Abstract
Rationale
Although patients with obstructive sleep apnea (OSA) have a higher risk for coronavirus disease (COVID-19) hospitalization, the causal relationship has remained unexplored.
Objectives
To understand the causal relationship between OSA and COVID-19 by leveraging data from vaccination and electronic health records, genetic risk factors from genome-wide association studies, and Mendelian randomization.
Methods
We elucidated genetic risk factors for OSA using FinnGen (total N = 377,277), performing genome-wide association. We used the associated variants as instruments for univariate and multivariate Mendelian randomization (MR) analyses and computed absolute risk reduction against COVID-19 hospitalization with or without vaccination.
Results
We identified nine novel loci for OSA and replicated our findings in the Million Veteran Program. Furthermore, MR analysis showed that OSA was a causal risk factor for severe COVID-19 (P = 9.41 × 10−4). Probabilistic modeling showed that the strongest genetic risk factor for OSA at the FTO locus reflected a signal of higher body mass index (BMI), whereas BMI-independent association was seen with the earlier reported SLC9A4 locus and a MECOM locus, which is a transcriptional regulator with 210-fold enrichment in the Finnish population. Similarly, multivariate MR analysis showed that the causality for severe COVID-19 was driven by BMI (multivariate MR P = 5.97 × 10−6, β = 0.47). Finally, vaccination reduced the risk for COVID-19 hospitalization more in the patients with OSA than in the non-OSA controls, with respective absolute risk reductions of 13.3% versus 6.3%.
Conclusions
Our analysis identified novel genetic risk factors for OSA and showed that OSA is a causal risk factor for severe COVID-19. The effect is predominantly explained by higher BMI and suggests BMI-dependent effects at the level of individual variants and at the level of comorbid causality.
Keywords: sleep apnea; GWAS; COVID-19; Mendelian randomization
Obstructive sleep apnea (OSA) affects ⩾15% of the population and is considered a sleep disorder that connects directly with cardiometabolic health (1, 2). In OSA, the airways collapse during sleep, which causes acute breathing cessations, an acute decrease in oxygen saturation levels, acute and long-term increase in blood pressure, and prolonged low-grade systemic inflammation (3, 4). Furthermore, high body mass index (BMI) and cardiometabolic diseases are the canonical risk factors or comorbidities of OSA (3). Similarly, a severe or hospitalization-requiring coronavirus disease (COVID-19) infection shares many risk factors and consequences with OSA, most notably lowered oxygen saturation (5). In addition, COVID-19 and OSA are connected by shared comorbidities and underlying conditions such as obesity, type 1 or type 2 diabetes, and coronary heart disease (6, 7).
Since the beginning of the COVID-19 pandemic, the association between OSA and COVID-19, and severe COVID-19 requiring hospital treatment in particular, has been under active research, not only because these diseases share several common risk factors but also because they may worsen each other’s symptoms (8–11). Furthermore, relatively early in the pandemic, patients with OSA were identified as being at high risk for a severe form of COVID-19, and several countries prioritized individuals with OSA for COVID-19 vaccination. In addition, since the deployment of vaccines, the risk for severe COVID-19 overall has decreased. Yet the risk reduction vaccinations provide for hospitalization for COVID-19 has remained elusive in individuals with OSA (12, 13).
The early analyses of the relationship between OSA and severe COVID-19 have been based on epidemiological settings and curated clinical samples and were not designed to assess causality (8–11). In this study, we first estimated the causal relationship between OSA and severe COVID-19 outcomes using univariate Mendelian randomization (MR) (14) and multivariate MR (MVMR) (15). Second, we investigated the risk for hospitalization with COVID-19 among individuals with a previous OSA diagnosis and compared the risk for hospitalization in vaccinated and nonvaccinated individuals with OSA. Similarly, we investigated these risks in a population without an OSA diagnosis.
Methods
Study Population
FinnGen is a large public–private partnership that combines genotype data from Finnish biobanks with digital health record data from Finnish health registries. It planned to collect data from 500,000 individuals by the end of 2023 (16).
FinnGen Release 9 included 377,277 individuals; of these, 38,998 had OSA (Table E1 in the data supplement) and 54,068 had confirmed COVID-19 cases. A total of 603 patients with OSA and 5,006 individuals without an OSA diagnosis had contracted COVID-19. A total of 319,836 individuals had received a COVID-19 vaccine (81.7%). There were no age restrictions in the data.
The OSA diagnosis is based on International Classification of Diseases (ICD), 10th Revision) code G47.3 and ICD, 9th Revision code 3472 (Finnish national version of ICD codes). These diagnoses are validated with individual-level data showing very high accuracy (17). COVID-19 is based on ICD, 10th Revision code U07.1, which requires a positive polymerase chain reaction or antigen test result. Hospitalization data were retrieved as part of the data that were provided for COVID-19–related events, including hospitalization.
Ethics Approval
Patients and control subjects in FinnGen provided informed consent for biobank research, based on the Finnish Biobank Act. Alternatively, separate research cohorts collected before the Finnish Biobank Act came into effect (September 2013) and the start of FinnGen (August 2017) were collected based on study-specific consents and later transferred to the Finnish Biobanks after approval by Fimea (Finnish Medicines Agency), the National Supervisory Authority for Welfare and Health. Recruitment protocols followed the biobank protocols approved by Fimea. The coordinating ethics committee of the Hospital District of Helsinki and Uusimaa statement number for the FinnGen study is Nr HUS/990/2017. Further information is provided at the end of this article.
Outcome Definitions
In the epidemiological analysis, we computed how vaccination affects hospitalization for COVID-19 in those with or without an OSA diagnosis. In addition, we used information about severe COVID-19 in genetic analyses. Severe COVID-19 was defined by hospitalization with laboratory-confirmed COVID-19 infection and death or respiratory support and hospitalization with COVID-19 as the primary reason for admission.
Other Data Sets
The COVID-19 host genetics initiative (https://www.covid19hg.org/) was founded to bring together the human genetic community to generate, share, and analyze data with the goal to learn the genetic determinants of COVID-19 susceptibility, severity, and outcome. The COVID-19 host genetics initiative collects biobank-based data internationally from several sources. The meta-analysis of the COVID-19 host genetics initiative in release 7 covers 18,152 COVID-19–positive patients and 1,145,546 controls. For our MR analysis, we used publicly available summary statistics of the meta-analysis with FinnGen left out from the meta-analysis to avoid sample overlapping.
Because a high BMI indicating obesity (BMI >30 kg/m2) is strongly associated with OSA and severe COVID-19 outcomes, we also used publicly available summary statistics released by the Genetic Investigation of ANthropometric Traits (GIANT) Consortium. This BMI meta-analysis included association results for as many as 339,224 individuals (18).
We replicated our genome-wide significant loci from FinnGen Freeze 9 in an independent cohort using the most recent OSA genome-wide association study (GWAS) from the Million Veteran Program cohort (19).
