Abstract
Human T‐cell lymphotropic virus type 1 is vertically transmitted in neonatal life and is causatively associated with adult T‐cell leukemia (ATL) and HTLV‐1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP) in adults. Persistence of HTLV‐1 in host T cells, clonal expansion of the HTLV‐1 carrying T cells, and emergence of malignantly transformed T cells are in accord with the multistep model of human cancer and roles for continuous interaction between host genes and environmental factors. This article reviews two lines of HTLV‐1 investigation, one regarding worldwide surveillance of HTLV‐1 infection foci by serological testing and molecular analysis of HTLV‐1 isolates, and the other focusing on genetics of the human leukocyte antigen (HLA) that determines the ethnic background of HTLV‐1 permissiveness and susceptibility to ATL or HAM/TSP. The serological surveillance revealed transcontinental dispersal of HTLV‐1 in the prehistoric era that started out of Africa, spread to Austro‐Melanesia and the Asian continent, then moved to North America and through to the southern edge of South America. This was highlighted by an Andean mummy study that proved ancient migration of paleo‐mongoloid HTLV‐1 from Asia to South America. Phylogenetic analysis of HLA alleles provided a basis for ethnic susceptibility to HTLV‐1 infection and associated diseases, both ATL and HAM/TSP. Ethnicity‐based sampling of peripheral blood lymphocytes has great potential for genome‐wide association studies to illuminate ethnically defined host factors for viral oncogenesis with reference to HTLV‐1 and other pathogenic elements causatively associated with chronic disease and malignancies. (Cancer Sci 2011; 102: 295–301)
Ethnoepidemiology of HTLV‐1 is a new paradigm in cancer research
Cancer is caused by chronic stimuli by chemical, physical, and biological agents( 1 , 2 , 3 , 4 ) which interact with the internal tissue milieu of the human body. Human T‐cell lymphotropic virus type 1( 5 ) is an oncornavirus that causes the T cell malignancy known as adult T cell leukemia (ATL) in a subgroup of HTLV‐1 infected people.( 6 , 7 , 8 , 9 ) It also gives rise to neurological, ophthalmological, and dermatological disorders called HTLV‐1‐associated myelopathy/tropical spastic paraparesis (HAM/TSP), HTLV‐1 uveitis, and infective dermatitis in other groups of HTLV‐1 infection.( 10 , 11 , 12 ) Here is an intriguing paradox in that one pathogen (HTLV‐1) causes two diseases (ATL and HAM/TSP), contradicting the classical theory of medical microbiology (Koch’s postulate of one pathogen, one disease). We thus hypothesized the presence of host factor(s) relevant to HTLV‐1 permissiveness and disease susceptibility that are determined by the genetic background of HTLV‐1 infected people of different ethnic groups.( 13 , 14 )
Molecular studies have revealed that 5′‐LTR genes of HTLV‐1 regulate encoding of Tax/Rex proteins that are responsible for HTLV‐1 transactivation,( 15 ) whereas 3′‐LTR genes encode HTLV‐1 basic leucine zipper (HBZ) proteins essential for maintenance of malignant features of HTLV‐1 infected T cells.( 16 , 17 , 18 ) However, it is yet unknown why most patients with ATL or HAM/TSP are late‐onset at adult age and only account for a very small minority of HTLV‐1 infected people, with an annual incidence <0.1%.( 19 ) It is likely that HTLV‐1 must remain a long time in the body of an infected person awaiting a rare event for malignant transformation of HTLV‐1 infected T‐cells to ATL, whereas other events in the central nervous system are responsible for the neuropathology of HAM/TSP.( 20 ) Persistence of HTLV‐1 in the host’s T cells, clonal expansion of HTLV‐1 positive T cells, and emergence of the malignantly transformed T cells are in accord with the multistep model of human cancer resulting from continuous interaction between environmental factors and host‐responsive elements, the former including various pathogens, and the latter varying with the genetic make‐up.( 21 ) Contemporary human beings form more than 60 000 ethnic groups that have developed over the last 200 000 years by surviving various physical, chemical, and biological impacts.( 22 )
This article reviews the global prevalence of HTLV‐1 and related diseases in the context of ethnicity‐based “ethnoepidemiology”, a new paradigm for cancer research to study host factor interaction with exogenous carcinogens.( 23 )
Ethnic clustering of HTLV‐1 infection and diseases
Human T‐cell lymphotropic virus type 1 is known to be vertically transmitted from mothers to infants and the virus is maintained within the infant’s family/ethnic group where maternal anti‐HTLV‐1 antibodies are known to inhibit milk‐borne infection of HTLV‐1 in early life.( 24 , 25 , 26 , 27 ) Serological surveillance of HTLV‐1 antibodies has enabled us to pinpoint HTLV‐1 infection foci in Japan and other countries in the world.( 23 ) Geographical clusters of HTLV‐1 include tribes of contemporary South Amerindians and the ancient Andeans (Fig. 1). Phylogenetic analysis revealed that South Amerindian and Andean mummy HTLV‐1 forms belong to a transcontinental subgroup that is related to Asian mongoloid HTLV‐1 (Fig. 2).( 28 , 29 )
Figure 1.
