Abstract
Although KIT and EGFR overexpressions are reported to occur in breast cancer, their pathological significance is still unclear. We examined KIT, EGFR, and c‐erbB‐2 overexpressions immunohistochemically in 150 cases of surgically resected breast cancer and their correlation with the histological type and grade and mesenchymal and/or myoepithelial immunophenotype of primary tumors. To facilitate the analysis, we constructed a tissue microarray comprising 2‐mm diameter tissues cored from the representative tissue block of each tumor. KIT, EGFR, and c‐erbB‐2 overexpressions were detected in 15 (10%), 12 (8%), and 23 (15%), respectively. The KIT was more frequent in the group comprising comedo‐type ductal carcinoma in situ and invasive ductal carcinomas (IDCs) of the solid‐tubular subtype than in the group of other histological types (P = 0.027), and the EGFR was more frequent in IDCs of solid‐tubular type than in other histological types (P < 0.05). KIT and EGFR overexpressions were correlated with nuclear grade 3 (P = 0.0095 and 0.0005) and tended to be concurrent (P = 0.005). KIT overexpression was correlated with vimentin and S‐100 expression (P = 0.003 and P = 0.005), and EGFR overexpression was correlated with S100 expression (P = 0.0001). These correlations with grade and mesenchymal/myoepithelial markers were not observed for c‐erbB‐2 overexpression. KIT and EGFR appeared to be indicators of high‐grade breast carcinoma groups that often contain the carcinomas with mesenchymal and/or myoepithelial differentiation, which are distinct from the group with c‐erbB‐2 overexpression. (Cancer Sci 2005; 96: 48 –53)
The KIT proto‐oncogene encodes a growth factor receptor with tyrosine kinase activity and is involved in the growth and development of mast cells and of premature stromal cell or interstitial cell of Cajal. 1 , 2 , 3 , 4 , 5 Among human tumors, the mutational activation of the KIT proto‐oncogene is frequent in gastrointestinal stromal tumors (GIST), which are suggested to originate from a premature stromal cell. 6 , 7 , 8 KIT activation is reported to occur in a very restricted subset in other common human cancers (i.e. small‐cell and large‐cell lung carcinomas). (9) In female breast cancer, the incidence of KIT expression has been reported to vary from 1 to 82%, but its biological and clinicopathological significance is unclear. 10 , 11 , 12
The EGFR (epidermal growth factor receptor, also called c‐erbB‐1 or HER‐1) and c‐erbB‐2 (or HER‐2/neu) proto‐oncogenes also encode growth factor receptors with tyrosine kinase activity. The EGFR and c‐erbB‐2 oncoproteins are overexpressed in 27–36% and 15–20% of primary breast cancers, respectively, and their overexpression was shown to be correlated with high grade and hormone‐receptor‐negative tumors and poorer patient prognosis. 13 , 14 , 15 , 16 Although concurrent overexpression of the EGFR and c‐erbB‐2 is reported to be correlated with much worse patient prognosis, it is unknown whether the EGFR and c‐erbB‐2 overexpressions occur in high‐grade breast cancers of a similar histological type or tend to occur in an alternative manner between high‐grade breast cancers of different histological types. In 62% of high‐grade breast cancers of common histological types, bimodal differentiation toward epithelial (glandular epithelial and myoepithelial) and mesenchymal phenotypes was reported to occur. 17 , 18
Tissue microarray (TMA) is a recently developed technique for high‐throughput evaluation of protein expression in a large number of archival tissue blocks used for routine histopathological diagnosis. A cohort of tissue core specimens obtained from these tissue blocks is arranged into a single recipient paraffin block. (19) The utility of TMAs has been proved in a number of immunohistochemical studies of various cancer types. 20 , 21 , 22 , 23
In the present study, to reveal the histopathological implication of the overexpressions of the KIT, EGFR, and c‐erbB‐2 oncoproteins, we examined the overexpressions and several mesenchymal and/or myoepithelial markers in 150 cases of breast carcinoma by TMA and immunohistochemistry (IHC).
Materials and methods
Cases. This study was approved by the institutional review board of the National Defense Medical College. We reviewed hematoxylin‐eosin‐stained tissue sections of breast carcinomas that were resected from patients who received a mastectomy or partial breast resection at the National Defense Medical College Hospital between 1995 and 1997. All cases were histologically classified according to the World Health Organization criteria. (24) Invasive ductal carcinoma (IDC) was further subclassified into papillo‐tubular, solid‐tubular, and scirrhous subtypes, according to the Japanese Breast Cancer Society (JBCS). (25) In papillo‐tubular, solid‐tubular, and scirrhous subtypes, tumor cells formed a mainly papillary or tubular structure, a solid nest structure, and a strand or trabecular structure, respectively. Cases of ductal carcinoma in situ (DCIS) were subclassified into comedo and non‐comedo subtypes, the latter comprising cribriform, solid, papillary, and low‐papillary ones.
From the viewpoint of nuclear grade, 150 carcinomas were classified into 17 cases of Grade 1, 84 cases of Grade 2, and 49 cases of Grade 3 by a nuclear grading system according to the JBCS. (25)
Ipsilateral axillary lymph node dissection was performed in 129 patients, and metastases were not detected in 77 cases. Metastases were detected in one to three lymph nodes in 15 patients and in four or more lymph nodes in 37 patients. In 21 cases, excisional biopsy only was performed, and lymph node dissection was not performed in the hospital.
