Abstract
Prostate cancer is usually androgen‐dependent and responds well to androgen ablation therapy based on castration. However, at a certain stage some prostate cancers eventually acquire a castration‐resistant phenotype where they progress aggressively and show very poor response to any anticancer therapies. To characterize the molecular features of these clinical castration‐resistant prostate cancers, we previously analyzed gene expression profiles by genome‐wide cDNA microarrays combined with microdissection and found dozens of trans‐activated genes in clinical castration‐resistant prostate cancers. Among them, we report the identification of a new biomarker, stanniocalcin 2, as an overexpressed gene in castration‐resistant prostate cancer cells. Real‐time polymerase chain reaction and immunohistochemical analysis confirmed overexpression of stanniocalcin 2, a 302‐amino‐acid glycoprotein hormone, specifically in castration‐resistant prostate cancer cells and aggressive castration‐naïve prostate cancers with high Gleason scores (8–10). The gene was not expressed in normal prostate, nor in most indolent castration‐naïve prostate cancers. Knockdown of stanniocalcin 2 expression by short interfering RNA in a prostate cancer cell line resulted in drastic attenuation of prostate cancer cell growth. Concordantly, stanniocalcin 2 overexpression in a prostate cancer cell line promoted prostate cancer cell growth, indicating its oncogenic property. These findings suggest that stanniocalcin 2 could be involved in aggressive phenotyping of prostate cancers, including castration‐resistant prostate cancers, and that it should be a potential molecular target for development of new therapeutics and a diagnostic biomarker for aggressive prostate cancers. (Cancer Sci 2009; 100: 914–919)
Prostate cancer (PC) is the most common malignancy in males and the second‐leading cause of cancer‐related death in United States and Europe.( 1 ) The incidence of PC has been increasing significantly in most developed countries, probably due to the prevalence of western lifestyle and the explosion of the aging population.( 1 , 2 ) Surgical and radiation therapies are effective to the localized disease, but nearly 30% of treated PC patients still suffer from relapse.( 3 , 4 , 5 ) Most of the patients with relapsed or advanced disease respond well to androgen‐ablation therapy (medical or surgical castration) because PCs are usually androgen‐dependent at a relatively early stage. However, they often acquire a castration‐resistant phenotype where they progress aggressively and finally kill PC patients. Development of new therapies on the basis of the molecular mechanisms of prostate carcinogenesis or castration‐resistant prostate cancer (CRPC) is urgently required.
We previously carried out a genome‐wide expression profile analysis of CRPCs and castration‐naïve prostate cancers (CNPCs) using cDNA microarray in combination with microdissection to enrich populations of cancer cells.( 6 ) Among dozens of genes being trans‐activated commonly in CRPC cells, we focus here on stanniocalcin 2 (STC2), an overexpressing gene in CRPC cells. STC was first identified as a glycoprotein hormone secreted from specific endocrine glands (corpuscle of Stannius) in the kidney region of bony fish that is involved in calcium and phosphate homeostasis.( 7 , 8 ) STC is released into the blood in response to rising serum calcium levels to regulate the Ca2+ and phosphate uptake in different target organs.( 7 , 8 ) In mammals that lack a specific corpuscle of Stannius gland, two related mammalian genes have been identified, STC1 and STC2, both of which are predicted to be secreted glycosylated proteins.( 9 , 10 ) However, their physiological and pathological functions in human beings and human cancers have not been clearly elucidated.
In this study, we validated STC2 overexpression in CRPCs and highly aggressive PCs with high Gleason scores (GS). We also showed its positive involvement in proliferation or viability of PC cells. Our data could provide new insights into the molecular mechanisms of PC progression and some clues to developing new therapeutic strategies or diagnostic biomarkers against aggressive PCs and CRPCs.
Materials and Methods
Cell lines. Human PC cell lines LNCaP, 22Rv1, DU‐145, and PC‐3 were obtained from American Type Culture Collection (Rockville, MD, USA). LNCaP‐derived CRPC cell line C4‐2B was purchased from ViroMed Laboratories (Minnetonka, MN, USA). All of the cell lines were cultured as monolayers in the following medium: DMEM (Dulbecco's Modified Eagle Medium. Sigma‐Aldrich, St. Louis, MO) for 22Rv1, LNCaP, C4–2B and DU‐145; and F‐12 (Gibco, Carlsbad, CA) for PC‐3 with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (Sigma‐Aldrich). Cells were maintained in incubators containing humidified air with 5% CO2 at 37°C.