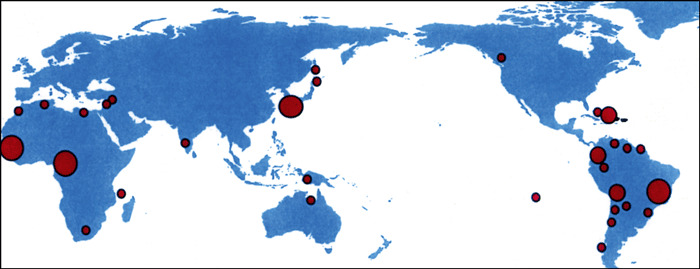
Worldwide distribution of HTLV‐1 carriers (modified from reference 14).
Figure 2.
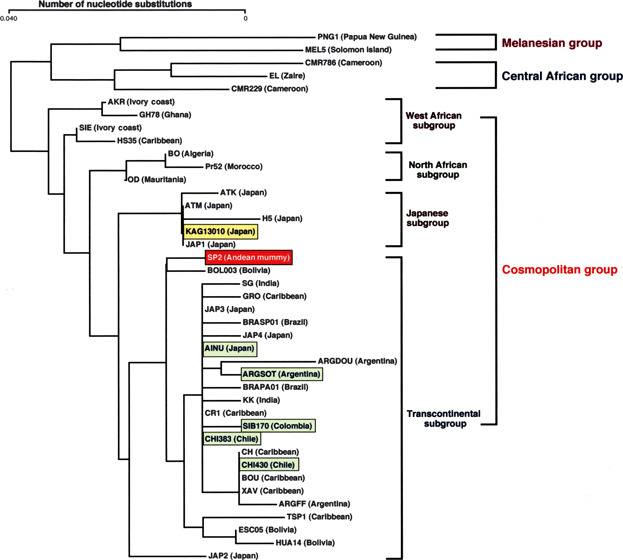
Phylogenetic relationship of HTLV‐1 isolates in the world (modified from references 28 and 29).
We obtained informed consent from all study participants to draw peripheral blood and to preserve lymphocytes and sera for laboratory analysis.( 30 ) We carried out the serological testing for HTLV‐1 manually in the field setting, often without any electricity, which was highly appreciated by the local people and the public health workers( 31 ). In fact, the sera were separated by hand‐driven centrifuge for HTLV‐1 agglutination testing, so that the villagers could be quickly informed of the HTLV‐1 positive or negative status within 60–90 min of blood collection. The screening test was followed by standard ELISA and Western blot analysis (World Health Organization criteria, 1990) for HTLV‐1 antibodies and further molecular detection of HTLV‐1 DNA.( 32 , 33 )
Ethnic factors impacting on susceptibility to ATL and HAM/TSP
It is known that HTLV‐1 foci contain certain numbers of patients with ATL or HAM/TSP segregated among families of Japanese HTLV‐1 carriers.( 34 , 35 ) Similar segregation of ATL and HAM/TSP was observed among Afro‐Caribbean and Afro‐Colombian patients whose clinical and hematological features were exactly the same as their Japanese counterparts.( 36 , 37 ) In Trinidad and Tobago, HAM/TSP was found to predominate in one group of Black people and ATL in another.( 38 ) In Zaire, HAM/TSP primarily affects a minor group of the Mundunga tribe.( 39 ) Thus, some ethnically defined factors are likely to be associated with HTLV‐1 persistence and development of ATL or HAM/TSP among HTLV‐1 endemic populations.
To approach ethnic factors involved in HTLV‐1 clustering and disease segregation of ATL and HAM/TSP, we investigated the genetic background of affected patients by analyzing polymorphic determinants of human leukocyte antigens (HLA) alleles and their immune responsiveness to HTLV‐1.( 14 , 40 ) We also identified HLA alleles elevating risk in Japanese patients,( 41 , 42 ) and confirmed their importance in Afro‐Caribbean( 43 , 44 , 45 ) and Iranian patients( 46 ) (Table 1). Phylogenic analysis revealed ATL‐associated HLA alleles (HLA‐A*26 and HLA‐A*36) to be segregated into two lineages within contemporary human populations whose ancestral genes come from the primate MHC, including gorilla, chimpanzee, and apes (Fig. 3).( 47 ) It is thus likely that HLA‐A*26 and HLA‐A*36 alleles might have evolved more than 50 million years before the present and that HLA holders became natural hosts of HTLV‐1 infection by virtue of a low immune responsiveness to the virus. This is a unique virus–host relationship clearly contrasting with conventional viruses, such as the Epstein–Barr virus, that usually survive host immune attacks by changing their genetic code (through mutation) and establishing a new virus–host relationship.( 48 ) It is likely that genetic polymorphisms of HLA alleles determine outcome regarding HTLV‐1 related disease in human hosts, one extreme being permissive to HTLV‐1 without any positive immune response due to low immune responder HLA alleles, and the other being non‐permissive to HTLV‐1 because of high immune responder HLA alleles that exacerbate inflammatory reactions in the host. The former predisposes to leukemogenesis and the latter to HAM/TSP immunopathology as described earlier.( 40 , 42 )
Table 1.