Tissue microarray construction. For each of the 150 cases of breast cancer, a representative hematoxylin and eosin (HE)‐stained section was selected by reviewing routine histopathological sections microscopically, and the corresponding tissue blocks stored in the hospital were used for this study. In order to construct TMA blocks, a single tissue core was taken from a cancer cell‐rich area of a donor block of each tumor, and the core specimens were transferred to a recipient block using a Tissue Microarrayer (Beecher Instruments, Silver Spring, MD) (Fig. 1).
Figure 1.
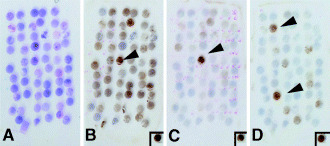
Detection of KIT, EGFR, and c‐erbB‐2 overexpression in breast carcinoma using tissue maicroarray. This TMA block contains 58 tissue cores 2.0 mm in diameter. (A) Hematoxylin and eosin stain (H&E). (B) KIT overexpression. Several cases, including case no. 33 (arrowhead), show KIT overexpression. (C) EGFR overexpression. Case no. 33 (arrowhead) shows EGFR overexpression, which is concurrent with KIT overexpression. (D) c‐erbB‐2 overexpression. Cases no. 14 and 50 (arrowheads) show c‐erbB‐2 overexpression with a score of 3+. (Insets) A case of gastrointestinal stromal tumor with KIT overexpression in (B), a stomach cancer with EGFR gene amplification (>10‐fold per haploid) in (C), and another case of stomach cancer with c‐erbB‐2 gene amplification (>10‐fold per haploid) in (D) were used as positive controls. Immunoperoxidase stain. × 7.
On constructing TMA in invasive carcinomas, we selected areas of invasive component with the highest nuclear grade. We used cores 2.0 mm in diameter and arranged them 0.7–0.8 mm apart in a recipient block. One TMA block contained a maximum of 66 tissue cores, and three TMA sets, comprising 150 core specimens, were prepared for the present study.
For the verification study, whole‐tissue sections of representative cancer tissue blocks were prepared from 10 cases (5 positive and 5 negative cases on TMA sections), and concordance in the rate of KIT, EGFR, and c‐erbB‐2 overexpression was examined between TMA sections and the corresponding whole‐tissue sections.
Immunohistochemistry. The expressions of KIT, EGFR, c‐erbB‐2, vimentin, S‐100, α‐smooth muscle actin (SMA), and CD34 were examined by IHC in the 150 breast carcinomas. TMA blocks were cut into 4‐µm‐thick sections. The antibodies used were polyclonal rabbit antihuman‐c‐KIT (1 : 50, Dako, Grostrup, Denmark), the PharmDx EGFR kit (Dako), the HercepTest kit for the c‐erbB‐2 (Dako), mouse monoclonal antivimentin (clone V9) (1 : 200, Dako), rabbit polyclonal anticow S‐100 (1 : 2000, Dako), mouse monoclonal antiα‐SMA (clone 1A4) (1 : 15, Shandon‐Lipshaw), and mouse monoclonal anti‐CD34 (clone QBent 10) (1 : 50, Dako). Estrogen receptor (ER) and progesterone receptor (PgR) were also immunohistochemically studied using monoclonal anti‐ER (clone 1D5, Dako) and monoclonal anti‐PgR (clone PgR636, Dako), respectively.
Antigen retrieval of the tissue sections was performed by the incubation of tissue sections in 10 mM sodium citrate (pH 6.0) with 0.1% Tween 40 at 95°C before the analysis using the anti‐KIT antibody (for 20 min). The sections were subjected to microwave treatment at 95°C for 15 min in 10 mM sodium citrate (pH 6.0) before analyzes using the anti‐CD34 antibody, and to autoclave at 120°C for 15 min in 10 mM sodium citrate (pH 6.0) before the analyses using the anti‐ER or anti‐PgR antibody.
After the antigen retrieval for the detection of KIT, EGFR, c‐erbB‐2, CD34, ER, and PgR, or without an antigen retrieval procedure for vimentin, S‐100, and α‐SMA, tissue sections were incubated in 0.3% hydrogen peroxide in methanol for 30 min, reacted with the primary antibody for 1–3 h, incubated with the dextran polymer reagent conjugated with peroxidase and secondary antibody (envision+, Dako) for 1 h, and subsequently reacted with 3,3′‐diaminobenzidine tetrahydrochloride‐hydrogen peroxide as a chromogen.
The KIT expression level was scored as 1+ if the cytoplasm was discretely and weakly to moderately stained and as 2+ if the cytoplasm was strongly stained with or without membrane staining in 10% or more of the constituent carcinoma cells. If no staining was observed or staining was observed in less than 10% of the constituent carcinoma cells, a score of 0 was given. Cases with a score of 2+ were judged as overexpression.
The EGFR and c‐erbB‐2 expressions were scored as 2+ and 3+ if the entire circumference of the cell membrane was weakly or moderately stained and strongly stained, respectively, in 10% or more of the constituent carcinoma cells. A score of 1+ was given if incomplete membrane staining was observed in 10% or more of the carcinoma cells, and a score of 0 was given if there was membrane staining in less than 10% of constituent cells or there was no membrane staining. Cases with a score of 2+ or 3+ were judged as overexpression.
Vimentin, S‐100, α‐SMA, and CD34 were judged as expressed if the cytoplasm of tumor cells was moderately to strongly stained in 10% or more of the tumor cells.