Semiquantitative reverse transcription–polymerase chain reaction (RT‐PCR). Purification of PC cells and normal prostatic epithelial cells from frozen PC tissues was described previously.( 6 ) Total RNA was extracted using RNeasy Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, treated with DNase Ι (Roche Diagnostics, Mannheim, Germany), and reversely transcribed to single‐stranded cDNA using random hexamer or oligo d(T)12–18 primer with Superscript reverse transcriptase ΙΙ (Invitrogen, Carlsbad, CA, USA). We prepared appropriate dilutions of each single‐stranded cDNA followed by normalizing cDNA content using β‐actin (ACTB) as a quantitative control, demonstrating the PCR reaction using single‐stranded cDNA as PCR templates. Primer sequences were: ACTB (forward, 5′‐TTGGCTTGACTCAGGATTTA‐3′; reverse, 5′‐ATGCTATCACCTCCCCTGTG‐3′), and STC2 (forward, 5′‐TTACTCCATGAGCCTTCCTTTG‐3′; reverse, 5′‐ TCCTGTTGCCTAAATCCGTAGTA‐3′). The conditions for PCR were initial denaturation at 95°C for 5 min, 23 cycles for ACTB and 30 cycles for STC2 of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s on a GeneAmp PCR system 9700 (Applied Biosystems, Foster, CA, USA). We also carried out quantitative real‐time PCR using a a Prism 7700 sequence detector with the SYBR Premix ExTaq (Takara, Kyoto, Japan) in accordance with the manufacturer's instructions. The primers for real‐time quantitative PCR were the same as those described above.
Northern blot analysis. We extracted total RNA from five PC cell lines using the RNeasy Kit (Qiagen), and their mRNAs were purified using an mRNA Purification Kit (GE Healthcare Bioscience, Piscataway, NJ, USA), according to the manufacturer's protocols. A 1 µg aliquot of each mRNA from the PC cell lines, as well as those isolated from normal human brain, heart, kidney, liver, lung, prostate, and testis (BD Biosciences, Palo Alto, CA, USA), were separated on 1% denaturing agarose gels and transferred onto nylon membranes. The membranes were hybridized for 20 h with 32P‐labeled STC2 cDNA that was labeled by Megaprime DNA labeling system (GE Healthcare Bioscience). Probe cDNA of STC2 was prepared as a 551‐bp PCR product using the following primers: forward, 5′‐AACATTTACCATTAGAGAGGGGG‐3′; reverse 5′‐CACATAGAAATGACACTCCTCCC‐3′. Prehybridization, hybridization, and washing were carried out according to the manufacturer's instructions. The blots were autoradiographed at –80°C for 7 days.
Generating antibody to STC2 and immunohistochemical analysis. Monoclonal mouse antibody to STC2 was generated by immunizing recombinant STC2 proteins at Medical and Biological Laboratories (Nagoya, Japan). CNPC tissues were obtained from the patients who underwent prostectomy in Kochi University Medical School (Nankoku, Japan). CRPC tissues were obtained from Kochi University Medical School, Iwate Medical College (Morioka, Japan), and Okayama University Medical School (Okayama, Japan) with appropriate informed consent. Immunohistochemical studies were carried out using the Ventana automated immunohistochemical systems (Discovery; Ventana Medical Systems, Tucson, AZ, USA). Sections were incubated with a 1:100 diluted solution of purified anti‐STC2 monoclonal antibody (0.75 mg/mL) for 16 min. The automated protocol is based on an indirect biotin–avidin system using a biotinylated universal secondary antibody and diaminobenzidine substrate with hematoxylin counterstaining. The specificity of the binding was confirmed by negative staining using mouse non‐immune serum as a primary antibody.