Distribution of susceptible level to adult T‐cell leukemia (ATL) or HTLV‐1‐asociated myelopathy/tropical spastic paraparesis (HAM/TSP) by human leukocyte antigen alleles
| ATL (ethnicity) | HAM/TSP (ethnicity) | |
|---|---|---|
| A*24 | + | +++ (Japanese) |
| A*26 | +++ (Japanese) | + |
| A*36 | +++ (Afro‐Caribbean) | + |
| B*07 | + | +++ (Japanese) |
| B*4002/6 | +++ (Japanese) | + |
| B*4801 | +++ (Japanese) | + |
| DRB1*0101 | + | +++ (Japanese, Iranian) (Brazilian, Afro‐Caribbean) |
| DRB1*0901 | +++ (Japanese) |
Figure 3.
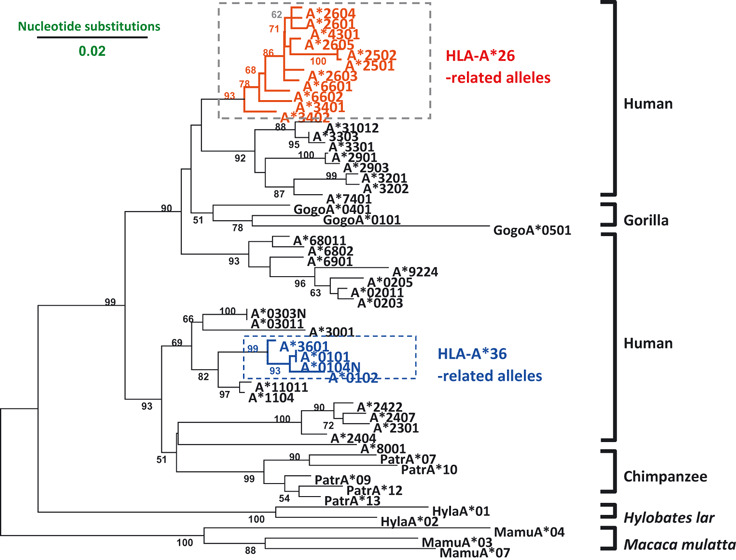
Phylogenetic tree of human leukocyte antigen A (HLA‐A) alleles (modified from references 42, 46, and 48).
Transcontinental dispersal of HTLV‐1 with human migration
Human T‐cell lymphotropic virus type 1 is transmissible from mother to infants through breast milk, from male to female through semen, and to blood recipients through HTLV‐1 carrier’s lymphocytes. All the transmission routes efficiently localize HTLV‐1 infection foci within particular family/ethnic groups.( 49 , 50 , 51 ) Efficacy of vertical transmission of HTLV‐1 was confirmed by recent studies of HLA concordance between mothers and infants, likely mediated by either immunological or virological synapse of HTLV‐1‐infected T cells.( 52 , 53 , 54 ) Thus, serological and virological surveillance allowed location of HTLV‐1 positive ethnic groups across the world, as depicted in Figure 1. By virtue of the genomic stability of HTLV‐1, we could trace the transcontinental dispersal by phylogenetic relationships of viral isolates from various ethnic groups.( 28 ) The early transmission of HTLV‐1 to South America was highlighted by the discovery of ancient HTLV‐1 DNA preserved within an Andean mummy.( 29 ) The mummy was buried 1500 years ago in the desert of Atacama, North Chile, and the body was kept intact in the dry climate with salty and alkaline soil. When bone marrow samples of mummy femur were processed to extract DNA by a glass meal method, followed by PCR amplification of HTLV‐1 DNA, nucleotide sequences of the mummy’s HTLV‐1 Tax and LTR DNA were very similar to HTLV‐1 isolated from the contemporary Atacama tribe and the nearest neighbor Chilean indigenous people. Of particular interest, they were closely related to HTLV‐1 of Ainu, southern Japanese, and some Asian mongoloid subgroups. This phylogenic relationship suggested that the Andean mummy’s HTLV‐1 might have come from an Asian paleo‐mongoloid, as mentioned above (Fig. 2).
The transcontinental migration of HTLV‐1 was supported by studies showing two lineages of HLA‐A alleles in association with HTLV‐1 endemic populations in the world, one being HLA‐A*26 and the other HLA‐A*36 and both originally evolving in African blacks and becoming dispersed to Melanesians, Israeli Jews, Asian Indians, southwestern Japanese, Sakharin natives, and North/South Amerindians, and carried by Carib/South American Black peoples (Fig. 4). It is likely that ancient HTLV‐1 passing peoples in Africa moved to the Asian continent, then passing the Bering Strait to North America and further to the southern edge of South America, as depicted in Figure 5. This transcontinental dispersal of HTLV‐1 partially overlapped with the migration pattern of South‐East Asian mongoloids.( 55 ) We speculate that Japanese HTLV‐1 carrying people are descended from the paleo‐mongoloids who reached the Japan archipelago before South‐East Asian mongoloids.
Figure 4.
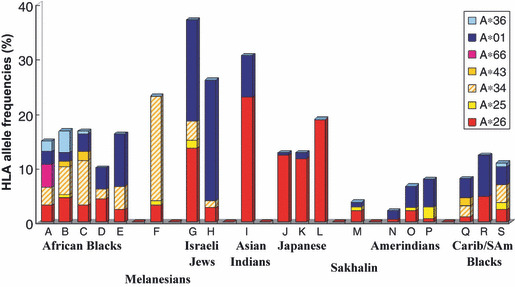
Distribution of human leukocyte antigen A (HLA‐A) alleles associated with worldwide HTLV‐1 carrier populations (modified from reference 40). Carib, Caribbean; SAm, South American.