Genetic Analyses
We performed a GWAS with 38,998 individuals with OSA and 336,659 controls in FinnGen Release 9 using Regenie (20). See data supplement for genotyping and imputation protocol. The GWAS model was adjusted for sex, age, the 10 first principal components, genotype chip, and genetic relatedness matrix, and each locus was fine-mapped using the sum of single effects model (21).
In the MR analyses, we selected independent lead variants for the genetic loci, and the association statistics for each instrumental (lead) variant are shown in Table 1. See data supplement for the respective outcome variant statistics. The outcome was defined as hospitalization with laboratory-confirmed COVID-19 infection and death or respiratory support and hospitalization with COVID-19 as the primary reason for admission. Univariate MR analysis was conducted using the TwoSampleMR R package (version 4.1.3), and instrument single-nucleotide polymorphisms (SNPs) were extracted from the outcome GWAS. We excluded palindromic variants and multiallelic variants. We then harmonized variants for consistency, resulting in 19 SNPs (Table E2 in the data supplement). Two methods were evaluated: MR inverse variance weighted and MR Egger (22). Also, leave-one-out analyses (i.e., remove one variant from the analysis and reestimate the causal effect) were assessed to estimate the reliance of an MR analysis on a particular variant (23).
Table 1.
Characterization of 23 genome-wide significant obstructive sleep apnea loci
| Chr | Position | Ref | Alt | RSIDS | Nearest Genes | Consequence | Info | Fin.enr* | MAF | MAF Cases | MAF Control | β | P Value | BMI-adjusted P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 96567092 | C | CT | rs34013490 | PTBP2 | Intergenic | 0.99 | 0.97 | 0.66 | 0.67 | 0.66 | 0.05 | 6.33 × 10−9 | 3.91 × 10−7 |
| 2 | 102507453 | A | C | rs76229479 | SLC9A4 | Intron | 0.99 | 0.70 | 0.10 | 0.09 | 0.10 | −0.08 | 2.64 × 10−9 | 4.97 × 10−10 |
| 3 | 132779050 | C | T | rs114106239 | NPHP3-ACAD11 | Intron | 0.98 | 2.43 | 0.04 | 0.03 | 0.04 | −0.12 | 1.19 × 10−8 | 1.40 × 10−4 |
| 3 | 169159948 | A | T | rs181010833 | MECOM | Intron | 0.99 | 210.62 | 0.04 | 0.03 | 0.04 | −0.12 | 1.52 × 10−8 | 2.69 × 10−4 |
| 4 | 47311502 | C | CA | rs34925548 | GABRB1 | Intron | 0.98 | 1.19 | 0.70 | 0.69 | 0.70 | −0.05 | 1.12 × 10−8 | 7.15 × 10−4 |
| 6 | 7746430 | T | C | rs60700772 | BMP6 | Intron | 0.99 | 1.02 | 0.22 | 0.23 | 0.22 | 0.05 | 2.20 × 10−8 | 1.18 × 10−5 |
| 7 | 69980355 | A | G | rs11981973 | AUTS2 | Intron | 1.00 | 1.03 | 0.18 | 0.19 | 0.18 | 0.06 | 4.72 × 10−10 | 3.94 × 10−5 |
| 7 | 75029963 | A | C | rs17148727 | RCC1L | Intron | 0.97 | 1.00 | 0.68 | 0.67 | 0.68 | −0.05 | 1.02 × 10−8 | 2.40 × 10−3 |
| 9 | 76652155 | G | A | rs679880 | PRUNE2 | Noncoding transcript exon | 0.99 | 0.92 | 0.75 | 0.75 | 0.74 | 0.05 | 2.53 × 10−8 | 4.36 × 10−5 |
| 9 | 125375595 | C | T | rs10986730 | GAPVD1 | Upstream gene | 0.99 | 1.23 | 0.52 | 0.51 | 0.53 | −0.05 | 1.16 × 10−9 | 3.12 × 10−6 |
| 10 | 18302297 | G | A | rs113955098 | CACNB2 | Intron | 0.98 | 3.26 | 0.07 | 0.06 | 0.07 | −0.10 | 3.38 × 10−10 | 1.01 × 10−4 |
| 10 | 100866753 | G | T | rs61873510 | PAX2 | Intergenic | 0.98 | 0.86 | 0.30 | 0.31 | 0.30 | 0.05 | 3.28 × 10−8 | 1.12 × 10−3 |
| 11 | 28365912 | G | A | rs6484367 | METTL15 | Intron | 0.99 | 0.99 | 0.50 | 0.51 | 0.50 | 0.05 | 1.13 × 10−9 | 4.00 × 10−6 |
| 12 | 56632124 | A | C | rs59333125 | BAZ2A | Intron | 0.99 | 1.13 | 0.08 | 0.08 | 0.08 | −0.08 | 1.32 × 10−8 | 1.28 × 10−5 |
| 12 | 97359374 | C | T | rs10507084 | NEDD1 | Intergenic | 0.99 | 3.03 | 0.18 | 0.19 | 0.18 | 0.06 | 8.23 × 10−11 | 1.22 × 10−7 |
| 12 | 107586546 | C | T | rs2016950 | BTBD11 | Intron | 0.99 | 1.33 | 0.16 | 0.15 | 0.16 | −0.06 | 4.13 × 10−8 | 4.48 × 10−5 |
| 14 | 79424334 | C | T | rs2370982 | NRXN3 | Intron | 1.00 | 1.12 | 0.24 | 0.25 | 0.24 | 0.05 | 7.97 × 10−9 | 8.14 × 10−3 |
| 16 | 53771295 | C | A | rs11075985 | FTO | Intron | 1.00 | 0.98 | 0.43 | 0.45 | 0.43 | 0.08 | 2.15 × 10−26 | 2.59 × 10−5 |
| 16 | 73774720 | C | G | rs13333522 | PSMD7 | Intron | 0.99 | 1.08 | 0.53 | 0.53 | 0.53 | 0.04 | 4.06 × 10−8 | – |
| 19 | 45679046 | T | A | rs10423928 | GIPR | Intron | 1.00 | 1.19 | 0.26 | 0.25 | 0.26 | −0.05 | 7.55 × 10−9 | 3.20 × 10−1 |
| 20 | 52363114 | G | C | rs4809902 | ZFP64 | Intron | 1.00 | 0.78 | 0.23 | 0.22 | 0.23 | −0.06 | 1.51 × 10−9 | 2.59 × 10−5 |
| 20 | 59073311 | C | T | rs140896965 | PRELID3B | Regulatory region | 0.99 | 9.68 | 0.05 | 0.04 | 0.05 | −0.11 | 1.11 × 10−9 | 6.40 × 10−2 |
| 23 | 155574640 | G | A | rs5940397 | TMLHE | Intron | 1.00 | 1.01 | 0.75 | 0.77 | 0.75 | 0.05 | 2.05 × 10−10 | 9.70 × 10−7 |
Definition of abbreviations: Alt = alternate allele; Chr = chromosome; POS = genomic position in build hg38; Ref = reference allele; Info = imputation quality score; MAF = minor allele frequency.