Scoring of immunohistochemical staining. To evaluate both the intensity of staining and proportion of positive‐stained cells, we used a scoring method reported previously.( 11 ) Positive staining of anti‐STC2 antibody was defined as follows: score 1, variable weak cytoplasmic staining; score 2, segmental and apical granular cytoplasmic staining; and score 3, diffuse continuous and intense cytoplasmic staining. For each score, the proportion of cells with the score was estimated visually. A combined weighted score (STC2 IHC score) consisting of the sum of the proportion of cells with each score was calculated for each sample as described previously.( 11 ) For example, a case with 70% score 3 staining, 20% score 2 staining, and 10% score 1 staining would be scored as follows: 70 × 3 + 20 × 2 + 10 × 1 = 260. The maximum score should be 300. All sections were scored, blinded and independently, by a clinical pathologist (M.F.). Comparisons of STC2 expression in three groups (GS 2–6, 7, and 8–10) were analyzed using the Kruskal–Wallis test for multiple comparisons. P < 0.01 was considered significant. Additional post‐testing was carried out using Mann–Whitney U‐test with the Bonferroni method to adjust for multiple pair‐wise differences. P < 0.003 between three groups was considered significant with the Bonferroni method. All statistical calculations were done with Statview software (version 5.0; SAS Institute, Cary, NC, USA).
Construction of short hairpin RNA (shRNA)‐expressing vectors and cell viability assay. To investigate the biological function of STC2 in PC cells, we used psiU6BX3.0 vector for expression of shRNA against a target gene as described previously.( 6 ) Plasmids designed to express shRNA were prepared by cloning of double‐stranded oligonucleotides into psiU6BX vector. The oligonucleotide sequences of target sequences for STC2 are: sense strand sequence for si1, 5′‐CAACTCTTGTGAGATTCGG‐3′; si2, 5′‐GACGAACAGTCTGAGTATT‐3′; si3, 5′‐GCAGGAGCTGGTATTGTAG‐3′; and siEGFP, 5′‐GAAGCAGCACGACTTCTTC‐3′ as a negative control. PC‐3 cells (2 × 106), which expressed STC2 at a high level, were seeded on 10‐cm dishes, transfected with psiU6‐STC2 (si1‐3), or psiU6‐siEGFP using FuGene6 (Roche Diagnostics) according to the manufacturer's instructions, then cultured in appropriate medium containing 800 µg/mL of Geneticin (Sigma‐Aldrich) for 14 days. The cells were fixed with 100% methanol, stained with 0.1% of crystal violet‐H20 for colony formation assay. In 3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide (MTT) assay, cell viability was measured using Cell Counting Kit‐8 (Dojindo Laboratories, Kumamoto, Japan) at 10 days after transfection. Absorbance was measured at 490 nm, and at 630 nm as a reference, with a Microplate Reader 550 (Bio‐Rad, Hercules, CA). Preliminarily, knockdown effects of these shRNA‐expression vectors on endogenous STC2 expression were validated 7 days after transfection by RT‐PCR using the primers for semiquantitative RT‐PCR.
Generation of STC2‐overexpressing cells and in‐vitro growth assay. Full‐length human STC2 cDNA was amplified using primers that were designed to contain HA‐tag sequences at the COOH terminus, and cloned into the pIRESneo3 vector (Clontech, Mountain View, CA). Human PC cell line 22Rv1 cells were seeded into a 100‐mm dish (5 × 105 cells per a dish) and transfected with 6 µg of pIRESneo3 empty vector or pIRESneo3‐STC2‐HA expression vector using FuGENE6 reagent (Roche Diagnostics) according to the manufacturer's instructions. Cells were selected with appropriate medium containing 400 µg/mL Geneticin (Sigma‐Aldrich) for 14 days when discrete colonies were isolated. All clones were maintained in selective medium. Each clone was assayed for STC2 protein expression by Western blot analysis using anti‐HA tag antibody (Roche Diagnostics). Proliferation of 22Rv1 cells that stably expressed STC2 (22Rv1‐STC2) or those transfected with pIRESneo3 empty vector (22Rv1‐mock clone mixture) were examined by the Cell Counting Kit‐8 (Dojindo Laboratories). Both 22Rv1‐STC2 and 22Rv1‐mock cells were seeded at the concentration of 3 × 103 cells per well using 48‐well plates. The assay was carried out every 48 h for 9 days, according to the manufacturer's instructions.
