Abstract
This study aimed to evaluate the diagnostic value of SYT‐SSX detected by reverse transcriptase–polymerase chain reaction (RT‐PCR) and fluorescence in situ hybridization (FISH) for synovial sarcoma (SS) in known and potential cases. SYT‐SSX was analyzed in formalin‐fixed, paraffin‐embedded tissues of 62 known SS, 60 non‐SS and 133 potential SS by RT‐PCR and FISH. FISH was mainly performed on a tissue microarray with some modifications. SYT‐SSX was detected in 94.7% (54/57) of known SS and 70.5% (86/122) of potential SS by RT‐PCR and in 96.7% (58/60) of known SS and 78.1% (100/128) of potential SS by FISH. Moreover, SYT‐SSX was negative in 100% (58/58) of non‐SS by RT‐PCR and in 100% (59/59) of non‐SS by FISH. Accordingly, SYT‐SSX was detected in 106 potential SS by RT‐PCR or FISH, including 80 cases manifested by both methods, 20 specimens verified only by FISH and 6 samples confirmed only by RT‐PCR. Clinical findings and immunohistochemistry data were analyzed in potential SS with final molecular diagnosis. The positive ratio of cytokeratin (CK) and epithelial membrane antigen (EMA) in finally diagnosed SS was 51.9% (55/106) and 61.3% (65/106), respectively. Except EMA, clinical parameters (age, sex, tumor size, tumor sites) and other immunohistochemistry indexes (CK, S‐100, neurone specific enolase (NSE), CD99, myoglobin, smooth muscle actin (SMA), cluster of differentiation (CD) 68 and mesothelial cell) had no significant difference between finally diagnosed SS and non‐SS. It is indicated that the efficiency of FISH is comparable to or even higher than that of RT‐PCR for SYT‐SSX detection. The detection of SYT‐SSX by RT‐PCR or FISH is very useful for the final diagnosis of potential synovial sarcomas. (Cancer Sci 2008; 99: 1355–1361)
Synovial sarcoma (SS) accounts for approximately 7–10% of soft tissue sarcomas and the diagnosis is based mainly on clinical context, histological aspect and immunohistochemistry.( 1 ) SS occur mainly in the extremities, particularly in the vicinity of large joints in adolescents and young adults.( 1 , 2 , 3 , 4 ) In the past few years, an increasing number of primary SS have been detected in unexpected sites,( 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 ) which sets back the diagnosis. Histological diagnosis of SS principally relies on the recognition of concurrent epithelial and spindle cell differentiation, but it is problematic when the tumor presents as the monophasic type or poorly differentiated type. In this case, immunohistochemistry staining is useful because tumor cells in SS are usually positive for epithelial markers and negative for some markers specific for other tumors.( 15 , 16 , 17 , 18 , 19 ) Nevertheless, positive reaction for S‐100 protein( 15 , 17 ) and cluster of differentiation (CD) 99( 17 , 20 ) has been reported in 27–43% and in 40–100% of SS, respectively, which renders the differential diagnosis with malignant peripheral nerve sheath tumors (MPNST) and Ewing's sarcoma/peripheral neuroectodermic tumors (PNET) difficult. The specific immunohistochemistry marker for SS has not been found so far.
Several studies have indicated that the t(X;18)(p11.2;q11.2) and consequently SYT‐SSX gene fusion arise exclusively in SS,( 21 , 22 , 23 , 24 , 25 ) therefore they can be used as excellent diagnostic hallmarks for this malignancy. Since Lasota et al.( 26 ) developed an RT‐PCR assay capable of detecting SYT‐SSX chimeric RNA from formalin‐fixed, paraffin‐embedded tissue, this new technique has been extensively used to differentiate SS from other tumors. Nevertheless, RT‐PCR analysis can not remove the influence of non‐neoplastic areas that may provoke false‐negative results. FISH allows the detection of molecular features in synopsis with histomorphology and non‐neoplastic areas can be excluded. However, Amary et al.( 27 ) reported that analysis with FISH is tedious and problematic. Furthermore, the diagnostic accuracy of RT‐PCR or FISH assay for SS has been demonstrated on preselected specimens of well‐established histologic types,( 21 , 22 , 23 , 24 , 25 , 26 , 28 ) but few studies focus on the simultaneous detection by FISH and RT‐PCR and their diagnostic utility in a series of potential SS.
In the present study, the SYT‐SSX fusion gene was analyzed by FISH mainly on tissue microarray (TMA) and the SYT‐SSX transcript was detected by RT‐PCR in 62 known SS, 60 non‐SS and 133 potential SS. The aims of the present study are to assess the diagnostic values of FISH and RT‐PCR for SS and to improve the techniques for extensive use in the future. There are four features in the present study. (1) It includes a review study in certainly diagnosed specimens and a prospective study in potential SS. (2) Two hundred and fifty‐five paraffin‐embedded samples were analyzed by FISH and RT‐PCR in a single‐blind setting and the results of two molecular methods were compared. (3) Clinical and immuohistochemistry data were analyzed in potential SS after the final molecular diagnosis was conducted so that we could compare the diagnostic value of molecular technique and traditional methods. (4) FISH was performed on TMA with some modifications in operation and assessment.