Effect sizes and allele frequencies are reported in terms of alternate allele.
Finnish enrichment, computed using the Genome Aggregation Database data comparing Finnish versus other European populations.
In addition, we conducted an MVMR analysis to estimate and evaluate possible pleiotropic pathways, as the method allows multiple traits as exposures (15). We included SNPs that were genome-wide significant in the FinnGen’s OSA GWAS or the GIANT Consortium’s BMI GWAS (18). We applied the method to estimate the effect of OSA on severe COVID-19 outcome when the model was adjusted for BMI (Table E3 in the data supplement).
To further investigate the independent and shared loci of OSA and BMI, we used the line model (24). This analysis allows a probabilistic clustering of variables based on their observed effect sizes on two outcomes.
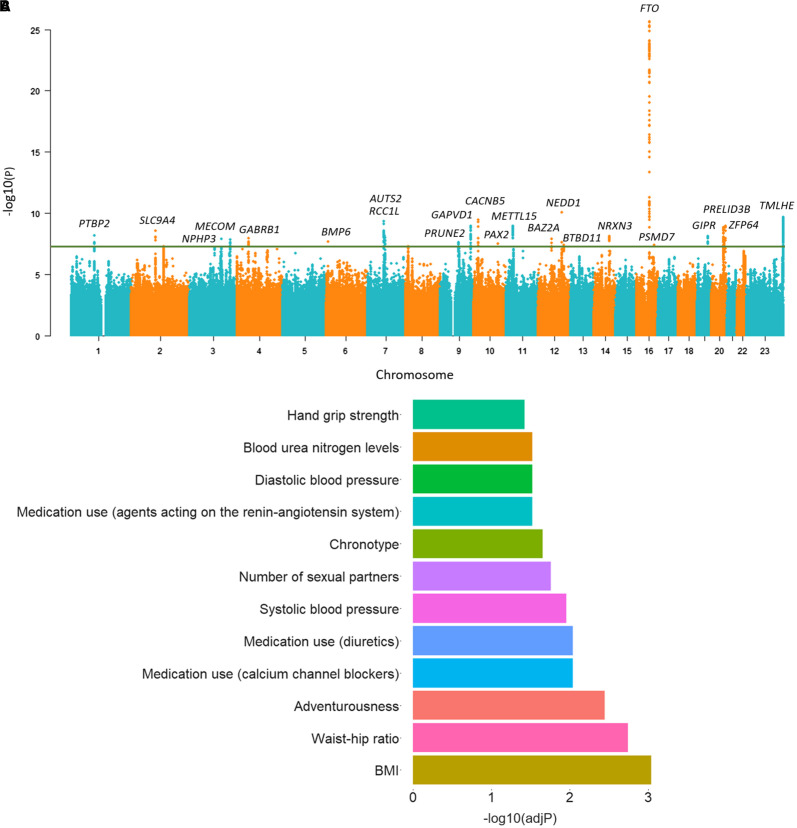
The 23 OSA-related genes (Figure 1A and Table 1) resulting from our OSA GWAS were analyzed using the FUMA web application to obtain information on the biological context in which these proteins are inserted (25). FUMA is an integrative web-based platform that provides pathway enrichment results by performing hypergeometric tests to evaluate whether genes of interest are overrepresented in any of the predefined sets from different categories (25).
Figure 1.
(A) Manhattan plot for the genome-wide association study of obstructive sleep apnea (OSA) including 38,998 patients with OSA and 336,659 controls. For each genetic variant, the x-axis shows chromosomal position and the y-axis shows the −log10(P) value. The horizontal line indicates the genome-wide significance threshold of P = 5 × 10−8. (B) Enrichment analysis of the OSA-related genes into gene sets using the FUMA web application resulted in 28 significantly overrepresented pathways after false discovery rate correction, with one trait per category or disease shown here. BMI = body mass index.
We used the stratified linkage disequilibrium score regression as implemented in LDSC software (26) to explore the tissue enrichment OSA risk variants in a genome-wide setting. We computed the stratified scores for ENCODE (27) (https://www.encodeproject.org/) tissue-specific methylation signals and corrected enrichment P values by false discovery rate (Table E4 in the data supplement). Furthermore, we examined the association of individual significant associations from the OSA GWAS using data from GTEx version 8 (28) as implemented on www.gtexportal.org using correction for the number of variants (n = 24) and tissues (n = 49) tested, whereby a Bonferroni-corrected P value <0.05 corresponds to an uncorrected P value <4.25 × 10−5 (Table E5 in the data supplement).
Epidemiological Analyses
We also analyzed the risk for hospitalization due to COVID-19 among individuals who had a previous OSA diagnosis and compared the risk for hospitalization in vaccinated and nonvaccinated individuals with OSA using the FinnGen data. Individuals who had died or moved abroad before January 1, 2020, were removed from our analyses, reducing the sample size to 336,867 individuals. We had information on hospitalization and vaccination status for the whole population in FinnGen, and the COVID-19, vaccination data, and hospitalization data following COVID-19 were available from February 2020 to May 2022. In these data, there were a total of 603 patients with OSA who had contracted COVID-19 and 5,006 individuals without an OSA diagnosis who had contracted COVID-19. These study participants also had information about vaccination. Individuals were divided into vaccination status groups according to whether the vaccination was received before or after contracting COVID-19 or if the vaccination had not been taken.
To study the effect of vaccination on hospitalization among patients with OSA and those without OSA, we calculated absolute risk reduction (ARR) implemented as the arithmetic difference between the hospitalization rates in the vaccinated and unvaccinated groups. Similarly, we compared the relative risk reduction (RRR) using the grouping as hospitalization rates in the vaccinated and unvaccinated groups. We compared baseline characteristics using the χ2 test for categorical variables and the t test for continuous variables (Tables 2 and 3). In addition, we assessed a multivariate logistic regression model to estimate the adjusted effect of the vaccine to prevent COVID-19–related hospital care. In this analysis, model 1 was adjusted for sex and age; model 2 was adjusted for age, sex, and BMI; and model 3 was adjusted for diabetes, hypertension, coronary heart disease, asthma, and chronic obstructive pulmonary disease in addition to the aforementioned covariates (Tables 4 and 5).
Table 2.
Baseline characteristics of individuals with an OSA diagnosis
| Characteristic | All (n = 603) | Vaccinated (n = 307) | Nonvaccinated (n = 296) | P Value |
|---|---|---|---|---|
| Male sex | 338 (56.1%) | 169 (55.0%) | 169 (57.1%) | 0.672 |
| Age, yr | 59.2 ± 12.6 | 59.5 ± 13.1 | 58.9 ± 12.0 | 0.553 |
| BMI, kg/m2 | 32.9 ± 7.3 | 32.3 ± 6.8 | 33.4 ± 7.7 | 0.093 |
| Diabetes | 179 (29.7%) | 97 (31.6%) | 82 (27.7%) | 0.339 |
| Hypertension | 246 (40.8%) | 131 (42.7%) | 115 (38.9%) | 0.384 |
| CHD | 57 (9.5%) | 33 (10.7%) | 24 (8.1%) | 0.333 |
| Asthma | 114 (18.9%) | 54 (17.6%) | 60 (20.3%) | 0.462 |
| COPD | 28 (4.6%) | 14 (4.6%) | 14 (4.7%) | 1 |
| Hospitalization | 110 (18.2%) | 36 (11.7%) | 74 (25.0%) | 3.89 × 10−5 |
Definition of abbreviations: BMI = body mass index; CHD = coronary heart disease; COPD = chronic obstructive pulmonary disease; OSA = obstructive sleep apnea; SD = standard deviation.
