Abstract
The protein product of the pseudorabies virus (PRV) Us9 gene is a phosphorylated, type II membrane protein that is inserted into virion envelopes and accumulates in the trans-Golgi network. It is among a linked group of three envelope protein genes in the unique short region of the PRV genome which are absent from the attenuated Bartha strain. We found that two different Us9 null mutants exhibited no obvious phenotype after infection of PK15 cells in culture. Unlike those of gE and gI null mutants, the plaque size of Us9 null mutants on Madin-Darby bovine kidney cells was indistinguishable from that of wild-type virus. However, both of the Us9 null mutants exhibited a defect in anterograde spread in the visual and cortical circuitry of the rat. The visual system defect was characterized by restricted infection of a functionally distinct subset of visual projections involved in the temporal organization of behavior, whereas decreased anterograde spread of virus to the cortical projection targets was characteristic of animals receiving direct injections of virus into the cortex. Spread of virus through retrograde pathways in the brain was not compromised by a Us9 deletion. The virulence of the Us9 null mutants, as measured by time to death and appearance of symptoms of infection, also was reduced after their injection into the eye, but not after cortical injection. Through sequence analysis, construction of revertants, measurement of gE and gI protein synthesis in the Us9 null mutants, and mixed-infection studies of rats, we conclude that the restricted-spread phenotype after infection of the rat nervous system reflects the loss of Us9 and is not an indirect effect of the Us9 mutations on expression of glycoproteins gE and gI. Therefore, at least three viral envelope proteins, Us9, gE, and gI, function together to promote efficient anterograde transneuronal infection by PRV in the rat central nervous system.
Pseudorabies virus (PRV) is a neurotropic alphaherpesvirus of the Herpesviridae family that is able to infect neurons of both the peripheral and central nervous systems of a wide variety of mammals and birds (4, 39). When PRV replicates and spreads in the nervous system, it acts as a self-amplifying tracer of synaptically connected neurons (10, 16). Integral to this process is the demonstrated ability of the virus to exploit the polarized cellular architecture of neurons and move throughout the brain of a host organism. Thus, assays of the spread of virus through the brain naturally incorporate a determination of the direction of viral transport through a circuit. Direction in the nervous system is defined by the terms “anterograde” and “retrograde,” which can be used to describe directional movement of virus inside a cell as well as the spread of virus between synaptically connected neurons. We use the terms anterograde spread and retrograde spread in this report to describe movement of virus between synaptically connected neurons. Spread from the primary infected neuron to the second-order uninfected neuron in the direction of the nerve impulse is taken to be anterograde spread. Spread in the opposite direction, against the direction of the nerve impulse, is described as retrograde spread. In this report, both of these terms are used to describe the direction of transynaptic viral transport through two model circuits that we have developed to study the role of PRV-encoded envelope proteins in directional spread of virus in the rat nervous system.
Hints that viral gene products affect the direction of spread in herpesvirus neuronal infections come from several studies. For instance, differential-spread patterns for two strains of herpes simplex virus type 1 (HSV-1) have been noted both in the primate motor system and in the murine optic pathway (17, 46). However, neither of these phenotypes has yet been associated with specific HSV-1 gene products. Studies in our laboratory have demonstrated that an attenuated PRV strain called PRV Bartha selectively infects a restricted subset of neurons in the rat visual system (12) and the rat prefrontal cortex (PFC) (9). Similarly, Rotto-Percelay and colleagues (40) demonstrated a restricted invasion by PRV Bartha of neurons innervating the gastrocnemius muscle of rats. In that analysis, virus replicated efficiently in motor and autonomic neurons innervating the muscle but did not replicate in neurons in the dorsal root ganglia (DRG) that provide the sensory innervation of the muscle. Importantly, PCR analysis in that study demonstrated PRV DNA in the sensory neurons. We have further demonstrated that wild-type virus (PRV Becker), but not PRV Bartha, injected into neck musculature of a cat replicates efficiently in DRG neurons serving the inoculated muscles (8). Several laboratories have demonstrated that the absence of the gE and gI genes from PRV Bartha accounts for the restricted spread in neurons in a variety of animal models (1, 3, 10, 12, 22, 25, 26, 29, 31, 45).