Results
STC2 overexpression in CRPC cells. We previously reported the genome‐wide expression profiles of CRPC cells and CNPC cells purified from clinical PC tissues.( 6 ) Of a number of trans‐activated genes in CRPC cells compared with normal prostatic epithelial cells, we focused on STC2 in this study. Semiquantitative RT‐PCR (Fig. 1a) and real‐time quantitative RT‐PCR (Fig. 1b) confirmed the elevated expression of STC2 in 6 out of 7 clinical CRPC cells, compared with CNPC cells and normal prostatic epithelial cells. Northern blot analysis using five PC cell lines and normal adult tissues confirmed the elevated expression of STC2 in all PC cell lines, compared with normal prostate and adult vital organs including brain, heart, kidney, liver, and lung (Fig. 1c). Multiple tissue northern blot analysis also revealed no or very limited expression of STC2 in normal adult organs, except for the pancreas (Fig. 1d).
Figure 1.
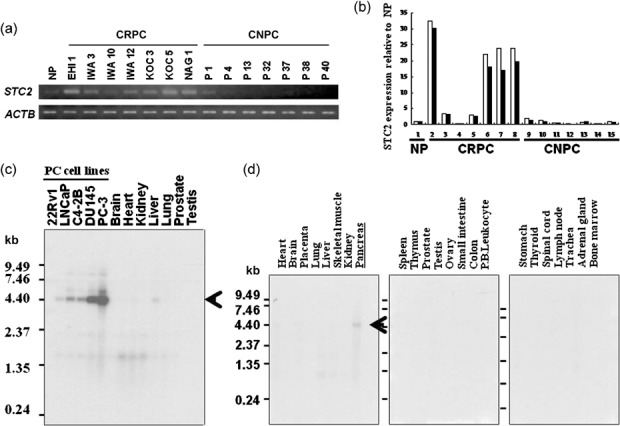
Stanniocalcin 2 (STC2) overexpression in castration‐resistant prostate cancer (CRPC) cells. (a) Semiquantitative reverse transcription–polymerase chain reaction (RT‐PCR) validated that STC2 was overexpressed in the microdissected CRPC cells, compared with castration‐naïve prostate cancer (CNPC) cells and normal prostatic epithelial (NP) cells, which were also microdissected. β‐actin (ACTB) was used to quantify the cDNA contents. (b) Real‐time quantitative RT‐PCR showed overexpression of STC2 transcript in CRPC cells (samples 2–8) compared with that of CNPC cells (samples 9–15) and NP cells (sample 1). ACTB was used to quantify each of the cDNA contents, and the relative quantity (y axis) was calculated so that the expression in NP cells was one. Real‐time quantitative RT‐PCR was duplicated for each sample (white and black columns). (c) Northern blot analysis showed a high level of STC2 expression in five prostate cancer cell lines (lanes 1–5), whereas its expression was hardly detectable in adult normal organs, including brain, heart, kidney, liver, lung, prostate, and testis (lanes 6–12). (d) Multiple tissue northern blot analysis indicated that STC2 was expressed only in normal pancreas, with no or very low expression in adult normal organs. The length of the STC2 transcript was approximately 5.4 kb. P.B.Leukocyte, peripheral blood leukocyte.
Immunohistochemical analysis of STC2 in clinical PCs and correlation with GS. To validate the overexpression of STC2 protein in CRPC cells, we carried out immunohistochemical analysis on clinical PC tissues using a monoclonal antibody specific to human STC2. As shown in Figure 2a, a strong immunochemical signal for STC2 was detected predominantly in the cytoplasm of cancer cells, especially in CRPC cases. Six of nine CRPCs we examined showed strong immunoreactivity to anti‐STC2 antibody. One CNPC with GS 10 (Fig. 2b) and five CNPCs with GS 9 also showed strong immunoreactivity, whereas most CNPCs with GS 7 showed no or very weak immunoreactivity to anti‐STC2 antibody (Fig. 2c). Adjacent normal prostatic epithelium in the same patient revealed very weak or no signal for STC2. Hormone ablation therapy is usually ineffective for PCs with high GS, which progress aggressively and are lethal, and these findings showed that STC2 was expressed specifically in CRPCs and aggressive PCs.