Data presented as mean ± standard deviation where applicable. P values were calculated with a χ2 test for categorical variables (diabetes, hypertension, CHD, asthma, COPD) and a t test for continuous variables (age and BMI). Hospitalization data were retrieved as part of the data that were provided for coronavirus disease (COVID-19)–related events, including hospitalization.
Table 3.
Baseline characteristics of individuals without an OSA diagnosis
| Characteristic | All (n = 5,006) | Vaccinated (n = 2,358) | Nonvaccinated (n = 2,648) | P Value |
|---|---|---|---|---|
| Male sex | 1777 (35.5%) | 842 (35.7%) | 935 (35.3%) | 0.791 |
| Age, yr | 50.7 ± 16.3 | 51.0 ± 16.4 | 50.5 ± 16.3 | 0.295 |
| BMI, kg/m2 | 27.0 ± 5.4 | 26.9 ± 5.3 | 27.1 ± 5.4 | 0.363 |
| Diabetes | 480 (9.6%) | 253 (10.7%) | 227 (8.6%) | 1.10 × 10−2 |
| Hypertension | 750 (15.0%) | 366 (15.5%) | 384 (14.5%) | 3.32 × 10−1 |
| CHD | 187 (3.7%) | 99 (4.2%) | 88 (3.3%) | 1.20 × 10−1 |
| Asthma | 542 (10.8%) | 252 (10.7%) | 290 (11.0%) | 0.799 |
| COPD | 72 (1.4%) | 40 (1.7%) | 32 (1.2%) | 1.84 × 10−1 |
| Hospitalization | 429 (8.6%) | 124 (5.3%) | 305 (11.5%) | 4.26 × 10−15 |
Definition of abbreviations: BMI = body mass index; CHD = coronary heart disease; COPD = chronic obstructive pulmonary disease; OSA = obstructive sleep apnea; SD = standard deviation.
Data presented as mean ± standard deviation where applicable. P values were calculated with a χ2 test for categorical variables (diabetes, hypertension, CHD, asthma, COPD) and a t test for continuous variables (age and BMI). Hospitalization data were retrieved as part of the data that were provided for coronavirus disease (COVID-19)–related events, including hospitalization.
Table 4.
Results of univariate MR analysis between OSA and severe COVID-19 outcome
| Method | SNPs | β | SE | P Value |
|---|---|---|---|---|
| MR Egger | 19 | 0.145 | 0.313 | 0.648 |
| Weighted median | 19 | 0.315 | 0.102 | 0.002 |
| Inverse variance weighted | 19 | 0.241 | 0.073 | 9.41 × 10−4 |
| Simple mode | 19 | 0.207 | 0.172 | 0.243 |
| Weighted mode | 19 | 0.417 | 0.132 | 0.005 |
Definition of abbreviations: COVID-19 = coronavirus disease; MR = Mendelian randomization; OSA = obstructive sleep apnea; SE = standard error; SNP = single-nucleotide polymorphism.
Table 5.
Results of BMI-adjusted multivariate MR analysis between OSA and severe COVID-19 outcome
| Exposure | Outcome | β | SE | P Value |
|---|---|---|---|---|
| BMI | COVID-19 | 0.473 | 0.097 | 5.95 × 10−6 |
| OSA | COVID-19 | 0.135 | 0.10 | 0.180 |
Definition of abbreviations: BMI = body mass index; COVID-19 = coronavirus disease; MR = Mendelian randomization; SE = standard error.
Results
Genome-Wide Association Scan of OSA
We identified 23 independent genome-wide significant (P < 5.0 × 10−8) SNPs (Figure 1A and Table 1). Although all lead variants localized at noncoding exons, introns, or intergenic regions, fine mapping identified six coding variants among the variant credible sets at PRUNE2, MECOM, BAZ2A, GIPR, and GAPVD1 loci (Table E6 in the data supplement). Furthermore, in BMI-adjusted analysis, an OSA association with SLC9A4 locus (P = 4.97 × 10−10) was still observed. Another study found an association with rs77375846 (P = 1.57 × 10−9) (29). This variant is in high linkage disequilibrium with the lead variant rs76229479 from FinnGen (r2 = 0.96), likely reflecting the same signal (FinnGen P = 9.10 × 10−9; BMI-adjusted OSA P = 1.46 × 10−9). In addition, we replicated 13 of the variants with P < 0.05 in the Million Veteran Program cohort (Table E7 in the data supplement). See data supplement for full variant association statistics, fine-mapping results, and instrumental variables used for MR analysis. The enrichment analysis of the variant associated genes into functional categories using FUMA (25) resulted in 28 significantly overexpressed pathways with several obesity-related categories such as BMI, waist/hip ratio, blood pressure, and blood pressure medication (representative traits are shown in Figure 1B, and full trait enrichments are shown in Table E8 in the data supplement).
Concordantly, we observed significant single-tissue expression quantitative trait loci tissues that are important in OSA, including adipose tissue (ACPP, GATSL2, GTF2IRD2, RCC1L, PMS2P5, GTF2IP1, TSHZ2); brain and neuronal tissues (GATSL2, GTF2IRD2, PMS2P5, RCC1L, GAPVD1, OLFML2A, PRPS1P2, RP11–179B2.2); whole blood (RCC1L, PMS2P5, GTF2IRD2, GTF2I, RABEPK, GAPVD1, TSHZ2); connective tissue including muscle, skin, fibroblasts, and esophagus (GATSL2, GTF2IRD2, PMS2P5, GTF2IP1, RCC1L, SPDYE5, GAPVD1, CACNB2, FTO, RCC1L, PMS2P5, GTF2I, SPDYE5, STAG3L2, TSHZ2, PTBP2, STAG3L2, PRPS1P2, METTL15, SNRPD2, RABEPK, SYMPK), heart tissue (CC1L, PMS2P5, GATSL2, GTF2IRD2, SPDYE5); and lung tissue (GTF2IRD2, GATSL2, SPDYE5, GAPVD1, PRPS1P2), with several variants associating with gene expression in more than one relevant tissue (see Table E5). The association of neuronal tissues in particular was seen with stratified linkage disequilibrium score regression, in which we observed enrichment across different brain regions and overall in the brain (fetal brain P = 3.26 × 10−4; FDR-corrected P < 0.05; see Table E4).
