |
| Mutation | 16 567 |
| Polymorphism | 2051 |
| Literature | 2306 |
Table 3.
Number of disease genes in each category of MutationView (1 December 2006)
| Disease category | No. genes † | Typical diseases |
|---|---|---|
| Eye | 108 | RP, glaucoma, corneal dystrophy |
| Ear | 56 | Deafness |
| Heart | 37 | Cardiomyopathy, heart dysmorphism |
| Muscle | 35 | DMD/BMD, MD Fukuyama type |
| Bone | 45 | Craniometaphyseal Dysplasia |
| Brain/Neuron | 74 | Familial Parkinsonism, Alzheimer's disease |
| Cancer‐related | 42 | Breast, retinoblastoma |
| Blood | 45 | CML, citrullinemia |
| Kidney | 20 | Bartter syndrome |
The number of diseases does not necessarily match with the number of genes because it is known that some diseases with different clinical symptoms are caused by different mutation types in the same gene and that other diseases with the same clinical phenotype are caused by different genes. BMD, Becker muscular dystrophy; CML, chronic myeloblastic leukenia; DMD, Duchenne muscular dystrophy; MD, muscular dystrophy; RP, retinitis pigmentosum.
KMcancerDB contains 42 genes related to 89 kinds of cancers (Fig. 1c). Table 4 shows a partial list of familial cancers, including hereditary non‐polyposis colorectal cancers and xeroderma pigmentosa as well as premature aging and increased cancer risk disorders such as Bloom and Werner syndromes.
Table 4.
Partial list of familial tumors and their causative genes
| Disease name | MIM number | Gene symbol † | MIM number | Locus |
|---|---|---|---|---|
| I. Autosomal Dominant | ||||
| 1. Tumor suppresor gene | ||||
| a. neuron, sensory organ, sukin, kidney, urinary organ | ||||
| Retinoblastoma (RB) | 180200 | RB1 | 180200 | 13q14.1‐q14.2 |
| Neurofibromatosis, type I (NF1),von Recklinghausen disease | 162200 | NF1 | 162200 | 17q11.2 |
| Neurofibromatosis, type II (NF2) | 101000 | NF2 | 101000 | 22q12.2 |
| Tuberous sclerosis (TS) | 191100 | TSC1 | 605284 | 9q34 |
| TSC2 | 191092 | 16p13.3 | ||
| Wilms tumor 1(WT1), nephroblastoma | 194070 | WT1 | 194070 | 11p13 |
| von Hippel‐Lindau Syndrome (VHL) | 193300 | VHL | 193300 | 3p26‐p25 |
| melanoma, cutaneous malignant, 2 (CMM2) | 155601 | p16/MTS1 (CDKN2A) | 600160 | 9p21 |
| Basal cell nevus syndrome (BCNS) (Gorlinsyndrome) | 109400 | PTCH | 601309 | 9q22.3 |
| PTCH2 | 603673 | 1p32 | ||
| b. endocrine | ||||
| Multiple endocrine neoplasia, type I (MEN 1) | 131100 | MEN1 | 131100 | 11q13 |
| c. extremity | ||||
| Exostoses, multiple, type I (EXT) | 133700 | EXT1 | 133700 | 8q24.11‐q24.3 |
| EXT2 | 133701 | 11p12‐p11 | ||
| d. digestive organ | ||||
| Gastric cancer | 137215 | ECAD (CDH1) | 192090 | 16q22.1 |
| Familial adenomatous polyposis (FAP) | 175100 | APC | 175100 | 5q21‐q22 |
| Gardner syndrome (GS) | 175100 | APC | 175100 | 5q21‐q22 |
| Turcot syndrome | 276300 | APC | 175100 | 5q21‐q22 |
| MLH1 | 120436 | 3p21.3 | ||
| PMS2 | 600259 | 7p22 | ||
| Peutz‐Jeghers syndrome (PJS) | 175200 | STK11/LKB1 | 602216 | 19p13.3 |
| Polyposis, juvenile intestinal (PJI) | 174900 | PTEN | 601728 | 10q23 |
| SMAD4/DPC4 | 600993 | 18q21 | ||
| Cowden disease (CD) | 158350 | PTEN | 601728 | 10q23 |
| e. multiple organ | ||||
| Li‐Fraumeni syndrome (LFS) | 151623 | p53 (TP53) | 191170 | 17p13.1 |
| 2. Oncogene | ||||
| Multiple endocrine neoplasia, type II (MEN2) | 171400 | RET | 164761 | 10q11.2 |