The PRV Bartha strain carries a large deletion in the unique short region of the genome that removes not only the gE and gI coding sequences but also all of the Us9 gene and part of the Us2 coding sequences (27, 29, 34). While the phenotypes of individual null mutants of gE, gI, and Us2 have been analyzed, the phenotype of a PRV Us9 null virus in an otherwise wild-type background has never been reported. All previously characterized Us9 mutant viruses contained additional mutations, especially in the neighboring genes, the gI and gE genes (30, 32, 35–37, 43). Therefore, to determine the Us9 null phenotype in vitro and in vivo, we constructed two isogenic strains of PRV Becker containing defined mutations in the Us9 gene. PRV 160 contains a nonsense mutation at amino acid 4, and PRV 161 contains a deletion removing the majority of the Us9 gene open reading frame. In this report, we used these two mutants and their revertants to determine if the Us9 protein plays any role in infection of standard tissue culture cell lines and in directional transneuronal infection of the rodent nervous system by using two well-characterized models of PRV invasiveness. We demonstrate that while mutants lacking the PRV Us9 gene have no obvious phenotypes in tissue culture cells, the mutants are defective in anterograde spread in visual and cortical circuitry.
MATERIALS AND METHODS
Virus strains and cells.
Pig kidney (PK15) cells and Madin-Darby bovine kidney (MDBK) cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All viruses used in this study were propagated on PK15 cells in DMEM containing 2% FBS as previously described (45). PRV 91 contains a deletion removing the gE gene open reading frame and has been described previously (11). PRV 98 contains a deletion removing the gI gene open reading frame (45).
Antisera.
The rabbit polyvalent and mouse monoclonal Us9 antisera have been described previously (5, 6). The gE monoclonal pool (M133, M138, M156) was received from T. Ben-Porat. The rabbit polyvalent gE and gI antisera were kindly provided by K. Bienkowska-Szewczyk (University of Gdansk). Rb133 is a rabbit polyvalent antiserum against acetone-inactivated PRV Becker and recognizes all of the major envelope glycoproteins (11). The polyvalent goat PRV gB (Ab 284) antiserum has been described previously (38).
Virus construction and characterization. (i) Construction of PRV 160 and PRV 161.
PRV 160 is an isogenic strain of PRV Becker which does not express the Us9 protein due to insertion of a nonsense codon at amino acid 4 in the Us9 open reading frame (Fig. 1). To construct PRV 160, a transfer vector in which the phenylalanine (TTC) codon at position 4 of the Us9 open reading frame was changed to a nonsense stop codon (TAA) by site-directed mutagenesis was first engineered. Specifically, oligonucleotide mutagenesis was performed with the Altered Sites kit (Promega) on plasmid pRS3 containing the SphI-MluI region of the PRV BamHI 7 fragment (41). The oligonucleotide (Us9 AflII-1) used to introduce the nonsense stop codon at position 4 also introduced a unique AflII restriction enzyme site at position 3 of the Us9 open reading frame. Addition of the AflII site facilitated the screening of mutated clones and the confirmation of recombinant viruses. The mutagenized pRS3 plasmid was digested with restriction enzymes SphI and MluI to release a 1,004-bp fragment containing the Us9 gene with the nonsense stop codon at position 4. This fragment was then used to replace the SphI-MluI fragment of pPH2, and the resulting plasmid was named pAB24. pPH2 contains the SalI-MluI region of the PRV Becker BamHI 7 fragment encompassing a portion of the gD gene; the gI, gE, and Us9 genes; and the 5′ end of the Us2 gene. The nucleotide sequence of the mutagenized SphI-MluI fragment was confirmed by DNA sequencing with Sequenase (United States Biochemical). The pAB24 transfer vector was cotransfected by the calcium phosphate method (18) with PRV 91 viral DNA, in which the gE sequences are deleted, and the infected cells and medium were collected when total cytopathic effect was observed. The virus stock was replated onto PK15 cells, and recombinant viruses were screened for gE expression by an immunoreactivity assay with a gE monoclonal pool. gE immunoreactive plaques were picked and plaque purified five times.
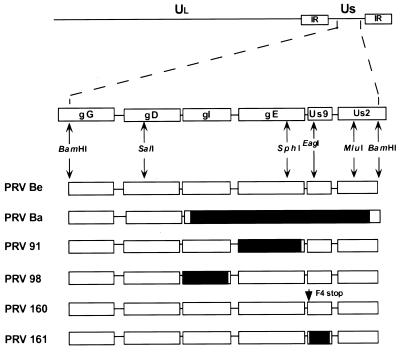
FIG. 1.
PRV genome and map of virus strains used in this study. The BamHI 7 fragment of the unique short (Us) region is expanded to show the genes contained in this fragment. The Us deletion of PRV Bartha (Ba) is indicated by a black box. PRV 91 contains a deletion removing the gE coding sequences. PRV 98 is a gI null virus. PRV 160 contains a nonsense stop mutation (stop) at position 4 in place of a phenylalanine residue (F) in the Us9 open reading frame. PRV 161 contains a 258-bp deletion in the Us9 open reading frame. Relevant restriction enzyme sites used in construction of the various recombinant viruses are indicated. Ul, unique long region; PRV Be, PRV Becker; IR, internal repeat.