A case of GIST was used as a positive control for the expressions of KIT and vimentin. A stomach cancer with EGFR gene amplification (>10‐fold per haploid) and another case of stomach cancer with c‐erbB‐2 gene amplification (>10‐fold per haploid), detected by fluorescence in situ hybridization, were used as positive controls of EGFR and c‐erbB‐2 overexpression, respectively. For the internal control of S‐100, α‐SMA, and CD34, peripheral nerve, smooth muscle, and endothelial cells were used, respectively. As negative controls, sections without loading the primary antibody were used in each assay.
Evaluation of interobserver and intraobserver agreement. For the evaluation of interobserver agreement, immunohistochemical results were evaluated by two observers (H.T. and Y.O.) independently, and cases with discrepant judgments were re‐evaluated with discussion. Consensus judgments were acquired as the final ones. For the assessment of intraobserver agreement level, one observer (H.T.) judged twice all the cases blindly at an interval of 2 months. The degree of interobserver or intraobserver agreement for evaluating the KIT, EGFR, and c‐erbB‐2 was computed using the generalized κ‐test for two or more observers. (26) In accordance with the criteria of Landis and Koch, (27) κ‐values were assigned to a scale of strength of agreement. When the κ‐value was <0.00, 0.00–0.20, 0.21–0.40, 0.41–0.60, 0.61–0.80, and 0.81–1.00 the strength of agreement was judged as poor, slight, fair, moderate, substantial, and almost perfect, respectively.
Statistical analysis. Statistical differences were analyzed by the χ2 test or Fisher's exact test. The concordance between TMA sections and corresponding whole‐tissue sections in the 10 cases for the KIT, EGFR, and c‐erbB‐2 was also evaluated by the κ‐test.
Results
Verification of TMA. The percent interobserver agreements in the evaluation of KIT, EGFR, and c‐erbB‐2 immunostaining were 96%, 99%, and 99%, respectively, between the tumor group with overexpression (2+ or 3+) and that without overexpression (0 or 1+). In all discrepant cases, agreement was finally reached upon re‐evaluation by the two observers using a discussion microscope. The κ‐values for KIT, EGFR, and c‐erbB‐2 were 0.81, 0.96, and 0.95, respectively, and the agreement levels of all these were almost perfect.
The percent intraobserver agreements in the evaluation of KIT, EGFR, and c‐erbB‐2 immunostaining were 92%, 97%, and 99%, respectively, between the tumor group with overexpression (2+ or 3+) and that without overexpression (0 or 1+). The κ‐values for KIT, EGFR, and c‐erbB‐2 were 0.66, 0.84, and 0.95, respectively, and the agreement level was substantial for KIT, and almost perfect for EGFR and c‐erbB‐2.
The score of immunohistochemistry (0 or 1+ vs 2+ for KIT, and 0 or 1+ vs 2+ or 3+ for EGFR and c‐erbB‐2) was concordant between a TMA section and the corresponding whole‐tissue section in 7 (70%) of 10 cases for KIT, 9 (90%) of 10 cases for EGFR, and 10 (100%) of 10 cases for c‐erbB‐2. The agreement levels for KIT, EGFR, and c‐erbB‐2 calculated by κ‐test were 0.40, 0.80, and 1.00, respectively, and the levels were fair for KIT, and almost perfect for EGFR and c‐erbB‐2. Because the total number of cases for the κ‐test was only 10, the reliability of the estimated κ‐values was unclear.
KIT overexpression. The staining of KIT 2+ cancer cells was stronger than that of normal mammary glands, but the staining of KIT 1+ was similar to or weaker than that of normal glands. The cytoplasm and membrane staining of the KIT in the control GIST case was categorized as score 2+. KIT overexpression was detected in 15 cases (10%) of breast carcinomas (Fig. 2). In each histological type, KIT overexpression was detected in 10% (13 of 130) of IDCs: 22% (9 of 41) of the solid‐tubular subtype, 9% (3 of 35) of the papillo‐tubular subtype, and 2% (1 of 54) of the scirrhous subtype. KIT overexpression was detected in 2 (13%) of DCIS, comprising 40% (2 of 5) of the comedo subtype but 0% (0 of 10) of the non‐comedo type. Five cases of ILC did not show KIT overexpression. Therefore, KIT was detected more frequently in the group comprising IDC of the solid‐tubular subtype and comedo‐type DCIS than in the group of other histological types (P = 0.027) (Table 1).
Figure 2.
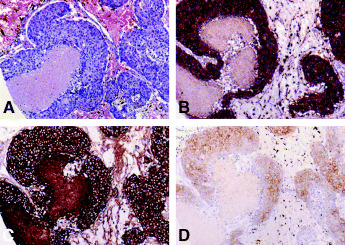
A case of invasive ductal carcinoma (Case no. 33) with KIT and EGFR co‐overexpression. (A) Histologically, Grade 3, solid‐tubular subtype. H&E stain. (B) KIT overexpression, and (c) EGFR overexpression. (D) c‐erbB‐2 expression was scored as 2+. (A) × 100. (B, C, D) Immunoperoxidase stain. × 100.
Table 1.