Estimating Causality between OSA and COVID-19
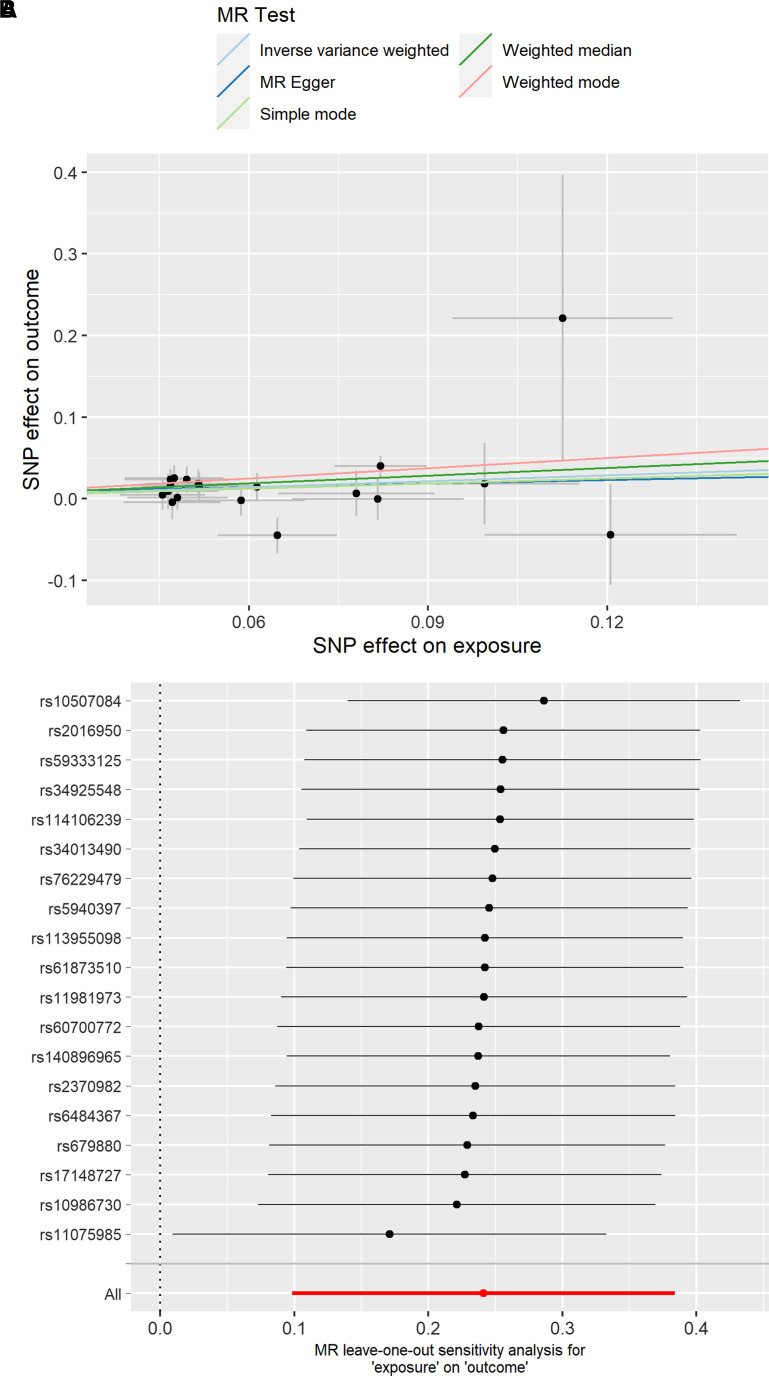
Our aim was to examine the potential causal relationship between OSA and severe COVID-19. After harmonization of OSA and COVID-19 GWAS data, 19 independent SNPs were qualified as instrumental variables for univariate MR analysis (14) to predict the COVID-19 outcome (see Table E2). Our analysis suggests that OSA is a causal risk factor for severe COVID-19 (inverse variance weighted β = 0.241; P = 9.41 × 10−4; Figure 2A and Table 4).
Figure 2.
(A) Univariate Mendelian randomization estimates of the causal effect between obstructive sleep apnea (exposure) and severe coronavirus disease (COVID-19) (outcome) using genome-wide association study results from FinnGen and the COVID-19 Host Genetics Initiative with 17 single-nucleotide polymorphisms (inverse variance weighted β = 0.241; P = 9.41 × 10−4). (B) Leave-one-out analysis removes one variant from the Mendelian randomization analysis and reestimates the causal effect between obstructive sleep apnea and COVID-19. MR = Mendelian randomization; SNP = single-nucleotide polymorphism.
Consequently, to estimate the robustness of our findings and to account for any high effect variant that might be shared between OSA and cardiometabolic traits such as high BMI, we computed leave-one-out analysis by individual variants (23). Exclusion of any single variant did not remove the causal association signal between OSA and severe COVID-19 (Figure 2B). Furthermore, we estimated potential pleiotropy with the MR Egger intercept and did not observe confounding through pleiotropy (Egger intercept P > 0.05).
It is notable that the variant that had the strongest association with OSA and COVID is located at the FTO locus and is well known for its association with BMI (30). Furthermore, our scan identified several other variants that had smaller but significant effects on BMI and OSA. Although OSA is not caused exclusively by high BMI and the effect of craniofacial structures in the predisposition to OSA is well established, it is likely that a significant proportion of the causality on COVID-19 susceptibility is mediated through BMI.
To formally test the connection between OSA and BMI, we performed the following analyses. We first computed genetic correlation between OSA and BMI using data from GIANT (18) and FinnGen and discovered a relatively strong positive correlation between BMI and OSA in both cohorts (GIANT genetic correlation rg = 0.60; P = 6.78 × 10−174, FinnGen rg = 0.50; P = 6.22 × 10−93). Furthermore, using MR, we showed that BMI is also a risk factor for OSA (inverse variance weighted β = 0.60; P = 3.24 × 10−21; MR Egger β = 0.31, P = 9.84 × 10−2). Finally, we examined if some variants that associate with OSA fully reflect the signal from higher BMI using probabilistic modeling (24). We discovered that, although many variants had similar effect sizes between OSA and BMI, two variants were estimated to have a probability greater than 0.95 to be specific to BMI or OSA. The first variant was, perhaps unsurprisingly, the FTO variant that, according to our analysis, reflects signal from BMI and likely contributes to OSA through BMI directly. The second variant was located in the MECOM locus and had a probability of more than 0.95 of reflecting signal only from OSA (BMI-adjusted P = 2.69 × 10−4). These findings are in line with earlier work by our group and others on the genetic etiology of OSA (17, 19, 29).
Given the connection between BMI and OSA, we also estimated the effect of BMI and OSA together on severe COVID-19 risk using MVMR (15) (see Table E2). The effect of BMI on severe COVID-19 infection was strong and significant (β = 0.47; inverse variance weighted P = 5.97 × 10−6). MR Egger was not significant, although the β estimate was consistent in the same direction (MR Egger causal β = 0.07; P = 0.84). Finally, the BMI-independent effect of OSA was not statistically significant, as expected (β = 0.13; P = 0.180). Additionally, we tested MVMR F-statistics for weak instruments (BMI, F = 8.76; OSA, F = 4.79) and horizontal pleiotropy, showing a nonsignificant pleiotropy estimate (Q-statistic for instrument validity, 85.35 on 70 degrees of freedom; P = 0.102). Our multivariate MR analysis suggests that the majority of causal signal from OSA to severe COVID-19 was explained by BMI (Table 5).