Figure 5.
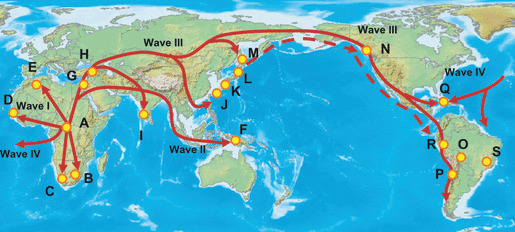
Transcontinental dispersal of HTLV‐1 out of Africa to South America. A–E, African Blacks; F, Melanesians; G, H, Israeli Jews; I, Asian Indians; J–L, Japanese; M, Sakhalin; N–P, Amerindians; Q–S, Caribbean/South American Blacks.
Ethnic segregation of HTLV‐1 and other human T‐cell lymphotropic viruses
Human T‐cell lymphotropic virus type 1 was first discovered as a human retrovirus cause of ATL.( 7 , 8 ) Human T‐ell lymphotropic virus type 2 was subsequently discovered among Amerindians of North, Central, and South America( 56 , 57 ) and Pygmies in central Africa.( 58 ) Recently, two further prototypic human retroviruses (HTLV‐3 and HTLV‐4) were found to be prevailing among bushmen living in the deep forest of Central Africa.( 59 , 60 ) We also identified a new focus of HTLV‐2 among South American natives of Orinoco, Colombia.( 61 ) In addition, HTLV‐1 positive Andes highlanders could be distinguished from HTLV‐1 positive Amazon lowlanders by segregating their HLA class II haplotypes.( 62 )
Human T‐cell lymphotropic virus type 2 associated disease remains to be confirmed, although a chronic neurodegenerative syndrome has been documented in association with HTLV‐2 infection.( 63 ) Virulence of HTLV‐2 appears be highly attenuated by natural adaptation to the human host, as with HTLV‐3 and HTLV‐4. It is thus hypothesized that the chronological order of virus emergence and length of coexistence with human hosts may correlate with morbidity due to human T cell lymphotropic viruses. Ethnoepidemiology of HTLV‐1, ‐2, ‐3, and ‐4 warrants further investigation to generate a better understanding of HTLV‐1‐associated T cell leukemia (ATL) and neurological disorder (HAM/TSP) with reference to interaction with ethnic factors in contemporary human hosts.
Beyond HTLV‐1 ethnoepidemiology
Ethnic clustering of HTLV‐1 infection foci was first noted in relation to the origin of Japanese HTLV‐1,( 64 ) which was followed by our worldwide study on the ethnic background of HTLV‐1 related diseases.( 14 , 29 ) Long‐lasting interaction between HTLV‐1 and the host’s responsive elements provides a typical model of human cancer. In the case of HTLV‐1‐associated T‐cell malignancy, monoclonal expansion of HTLV‐1‐infected T cells is causatively associated with ATL,( 65 , 66 ) whereas a polyclonal status permits an asymptomatic state and reduces risk for ATL, as typically shown in HAM/TSP patients.( 67 , 68 )
Neonatal infection with HTLV‐1 is preventable through short‐term breastfeeding for <3–6 months after birth and/or bottle feeding,( 26 , 27 ) as confirmed in an Afro‐Caribbean study.( 50 ) Recent surveys in Kyushu, Japan, have revealed a decreasing trend of HTLV‐1 positive blood donors, suggesting that controlled breastfeeding of infants born to HTLV‐1 carrier mothers and exclusion of HTLV‐1 contaminated blood has successfully inhibited HTLV‐1 transmission.( 20 )
Studies on chemoprevention of ATL have shown that interference with monoclonal outgrowth of ATL cells is possible using biological products such as dehydroxymethylepoxyquinomicin (DHMEQ)( 69 ) and fucoidin.( 70 ) We have studied green tea polyphenols that induce apoptosis in both ATL and HTLV‐1 infected T cells, eventually diminishing HTLV‐1 viral load in HTLV‐1 carriers’ peripheral blood lymphocytes.( 71 ) Thus, daily drinking of green tea and its products is a feasible approach for suppressing outgrowth of HTLV‐1 infected T cells and reducing risk of ATL and HAM/TSP.
Experimental therapeutics for ATL patients are now under investigation using hematopoietic stem cell transplantation( 72 ) and molecular targeting by CCR4 antibodies.( 73 ) Another therapeutic approach is to mark the HTLV‐1 Tax protein( 74 ) and the HBZ protein,( 75 ) both of which confer a strong CTL response. However, constitutive expression of Tax and HBZ antigenic epitopes on ATL cells is fairly limited to generating a positive CTL response,( 76 ) probably due to altered HLA phenotypes of ATL cells.( 77 )
Finally, we would like to stress accumulating evidence regarding functional diversity of NKG2D natural killer cells( 78 ) and pattern recognition receptors (PRRs) in the innate immune system.( 79 ) Both NKG2D and PRRs are thought to enforce the first line of host defense against microbes, so they must play an important role in HTLV‐1 infection and related diseases. However, it is unclear whether genes for NKG2D and PRRs are polymorphic in this regard. In order to address this important issue, ethnicity‐based collection of peripheral blood lymphocytes (the Sonoda–Tajima Collection of the Rikken Bio‐Resource Center) should provide a useful resource for genome‐wide association scans to illuminate ethnic variation of human genes and susceptibility to exogenous pathogens with reference to HTLV‐1 and other pathogenic elements causatively associated with chronic disease and malignancies.