| Renal cell carcinoma, papillary, 1 (PRCC) | 179755 | MET | 164860 | 7q31 |
| Gastrointestinal stromal tumor (GIST) | 164920 | KIT | 164920 | 4q21 |
| 3. DNA repair‐related gene | ||||
| Colorectal cancer, hereditary non‐polyposis, type 1 (HNPCC1) | 120435 | MSH2 | 120435 | 2p21 |
| Colorectal cancer, hereditary non‐polyposis, type 2 (HNPCC2) | 120436 | MLH1 | 120436 | 3p21.3 |
| Colorectal cancer, hereditary non‐polyposis, type 5 (HNPCC5) | 600678 | MSH6 | 600678 | 2p16 |
| Colorectal cancer, hereditary non‐polyposis, type 3 (HNPCC3) | 600258 | PMS1 | 600258 | 2q31‐q33 |
| Colorectal cancer, hereditary non‐polyposis, type 4 (HNPCC4) | 600259 | PMS2 | 600259 | 7p22 |
| Breast cancer, familial (FBC) | 114480 | BRCA1 | 113705 | 17q21 |
| BRCA2 | 600185 | 13q12.3 | ||
| II. Autosomel Recessive | ||||
| Xeroderma pigmentosum | XPA | 278700 | 9q34.1 | |
| XPB (ERCC3) | 133510 | 2q21 | ||
| XPC | 278720 | 3p25 | ||
| XPD (ERCC2) | 278730 | 19q13.2 | ||
| XPE (DDB2) | 278740 | 11p12‐p11 | ||
| XPF (ERCC4) | 278760 | 16p13 | ||
| XPG (ERCC5) | 133530 | 13q33 | ||
| Ataxia‐telangiectasia (AT) | 208900 | ATM | 208900 | 11q22.3 |
| Fanconi anemia | FAA (FANCA) | 227650 | 16q24.3 | |
| FAC (FANCC) | 227645 | 9q22.3 | ||
| Bloom syndrome (BLM) | 210900 | BLM (RECQ2) | 604610 | 15q26.1 |
| Werner syndrome (WRN) | 277700 | WRN (RECQ3) | 604611 | 8p12‐11.2 |
| Chediak‐Higashi syndrome (CGS1) | 214500 | LYST (CHS1) | 214500 | 1q42.1‐q42.2 |
| III. X‐linked | ||||
| Lymphoproliferative disease, X‐linked (XLP) | 308240 | SH2D1A | 308240 | Xq25 |
| Wiskott‐Aldrich syndrome (WAS) | 301000 | WAS | 301000 | Xp11.23‐p11.22 |
)The genes which has been already entered in MutationView are shown in bold character.
Prospects. We are expanding the contents of KMcancerDB (Table 4) to establish a LSDB with a complete list of familial tumor genes. Furthermore, we are making efforts to create software to manage search queries across multiple genes, which will add a CMDB‐like capability to MutationView. Hyperlinks to a variety of other databases are in progress. MutationView is evolving into a next‐generation knowledge base to deal with the more complex nature of cancer‐related genes.
Accessibility and availability. Users can have access to MutationView/KMcancerDB without an ID and password. However, this database is strictly for academic research purposes and is not intended for commercial use. Most internet browser software including Internet Explorer, Netscape, Mozilla and Safari can be used. The software of the MutationView disease server and main server are made available to LSDB curators in the spirit of cooperative database development. For inquiries, contact Nobuyoshi Shimizu (shimizu@dmb.med.keio.ac.jp) or Shinsei Minoshima (mino@hama-med.ac.jp).
Acknowledgments
We thank Dr Takashi Kawamura and Mr Susumu Mitsuyama at the Keio University School of Medicine, Professor Fumiaki Ito and Ms Sachiko Ito at Stsunan University, and Dr Shin‐ichi Moriwaki at Osaka Medical University for their collaborative efforts to construct the database. We also thank Chi Co. Ltd for long‐lasting collaboration for computer programming and database construction. This work was supported in part by a Grant‐in‐Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and a Grant‐in‐Aid for Publication of Scientific Research Results from the Japan Society for the Promotion of Science (JSPS).