Materials and Methods
Specimens. One hundred and ninety‐five tumor specimens diagnosed as SS or potential SS at the Cancer Hospital of Tianjin Medical University from 1974 to 2005 were reviewed by three pathologists. Immunohistochemical staining was supplied for those cases without immunohistochemistry data at initial diagnosis. Sixty‐two cases were certainly diagnosed as SS and served as our known SS sample set because they displayed typical clinical context, histologic aspect and immunohistochemical profile. Another 133 tumors were considered as potential SS (SS was either considered the primary diagnosis or included as part of the main differential diagnosis). In addition, 60 specimens representing other neoplasmas included in the differential diagnosis of SS (10 fibrosarcomas, 10 leiomyosarcomas, 10 Ewing's sarcomas/PNET, 10 MPNST, 10 malignant fibrohistiocytomas and 10 hemangio‐peritheliomas) were considered non‐SS. SYT‐SSX fusion was analyzed prospectively on these formalin‐fixed, paraffin‐embedded specimens as a systematic test. The study of the tumor material was approved by a national ethical committee.
Immunohistochemistry. Immunohistochemistry staining of cytokeratin (CK) (AE1/AE3, 1:100 dilution, Zymed Laboratory Inc., S. San Francisco, CA, USA), epithelial membrane antigen (EMA) (ZCE113, 1:100 dilution, Zymed) and vimentin (V9, 1:200 dilution, Zymed) was performed systematically; whereas the staining of smooth muscle actin (SMA) (1A4, Zymed), myoglobin (Z001, Zymed), CD68 (KP1, Zymed), CD99 (O13, Zymed), mesothelial cell (HBME‐1, Zymed), S‐100 (4C4.9, Zymed) and neurone specific enolase (NSE) (NSE‐1 G4, Zymed) was conducted according to differential diagnosis. The streptavidinbiotin–peroxidase method was used. After being subjected to heating for 15 min in 0.1 M citrate buffer (pH 6.0) or to digestion in 0.1% trypsin for 15 min at 37°C, the sections were incubated with antibodies overnight at 4°C. The sections were then immunostained using the streptavidinbiotin‐peroxidase (SP) kit (Zymed) and the signals were revealed using diaminobenzidine as substrate. Positive and negative controls were included in each immunohistochemistry run.
Tissue microarray. A TMA was constructed using a manual tissue arrayer (Beecher Instruments Inc, Sun Prarie, WI, USA). Two typical areas from different situations of each case were selected according to hematoxylin and eosin (HE) slides and marked in the paraffin blocks. Two 0.6‐mm cores from the two areas of every case were taken for the array. Moreover, two positive (SS with SYT‐SSX fusion gene confirmed by sequencing) and two negative (normal colon mucosa without SYT‐SSX confirmed by sequencing) controls were included in the TMA. The cases were ordered according to the pathological number instead of categories (known SS, non‐SS or potential SS).
RT‐PCR. Ten 5‐µm sections from the paraffin‐embedded tissue were treated with xylene and ethanol and incubated in protein digestion buffer (20 mmol/L TRIS‐HCl [pH 8.0], 200 mM NaCl, 1.5 mM CaCl2, 0.5 mg/mL proteinase K) overnight at 55°C. RNA was then extracted using TRIZOL (Invitrogen Inc., Karlsruhe, Germany), precipitated with isopropanol and redissolved in RNase‐free water.
RT‐PCR was performed using RNA PCR Kit ver. 3.0 (Takara Bio Inc., Kyoto, Japan). A 20‐µL volume of reverse transcriptase (RT) reaction contained 5 U avian myeloblastosis virus (AMV) RT, 20 U RNase inhibitor, 20 mmol deoxyribonucleotide triphosphate (dNTP), 50 pmol random primers and 1 µg RNA. RNA was reverse‐transcribed into complementary DNA (cDNA) at 42°C for 50 min. Polymerase chain reaction (PCR) was performed in a 20‐µL volume system with 1.0 U Taq DNA polymerase, 10 × PCR buffer (including 1.5 mmol/L MgCl2), 4 pmol each of forward and reverese primers (SYT‐fw: 5′‐CCAGCAGAGGCCTTATGGATA‐3′, SSX‐rv: 5′‐TTTGTGGGCCAGATGCTTC‐3′) and 2 µL cDNA. The amplification profile of the PCR consisted of 40 cycles of denaturation at 94°C for 50 s, annealing at 58°C for 30 s and extension at 72°C for 1 min. As positive controls for the integrity of messenger RNA (mRNA) in each sample, PCR for ubiquitously expressed glyzeraldehyde‐3‐phosphate dehydrogenase (GAPDH) was performed (GAPDH‐fw: 5′‐GAAGGTGAAGGTCGGAGTC‐3′, GAPDH‐rv: 5′‐GAAGATGGTGATGGGATTTC‐3′). A negative (no template) and positive control (SYT‐SSX fusion gene confirmed by sequencing) were used for each experiment.
An aliquot of the PCR products was electrophoresed on a 3% agarose gel and stained with ethidium bromide. Moreover, the PCR products were cloned into a pMD18‐T vector (Takara Bio Inc.) and sequenced using ABI PRISM 3730 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Assessment of the cases was performed without knowledge of the categories.