Figure 2.
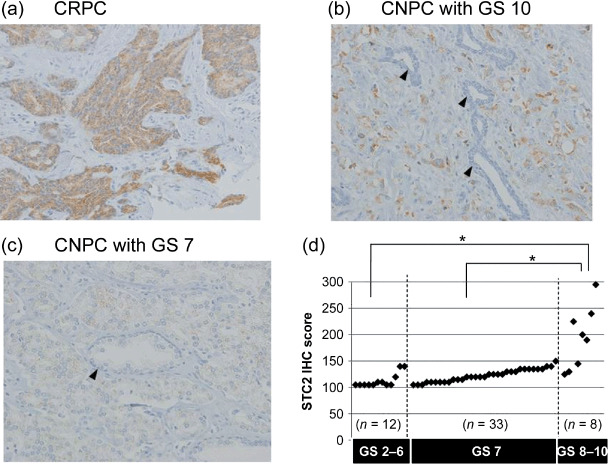
Immunohistochemical analysis of stanniocalcin 2 (STC2) in prostate cancer tissues. Immunoreactivity with anti‐STC2 antibody was observed in castration‐resistant prostate cancers (CRPCs) and castration‐naïve prostate cancers (CNPCs) with high Gleason scores (GS), exhibiting strong positive immunostaining in the cytoplasm of prostate cancer cells. (a) The representative picture of CRPC (×200); (b) the representative picture of CNPC with GS 10 (×200); (c) the representative picture of CNPC with GS 7 (×200). Adjacent normal prostatic epithelium revealed very weak or no signal for STC2 ([b,c] arrowheads). (d) Correlation between STC2 immunohistochemical (IHC) score and GS in CNPCs. There were significant differences in STC2 IHC scores between the GS 2–6 group (n = 12) and the GS 8–10 group (n = 8), and between the GS 7 (n = 33) and GS 8–10 groups (Mann–Whitney U‐test with Bonferroni method; P = 0.0006 and P = 0.0004, respectively).
To further investigate the clinic–pathologic significance of STC2 expression in PC tissues, we analyzed the relationship between the calculated immunohistochemical score for STC2 and GS in 53 CNPC tissues with various GS. As each PC specimen apparently showed a different degree of staining intensity and different proportion of positively stained cells, indicating a heterogeneous pattern as well as the GS system, we took this heterogeneity into consideration and employed the immunohistochemical scoring system. A combined weighted score was given by the sum of the proportion (0–100%) of stained cells for which the score 1, 2, or 3 was given, according to the signal STC2 immunohistochemical scores and intensity, as described previously.( 11 ) Compared to STC2 expression, there were significant differences between the GS 2–6 group (n = 12) and the GS 8–10 group (n = 8), and between the GS 7 group (n = 33) and the GS 8–10 group (Mann–Whitney U‐test with Bonferroni method; P = 0.0006 and P = 0.0004, respectively). No significant difference was observed between the GS 2–6 and GS 7 groups (P = 0.0110). We confirmed that the STC2 immunohistochemical score revealed a strong correlation with high GS (Fig. 2d).
Knockdown of STC2 expression by shRNA‐attenuated PC cell growth. To examine the biological roles of STC2 overexpression in PC cells, we constructed three vectors designed to express shRNA specifically to STC2, and transfected them into the PC‐3 cell line, which expressed endogenous STC2 at a high level. Among the three shRNA‐expression vectors, si2 and si3 showed significant knockdown effects on endogenous STC2 transcripts (Fig. 3a), and this transfection resulted in reduction in the numbers of colonies (Fig. 3b), as well as those of the viable cells measured by MTT assay for PC‐3 cells (Fig. 3c). In contrast, the transfection of si1 and a negative control (siEGFP) showed little or no knockdown effect on STC2 expression and did not affect the viability of PC‐3 cells. These findings indicated that STC2 overexpression could play important roles in PC cell growth or viability.
Figure 3.