Incidence of KIT, EGFR, and c‐erbB‐2 overexpressions in various histological types of breast carcinoma
| Histological type | Number of cases (%) | |||
|---|---|---|---|---|
| Total | KIT over‐expression* | EGFR over‐expression † | c‐erbB‐2 over‐expression ‡ | |
| A. Invasive carcinoma | ||||
| Invasive ductal carcinoma | 130 | 13 (10) | 11 (8) | 18 (14) |
| Solid‐tubular subtype | 41 | 9 (22) | 7 (17) | 7 (17) |
| Papillo‐tubular subtype | 35 | 3 (9) | 1 (3) | 4 (11) |
| Scirrhous subtype | 54 | 1 (2) | 3 (6) | 7 (13) |
| Invasive lobular carcinoma | 5 | 0 (0) | 0 (0) | 0 (0) |
| B. Ductal carcinoma in situ (DCIS) | ||||
| Comedo subtype | 5 | 2 (40) | 0 (0) | 3 (60) |
| Non‐comedo subtype | 10 | 0 (0) | 1 (10) | 2 (20) |
| Total | 150 | 15 (10) | 12 (8) | 23 (15) |
P = 0.027 between the group comprising solid‐tubular type and comedo subtype and the group of other histological types;
P < 0.05 between solid‐tubular type and other histological types;
P < 0.05 between the group of DCIS and the group of invasive carcinomas.
Considering the nuclear grade of breast carcinoma, the KIT was overexpressed in 0% (0 of 17), 6% (5 of 84), and 20% (10 of 49) of cases of Grades 1, 2, and 3, respectively (P = 0.0095) (Table 2). KIT overexpression was inversely correlated with ER expression (P < 0.0001) and with PgR expression (P = 0.0002), but was not correlated with lymph node status (Table 2).
Table 2.
Correlation of KIT, EGFR, and c‐erbB‐2 overexpressions in breast carcinoma with nuclear grade, hormone receptor statuses, and lymph node metastasis
| Parameter | Number of cases (%) | |||
|---|---|---|---|---|
| Total | KIT over‐ expression | EGFR over‐ expression | c‐erbB‐2 over‐ expression | |
| Nuclear grade | ||||
| Grade 1 | 17 | 0 (0)* | 0 (0) † | 2 (12) |
| Grade 2 | 84 | 5 (6) | 2 (2) | 10 (12) |
| Grade 3 | 49 | 10 (20) | 10 (20) | 11 (22) |
| Estrogen receptor | ||||
| 0 | 45 | 12 (27) ‡ | 12 (27) ‡ | 17 (38) ‡ |
| 1+ | 28 | 0 (0) | 0 (0) | 4 (14) |
| 2+ | 77 | 3 (4) | 0 (0) | 2 (3) |
| Progesterone receptor | ||||
| 0 | 57 | 13 (23) § | 11 (19)¶ | 17 (30)** |
| 1+ | 6 | 0 (0) | 0 (0) | 0 (0) |
| 2+ | 87 | 2 (2) | 1 (1) | 6 (7) |
| Number of metastatic axillary lymph nodes | ||||
| 0 | 77 | 7 (9) | 4 (5) | 13 (17) |
| 3 | 15 | 1 (7) | 3 (20) | 2 (13) |
| ≥4 | 37 | 6 (16) | 3 (8) | 7 (29) |
| No data | 21 | 1 (5) | 4 (19) | 1 (5) |
| Total | 150 | 15 (10) | 12 (8) | 23 (15) |
P = 0.0095 among the cases of Grades 1, 2, and 3;
P = 0.0005 between the cases of Grade 1 or 2 and the cases of Grade 3;
P < 0.0001 among the cases of ER 0, 1+, and 2+;
P = 0.0002,
P = 0.0003, and
P = 0.0005 among the cases of PgR 0, 1+, and 2+.
EGFR overexpression. EGFR overexpression was detected in 12 cases (8%) of breast carcinoma (Fig. 2). The staining of the EGFR in the control stomach cancer case was categorized as 3+. The incidence of EGFR overexpression was 8% (11 of 138) of IDC: 17% (7 of 41) of the solid‐tubular subtype (Fig. 2b), 6% (3 of 54) of the scirrhous subtype, and 3% (1 of 35) of the papillo‐tubular subtype. EGFR overexpression was detected in only one (7%) of 14 cases of DCIS and in 0 of five cases of ILC. Therefore, EGFR tended to occur more frequently in IDC of the solid‐tubular subtype than in the other histological types (P < 0.05) (Table 1). EGFR overexpression was detected in 0% (0 of 17), 2% (1 of 84), and 20% (10 of 49) of carcinomas of Grades 1, 2, and 3, respectively (P = 0.0005) (Table 2). EGFR overexpression was inversely correlated with ER expression (P < 0.0001) and with PgR expression (P = 0.0003), but was not correlated with lymph node status (Table 2).
c‐erbB‐2 overexpression. The c‐erbB‐2 was overexpressed in 23 cases (15%) (Fig. 2). The staining of the c‐erbB‐2 in the control stomach cancer case was categorized as 3+ (Fig. 1c). Of 130 cases of IDC, the c‐erbB‐2 was overexpressed in 18 (14%) (Table 1): 17% (7 of 41) of the solid‐tubular subtype, 13% (7 of 54) of the scirrhous subtype, and 11% (4 of 35) of papillo‐tubular subtype. C‐erbB‐2 overexpression was detected in five (33%) of 15 cases of DCIS, comprising 60% (3 of 5) of the comedo subtype but 20% (2 of 10) of the non‐comedo type. Five cases of ILC did not show EGFR overexpression (Table 1). Therefore, the c‐erbB‐2 was more frequently overexpressed in DCIS (5 of 15, 33%) than in invasive carcinomas (18 of 135, 13%) (P < 0.05). C‐erbB‐2 overexpression was detected in 12% (2 of 17), 12% (10 of 84), and 22% (11 of 49) of carcinomas of Grades 1, 2, and 3, respectively, and was not correlated with the nuclear grades (Table 2). C‐erbB‐2 overexpression was inversely correlated with ER expression (P < 0.0001) and with PgR expression (P = 0.0005), but was not correlated with lymph node status (Table 2).