FISH. Four 5‐µm sections of the TMA were transferred to positively charged slides and baked overnight at 60°C. These slides were deparaffinized by three 10‐min xylene washes and two 5‐min 100%‐ethanol washes. After pretreated by immersion in 1 mol/L sodium thiocyanate at 80°C for 20 min and a 3‐min distilled water wash, four TMA slides were incubated in 4 mg/mL pepsin solution (in 0.2 N HCl, pH 1.0) at 37°C for 15, 30, 45 and 60 min, respectively. The slides were washed in distilled water for 3 min, and then air dried for 5 min. A 20‐µL location specific insite (LSI) SYT probe (Vysis, Downers Grove, IL, USA) mixture (probe : hybridization buffer : purified H2O = 1:7:2) was added to every slide and sealed under a coverslip with rubber cement. The slides were then incubated at 85°C for 5 min to co‐denature, followed by hybridization with the probes in a humidified chamber (HYBrite, Vysis) at 37°C overnight. After coverslips were removed, the slides were soon immersed in posthybridization buffer (2 × SSC with 0.3% NP40) at 73°C for 2 min, followed by a 2‐min wash in 2 × SSC/0.1% NP40 at room temperature. Finally, the slides were air dried, counterstained with 20 µL of 4′,6‐diamidino‐2‐phenylindole (DAPI) II reagent (Vysis) and coverslipped.
Fluorescence signals were analyzed using a Nikon Eclipse 90i fluorescence microscope (Nikon, Tokyo, Japan) equipped with an appropriate filter set. Among four TMA slides, the best one for each case (according to DAPI counterstaining and fluorescence signals) was selected for evaluation. One hundred non‐overlapping nuclei, which were clearly identified and contained unequivocal signals, were counted for each case. If a case could not be analyzed on TMA because of tissue shedding, obscured signals or insufficient nuclei, it was then performed and assessed on a traditional slide. A probe was considered to be break‐apart when a pair of orange and green signals was separated by a distance greater than the size of one hybridization signal (spectrum orange 650 kb, spectrum green 1044 kb): this cutoff was chosen because the distance between the non‐split signals is 56 kb. Cells positive for SYT‐SSX showed at least one pair of break‐apart signals per nucleus. The ratio of SYT‐SSX‐positive cells was calculated for each case. The mean and standard deviation of the ratio of SYT‐SSX‐positive cells in non‐SS were used to determine a cutoff value that would optimally divide the known SS and non‐SS with respect to test sensitivity (the percentage of known SS above the cutoff) and specificity (the percentage of non‐SS below the cutoff). This cutoff value was then applied to the test samples to determine if a SYT‐SSX fusion was present. Assessment of the cases was performed without knowledge of the categories and RT‐PCR results. The main procedure of FISH was summarized in Figure 1 and the modifications of operation and assessment are shown in red bold italic.
Figure 1.
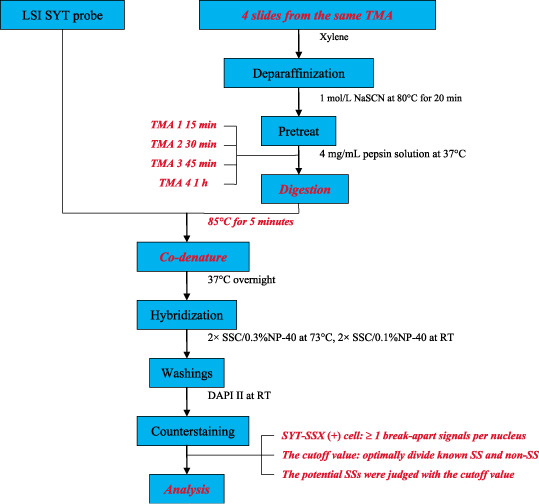
A schematic drawing of fluorescence in situ hybridization (FISH). The modifications of operation and assessment in the present study are shown in red bold italic. TMA, tissue microarray; RT, room temperature
Statistical analysis. Results were analyzed using SPSS 11.5 for windows software (SPSS Inc., Chicago, IL, USA). The Independent Samples T‐test was used to analyze the difference of ages between finally diagnosed SS and non‐SS. Then the difference of sex, tumor size, tumor site and expression of immunohistochemistry indexes between groups was analyzed by χ2 test or Fisher's exact test. A two‐tailed P‐value of less than 0.05 was considered significant.
Results
RT‐PCR. Two non‐SS (3.3%), five known SS (8.1%) and 11 potential SS (8.3%) could not be evaluated by RT‐PCR because of poor RNA: a PCR product for the housekeeping gene, GAPDH, failed to be generated in these cases. In the non‐SS detected by RT‐PCR, 100% (58/58) of cases were negative for SYT‐SSX transcript. Otherwise, 94.7% (54/57) of known SS were positive for SYT‐SSX. Furthermore, SYT‐SSX was detected in 70.5% (86/122) of potential SS. The electrophoresis and sequencing images of the PCR products are shown in Figure 2.
Figure 2.
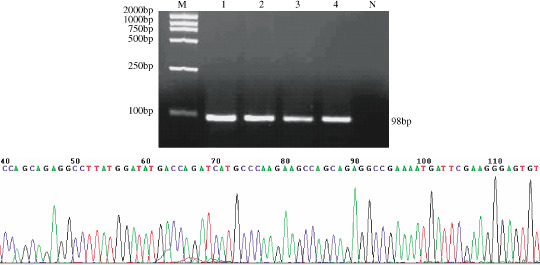
(a) Electrophoresis and (b) sequencing image of reverse transcriptase–polymerase chain reaction (RT‐PCR) products of SYT‐SSX. M: DNA Marker; 1: RT‐PCR product of the positive control; 2–4: RT‐PCR products from paraffin‐embedded tissue of three synovial sarcomas; N: RT‐PCR product of the negative control. The arrow in (b) shows the fusion location of SYT and SSX.