Acknowledgments
We greatly thank Drs. V. Zaninovic, A. Blank, M. Blank (Colombia), L. Cartier, L.Ramirez, L Nunez (Chile), L. Hurtado, L.V. Hurtado, R. Andrade (Bolivia), B. Hanchard, O. Morgan, B. Cranston (Jamaica), A. Manns, W. Blattner, P. Levine, N. Mueller, M. Hisada, R. Biggar, J. Goedert, V. Franchini, S. Jacobson, W. Harrington Jr., J. Byrnes, M. Hojo (USA), G. de The, A. Gessain (France), B. Kitze (Germany), W. Hall (Ireland), K.Takahashi, Y.Nagata, K. Kawakami, T. Takezaki, N. Arima, K. Usuku, S. Yashiki, T. Fujiyoshi, L. Hong, M. Osame, A. Utsunomiya, S. Kashiwagi, T. Miura, M. Hayami, S. Horai, and M. Moore (Japan) for their kind collaboration in our field studies and laboratory work, as well as in preparation of this article. We highly appreciate Drs. I. Danjoh, Y. Nakamura, and Y. Obata, the Rikken Bio‐Resource Center of Japan for their expertise in preserving peripheral blood lymphocytes collected in our field studies and in establishing B cell lines for future studies on human genome diversity and genome‐wide scans of disease‐susceptible genes. The present investigation was supported by the Monbusho International Collaborative Study of Cancer Research and by Grants‐in‐Aid from the Ministry of Science, Education, Sports and Technology of Japan.
References
- 1. Sugimura T. Studies on environmental chemical carcinogenesis in Japan. Science 1986; 233(4761): 312–8. [DOI] [PubMed] [Google Scholar]
- 2. Doll R. Hazards of ionizing radiation: 100 years of observations on man. Br J Cancer 1995; 72(6): 1339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J Exp Med 1911; 13(4): 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1964; 1(7335): 702–3. [DOI] [PubMed] [Google Scholar]
- 5. Popovic M, Reitz MS Jr, Sarngadharan MG et al. The virus of Japanese adult T‐cell leukaemia is a member of the human T‐cell leukaemia virus group. Nature 1982; 300(5887): 63–6. [DOI] [PubMed] [Google Scholar]
- 6. Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T‐cell leukemia: clinical and hematologic features of 16 cases. Blood 1977; 50(3): 481–92. [PubMed] [Google Scholar]
- 7. Hinuma Y, Nagata K, Hanaoka M et al. Adult T‐cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci U S A 1981; 78(10): 6476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T‐cell leukemia and its implication in the disease. Proc Natl Acad Sci U S A 1982; 79(6): 2031–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poiesz BJ, Ruscetti FW, Gazdar AF et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T‐cell lymphoma. Proc Natl Acad Sci U S A 1980; 77(12): 7415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Román GC, Osame M. Identity of HTLV‐I‐associated tropical spastic paraparesis and HTLV‐I‐associated myelopathy. Lancet 1988; 1(8586): 651. [DOI] [PubMed] [Google Scholar]
- 11. Mochizuki M, Watanabe T, Yamaguchi K et al. HTLV‐I uveitis: a distinct clinical entity caused by HTLV‐I. Jpn J Cancer Res 1992; 83(3): 236–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LaGrenade L, Hanchard B, Fletcher V, Cranston B, Blattner W. Infective dermatitis of Jamaican children: a marker for HTLV‐I infection. Lancet 1990; 336(8727): 1345–7. [DOI] [PubMed] [Google Scholar]
- 13. Tajima K, Tominaga S, Shimizu H et al. A hypothesis on the etiology of adult T‐cell leukemia/lymphoma. Gann 1981; 72(5): 684–91. [PubMed] [Google Scholar]
- 14. Sonoda S, Fujiyoshi T, Yashiki S. Immunogenetics of HTLV‐I/II and associated diseases. J Acquir Immune Defic Syndr Hum Retrovirol 1996; 13 (Suppl 1): S119–23. [DOI] [PubMed] [Google Scholar]
- 15. Yoshida M, Inoue J, Fujisawa J, Seiki M. Molecular mechanisms of regulation of HTLV‐1 gene expression and its association with leukemogenesis. Genome 1989; 31(2): 662–7. [DOI] [PubMed] [Google Scholar]
- 16. Gaudray G, Gachon F, Basbous J et al. The complementary strand of the human T‐cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down‐regulates viral transcription. J Virol 2002; 76(24): 12813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Satou Y, Yasunaga J, Yoshida M et al. HTLV‐I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci U S A 2006; 103(3): 720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukumoto R, Andresen V, Bialuk I et al. In vivo genetic mutations define predominant functions of the human T‐cell leukemia/lymphoma virus p12I protein. Blood 2009; 113(16): 3726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tajima K, Kuroishi T. Estimation of rate of incidence of ATL among ATLV (HTLV‐I) carriers in Kyushu, Japan. Jpn J Clin Oncol 1985; 15(2): 423–30. [PubMed] [Google Scholar]