Of 21 solid‐tubular subtype IDCs, Grade 3, overexpressions of the KIT, EGFR, and c‐erbB‐2 were detected in six (29%), six (29%), and three (14%), respectively. As cases with special histological features in this group, two had a feature of metaplastic carcinoma of the spindle‐cell type, and one of these cases showed EGFR overexpression. Another case showed a feature of matrix‐producing carcinoma, and that case concurrently showed KIT and EGFR overexpressions.
Correlation between KIT, EGFR, and c‐erbB‐2 overexpression. Of the 15 cases expressing the KIT, four (27%) co‐overexpressed EGFR (Figs 2b,c). In contrast, of the 135 cases that did not show KIT overexpression, only eight (6%) overexpressed EGFR. There was a correlation between KIT overexpression and EGFR overexpression (P = 0.005) (Table 3). All four cases with co‐overexpression of the KIT and EGFR, including two cases accompanied with co‐overexpression of the c‐erbB‐2, were IDC of the solid‐tubular subtype and Grade 3.
Table 3.
Correlation of KIT overexpression with EGFR overexpression and with c‐erbB‐2 overexpression in breast carcinomas
| Number of cases (%) | |||
|---|---|---|---|
| Total | EGFR overexpression | c‐erbB‐2 overexpression | |
| A. KIT overexpression | |||
| Present | 15 | 4 (27)* | 6 (40) † |
| Absent | 135 | 8 (6) | 17 (13) |
| B. EGFR overexpression | |||
| Present | 12 | 4 (33) ‡ | |
| Absent | 138 | 19 (14) | |
| Total | 150 | 12 (8) | 23 (15) |
P = 0.005;
P = 0.050 (only marginally significant);
P = 0.071 (not significant).
C‐erbB‐2 overexpression was detected in 40% (6 of 15) of KIT‐overexpressing cases and 13% (17 of 135) of KIT‐non‐overexpressing cases (P = 0.050), the difference being only marginally significant. C‐erbB‐2 overexpression was detected in 33% (4 of 12) of carcinomas with EGFR overexpression and 14% (19 of 138) without EGFR overexpression, the difference being not significant (P = 0.071) (Table 3). Concurrence of KIT and c‐erbB‐2 overexpressions, without EGFR overexpression, was observed in four cases, which comprised two cases of comedo‐type DCIS (one Grade 3 and one Grade 2), one case of IDC of the solid‐tubular subtype, Grade 3, and one case of IDC of the papillo‐tubular subtype, Grade 2. Concurrence of EGFR and c‐erbB‐2 overexpressions, without KIT overexpression, was observed in a case of IDC of the scirrhous subtype, Grade 3.
Expression of mesenchymal and myoepithelial markers. Vimentin, CD34, S‐100 protein, and α‐SMA were expressed in seven (5%), one (0.7%), six (4%), and one (0.7%) of the 150 cases, respectively (Fig. 3). Vimentin was expressed in 5% (6 of 130) of IDC: 10% (4 of 41) of the solid‐tubular subtype, 4% (2 of 54) of the scirrhous subtype, and 0% (0 of 35) of the papillo‐tubular subtype (Fig. 3b). Vimentin expression was detected in one (7%) of 15 cases of DCIS, comprising 20% (1 of 5) of the comedo subtype, but 0% (0 of 10) of the non‐comedo type. Five cases of ILC did not show vimentin expression. Vimentin expression was more frequent in IDC of the solid‐tubular subtype than in other histological types (P = 0.044). Vimentin expression was detected in 0% (0 of 17), 4% (3 of 84), and 8% (4 of 49) of carcinomas of Grades 1, 2, and 3, respectively, and the difference was not significant.
Figure 3.
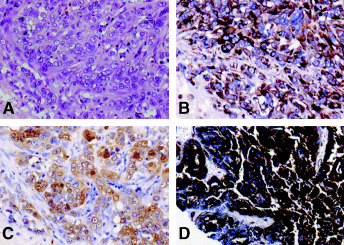
Breast carcinomas with mesenchymal or myoepithelial immunophenotype (A to C) and with CD34 expression (D). (A–C) Case no. 109. (A) Histologically, invasive ductal carcinoma (IDC) solid‐tubular subtype Grade 3. H&E stain. × 200. Vimentin (B) and S‐100 protein (C) expressions are diffusely positive. This case showed concurrent EGFR overexpression. (B, C) Immunoperoxidase stain. × 200. (D) Case no. 146. Histologically, IDC papillotubular subtype, Grade 2. CD34 is strongly and diffusely positive. Immunoperoxidase stain. × 40.
S‐100 was expressed in 5% (6 of 130) of IDC: 10% (4 of 41) of the solid‐tubular subtype, 4% (2 of 54) of the scirrhous subtype, and 0% (0 of 35) of the papillo‐tubular subtype (Fig. 3c). S‐100 expression was not detected in 15 cases of DCIS or five cases of ILC. S‐100 expression was more frequent in IDC of the solid‐tubular subtype than in other histological types (P = 0.038). S‐100 expression was detected in 0% (0 of 17), 0% (0 of 84), and 12% (6 of 49) of carcinomas of Grades 1, 2, and 3, respectively (P = 0.007). α‐SMA was expressed in only one case, an IDC of the solid‐tubular subtype, Grade 3. CD34 was diffusely and strongly expressed in only one case, IDC of the papillo‐tubular subtype, Grade 2 (Fig. 3d).