FISH. Seven non‐SS (11.7%), seven known SS (11.3%) and 20 potential SS (15.0%) on TMA could not be interpreted owing to tissue shedding, obscured signals or insufficient nuclei. Among them, six non‐SS, five known SS and 15 test cases were evaluated successfully by FISH on traditional slides. At last, 1.7% of non‐SS (1/60), 3.2% of known SS (2/62) and 3.8% of potential SS (5/133) could not be assessed by FISH. In 247 cases analyzed by FISH, two pairs of fluorescence signals (diploid) were found in most tumor cells in 164 specimens (66.4%), and more than two pairs of fluorescence signals (multiploid) were present in some tumor cells in 83 cases (33.6%), including 20 non‐SS, 18 known SS and 45 potential SS.
The FISH images of one Ewing's sarcoma, a known SS and one potential SS are shown in Figure 3. The ratio of SYT‐SSX‐positive cells in 59 non‐SS was 1–16% (7.81 ± 2.86%), and the ratio in 60 known SS was 13–94% (73.56 ± 23.87%). The optimal cutoff for sorting SYT‐SSX‐positive and negative cases was determined by adding multiples of the standard deviation to the mean of non‐SS until the best compromise between sensitivity and specificity was achieved to divide the known SS and non‐SS (Table 1). The optimal cutoff was determined to be 16.39%, that is to say, samples with the ratio equal to or above 16.39% would be considered to be positive for SYT‐SSX, whereas other cases were considered negative.
Figure 3.
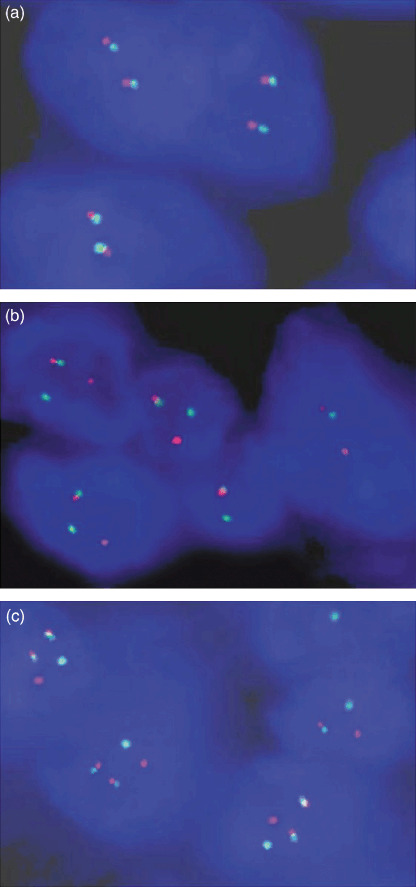
The fluorescence in situ hybridization (FISH) images in (a) one Ewing's sarcoma, (b) one known synovial sarcoma (SS) and (c) one potential SS. There were two pairs of fusion signals in every nucleus in (a), one pair of fusion signals and one pair of break‐apart signals in most tumor cells in (b) and two pairs of fusion signals and one pair of break‐apart signals in most tumor cells in (c).
Table 1.
The sensitivity and specificity of fluorescence in situ hybridization (FISH) at different cutoff values
| Cutoff (+SD) | Known synovial sarcomas | Non‐synovial sarcomas |
|---|---|---|
| 10.67% (1) | 100% (60/60) | 78.0% (46/59) |
| 13.53% (2) | 100% (60/60) | 88.1% (52/59) |
| 16.39% (3) | 96.7% (58/60) | 100% (59/59) |
| 19.25% (4) | 91.7% (57/60) | 100% (59/59) |
Furthermore, the ratio of SYT‐SSX‐positive cells of 128 potential SS was 3–95% (56.55 ± 31.04%). According to the above cutoff value, 78.1% (100/128) of potential SS were considered to be positive for SYT‐SSX, including 35 cases with multiple signals in more or less tumor cells.
Molecular diagnosis in known SS and non‐SS. Of the 62 known SS, 57 cases could be analyzed by both FISH and RT‐PCR, three could be detected only by FISH and two had neither FISH nor RT‐PCR results (Table 2). The sensitivity of FISH and RT‐PCR were 96.7% (58/60) and 94.7% (54/57), respectively, and the consistency of the two methods was 94.7% (54/57). However, one known SS was recorded as negative for SYT‐SSX with both methods.
Table 2.
The results of reverse transcriptase–polymerase chain reaction (RT‐PCR) and fluorescence in situ hybridization (FISH) in 62 known synovial sarcomas
| FISH positive | FISH negative | FISH no result | Total number | |
|---|---|---|---|---|
| RT‐PCR positive | 53 | 1 | 0 | 54 |
| RT‐PCR negative | 2 | 1 | 0 | 3 |
| RT‐PCR no result | 3 | 0 | 2 | 5 |
| Total number | 58 | 2 | 2 | 62 |
SYT‐SSX could not be detected by either RT‐PCR or FISH in 58 non‐SS. Moreover, one case without RT‐PCR result was indicated to be negative for SYT‐SSX by FISH. In addition, one non‐SS had no molecular result (Table 3). Both RT‐PCR and FISH had specificity of 100% (59/59 and 58/58, respectively).