References
- 1. HUGO MDI. HUGO‐mutation database initiative newsletter no. 18. 2001. [Cited 27 November 2001.] Available from URL: http://www.hgvs.org/newsletters/news18.html
- 2. Genomic Disorders Research Centre. The Human Variome Project meeting − International collection of human gene variation. 2006. [Cited 20 June 2006.] Available from URL: http://www.humanvariomeproject.org/
- 3. Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 Database: new online mutation analysis and recommendations to users. Hum Mutat 2002; 19: 607–14. [DOI] [PubMed] [Google Scholar]
- 4. Beroud C, Collod‐Beroud G, Boileau C, Soussi T, Junien C. UMD (universal mutation database): a generic software to build and analyze locus‐specific databases. Hum Mutat 2000; 15: 86–94. [DOI] [PubMed] [Google Scholar]
- 5. Beroud C, Soussi T. The UMD‐p53 database: new mutations and analysis tools. Hum Mutat 2003; 21: 176–81. [DOI] [PubMed] [Google Scholar]
- 6. Forbes S, Clements J, Dawson E et al. COSMIC 2005. Br J Cancer 2006; 94: 318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huret JL, Senon S, Bernheim A, Dessen P. An atlas on genes and chromosomes in oncology and haematology. Cell Mol Biol 2004; 50: 805–7. [PubMed] [Google Scholar]
- 8. Strausberg RL. The cancer genome anatomy project: new resources for reading the molecular signatures of cancer. J Pathol 2001; 195: 31–40. [DOI] [PubMed] [Google Scholar]
- 9. Steffen DL, Levine AE, Yarus S, Baasiri RA, Wheeler DA. Digital reviews in molecular biology: approaches to structured digital publication. Bioinformatics 2000; 16: 639–49. [DOI] [PubMed] [Google Scholar]
- 10. Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2002; 30: 52–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stenson PD, Ball EV, Mort M et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 2003; 21: 577–81. [DOI] [PubMed] [Google Scholar]
- 12. Minoshima S, Mitsuyama S, Ohno S, Kawamura T, Shimizu N. Keio Mutation Database (KMDB) for human disease gene mutations. Nucleic Acids Res 2000; 28: 364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Minoshima S, Mitsuyama S, Ohtsubo M et al. KMDB/MutationView: a mutation database for human disease genes. Nucleic Acids Res 2001; 29: 327–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gnarra JR, Duan DR, Weng Y et al. Molecular cloning of the von Hippel‐Lindau tumor suppressor gene and its role in renal carcinoma. Biochim Biophys Acta 1996; 1242: 201–10. [DOI] [PubMed] [Google Scholar]
- 15. Valiaho J, Riikonen P, Vihinen M. Novel immunodeficiency data servers. Immunol Rev 2000; 178: 177–85. [DOI] [PubMed] [Google Scholar]
- 16. Albrecht P, Ansperger‐Rescher B, Schuler A, Zeschnigk M, Gallie B, Lohmann DR. Spectrum of gross deletions and insertions in the RB1 gene in patients with retinoblastoma and association with phenotypic expression. Hum Mutat 2005; 26: 437–45. [DOI] [PubMed] [Google Scholar]
- 17. Sedlacek Z, Kodet R, Poustka A, Goetz P. A database of germline p53 mutations in cancer‐prone families. Nucleic Acids Res 1998; 26: 214–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cariello NF, Douglas GR, Soussi T. Databases and software for the analysis of mutations in the human p53 gene, the human hprt gene and the lacZ gene in transgenic rodents. Nucleic Acids Res 1996; 24: 119–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fokkema IFAC, Den Dunnen JT, Taschner PEM. LOVD: easy creation of a locus‐specific sequence variation database using an ‘LSDB‐in‐a‐Box’ approach. Hum Mutat 2005; 26: 63–8. [DOI] [PubMed] [Google Scholar]
- 20. Kato K, Yamashita R, Matoba R et al. Cancer gene expression database (CGED): a database for gene expression profiling with accompanying clinical information of human cancer tissues. Nucleic Acids Res 2005; 33: D533–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cuticchia AJ. Future vision of the GDB human genome database. Hum Mutat 2000; 15: 62–7. [DOI] [PubMed] [Google Scholar]
- 22. Frezal J. Genatlas database, genes and development defects. C R Acad Sci III 1998; 321: 805–17. [DOI] [PubMed] [Google Scholar]
- 23. Safran M, Solomon I, Shmueli O et al. GeneCards 2002: towards a complete, object‐oriented, human gene compendium. Bioinformatics 2002; 18: 1542–3. [DOI] [PubMed] [Google Scholar]
- 24. Fredman D, Munns G, Rios D et al. HGVbase: a curated resource describing human DNA variation and phenotype relationships. Nucleic Acids Res 2004; 32: D516–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirakawa M. HOWDY: an integrated database system for human genome research. Nucleic Acids Res 2002; 30: 152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christoffels A, Van Gelder A, Greyling G, Miller R, Hide T, Hide W. STACK: Sequence tag alignment and consensus knowledgebase. Nucleic Acids Res 2001; 29: 234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