Of 21 solid‐tubular subtype IDCs, Grade 3, expressions of vimentin, S‐100, α‐SMA, and CD34 were detected in three (14%), three (14%), one (5%), and 0 (0%), respectively. Two of these cases showing a feature of metaplastic carcinoma of the spindle‐cell type expressed vimentin and S‐100 protein. Another case with a feature of matrix‐producing carcinoma showed the expression of S‐100 protein.
The incidences of expressions of vimentin and S‐100 protein were 20% (3 of 15) and 13% (2 of 15) in KIT‐overexpressing cases but only 3% (4 of 135) and 3% (4 of 135) in KIT‐non‐overexpressing cases, respectively (P = 0.003 and P = 0.005, respectively) (Table 4). A similar tendency of such correlations was seen between EGFR‐overexpressing and EGFR‐non‐overexpressing cases. The incidences of expressions of vimentin and S‐100 protein were 8% (1 of 12) and 25% (3 of 12) in EGFR‐overexpressing cases, but only 4% (6 of 138) and 2% (3 of 138) in EGFR‐non‐overexpressing cases, respectively. S‐100 protein was expressed more frequently in EGFR‐overexpressing cases than in EGFR‐non‐overexpressing ones (P = 0.0001) (Table 4).
Table 4.
Correlation of KIT, EGFR, and c‐erbB‐2 overexpressions with the expression of markers of mesenchymal or myoepithelial differentiation in breast carcinomas
| Total | Expression of mesenchymal or myoepithelial markers | ||||
|---|---|---|---|---|---|
| Number of cases (%) | |||||
| Vimentin | S‐100 protein | α‐smooth muscle actin | CD34 | ||
| A. KIT overexpression | |||||
| Positive | 15 | 3 (20)* | 2 (13) † | 1 (7) | 1 (7) |
| Negative | 135 | 4 (3) | 4 (3) | 0 (0) | 0 (0) |
| B. EGFR overexpression | |||||
| Positive | 12 | 1 (8) | 3 (25) ‡ | 0 (0) | 0 (0) |
| Negative | 138 | 6 (4) | 3 (2) | 1 (0.7) | 1 (0.7) |
| c‐erbB‐2 overexpression | |||||
| Positive | 23 | 1 (4) | 0 (0) | 0 (0) | 0 (0) |
| Negative | 127 | 6 (5) | 6 (5) | 1 (0.7) | 1 (0.8) |
| Total | 150 | 7 | 6 | 1 | 1 |
P = 0.003;
P = 0.005;
P = 0.0001.
On the other hand, the expression of vimentin and S‐100 were not correlated with c‐erbB‐2 overexpression. The incidences of vimentin and S‐100 protein expressions were 4% (1 of 23) and 0% (0 of 23) in c‐erbB‐2‐overexpressing cases but 5% (6 of 127) and 5% (6 of 127) in c‐erbB‐2‐non‐overexpressing ones, respectively (Table 4).
Discussion
We examined the histopathological characteristics of breast carcinomas with KIT, EGFR, and c‐erbB‐2 overexpression. KIT overexpression was more frequent in the group comprising the solid‐tubular subtype IDC and comedo subtype DCIS than in the group of other histological types, and EGFR overexpression was more frequent in IDC of the solid‐tubular subtype than in other histological types. In contrast, c‐erbB‐2 overexpression was more frequent in DCIS than in invasive carcinomas. KIT and EGFR were more frequent in Grade 3 carcinomas than in Grade 1 or 2 carcinomas and tended to concur. Deduced from their combination and histological type of tumors, the concurrent KIT and EGFR overexpressions were suspected to be related with the development of IDC of the solid‐tubular subtype, Grade 3.
From these results, KIT and EGFR overexpressions appear to be correlated with similar spectra of histological types. C‐erbB‐2 overexpression was reported to be correlated with high histological and nuclear grades, but, in the present study, KIT and EGFR overexpressions were more correlated with Grade 3 than c‐erbB‐2 overexpression. The carcinoma group with KIT and/or EGFR overexpression might be less differentiated than the carcinoma group with c‐erbB‐2 overexpression. KIT, EGFR, and c‐erbB‐2 were inversely correlated with ER or PgR expression. These results might indicate association between increased tyrosine kinase pathway and tamoxifen resistance in breast cancer.
From the viewpoint of mesenchymal and myoepithelial differentiation, KIT‐overexpressing carcinoma tended to coexpress vimentin and S‐100 protein more frequently than carcinomas without KIT overexpression. EGFR‐overexpressing carcinomas tended to be concurrent with S‐100 protein expression. In contrast, c‐erbB‐2 overexpression was not correlated with mesenchymal or myoepithelial differentiation. Mesenchymal and/or myoepithelial differentiation appeared to be characteristic features of breast carcinomas with KIT and/or EGFR overexpression.