Table 3.
The results of reverse transcriptase–polymerase chain reaction (RT‐PCR) and fluorescence in situ hybridization (FISH) in 60 non‐synovial sarcomas
| FISH positive | FISH negative | FISH no result | Total number | |
|---|---|---|---|---|
| RT‐PCR positive | 0 | 0 | 0 | 0 |
| RT‐PCR negative | 0 | 58 | 0 | 58 |
| RT‐PCR no result | 0 | 1 | 1 | 2 |
| Total number | 0 | 59 | 1 | 60 |
Molecular diagnosis of potential SS. One hundred 29 (97.0%) potential SS could be analyzed by RT‐PCR or FISH, including 121 cases with results by both, seven specimens with only FISH assessment and one sample with only RT‐PCR result (Table 4). In the 121 potential SS tested by two methods, 102 samples (84.3%) had concordant results, including 80 positive and 22 negative cases. Furthermore, 19 potential SS had incompatible results about the presence of SYT‐SSX. In the five specimens with positive RT‐PCR but negative FISH results, the ratio of SYT‐SSX‐positive cells for FISH was 11–16%, and there were two cases with multiple signals. Otherwise, among the 14 samples with positive FISH but negative RT‐PCR result, the ratio of SYT‐SSX‐positive cells of FISH was 64–92%.
Table 4.
The results of reverse transcriptase–polymerase chain reaction (RT‐PCR) and fluorescence in situ hybridization (FISH) in 133 potential synovial sarcomas
| FISH positive | FISH negative | FISH no result | Total number | |
|---|---|---|---|---|
| RT‐PCR positive | 80 | 5 | 1 | 86 |
| RT‐PCR negative | 14 | 22 | 0 | 36 |
| RT‐PCR no result | 6 | 1 | 4 | 11 |
| Total number | 100 | 28 | 5 | 133 |
Altogether, 106 potential cases were diagnosed as SS by molecular analysis for the positive results of RT‐PCR or FISH. Twenty‐three cases were eliminated from SS in molecular diagnosis, including 18 with negative results of RT‐PCR and FISH, three with negative RT‐PCR but without FISH result and two with negative FISH but without RT‐PCR result. In the 23 samples negative for SYT‐SSX, six were diagnosed as Ewing's sarcoma/PNET, desmoplastic small round cell tumor or clear cell sarcoma, as EWS rearrangement was detected by FISH (data not shown). Among them, five cases were positive for CD99 and S‐100 and three cases were positive for NSE. Although similar to SS in histological feature, three, one and one tumors were considered as malignant fibrous histiocytoma, rhabdomyosarcoma and leiomyosarcoma for positive expression of CD68, myoglobin and SMA, respectively. Moreover, MPNST was the first diagnoses for four cases in which S‐100 and NSE were a positive expression. Another specimen was firstly considered as mesothelioma, since it was present on pleura and showed very extensive expression of CK in the spindle cell areas, though mesothelial cell was negative in it. Furthermore, fibrosarcoma could not be excluded in four samples on the basis of histological findings. In addition, three cases were diagnosed as sarcoma, but the specific classifications were unclear.
Clinical findings and immunohistochemistry data of potential SS with final molecular diagnosis. The mean ages of SS and non‐SS manifested by molecular analysis were 37 and 40 years old, respectively. There was no significant difference between them (t = 0.874, P = 0.384). Furthermore, there was no difference in sex, tumor size or tumor site between finally diagnosed SS and non‐SS (Table 5).
Table 5.
The clinical and immunohistochemistry data of finally diagnosed synovial sarcomas (SS) and non‐synovial sarcomas (non‐SS) from potential cases
| Parameter | Final diagnosis | χ2 | P | ||
|---|---|---|---|---|---|
| SS | Non‐SS | ||||
| Sex | Male | 58 | 11 | 0.361 | 0.548 |
| Female | 48 | 12 | |||
| Tumor size | < 5 cm | 37 | 10 | 0.600 | 0.439 |
| ≥ 5 cm | 69 | 13 | |||
| Tumor site | Extremity | 82 | 18 | 0.009 | 0.925 |
| Trunk | 24 | 5 | |||
| CK | Positive | 55 | 8 | 2.213 | 0.137 |
| Negative | 51 | 15 | |||
| EMA | Positive | 65 | 8 | 5.418 | 0.020 |
| Negative | 41 | 15 | |||
| Vimentin | Positive | 106 | 23 | / | / |
| Negative | 0 | 0 | |||
| NSE | Positive | 24 | 7 | 1.392 | 0.238 |
| Negative | 42 | 6 | |||
| S‐100 | Positive | 36 | 10 | 2.236 | 0.135 |
| Negative | 30 | 3 | |||
| CD99 | Positive | 21 | 5 | 0.596 | 0.440 |
| Negative | 23 | 3 | |||
| Myoglobin | Positive | 0 | 1 | 3.529 | 0.060 |
| Negative | 12 | 2 | |||
| SMA | Positive | 0 | 1 | 1.780 | 0.182 |
| Negative | 5 | 3 | |||
| CD68 | Positive | 0 | 3 | 1.434 | 0.231 |
| Negative | 2 | 6 | |||
| Mesothelial cell | Positive | 0 | 0 | / | / |
| Negative | 2 | 4 | |||
CK, cytokeratin; EMA, epithelial membrane antigen; NSE, neurone specific enolase; CD, cluster of differentiation; SMA, smooth muscle antigen.