Understanding the Role of Vaccination as a Tool to Prevent COVID-19 Hospitalization among Individuals with OSA
We investigated hospitalizations in patients with OSA in two separate patient groups. The first group had received the COVID-19 vaccine, whereas the second group had not been vaccinated. Furthermore, we compared the risk for hospitalization in these groups versus the risk for COVID-19 hospitalizations in the general population in FinnGen without an OSA diagnosis. The baseline characteristics of the participants are presented in Table 3. Furthermore, there were no significant differences at baseline in medication use among the individuals with OSA, whereas metformin and thyroxine use differed at baseline in individuals without OSA (Table E9 in the data supplement).
First, we calculated the protection of the vaccine against hospitalization due to COVID-19 infection by calculating ARR in the OSA group. The number of patients requiring hospital treatment in the vaccinated group was 36 per 307 vaccinated individuals (11.7%), compared with 74 per 296 in the nonvaccinated group (25.5%). Hence, the ARR was 13.3% (95% confidence interval [CI], 7.2–19.4%) and the number needed to treat (NNT) was 8 (95% CI, 5.2–14.0). Similarly, we calculated the relative risk, showing a 53.1% (95% CI, 32.4–67.4%) RRR.
Correspondingly, we investigated how the ARR, NNT, and RRR appeared in the population without an OSA diagnosis. In these analyses, the number of patients requiring hospital treatment in the vaccinated group was 124 per 2,358 vaccinated individuals (5.3%), compared with 345 per 2,648 in the nonvaccinated group (11.5%). In this population, the ARR was 6.3 (95% CI, 4.8–7.8%), NNT was 16 (95% CI, 12.9–21.1), and RRR was 54.3% (95% CI, 44.2–62.7%).
In addition, sex, age, BMI, and other demographic factors affect COVID-19 severity (31), and the raw effect estimates such as ARR, NNT, or RRR do not account for these factors. Therefore, we built a multivariate logistic regression model (Table 6) and used the data of patients with OSA. The raw model (model 1) was adjusted for age and sex, and odds were calculated between hospitalized and nonhospitalized groups, showing a 65% reduced risk (odds ratio [OR] = 0.35; 95% CI = 0.22–0.55; P = 5.74 × 10−6) for hospitalization if a patient had received a vaccination. In addition, we adjusted our model (model 2) for BMI and showed a similar reduced reduction (61%) in the risk for hospitalization (OR = 0.39; 95% CI = 0.23–0.66; P = 5.49 × 10−4). Finally, we adjusted the analysis for comorbidities shared between OSA and COVID-19—diabetes, hypertension, coronary heart disease, asthma, and chronic obstructive pulmonary disease—and showed a 62% reduction in risk (OR = 0.38; 95% CI = 0.22–0.66; P = 0.00596). Similarly, we calculated corresponding models for the non-OSA population and showed significant risk reductions of 65% (OR = 0.35; 95% CI = 0.28–0.45; P < 2.0 × 10−16), 95% (OR = 0.41; 95% CI = 0.31–0.54; P = 5.33 × 10−10), and 66% (OR = 0.38; 95% CI = 0.29–0.51; P = 1.14 × 10−10], respectively.
Table 6.
ORs associating vaccination status with hospitalization among individuals with and without an OSA diagnosis
| Individuals with OSA Diagnosis |
Individuals without OSA Diagnosis |
|||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| Model 1 | 0.35 | 0.22–0.55 | 5.74 × 10−6 | 0.35 | 0.28–0.45 | <2.0 × 10−16 |
| Model 2 | 0.39 | 0.23–0.66 | 0.000549 | 0.41 | 0.31–0.54 | 5.33 × 10−10 |
| Model 3 | 0.38 | 0.22–0.66 | 0.00596 | 0.38 | 0.29–0.51 | 1.14 × 10−10 |
Definition of abbreviations: CI = confidence interval; OR = odds ratio; OSA = obstructive sleep apnea.
Model 1 is adjusted for age and sex; model 2 is adjusted for age, sex, and body mass index; and model 3 is adjusted for diabetes, hypertension, coronary heart disease, asthma, and chronic obstructive pulmonary disease status in addition to the aforementioned covariates.
Discussion
In this study, we performed a large GWAS for OSA using the FinnGen cohort. We discovered 23 loci, replicating previously defined risk loci and identifying 9 novel loci. Our results portray OSA as a multifactorial trait with significant correlation with cardiometabolic traits in particular. Furthermore, our findings support the previously reported connection between OSA and COVID-19. In particular, we examined the causal relationship between OSA and severe COVID-19 and estimated the protection of vaccination against hospitalization due to COVID-19. We discovered a causal relationship that was long suspected, but not previously shown, that OSA increases the risk for a severe COVID-19 outcome mainly through BMI. Our results indicate that BMI is the main driver between OSA and COVID-19 severity in individuals with OSA. Our findings support the earlier epidemiological literature that has shown the association between OSA and severe COVID-19 (8–11). In addition, we show that individuals with OSA benefit from vaccination, which offers particular protection from COVID-19 hospitalization. The ARR and NNT results indicated that the absolute benefit of vaccination in preventing hospitalization for COVID-19 is greater for individuals with OSA than for those without OSA.
Although the association between OSA and severe COVID-19 has been studied recently, their causal relationship has not been formally estimated (8–11). Large biobank-based datasets and MR as a method provide us with a basis to investigate this link without the traditional randomized experimental design (14). It is important to note that the causality we found was largely mediated by obesity. The robustness of BMI as one of the contributing factors was also observed from a genetic basis (17), on which FTO was associated with OSA at a genome-wide significance level and was also the strongest genetic instrument in the causality analysis for severe COVID-19.
Although we observed a significant causal effect of OSA on severe COVID-19 in the MR setting, the finding is primarily related to a higher BMI in the OSA population. This relationship is further supported by multivariate MR, in which primarily BMI was increasing the risk for severe COVID-19 and the causal effect of OSA on severe COVID-19 was not significant after controlling for BMI. These findings further highlight the strong disease burden of BMI in OSA.
Our analysis is an example of how causality analysis by MR can first define a causal relationship between two diseases and then begin elucidating deeper biological relationships. In the case of OSA, it is clear that, although OSA is a risk factor for severe COVID-19, this causal relationship is mediated primarily, if not fully, by BMI.