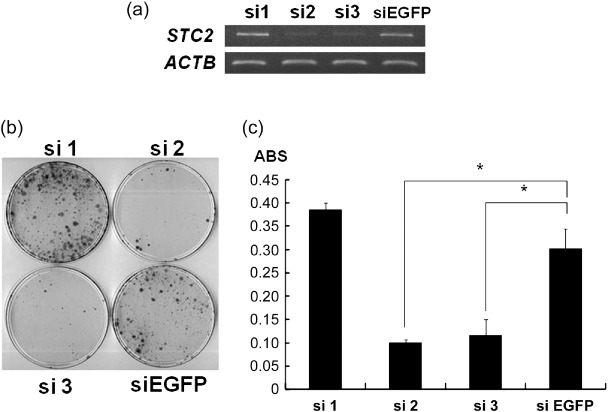
Knockdown of stanniocalcin 2 (STC2) expression by shRNA attenuated prostate cancer (PC) cell viability. (a) Knockdown effect of siRNA on STC2 in PC‐3 cells. Semiquantitative reverse transcription–polymerase chain reaction was carried out using cells transfected with each shRNA‐expressing vector to STC2 (si1‐3) as well as a negative control vector (siEGFP). β‐actin (ACTB) was used to quantify RNAs. (b) Colony formation assay of PC‐3 cells transfected with each of the indicated shRNA‐expressing vectors to STC2 (si1–3) and a negative control vector (siEGFP). Cells were visualized with 0.1% crystal violet staining after 14 days of incubation with Geneticin. (c) 3‐[4,5‐Dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide (MTT) assay of PC‐3 cells transfected with indicated shRNA‐expressing vectors to STC2 (si1–3) and a negative control vector (siEGFP). Each average is plotted with error bars indicating standard deviation after 10 days of incubation with Geneticin. These experiments were carried out in triplicate (Student's t‐test; *P < 0.01). ABS, absorbance at 490 nm, and at 630 nm as a reference, measured with a microplate reader.
STC2 overexpression promoted cancer cell growth. To further investigate the potential oncogenic function of STC2, we established three stable transformants (Clones 1–3) from 22Rv1 cells, in which exogenous STC2 expressed constitutively. We also prepared a control 22Rv1 cells transfected empty vector (Mock) and compared their proliferation. Western blot analysis (Fig. 4a) confirmed high levels of exogenous STC2 expression in three stable clones (Clones 1–3). MTT assay showed that the three STC2‐overexpressing clones grew more rapidly than the 22Rv1‐mock clone mixture (Student's t‐test; *P < 0.01, **P < 0.05), indicating that STC2 overexpression promoted PC cell growth (Fig. 4c).
Figure 4.
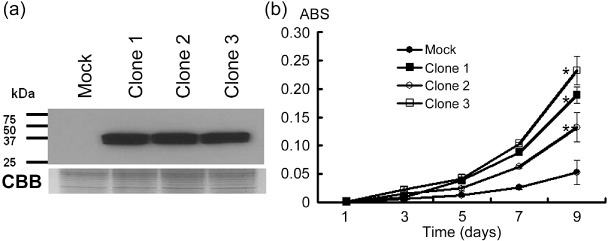
Stanniocalcin 2 (STC2) overexpression promoted cancer cell growth. (a) Western blot analysis with anti‐HA‐tag antibody validated constitutive expression of exogenous STC2 in three stable transformants (Clones 1–3). Mock was the 22Rv1‐mock clone mixture. Coomassie Brilliant Blue (CBB) stain served as a loading control. (b) In vitro growth curve by 3‐[4,5‐dimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide assay showed that three stable transformants (Clones 1–3) grew more rapidly than the 22Rv1‐mock clone mixture (Student's t‐test; *P < 0.01, **P < 0.05). ABS, absorbance at 490 nm, and at 630 nm as a reference, measured with a microplate reader.