Seventy‐three (68.9%) SS indicated by molecular analysis were positive for CK or EMA, and the positive ratio of CK and EMA in finally diagnosed SS was 51.9% (55/106) and 61.3% (65/106), respectively. There was no significant difference of CK expression between finally diagnosed SS and non‐SS. However, the expression of EMA was significantly different between them (Table 5). Moreover, all the potential SS, regardless of finally diagnosed SS or non‐SS, were positive for vimentin. In addition, immunohistochemistry staining of some antibodies were performed in certain cases for differential diagnosis. The positive ratio of S‐100, NSE, and CD99 in finally diagnosed SS was 54.5% (36/66), 36.4% (24/66) and 47.7% (21/44), respectively. Myoglobin, SMA, CD68 and mesothelial cell were negative in the molecular results of all SS. There was no significant difference of expression of S‐100, NSE, CD99, myoglobin, SMA and CD68 in finally diagnosed SS and non‐SS (Table 5).
Discussion
At present, in distinguishing SS from other tumors, SYT‐SSX detected by RT‐PCR often serves as a most helpful method.( 21 , 22 , 24 , 26 ) The advantages of RT‐PCR are the easily visible results and the high confidence established on sequencing. Guillous et al.( 21 ) reported that SYT‐SSX could be detected by RT‐PCR with a specificity of 100% and a sensitivity of 96%. In the clearly diagnosed cases of this study, the specificity and sensitivity of RT‐PCR were 100% and 94.7%, respectively. However, 3.3% of non‐SS, 8.1% of known SS and 8.3% of potential SS could not be evaluated by RT‐PCR owing to poor RNA, which possibly resulted from RNA degradation in tissue fixing, embedding, RNA extraction or RT‐PCR operation. The poor stability of RNA is one disadvantage of RT‐PCR. In addition, RT‐PCR can not show the molecular and histomorphological features simultaneously, thus it failed to exclude the non‐neoplastic areas that may produce false‐negative results. Even if this pitfall can be overcome by additional methods like laser capture microdissection,( 29 ) the latter procedure tends to be time consuming. Therefore, some other valid tools need to be established for SS diagnosis.
FISH works stably for diagnosis, as it detects specific DNA sequences by hybridization with complimentary DNA probes. Moreover, FISH requires relatively little tumor material that is suitable for fine needle biopsy. Most importantly, FISH analysis allows the detection of molecular features in synchrony with histomorphology because non‐neoplastic areas can be excluded from analysis. Accordingly, FISH has particular advantages in the detection of chromosome translocation and gene rearrangement. Friedrichs et al.( 30 ) recommended FISH as a sensitive screening tool followed by subsequent RT‐PCR for the t(11; 22) translocation and EWS rearrangement. However, there have been few investigations about the diagnostic value of RT‐PCR and FISH in a large study of SS at the same time. Amary et al.( 27 ) reported FISH was less sensitive than RT‐PCR and raised some problems in interpreting FISH results, which impeded the extensive application of FISH.
In the present study, we tried to improve the manipulation and assessment of FISH in order to enhance its sensitivity and specificity. As this was a large‐sample study and the SYT probe was expensive, we performed FISH on TMA instead. Terry et al.( 28 ) applied SYT two‐color break‐apart style FISH probe to analyze SYT‐SSX fusion gene in 74 tumors on TMA, but 17 cases (23.0%) could not be interpreted owing to high background, which might be caused partly by the insufficient digestion of protease. Given that the structure of tumor cells and the density of stroma are different among tumors, the most appropriate protease digestion for each case is different. However, it is difficult to use different digestion times on one TMA slide. In the present study, we cut four slides from every TMA, with a different digestion time for each slide, and selected the best slide for analysis for each sample. Also, analysis would be performed on a traditional slide by FISH if it could not be completed on TMA owing to tissue shedding, obscured signals or insufficient nuclei. Hence, the observation ratio of FISH in the present study reached 96.9% (247/255). Furthermore, co‐denature of probe and TMA was used in this study in order to reduce the procedure and decrease the influence of other factors. Most importantly, the assessment of FISH was improved. At first, a cell showing at least one pair of break‐apart signals was defined as positive for SYT‐SSX, which avoids the trouble of discriminating diploid and polyploid. Second, the cutoff value for determining SYT‐SSX‐positive cases was found by analyzing non‐SS and known SS with respect to sensitivity and specificity. Because FISH was used to distinguish SS from other diseases, the cutoff based on non‐SS included in the differential diagnosis of SS is valid. Moreover, it is conducive to solving the problem of the cutoff value being established subjectively.( 27 , 30 , 31 ) Through the improvement of FISH in operation and assessment, the observation rate, specificity and sensitivity of FISH were increased compared with former studies.( 25 , 27 , 28 )
In the present study, SYT‐SSX could be detected in 94.7% (54/57) of known SS and 70.5% (86/122) of potential SS by RT‐PCR, but in 96.7% (58/60) of known SS and 78.1% (100/128) of potential SS by FISH. SYT‐SSX was detected in 86 potential SS by RT‐PCR, but in 100 cases by FISH. These results demonstrated that, the efficiency of FISH was comparable to or even higher than that of RT‐PCR. Therefore, FISH is also a powerful aid in the diagnosis of SS. However, one known SS and five potential SS with negative FISH results were determined as positive for SYT‐SSX by RT‐PCR and the ratio of SYT‐SSX‐positive cells in the six case was 11–16%, which suggested that RT‐PCR is an efficient complement to FISH, especially for the cases with the ratio of SYT‐SSX‐positive cells in the vicinity of the cutoff value.