Several special types of breast carcinomas are known to show myoepithelial or mesenchymal differentiation (e.g. carcinoma with metaplasia of the spindle‐cell type [spindle‐cell carcinoma] and matrix‐producing carcinoma [a variant of carcinoma with cartilaginous metaplasia]). 28 , 29 In addition, even among common histological types, this kind of bimodal differentiation toward epithelial and mesenchymal phenotypes was reported to occur in 62% of Grade 3 IDCs. As subtypes of Grade 3 IDCs, IDC with a large central acellular zone and atypical medullary carcinoma, which could also be a variant of medullary carcinoma, frequently express vimentin, α‐SMA, and/or S‐100 protein. 30 , 31 , 32 In contrast, the coexistence of epithelial and mesenchymal features was infrequent in lower‐grade (Grade 1 or 2) IDCs. 17 , 18
It is well recognized that TMA is efficient for screening of molecular alterations in a large number of tumor cases. In contrast, the limitation and possible pitfalls of TMA analysis would consist in that the characteristics of sampled tissue do not always represent those of the whole tumor. It is shown that there are heterogeneity of KIT and EGFR immunolocalization in breast cancer. On constructing TMA in invasive carcinomas, we selected the areas of invasive component with the highest nuclear grade. We also preferred a 2.0‐mm diameter punch and did not use a 0.6‐mm diameter punch as was used in a majority of TMA examinations. 19 , 20 , 21 , 22 , 23 In this study, we showed that judgments of KIT, EGFR, and c‐erbB‐2 were concordant between TMA sections and whole‐tissue sections in 70%, 90%, and 100% of cases. By careful selection of area for TMA construction and the use of a 2.0‐mm diameter punch, a TMA block would be able to represent substantially the status of KIT, EGFR, and c‐erbB‐2 status in the whole tumor tissue.
The mechanism of KIT overexpression in breast carcinomas is still unknown. In GIST, acute myelogenous leukemia, and mastocytosis, the KIT is activated by somatic mutation. 33 , 34 In other cancers (e.g. small cell lung cancer and ovarian cancers) paracrine or autocrine activation is postulated. 33 , 34 As a novel anticancer drug, imatinib (gleebec or glivec), a tyrosine kinase inhibitor to inhibit the activity of KIT and BCR‐ABL, has been developed and is now routinely used. 35 , 36 It might be worth investigating if the inhibition of the active KIT is effective for the patients with KIT‐overexpressing breast cancers.
In summary, we found that KIT and EGFR overexpressions tended to occur concurrently and were correlated with IDC of the solid‐tubular subtype, Grade 3. The solid‐tubular subtype, Grade 3 may contain atypical medullary carcinoma, IDC with a large central acellular zone, and matrix‐producing carcinoma. We plan to examine KIT, EGFR, c‐erbB‐2 overexpressions in these breast carcinoma types, which frequently express mesenchymal and myoepithelial features. KIT and EGFR appeared to be indicators of high‐grade breast carcinoma groups that often contain the carcinomas with mesenchymal and/or myoepithelial differentiation, which are distinct from the group with c‐erbB‐2 overexpression.
Acknowledgments
This work was supported in part by Grants‐in‐Aid for Cancer Research (16–6) and for Scientific Research from the Ministry of Health, Labor, and Welfare, Japan; by a Grant‐in‐Aid for special research, National Defense Medical College, and by the Chugai Pharmaceutical Co., Japan, the Astrazeneca Co., Japan, and the Taiho Pharmaceutical Co., Japan.
References
- 1. Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfield JH, Ashman LK, Kanayama Y, Matsuzawa Y, Kitamura Y, Kanakura Y. Identification of mutations in the coding sequence of the proto‐oncogene c‐KIT in human mast cell leukemia cell line causing ligand‐independent activation of c‐KIT product. J Clin Invest 1993; 92: 1736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baselga J. The EGFR as a target for anticancer therapy – focus on cetuximab. Eur J Cancer 2001; 37: S16–S22. [DOI] [PubMed] [Google Scholar]
- 3. Isozaki K, Hirota S, Nakama A, Miyagawa J, Shinomura Y, Xu Z, Nomura S, Kitamura Y. Disturbed intestinal movement, bile reflux to the stomach, and deficiency of c‐kit‐expressing cells in Ws/Ws mutant rats. Gastroenterology 1995; 109: 456–64. [DOI] [PubMed] [Google Scholar]
- 4. Qiu FH, Ray P, Brown K, Barker PE, Jhanwar S, Ruddle FH, Besmer P. Primary structure of c‐kit: Relationship with the CSF‐1/PDGF receptor kinase family‐oncogene activation of v‐KIT involves deletion of extracellular domain and C terminus. EMBO J 1998; 7: 1003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yarden Y, Kuang WJ, Yang‐Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A. Human protooncogene c‐kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J 1987; 6: 3341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Tunio GM, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain‐of‐function mutations of c‐KIT in human gastrointestinal stromal tumors. Science 1998; 279: 577–80. [DOI] [PubMed] [Google Scholar]
- 7. Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen C‐J, Xiao S, Tuveson DA, Demetri GD, Fletcher CDM, Fletcher JA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res 2001; 61: 8118–21. [PubMed] [Google Scholar]
- 8. Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c‐KIT mutation on prognosis of gastrointestinal stromal tumors. Cancer Res 1999; 59: 4297–300. [PubMed] [Google Scholar]