- 20. Kaplan JE, Osame M, Kubota H et al. The risk of development of HTLV‐I‐associated myelopathy/tropical spastic paraparesis among persons infected with HTLV‐I. J Acquir Immune Defic Syndr 1990; 3(11): 1096–101. [PubMed] [Google Scholar]
- 21. Bjorgen PJ. MHC restriction in three dimensions: a view of T cell receptor/ligand interactions. Cell 1997; 89: 167–70. [DOI] [PubMed] [Google Scholar]
- 22. Cavalli‐Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, New Jersey: Princeton University Press, 1994. [Google Scholar]
- 23. Tajima K, Sonoda S. Ethnoepidemiology, a new paradigm, for studying cancer risk factors and prevention strategy. In: Tajima K, Sonoda S, eds. Ethnoepidemiology of Cancer. Gann Monograph No.44. Tokyo: Japanese Scientific Societies Press, 1996; 3–12. [Google Scholar]
- 24. Kinoshita K, Hino S, Amagaski T et al. Demonstration of adult T‐cell leukemia virus antigen in milk from three sero‐positive mothers. Gann 1984; 75(2): 103–5. [PubMed] [Google Scholar]
- 25. Kusuhara K, Sonoda S et al. Mother‐to‐child transmission of T‐cell leukemia virus type I(HTLV‐I): fifteen‐year follow‐up study in Okinawa, Japan. Int J Cancer 1987; 40: 755–7. [DOI] [PubMed] [Google Scholar]
- 26. Takahashi K, Takezaki T, Oki T et al. Inhibitory effect of maternal antibody on mother‐to‐child transmission of human T‐lymphotropic virus type I. The mother‐to‐child transmission study group. Int J Cancer 1991; 49: 673–7. [DOI] [PubMed] [Google Scholar]
- 27. Takezaki T, Tajima K, Ito M et al. Short‐term breast‐feeding may reduce the risk of vertical transmission of HTLV‐I. The Tsushima ATL Study Group. Leukemia 1997; 11 (Suppl 3): 60–2. [PubMed] [Google Scholar]
- 28. Miura T, Fukunaga T, Igarashi T et al. Phylogenetic subtypes of human T‐lymphotropic virus type I and their relations to the anthropological background. Proc Natl Acad Sci U S A 1994; 91(3): 1124–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li HC, Fujiyoshi T, Lou H et al. The presence of ancient human T‐cell lymphotropic virus type I provirus DNA in an Andean mummy. Nat Med 1999; 5(12): 1428–32. [DOI] [PubMed] [Google Scholar]
- 30. Fujiyoshi T, Li HC, Lou H et al. Characteristic distribution of HTLV type I and HTLV type II carriers among native ethnic groups in South America. AIDS Res Hum Retroviruses 1999; 15(14): 1235–9. [DOI] [PubMed] [Google Scholar]
- 31. Hurtado L. A progress report of Bolivian field study: In Monbusho International Scientific Research Program. Special Cancer Research in 1994–1998, Overall Evaluation by Foreign Counterpart, pp 93–95
- 32. Fujiyama C, Fujiyoshi T, Matsumoto D et al. Evaluation of commercial HTLV‐1 test kits by a standard HTLV‐1 serum panel. Bull World Health Organ 1995; 73(4): 515–21. [PMC free article] [PubMed] [Google Scholar]
- 33. Seiki M, Hattori S, Hirayama Y et al. Human adult T‐cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A 1983; 80(12): 3618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tajima K et al. The 4th nation‐wide study of adult T‐cell leukemia/lymphoma (ATL) in Japan: estimates of risk of ATL and its geographical and clinical features. The T‐ and B‐cell Malignancy Study Group. Int J Cancer 1990; 45(2): 237–43. [DOI] [PubMed] [Google Scholar]
- 35. Osame M, Janssen R, Kubota H et al. Nationwide survey of HTLV‐I‐associated myelopathy in Japan: association with blood transfusion. Ann Neurol 1990; 28(1): 50–6. [DOI] [PubMed] [Google Scholar]
- 36. Zamora T, Zaninovic V, Kajiwara M et al. Antibody to HTLV‐I in indigenous inhabitants of the Andes and Amazon regions in Colombia. Jpn J Cancer Res 1990; 81(8): 715–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blank A, Yamaguchi K, Blank M et al. Six Colombian patients with adult T‐cell leukemia/lymphoma. Leuk Lymphoma 1993; 9(4–5): 407–12. [DOI] [PubMed] [Google Scholar]
- 38. Bartholomew C, Jack N, Edwards J et al. HTLV‐I serostatus of mothers of patients with adult T‐cell leukemia and HTLV‐I‐ associated myelopathy/tropical spastic paraparesis. J Hum Virol 1998; 1(4): 302–5. [PubMed] [Google Scholar]