Discussion
This study identified STC2 as a novel target molecule for therapy development or a biomarker of aggressive PCs and CRPCs. PC shows relatively good prognosis and hormone‐ablation therapy or castration is usually effective in most relapsed or advanced PCs. However, once CRPC cells emerge or PCs are at an advanced stage with a high GS, there are very limited treatment options for these PC patients, such as docetaxel plus predonisone,( 12 , 13 ) which can still offer the minimum effect on PCs. Hence, it is vital to identify molecular targets for CRPCs or aggressive PCs and develop novel therapies to target those molecules. Their expression pattern in normal adult organs is one of the most important factors in determining molecular targets for therapy, and as our northern blot data and a previous report suggested, STC2 was expressed in the α cells in the islet of the pancreas.( 14 ) As shown here, due to its restrictive expression in adult normal organs and its critical roles in PC cell viability, STC2 could be a promising target for a novel therapeutic approach with a minimal risk of adverse effects. Furthermore, STC2 was established as a secreted protein by a previous study( 10 ) and our enzyme‐linked immunosorbent assay study using the culture media of cancer cell lines (data not shown). STC2 could function to promote PC cell viability in an autocrine/paracrine manner. It is possible that STC2 could be detected in the patient serum as a diagnostic biomarker predicting PC aggressiveness, and neutralization of STC2 by highly specific antibodies might have potential as one of the therapeutic strategies against CRPCs and aggressive PCs.
Our immunohistochemical study clearly indicated STC2 overexpression not only in CRPCs, but also in high‐grade CNPC with GS 8–10. High‐grade CNPCs, similar to CRPCs, respond poorly to androgen‐ablation therapy and show highly aggressive behavior and poor prognosis. The GS system is based on tumor microarchitecture and, although GS 8–10 cancer is described as poorly differentiated, the molecular and cellular characteristics of its phenotype are not well known. A recent study by True et al.( 15 ) provided new insight into molecular characteristics that distinguish high Gleason grade PCs from well‐differentiated low Gleason grade PCs. They and Peehl et al.( 16 ) showed a significant association between high levels of monoamine oxidase A expression and high‐grade PCs. High‐grade PC had some similar molecular profiles to CRPCs, which might reflect dedifferentiation and explain the clinical association of grade with prognosis.( 17 ) Our findings regarding STC2 expression in CRPCs and high‐grade CNPCs suggest STC2 to be a response marker of androgen‐ablation therapy and a prognostic marker.
Recent reports suggested that HIF‐1 could possibly regulate STC2 expression( 18 ) and some proportions of renal cell cancers (RCCs) overexpressed STC2, which was correlated with their aggressiveness and poor prognosis.( 19 ) We also confirmed that STC2 overexpression in clinical RCC cells by our microarray data( 20 ) and RT‐PCR, and our shRNA experiment showed that STC2 was also essential to RCC cell viability or cell growth (data not shown). Another in vitro study implied that STC2 could have potential to protect the cells from various cell stresses, especially endoplasmic reticulum stress.( 21 ) HIF‐1, the endoplasmic reticulum stress pathway, or other cell stress pathways might induce STC2 overexpression in CRPC cells, and STC2 overexpression in PC cells is likely to provide their ability to survive under castration, ischemia, and other various cell stresses. However, the detailed mechanism of its growth‐promoting or survival effect on PCs and the mechanism of STC2 overexpression in aggressive PCs are unknown and should be defined by further investigations.
In summary, the autocrine/paracrine signaling pathway by STC2 can be essential in the cell viability of CRPCs and aggressive PCs, although the detailed pathways and binding receptors involved remain unknown. The detection of STC2 and its inhibition could provide us with a novel promising approach for a biomarker or a molecular treatment of CRPCs and aggressive PCs.
Acknowledgments
We would like to thank Ms. Hitomi Uchida for her technical assistance. This work was supported in part by Grant‐in‐Aid for Scientific Research #18590323 (H. Nakagawa) and Research for the Future Program Grants #00L01402 (Y. Nakamura) from the Japan Society for the Promotion of Science.