Vaccinations have been shown to be very effective in preventing COVID-19 infection severe enough to require hospital care (32, 33). However, it remains to be determined whether vaccination is as effective in individuals with OSA as in the general population. In this cohort, approximately 11.5% of nonvaccinated individuals without OSA needed hospitalization. It is noteworthy that the percentage was higher (25.0%) in the OSA population. However, the number of individuals within the OSA population who were vaccinated and had COVID-19 was relatively small (n = 42 with one dose and n = 265 with two doses), and we did not see a significant association for hospitalization between one versus two vaccine doses in the individuals who also had OSA (P = 0.767). Our study shows that the ARR for hospitalization in patients with OSA was 13.3%, compared with 6.3% in the population without an OSA diagnosis. Correspondingly, the NNTs were 8 in the OSA group and 16 in the non-OSA population. The interpretation of this is that, if eight patients with OSA are vaccinated, one hospitalization is prevented. Similarly, in the non-OSA population, 16 individuals would need to be vaccinated to prevent one hospitalization.
The ARR, RRR, and OR of vaccinations on hospitalization reflect the phenomenon in slightly different ways. Thus, we also investigated the RRR in populations with and without OSA, showing reductions of 53.1% and 54.3%, respectively. We also showed, using the adjusted ORs, that vaccination was associated with a reduced risk of hospitalization, reducing the outcome by 61–65% compared with patients with OSA who did not receive the vaccination. This reduction was very similar in the non-OSA population (59–66%). Our results are well in line with previous publications in which vaccinations have been estimated to reduce the risk of hospitalization by 60% in the general population, especially in older age groups (32, 34). This is the first study to demonstrate this risk reduction in the OSA population.
Our research should be interpreted in the light of some limitations. Although our registry data cover a person’s entire life span, including the information concerning vaccination, it does not directly tell us about the severity of OSA. In addition, asymptomatic COVID-19 admissions are also possible. Because we used a control group, we anticipate similar numbers of asymptomatic admissions in the OSA and control populations. Furthermore, our data set is older and enriched with patients with clinical diagnoses, and therefore it does not reflect effects at the population level. Moreover, we did not collect data for OSA therapy. It would be interesting to study whether treating OSA could reduce the risk of severe COVID-19 infection.
A limitation of our study is the lack of information on compliance with continuous positive airway pressure (CPAP) use. However, patients with moderate or severe OSA in the study population were offered CPAP in accordance with national guidelines. Earlier studies in the same population show compliance rates of 65–67% and a mean CPAP use duration of at 5 hours, 20 minutes per day (35). Because CPAP is an efficient treatment for OSA, the compliance with treatment likely affects our findings, increasing heterogeneity in the population between those who use CPAP and those who do not.
In summary, we show that OSA is a causal risk factor for severe COVID-19 and that this risk is mainly mediated by obesity. The ARR was twice as high in the OSA population as in the general population when comparing hospitalizations due to COVID-19 in vaccinated and nonvaccinated groups. Our study suggests that prioritizing patients with OSA as a risk group in the early stage of the COVID-19 pandemic reduced the need for hospitalizations due to COVID-19 infection.
Acknowledgments
Acknowledgment
The authors thank the participants and investigators of FinnGen study. The following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank (www.auria.fi/biopankki), THL Biobank (www.thl.fi/biobank), Helsinki Biobank (www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (www.ita-suomenbiopankki.fi/en), Central Finland Biobank (www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (www.veripalvelu.fi/verenluovutus/biopankkitoiminta) and Terveystalo Biobank (www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi). Finnish Biobank Cooperative -FINBB (https://finbb.fi/) is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious services (https://site.fingenious.fi/en/) managed by FINBB. The authors thank the Million Veteran Program participants and staff. This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award BX004821. This publication does not represent the views of the Department of Veteran Affairs or the United States Government.
The FinnGen study is approved by the Finnish Institute for Health and Welfare (permit numbers: THL/2031/6.02.00/2017, THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019, THL/1721/5.05.00/2019 and THL/1524/5.05.00/2020), Digital and population data service agency (permit numbers: VRK43431/2017-3, VRK/6909/2018-3, VRK/4415/2019-3), the Social Insurance Institution (permit numbers: KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019, KELA 98/522/2019, KELA 134/522/2019, KELA 138/522/2019, KELA 2/522/2020, KELA 16/522/2020), Findata permit numbers THL/2364/14.02/2020, THL/4055/14.06.00/2020, THL/3433/14.06.00/2020, THL/4432/14.06/2020, THL/5189/14.06/2020, THL/5894/14.06.00/2020, THL/6619/14.06.00/2020, THL/209/14.06.00/2021, THL/688/14.06.00/2021, THL/1284/14.06.00/2021, THL/1965/14.06.00/2021, THL/5546/14.02.00/2020, THL/2658/14.06.00/2021, THL/4235/14.06.00/202, Statistics Finland (permit numbers: TK-53-1041-17 and TK/143/07.03.00/2020 (earlier TK-53-90-20) TK/1735/07.03.00/2021, TK/3112/07.03.00/2021) and Finnish Registry for Kidney Diseases permission/extract from the meeting minutes on 4th July 2019. The Biobank Access Decisions for FinnGen samples and data utilized in FinnGen Data Freeze 9 include: THL Biobank BB2017_55, BB2017_111, BB2018_19, BB_2018_34, BB_2018_67, BB2018_71, BB2019_7, BB2019_8, BB2019_26, BB2020_1, Finnish Red Cross Blood Service Biobank 7.12.2017, Helsinki Biobank HUS/359/2017, HUS/248/2020, Auria Biobank AB17-5154 and amendment #1 (August 17 2020), AB20-5926 and amendment #1 (April 23 2020) and it´s modification (Sep 22 2021), Biobank Borealis of Northern Finland_2017_1013, Biobank of Eastern Finland 1186/2018 and amendment 22 §/2020, Finnish Clinical Biobank Tampere MH0004 and amendments (21.02.2020 & 06.10.2020), Central Finland Biobank 1-2017, and Terveystalo Biobank STB 2018001 and amendment 25th Aug 2020.
Footnotes
Supported by the Finnish Medical Foundation, Suomen Hammaslääkäriseura Apollonia, Finnish Sleep Research Society (S.S.), Instrumentarium Science Foundation, Academy of Finland (no. 340539; H.M.O.), FinnGen has been supported by Maze Therapeutics, Sanofi US Services Inc, Bristol-Myers Squibb, Genentech Inc, Janssen Biotech Inc, Novartis, GlaxoSmithKline Intellectual Property Development Ltd, Merck Sharp & Dohme LCC, Boehringer Ingelheim International GmbH, Business Finland grants HUS 4685/31/2016 and UH 4386/31/2016, AstraZeneca, Biogen MA Inc, Pfizer, and AbbVie.
Data availability: Individual-level genotypes and register data from FinnGen participants can be accessed by approved researchers via the Fingenious portal (https://site.fingenious.fi/en/) hosted by the Finnish Biobank Cooperative FinBB (https://finbb.fi/en/). Data release to FinBB is timed to the biannual public release of FinnGen summary results, which occurs 12 months after FinGen consortium members can start working with the data.