- 39. Kayembe K, Goubau P, Desmyter J et al. A cluster of HTLV‐1 associated tropical spastic paraparesis in Equateur (Zaire): ethnic and familial distribution. J Neurol Neurosurg Psychiatry 1990; 53(1): 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sonoda S. HLA and HTLV: 12th International Histocompatibility Worshop Study. In: Charron D, ed. Genetic Diversity of HLA, Functional and Medical Implication. Paris: EDK Med Sci, Int.Publ, 1997; 429–36. [Google Scholar]
- 41. Yashiki S, Fujiyoshi T, Arima N et al. HLA‐A*26, HLA‐B*4002, HLA‐B*4006, and HLA‐B* 4801 alleles predispose to adult T cell leukemia: the limited recognition of HTLV type 1 tax peptide anchor motifs and epitopes to generate anti‐HTLV type 1 tax CD8(+) cytotoxic T lymphocytes. AIDS Res Hum Retroviruses 2001; 17(11): 1047–61. [DOI] [PubMed] [Google Scholar]
- 42. Yamano Y, Kitze B, Yashiki S et al. Preferential recognition of synthetic peptides from HTLV‐I gp21 envelope protein by HLA‐DRB1 alleles associated with HAM/TSP (HTLV‐I‐associated myelopathy/tropical spastic paraparesis). J Neuroimmunol 1997; 76(1): 50–60. [DOI] [PubMed] [Google Scholar]
- 43. White JD, Johnson JA, Nam JM et al. Distribution of human leukocyte antigens in a population of black patients with human T‐cell lymphotrophic virus type I‐associated adult T‐cell leukemia/lymphoma. Cancer Epidemiol Biomarkers Prev 1996; 5(11): 873–7. [PubMed] [Google Scholar]
- 44. LaGrenade L, Sonoda S, Miller W et al. HLA DRB1*DQB1* haplotype in HTLV‐I‐ associated familial infective dermatitis may predict development of HTLV‐I‐associated myelopathy/tropical spastic paraparesis. Am J Med Genet 1996; 61(1): 37–41. [DOI] [PubMed] [Google Scholar]
- 45. Goedert JJ, Li HC, Gao XJ et al. Risk of human T‐lymphotropic virus type I‐associated diseases in Jamaica with common HLA types. Int J Cancer 2007; 121(5): 1092–7. [DOI] [PubMed] [Google Scholar]
- 46. Sabouri AH, Saito M, Usuku K et al. Differences in viral and host genetic risk factors for development of human T‐cell lymphotropic virus type 1 (HTLV‐1)‐associated myelopathy/tropical spastic paraparesis between Iranian and Japanese HTLV‐1‐infected individuals. J Gen Virol 2005; 86: 773–81. [DOI] [PubMed] [Google Scholar]
- 47. Robinson J, Malik A, Parham P et al. IMGT/HLA database‐‐a sequence database for the human major histocompatibility complex. Tissue Antigens 2000; 55(3): 280–7. [DOI] [PubMed] [Google Scholar]
- 48. De Campos‐Lima PO, Gavioli R et al. HLA‐A11 epitope loss isolates of Epstein–Barr virus from a highly A11+ population. Science 1993; 260(5104): 98–100. [DOI] [PubMed] [Google Scholar]
- 49. Hino S, Doi H, Yoshikuni H, Sugiyama H et al. HTLV‐I carrier mothers with high‐titer antibody are at high risk as a source of infection. Jpn J Cancer Res 1987; 78(11): 1156–8. [PubMed] [Google Scholar]
- 50. Wiktor SZ, Pate EJ, Rosenberg PS et al. Mother‐to‐child transmission of human T‐cell lymphotropic virus type I associated with prolonged breast‐feeding. J Hum Virol 1997; 1(1): 37–44. [PubMed] [Google Scholar]
- 51. Nomura K, Utsunomiya A, Furushou H et al. A family predisposition to adult T‐cell leukemia. J Clin Exp Hematop 2006; 46(2): 67–71. [DOI] [PubMed] [Google Scholar]
- 52. Biggar RJ, Ng J, Kim N, Hisada M et al. Human leukocyte antigen concordance and the transmission risk via breast‐feeding of human T cell lymphotropic virus type I. J Infect Dis 2006; 193(2): 277–82. [DOI] [PubMed] [Google Scholar]
- 53. Tomaru U, Yamano Y, Nagai M et al. Detection of virus‐specific T cells and CD8+ T‐cell epitopes by acquisition of peptide‐HLA‐GFP complexes: analysis of T‐cell phenotype and function in chronic viral infections. Nat Med 2003; 9(4): 469–76. [DOI] [PubMed] [Google Scholar]
- 54. Igakura T, Stinchcombe JC, Goon PK et al. Spread of HTLV‐I between lymphocytes by virus‐induced polarization of the cytoskeleton. Science 2003; 299(5613): 1713–6. [DOI] [PubMed] [Google Scholar]
- 55. HUGO Pan‐Asian SNP Consotium . Mapping human genetic diversity in Asia. Science 2009; 326(5959): 1541–5. [DOI] [PubMed] [Google Scholar]
- 56. Kalyanaraman VS, Sarngadharan MG, Robert‐Guroff M et al. A new subtype of human T‐cell leukemia virus (HTLV‐II) associated with a T‐cell variant of hairy cell leukemia. Science 1982; 218(4572): 571–3. [DOI] [PubMed] [Google Scholar]
- 57. Lairmore MD, Jacobson S, Gracia F et al. Isolation of human T‐cell lymphotropic virus type 2 from Guaymi Indians in Panama. Proc Natl Acad Sci U S A 1990; 87(22): 8840–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goubau P, Desmyter J, Ghesquiere J et al. HTLV‐II among pygmies. Nature 1992; 359(6392): 201. [DOI] [PubMed] [Google Scholar]