References
- 1. Gronberg H. Prostate cancer epidemiology. Lancet 2003; 361: 859–64. [DOI] [PubMed] [Google Scholar]
- 2. Hsing AW, Devesa SS. Trends and patterns of prostate cancer: what do they suggest? Epidemiol Rev 2001; 23: 3–13. [DOI] [PubMed] [Google Scholar]
- 3. Feldman BJ, Feldman D. The development of androgen‐independent prostate cancer. Nat Rev Cancer 2001; 1: 34–45. [DOI] [PubMed] [Google Scholar]
- 4. Scher HI, Sawyers CL. Biology of progressive, castration‐resistant prostate cancer: directed therapies targeting the androgen‐receptor signaling axis. J Clin Oncol 2006; 23: 8253–61. [DOI] [PubMed] [Google Scholar]
- 5. Han M, Partin AW, Piantadosi S, Epstein JI, Walsh PC. Era specific biochemical recurrence‐free survival following radical prostatectomy for clinically localized prostate cancer. J Urol 2001; 166: 416–19. [PubMed] [Google Scholar]
- 6. Tamura K, Furihata M, Tsunoda T et al . Molecular features of hormone‐refractory prostate cancer cells by genome‐wide gene expression profiles. Cancer Res 2007; 67: 5117–25. [DOI] [PubMed] [Google Scholar]
- 7. Wagner GF, Milliken C, Friesen HG et al . Studies on the regulation and characterization of plasma stanniocalcin in rainbow trout. Mol Cell Endocrinol 1991; 79: 129–38. [DOI] [PubMed] [Google Scholar]
- 8. Wagner GF, Jaworski E. Calcium regulates stanniocalcin mRNA levels in primary cultured rainbow trout corpuscles of stannius. Mol Cell Endocrinol 1994; 99: 315–22. [DOI] [PubMed] [Google Scholar]
- 9. Chang AC, Janosi J, Hulsbeek M et al . A novel human cDNA highly homologous to the fish hormone stanniocalcin. Mol Cell Endocrinol 1995; 112: 241–7. [DOI] [PubMed] [Google Scholar]
- 10. Ishibashi K, Miyamoto K, Taketani Y et al . Molecular cloning of a second human stanniocalcin homologue (STC2). Biochem Biophys Res Commun 1998; 250: 252–8. [DOI] [PubMed] [Google Scholar]
- 11. Ashida S, Furihata M, Katagiri T et al . Expression of novel molecules, MICAL2‐PV (mical2 prostate cancer variants), increases with high Gleason score and prostate cancer progression. Clin Cancer Res 2006; 12: 2767–73. [DOI] [PubMed] [Google Scholar]
- 12. Tannock IF, de Wit R, Berry WR et al . Docetaxel plus predonisone or mitoxantrone plus predonisone for advanced prostate cancer. N Engl J Med 2004; 351: 1502–12. [DOI] [PubMed] [Google Scholar]
- 13. Petrylak DP, Tangen CM, Hussain MH et al . Docetaxel and estramustine compared with mitoxantrone and predonisone for advanced refractory prostate cancer. N Engl J Med 2004; 351: 1513–20. [DOI] [PubMed] [Google Scholar]
- 14. Moore EE, Keuetner RE, Conklin DC et al . Stanniocalcin 2: characterization of the protein and its localization to human pancreatic α cells. Horm Metab Res 1999; 31: 406–14. [DOI] [PubMed] [Google Scholar]
- 15. True L, Coleman I, Hawley S et al . A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci USA 2006; 103: 10991–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peehl DM, Coram M, Khine H et al . The significance of monoamine oxidase‐A expression in high grade prostate cancer. J Urol 2008; 180: 2206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tomlins SA, Mehra R, Rhodes DR et al . Integrative molecular concept modeling of prostate cancer. Nat Genet 2007; 39: 41–51. [DOI] [PubMed] [Google Scholar]
- 18. Law AY, Lai KP, Ip CK et al . Epigenetic and HIF‐1 regulation of stanniocalcin‐2 expression in human cancer cells. Exp Cell Res 2008; 314: 1823–30. [DOI] [PubMed] [Google Scholar]
- 19. Meyer HA, Tölle A, Jung M et al . Identification of stanniocalcin 2 as prognostic marker in renal cell carcinoma. Eur Urol 2008. published online 09 April 2008. [DOI] [PubMed]
- 20. Hirota E, Yan L, Tsunoda T et al . Genome‐wide gene expression profiles of clear cell renal cell carcinoma: identification of molecular targets for treatment of renal cell carcinoma. Int J Oncol; 29: 799–827. [PubMed] [Google Scholar]
- 21. Ito D, Walker JR, Thompson CS et al . Characterization of stanniocalcin 2, a novel target of the mammalian unfolded protein response with cytoprotective properties. Mol Cell Biol 2004; 24: 9456–69. [DOI] [PMC free article] [PubMed] [Google Scholar]