We also analyzed the clinical findings and immunohistochemistry data of potential SS after the final diagnosis was conducted according to molecular results. It showed that there was no significant difference in age and sex of patients, tumor size and tumor sites between SS and other tumors included in the differential diagnosis. Moreover, the expression of CK was not distinctly different between finally diagnosed SS and non‐SS. Although the expression of EMA was significantly different between the two groups, the positive ratio of EMA in finally diagnosed SS was only 61.3%. Similar to the previous reports,( 15 , 17 , 20 ) S‐100, NSE and CD99 were inappropriate for distinguishing SS from MPNST and Ewing's sarcoma/PNET. Myoglobin, SMA, CD68 and mesothelial cell should be positive in rhabdomyosarcoma, leiomyosarcoma, malignant fibrohistiocytomas and mesothelioma, respectively, and be negative in SS. Therefore, they were very helpful to differentiate SS and the above corresponding tumors. Nevertheless, most of potential SS are not typical and need to be differentiated with several tumors, so many immunohistochemistry indexes will be used in one case, which increases work load and decreases sensitivity. This is a reason why the diagnostic value of myoglobin, SMA, CD68 and mesothelial cell was limited in this study. Through a prospective investigation and a comparison of molecular results and traditional data in 133 potential SS, we recommended molecular techniques as valid means for the diagnosis of SS. Nevertheless, one known SS was recorded as negative for SYT‐SSX by both RT‐PCR and FISH. Therefore, the results of molecular genetics had to be interpreted in the light of clinical findings, histological features and immunohistochemical data. RT‐PCR or FISH should be mainly used in the final diagnosis of potential SS as an assistance tool.
Acknowledgments
The authors thank the National Natural Science foundation of China for the financial support. The authors thank the help of Yumei Feng in the Department of Biochemistry and Molecular Biology, Tianjin Cancer Hospital and Institute.
References
- 1. Enzinger FM, Weiss SW. Soft Tissue Tumors, 3rd edn. St. Louis: Mosby, 1995. [Google Scholar]
- 2. Bergh P, Meis‐Kindblom JM, Gherlinzoni F et al . Synovial sarcoma. Identification of low and high risk groups. Cancer 1999; 85: 2596–607. [DOI] [PubMed] [Google Scholar]
- 3. Trassard M, Le Doussal V, Hacène K et al . Prognostic factors in localized primary synovial sarcoma: a multicenter study of 128 adult patients. J Clin Oncol 2001; 19: 525–34. [DOI] [PubMed] [Google Scholar]
- 4. Weiss SW, Goldblum JR. Malignant soft tissue tumors of uncertain type. In: Weiss SW, Goldblum JR, eds. Enzingerand Weiss's Soft Tissue Tumors. St. Louis: Mosby, 2001; 1483–571. [Google Scholar]
- 5. Argani P, Faria PA, Epstein JI et al . Primary renal synovial sarcoma: molecular and morphologic delineation of an entity previously included among embryonal sarcomas of the kidney. Am J Surg Pathol 2000; 24: 1087–96. [DOI] [PubMed] [Google Scholar]
- 6. Fetsch JF, Meis JM. Synovial sarcoma of the abdominal wall. Cancer 1993; 72: 469–77. [DOI] [PubMed] [Google Scholar]
- 7. Iwasaki H, Ishiguro M, Ohjimi Y et al . Synovial sarcoma of the prostate with t (X;18)(p11.2;q11.2). Am J Surg Pathol 1999; 23: 220–6. [DOI] [PubMed]
- 8. Iyengar V, Lineberger AS, Kerman S et al . Synovial sarcoma of the heart. Correlation with cytogenetic findings. Arch Pathol Lab Med 1995; 119: 1080–2. [PubMed] [Google Scholar]
- 9. Kim DH, Sohn JH, Lee MC et al . Primary synovial sarcoma of the kidney. Am J Surg Pathol 2000; 24: 1097–104. [DOI] [PubMed] [Google Scholar]
- 10. Nicholson AG, Goldstraw P, Fisher C. Synovial sarcoma of the pleura and its differentiation from other primary pleural tumours: a clinicopathological and immunohistochemical review of three cases. Histopathology 1998; 33: 508–13. [DOI] [PubMed] [Google Scholar]
- 11. O’Connell JX, Browne WL, Gropper PT et al . Intraneural biphasic synovial sarcoma: an alternative glandular tumor of peripheral nerve. Mod Pathol 1996; 9: 738–41. [PubMed] [Google Scholar]
- 12. Roth JA, Enzinger FM, Tannenbaum M. Synovial sarcoma of the neck: a follow‐up study of 24 cases. Cancer 1975; 35: 1243–53. [DOI] [PubMed] [Google Scholar]
- 13. Shmookler BM. Retroperitoneal synovial sarcoma. A report of four cases. Am J Clin Pathol 1982; 77: 686–91. [DOI] [PubMed] [Google Scholar]