Author Contributions: Designed, conducted, and analyzed data: S.S., E.A., V.T., T.K., M.B., T.S., and D.J.G. Mentorship and intellectual contributions: J.K., A.B., A.P., T.P., S.R, and H.M.O. Wrote the manuscript: S.S. and H.M.O. Revised the manuscript: S.S., E.A., V.T., T.K., M.B., S.E.R., J.K., A.B., T.S., D.J.G., A.P., T.P., S.R., and H.M.O.
This article has a data supplement, which is accessible at the Supplements tab.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Benjafield AV, Ayas NT, Eastwood PR, Heinzer R, Ip MSM, Morrell MJ, et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med . 2019;7:687–698. doi: 10.1016/S2213-2600(19)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Senaratna CV, Perret JL, Lodge CJ, Lowe AJ, Campbell BE, Matheson MC, et al. Prevalence of obstructive sleep apnea in the general population: a systematic review. Sleep Med Rev . 2017;34:70–81. doi: 10.1016/j.smrv.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 3. Young T, Skatrud J, Peppard PE. Risk factors for obstructive sleep apnea in adults. JAMA . 2004;291:2013–2016. doi: 10.1001/jama.291.16.2013. [DOI] [PubMed] [Google Scholar]
- 4. Kohler M, Stradling JR. Mechanisms of vascular damage in obstructive sleep apnea. Nat Rev Cardiol . 2010;7:677–685. doi: 10.1038/nrcardio.2010.145. [DOI] [PubMed] [Google Scholar]
- 5. Pena Orbea C, Wang L, Shah V, Jehi L, Milinovich A, Foldvary-Schaefer N, et al. Association of sleep-related hypoxia with risk of COVID-19 hospitalizations and mortality in a large integrated health system. JAMA Netw Open . 2021;4:e2134241. doi: 10.1001/jamanetworkopen.2021.34241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jordan RE, Adab P, Cheng KK. Covid-19: risk factors for severe disease and death. BMJ . 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 7. Mitra AK, Bhuiyan AR, Jones EA. Association and risk factors for obstructive sleep apnea and cardiovascular diseases: a systematic review. Diseases . 2021;9:88. doi: 10.3390/diseases9040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller MA, Cappuccio FP. A systematic review of COVID-19 and obstructive sleep apnoea. Sleep Med Rev . 2021;55:101382. doi: 10.1016/j.smrv.2020.101382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cade BE, Dashti HS, Hassan SM, Redline S, Karlson EW. Sleep apnea and COVID-19 mortality and hospitalization. Am J Respir Crit Care Med . 2020;202:1462–1464. doi: 10.1164/rccm.202006-2252LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strausz S, Kiiskinen T, Broberg M, Ruotsalainen S, Koskela J, Bachour A, et al. FinnGen. Sleep apnoea is a risk factor for severe COVID-19. BMJ Open Respir Res . 2021;8:e000845. doi: 10.1136/bmjresp-2020-000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cariou B, Hadjadj S, Wargny M, Pichelin M, Al-Salameh A, Allix I, et al. CORONADO investigators Phenotypic characteristics and prognosis of inpatients with COVID-19 and diabetes: the CORONADO study. Diabetologia . 2020;63:1500–1515. doi: 10.1007/s00125-020-05180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnish Institute for Health and Welfare. https://thl.fi/en/web/infectious-diseases-and-vaccinations/what-s-new/coronavirus-covid-19-latest-updates/risk-groups-for-severe-coronavirus-disease
- 13. Yek C WS, Wiltz JL, Sun J, Adjei S, Mancera A, et al. Risk factors for severe COVID-19 outcomes among persons aged ⩾18 years who completed a primary COVID-19 vaccination series — 465 health care facilities, United States, December 2020–October 2021. MMWR Morb Mortal Wkly Rep . 2022;71:19–25. doi: 10.15585/mmwr.mm7101a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med . 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 15. Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol . 2019;48:713–727. doi: 10.1093/ije/dyy262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. 2022. https://www.medrxiv.org/content/10.1101/2022.03.03.22271360v1
- 17. Strausz S, Ruotsalainen S, Ollila HM, Karjalainen J, Kiiskinen T, Reeve M, et al. FinnGen Genetic analysis of obstructive sleep apnoea discovers a strong association with cardiometabolic health. Eur Respir J . 2021;57:57. doi: 10.1183/13993003.03091-2020. [DOI] [PubMed] [Google Scholar]
- 18. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature . 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sofer T, Kurniansyah N, Murray M, Ho YL, Abner E, Esko T, et al. Estonian Biobank Research Team; VA Million Veteran Program Genome-wide association study of obstructive sleep apnoea in the Million Veteran Program uncovers genetic heterogeneity by sex. EBioMedicine . 2023;90:104536. doi: 10.1016/j.ebiom.2023.104536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, Ziyatdinov A, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet . 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 21. Zou Y, Carbonetto P, Wang G, Stephens M. Fine-mapping from summary data with the “sum of single effects” model. PLoS Genet . 2022;18:e1010299. doi: 10.1371/journal.pgen.1010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res . 2023;4:186. doi: 10.12688/wellcomeopenres.15555.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corbin LJ, Richmond RC, Wade KH, Burgess S, Bowden J, Smith GD, et al. BMI as a modifiable risk factor for type 2 diabetes: refining and understanding causal estimates using mendelian randomization. Diabetes . 2016;65:3002–3007. doi: 10.2337/db16-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pirinen M. linemodels: clustering effects based on linear relationships. Bioinformatics . 2023;39:btad117. doi: 10.1093/bioinformatics/btad115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun . 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet . 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature . 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science . 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cade BE, Lee J, Sofer T, Wang H, Zhang M, Chen H, et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; TOPMed Sleep Working Group Whole-genome association analyses of sleep-disordered breathing phenotypes in the NHLBI TOPMed program. Genome Med . 2021;13:136. doi: 10.1186/s13073-021-00917-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med . 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Booth A, Reed AB, Ponzo S, Yassaee A, Aral M, Plans D, et al. Population risk factors for severe disease and mortality in COVID-19: a global systematic review and meta-analysis. PLoS One . 2021;16:e0247461. doi: 10.1371/journal.pone.0247461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.https://covid.cdc.gov/covid-data-tracker/#vaccine-effectiveness
- 33. Havers FP, Pham H, Taylor CA, Whitaker M, Patel K, Anglin O, et al. COVID-19-associated hospitalizations among vaccinated and unvaccinated adults 18 years or older in 13 US states, January 2021 to April 2022. JAMA Intern Med . 2022;182:1071–1081. doi: 10.1001/jamainternmed.2022.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steele MK, Couture A, Reed C, Iuliano D, Whitaker M, Fast H, et al. Estimated number of COVID-19 infections, hospitalizations, and deaths prevented among vaccinated persons in the US, December 2020 to September 2021. JAMA Netw Open . 2022;5:e2220385. doi: 10.1001/jamanetworkopen.2022.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kreivi HR, Maasilta P, Bachour A. Willingness score obtained after a short CPAP trial predicts CPAP use at 1 year. Sleep Breath . 2014;18:207–213. doi: 10.1007/s11325-013-0872-x. [DOI] [PubMed] [Google Scholar]