- 9. Matsuda R, Takahashi T, Nakamura S, Sekido Y, Nishida K, Seto M, Seito T, Sugiura T, Ariyoshi Y, Takahashi T, Ueda R. Expression of the c‐KIT protein in human solid tumors and in corresponding fetal and adult normal tissue. Am J Pathol 1993; 142: 339–46. [PMC free article] [PubMed] [Google Scholar]
- 10. Chui X, Egami H, Yamashita J, Kurizaki T, Ohmachi H, Yamamoto S, Ogawa M. Immunohistochemical expression of the c‐KIT proto‐oncogene product in human malignant and non‐malignant breast tissues. Br J Cancer 1996; 73: 1233–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palmu S, Söderström KO, Quazi K, Isola J, Salminen E. Expression of C‐KIT and HER‐2 tyrosine kinase receptors in poor‐prognosis breast cancer. Anticancer Res 2002; 22: 411–4. [PubMed] [Google Scholar]
- 12. Tsuura Y, Suzuki T, Honma K, Sano M. Expression of c‐KIT protein in proliferative lesions of human breast: Sexual difference and close association with phosphotyrosine status. J Cancer Res Clin Oncol 2002; 128: 239–46. [DOI] [PubMed] [Google Scholar]
- 13. Hayes DF, Thor AD. c‐erbB‐2 in breast cancer: Development of a clinically useful marker. Semin Oncol 2002; 29: 231–45. [DOI] [PubMed] [Google Scholar]
- 14. Revillion F, Bonneterre J, Peyrat JP. ERBB2 oncogene in human breast cancer and its clinical significance. Eur J Cancer 1998; 4: 791–808. [DOI] [PubMed] [Google Scholar]
- 15. Tsutsui S, Ohno S, Murakami S, Hachitanda Y, Oda S. Prognostic value of epidermal growth factor receptor (EGFR) and its relationship to the estrogen receptor status in 1029 patients with breast cancer. Breast Cancer Res Treat 2002; 71: 67–75. [DOI] [PubMed] [Google Scholar]
- 16. Walker RA, Dearing SJ. Expression of epidermal growth factor receptor mRNA and protein in primary breast carcinomas. Breast Cancer Res Treat 1999; 53: 167–76. [DOI] [PubMed] [Google Scholar]
- 17. Malzahn K, Mitze M, Thoenes M, Moll R. Biological and prognostic significance of stratified epithelial cytokeratins in infiltrating ductal breast carcinomas. Virchows Arch 1998; 433: 119–29. [DOI] [PubMed] [Google Scholar]
- 18. Wetzels RH, Holland R, Van Haelst UJ, Lane EB, Leigh IM, Ramaekers FC. Detection of basement membrane components and basal cell keratin 14 in noninvasive and invasive carcinomas of the breast. Am J Pathol 1989; 134: 571–9. [PMC free article] [PubMed] [Google Scholar]
- 19. Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high‐throughput molecular profiling of tumor specimens. Nat Med 1998; 4: 844–7. [DOI] [PubMed] [Google Scholar]
- 20. Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Laboratory Invest 2000; 80: 1943–9. [DOI] [PubMed] [Google Scholar]
- 21. Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi OP, Sauter G. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 2001; 159: 2249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoos A, Urist MJ, Stojadinovic A, Mastorides S, Dudas ME, Leung DH, Kuo D, Brennan MF, Lewis JJ, Cordon‐Cardo C. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol 2001; 158: 1245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hendriks Y, Franken P, Dierssen JW, De Leeuw W, Wijnen J, Dreef E, Tops C, Breuning M, Brocker‐Vriends A, Vasen H, Fodde R, Morreau H. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 2003; 162: 469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization. International Histological Classification of Tumors No. 2. Histological Typing of Breast Tumors. 2nd edn. World Health Organization, Geneva 1981; 17–25. [Google Scholar]
- 25. Japanese Breast Cancer Society. General Rules for Clinical and Pathological Recording of Breast Cancer, 15th edn. Kanehara Shuppan, Tokyo 2004.. (In Japanese.) [Google Scholar]
- 26. Fleiss JL. Measuring nominal scale agreement among many raters. Psychol Bull 1971; 76: 378–82. [Google Scholar]
- 27. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33: 159–74. [PubMed] [Google Scholar]
- 28. Wargotz ES, Deos PH, Norris HJ. Metaplastic carcinomas of the breast. II. Spindle cell carcinoma. Hum Pathol 1989; 20: 732–40. [DOI] [PubMed] [Google Scholar]
- 29. Wargotz ES, Norris HJ. Metaplastic carcinomas of the breast. I. Matrix‐producing carcinoma. Hum Pathol 1989; 20: 628–35. [DOI] [PubMed] [Google Scholar]
- 30. Rosen PP. Breast Pathology. Lippincott‐Raven, Philadelphia 1997; 355–95. [Google Scholar]
- 31. Tot T. The cytokeratin profile of medullary carcinoma of the breast. Histopathology 2000; 37: 175–81. [DOI] [PubMed] [Google Scholar]
- 32. Tsuda H, Takarabe T, Hasegawa F, Fukutomi T, Hirohashi S. Large central acellular zones indicating myoepithelial tumor differentiation in high‐grade invasive ductal carcinomas as markers of predisposition to lung and brain metastases. Am J Surg Pathol 2000; 24: 197–202. [DOI] [PubMed] [Google Scholar]
- 33. Heinrich MC, Blanke CD, Druker BJ, Corless CL. Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT‐positive malignancies. J Clin Oncol 2002; 20: 1692–703. [DOI] [PubMed] [Google Scholar]
- 34. Heinrich MC, Corless CL, Demetri GD, Blanke CD, Von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 2003; 21: 4342–9. [DOI] [PubMed] [Google Scholar]
- 35. Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno‐Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the Bcr‐Abl tyrosine kinase in chronic myeloid leukemia. N Engl J Med 2001; 344: 1031–7. [DOI] [PubMed] [Google Scholar]
- 36. Demetri GD, Von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 2002; 347: 472–80. [DOI] [PubMed] [Google Scholar]