- 14. Zeren H, Moran CA, Suster S et al . Primary pulmonary sarcomas with features of monophasic synovial sarcoma: a clinicopathological, immunohistochemical and ultrastructural study of 25 cases. Hum Pathol 1995; 26: 474–80. [DOI] [PubMed] [Google Scholar]
- 15. Guillou L, Wadden C, Kraus MD et al . S100 protein reactivity in synovial sarcomas. A potentially frequent diagnostic pitfall. Immunohistochemical analysis of 100 cases. Appl Immunohistochem 1996; 4: 167–75. [Google Scholar]
- 16. Lopes JM, Bjerkehagen B, Holm R et al . Immunohistochemical profile of synovial sarcoma with emphasis on the epithelial‐type differentiation: a study of 49 primary tumors, recurrences and metastases. Pathol Res Pract 1994; 190: 168–77. [DOI] [PubMed] [Google Scholar]
- 17. Pelmus M, Guillou L, Hostein I et al . Monophasic fibrous and poorly differentiated synovial sarcoma: immunohistochemical reassessment of 60t (X;18)(SYT‐SSX)‐positive cases. Am J Surg Pathol 2002; 26: 1434–40. [DOI] [PubMed] [Google Scholar]
- 18. Smith TA, Machen SK, Fisher C et al . Usefulness of cytokeratin subsets in distinguishing monophasic synovial sarcoma from malignant peripheral nerve sheath tumor. Am J Clin Pathol 1999; 112: 641–8. [DOI] [PubMed] [Google Scholar]
- 19. Wrba F, Fertl H, Amann G et al . Epithelial markers in synovial sarcoma: an immunohistochemical study on paraffin embedded tissues. Virchows Arch a Pathol Anat Histopathol 1989; 415: 253–8. [DOI] [PubMed] [Google Scholar]
- 20. Dei Tos AP, Wadden C, Calonje E. Immunohistochemical demonstration of glycoprotein p30/32MIC2 in synovial sarcoma: a potential cause of diagnostic confusion. Appl Immunohistochem 1995; 3: 168–73. [Google Scholar]
- 21. Guillou L, Coindre J, Gallagher G et al . Detection of the synovial sarcoma translocation t(X; 18)(SYT; SSX) in paraffin–embedded tissues using reverse transcriptase–polymerase chain reaction: a reliable and powerful diagnostic tool for pathologists. A molecular analysis of 221 mesenchymal tumors fixed in different fixatives. Hum Pathol 2001; 32: 105–12. [DOI] [PubMed] [Google Scholar]
- 22. Hiraga H, Nojima T, Abe S et al . Diagnosis of synovial sarcoma with the reverse transcriptase‐polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol 1998; 7: 102–10. [DOI] [PubMed] [Google Scholar]
- 23. Van De Rijn M, Barr FG, Collins MH et al . Absence of SYT‐SSX fusion products in soft tissue tumors other than synovial sarcoma. Am J Clin Pathol 1999; 112: 43–9. [DOI] [PubMed] [Google Scholar]
- 24. Willeke F, Mechtersheimer G, Schwarzbach M et al . Detection of SYT‐SSX1/2 fusion transcripts by reverse transcriptase–polymerase chain reaction (RT‐PCR) is a valuable diagnostic tool in synovial sarcoma. Eur J Cancer 1998; 34: 2087–93. [DOI] [PubMed] [Google Scholar]
- 25. Yang P, Hirose T, Hasegawa T et al . Dual‐colour fluorescence in situ hybridization analysis of synovial sarcoma. J Pathol 1998; 184: 7–13. [DOI] [PubMed] [Google Scholar]
- 26. Lasota J, Jasinski M, Debiec‐Rychter M et al . Detection of the SYT‐SSX fusion transcripts in formaldehyde‐fixed, paraffin‐embedded tissue: a reverse transcription polymerase chain reaction amplification assay useful in the diagnosis of synovial sarcoma. Mod Pathol 1998; 11: 626–33. [PubMed] [Google Scholar]
- 27. Amary MF, Berisha F, Bernardi Fdel C et al . Detection of SS18‐SSX fusion transcripts in formalin‐fixed paraffin‐embedded neoplasms: analysis of conventional RT‐PCR, qRT‐PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol 2007; 20: 482–96. [DOI] [PubMed] [Google Scholar]
- 28. Terry J, Barry TS. Horsman DE et al . Fluorescence in situ hybridization for the detection of t(X;18)(p11.2;q11.2) in a synovial sarcoma tissue microarray using a breakapart–style probe. Diagn Mol Pathol 2005; 14: 77–82. [DOI] [PubMed] [Google Scholar]
- 29. Kasai T, Shimajiri S, Hashimoto H. Detection of SYT‐SSX fusion transcripts in both epithelial and spindle cell areas of biphasic synovial sarcoma using laser capture microdissection. Mol Pathol 2000; 53: 107–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Friedrichs N, Kriegl L, Poremba C et al . Pitfalls in the detection of t (11;22) translocation by fluorescence in situ hybridization and RT‐PCR: a single‐blinded study. Diagn Mol Pathol 2006; 15: 83–9. [DOI] [PubMed] [Google Scholar]
- 31. Qian X, Jin L, Shearer BM et al . Molecular diagnosis of Ewing's sarcoma/primitive neuroectodermal tumor in formalin‐fixed paraffin‐embedded tissues by RT‐PCR and fluorescence in situ hybridization. Diagn Mol Pathol 2005; 14: 23–8. [DOI] [PubMed] [Google Scholar]