- 59. Mahieux R, Gessain A. The human HTLV‐3 and HTLV‐4 retroviruses: new members of the HTLV family. Pathol Biol (Paris) 2009; 57(2): 161–6. [DOI] [PubMed] [Google Scholar]
- 60. Switzer WM, Salemi M, Qari SH et al. Ancient, independent evo;ution and distinct molecular features of the novel human T‐lymphotropic virus type4. Retrovirology 2009; 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fujiyama C, Fujiyoshi T, Miura T et al. A new endemic focus of human T lymphotropic virus type II carriers among Orinoco natives in Colombia. J Infect Dis 1993; 168(4): 1075–7. [DOI] [PubMed] [Google Scholar]
- 62. Fujiyoshi T, Yashiki S, Fujiyama C et al. Ethnic segregation of HTLV‐I and HTLV‐II carriers among South American native Indians. Int J Cancer 1995; 63(4): 510–5. [DOI] [PubMed] [Google Scholar]
- 63. Hjelle B, Appenzeller O, Mills R et al. Chronic neurodegenerative disease associated with HTLV‐II infection. Lancet 1992; 339(8794): 645–6. [DOI] [PubMed] [Google Scholar]
- 64. Ishida T, Hinuma Y. The origin of Japanese HTLV‐I. Nature 1986; 322(6079): 504. [DOI] [PubMed] [Google Scholar]
- 65. Yoshida M, Seiki M, Yamaguchi K et al. Monoclonal integration of human T‐cell leukemia provirus in all primary tumors of adult T‐cell leukemia suggests causative role of human T‐cell leukemia virus in the disease. Proc Natl Acad Sci U S A 1984; 81(8): 2534–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wong‐Staal F, Gallo RC. Human T‐lymphotropic retroviruses. Nature 1985; 317(6036): 395–403. [DOI] [PubMed] [Google Scholar]
- 67. Kubota R, Fujiyoshi T, Izumo S et al. Fluctuation of HTLV‐I proviral DNA in peripheral blood mononuclear cells of HTLV‐I‐associated myelopathy. J Neuroimmunol 1993; 42(2): 147–54. [DOI] [PubMed] [Google Scholar]
- 68. Furukawa Y, Fujisawa J, Osame M et al. Frequent clonal proliferation of human T‐cell leukemia virus type 1 (HTLV‐1)‐infected T cells in HTLV‐1‐associated myelopathy. Blood 1992; 80(4): 1012–6. [PubMed] [Google Scholar]
- 69. Watanabe M, Ohsugi T, Shoda M et al. Dual targeting of transformed and untransformed HTLV‐1‐infected T cells by DHMEQ, a potent and selective inhibitor of NF‐kappaB, as a strategy for chemoprevention and therapy of adult T‐cell leukemia. Blood 2005; 106(7): 2462–71. [DOI] [PubMed] [Google Scholar]
- 70. Haneji K, Matsuda T, Tomita M et al. Fucoidan extracted from Cladosiphon okamuranus Tokida induces apoptosis of human T‐cell leukemia virus type 1‐infected T‐cell lines and primary adult T‐cell leukemia cells. Nutr Cancer 2005; 52(2): 189–201. [DOI] [PubMed] [Google Scholar]
- 71. Sonoda J, Koriyama C, Yamamoto S et al. HTLV‐1 provirus load in peripheral blood lymphocytes of HTLV‐1 carriers is diminished by green tea drinking. Cancer Sci 2004; 95(7): 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Utsunomiya A, Miyazaki Y, Takatsuka Y et al. Improved outcome of adult T cell leukemia/lymphoma with allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant 2001; 27(1): 15–20. [DOI] [PubMed] [Google Scholar]
- 73. Ishida T, Ueda R. CCR4 as a novel molecular target for immunotherapy of cancer. Cancer Sci 2006; 97(11): 1139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kannagi M, Harashima N, Kurihara K et al. Tumor immunity against adult T‐cell leukemia. Cancer Sci 2005; 96(5): 249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Matsuoka M, Green PL. The HBZ gene, a key player in HTLV‐1 pathogenesis. Retrovirology 2009; 6: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Suemori K, Fujiwara H, Ochi T et al. HBZ is an immunogenic protein, but not a target antigen for human T‐cell leukemia virus type 1‐specific cytotoxic T lymphocytes. J Gen Virol 2009; 90 (Pt 8): 1806–11. [DOI] [PubMed] [Google Scholar]
- 77. Sonoda S, Yashiki S, Takahashi K et al. Altered HLA antigens expressed on T and B lymphocytes of adult T‐cell leukemia/lymphoma patients and their relatives. Int J Cancer 1987; 40(5): 629–34. [DOI] [PubMed] [Google Scholar]
- 78. Hayashi T, Imai K, Morishita Y et al. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res 2006; 66(1): 563–70. [DOI] [PubMed] [Google Scholar]
- 79. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev 2009; 227(1): 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]