PRV 161 is an isogenic strain of PRV Becker which does not express the Us9 protein due to a 258-bp deletion (from a total of 294 bp in the Us9 gene open reading frame) removing the nucleotide sequence encoding amino acids 4 through 89 of the Us9 gene open reading frame (Fig. 1). To construct PRV 161, a transfer vector was constructed by site-directed mutagenesis on plasmid pRS3 as described above for the construction of PRV 160. As for PRV 160, oligonucleotide Us9 AflII-1 was used to introduce an AflII restriction site at position 3 followed by a nonsense codon at position 4 in the Us9 gene open reading frame. In addition, a second oligonucleotide (Us9 AflII-2) was used in the same mutagenesis reaction to introduce an additional AflII site near the 3′ end of the Us9 gene at position 89. The resulting mutagenized plasmid, therefore, contained an AflII restriction site at both the 5′ and 3′ ends of the Us9 gene. Digestion of the doubly mutagenized plasmid with AflII released a 258-bp fragment containing the majority of the Us9 gene. The AflII-digested plasmid was then self-ligated, resulting in a plasmid with base pairs 8 through 267 of the Us9 gene deleted. The SphI-MluI fragment of this plasmid was then used to replace the corresponding fragment of pPH2, thereby constructing the final transfer vector, pAB25. Plasmid AB25 was cotransfected with PRV 91 viral DNA, and gE-immunoreactive plaques were picked and plaque purified as described above for PRV 160.
(ii) Construction of PRV 160 and PRV 161 revertants.
PRV 160R and PRV 161R are isogenic strains of PRV 160 and PRV 161, respectively, in which the mutated Us9 gene was replaced with wild-type Us9 gene sequences by homologous recombination. PRV 160R and PRV 161R were created by cotransfection of either PRV 160 or PRV 161 viral DNA and a gel-purified SphI-MluI PRV Becker fragment. The 1,004-bp SphI-MluI fragment spans from the gE gene cytoplasmic tail to the beginning of the Us2 gene. Recombinant viruses were screened for with a black-plaque immunoreactivity assay with polyvalent Us9 antiserum. Both PRV 160R and PRV 161R were subjected to five rounds of plaque purification.
(iii) Southern blot analysis.
Southern blot analysis was used to confirm the desired mutations in all the viruses described above. Both the nonsense mutation in PRV 160 and the deletion in PRV 161 created a novel AflII site in the Us9 gene. For PRV 160, digestion of the BamHI 7 fragment with AflII converts a 6,613-bp fragment to two fragments of 5,708 and 905 bp. For PRV 161, the 6,355-bp BamHI 7 fragment is cleaved by AflII into a 5,708-bp fragment and a 647-bp fragment. PRV 160R and PRV 161R were examined by Southern blot analysis for the loss of the unique AflII site in the BamHI 7 fragment.
(iv) Black-plaque immunoreactivity assay.
Monolayers of PK15 cells were infected with virus to yield approximately 500 plaques per 10-cm-diameter dish and were overlayed with DMEM containing 1% Methocel and 2% FBS. After approximately 48 h, the Methocel solution was aspirated and the plaques were washed three times with phosphate-buffered saline (PBS). The plaques were then incubated in primary antibody for 1 h at room temperature on a rocking platform. For gE immunoreactivity assays, the gE monoclonal pool was first mixed 1:1:1 and then diluted 1:10 in PBS containing 3% bovine serum albumin (BSA). For Us9 immunoreactivity assays, the rabbit polyvalent Us9 antiserum was diluted 1:200 in PBS plus 3% BSA. After incubation with the primary antibody, the plaques were washed three times with PBS and incubated in a peroxidase-labeled goat anti-mouse (gE monoclonal pool) or anti-rabbit (Us9) secondary antibody (Kirkegaard and Perry) diluted 1:200 in PBS plus 3% BSA. Following a 1-h incubation in the secondary antibody, the plates were rinsed three times with PBS and the immunoreactive plaques were detected by the addition of a detection solution containing 10 mg of 4-chloro-1-naphthol (Sigma)/ml and 0.3% hydrogen peroxide. Positive plaques were picked and resuspended in 1 ml of DMEM plus 2% FBS.
(v) Plaque size determination.
Monolayers of MDBK cells in 60-mm-diameter dishes were infected with virus to yield approximately 200 plaques per dish. After approximately 48 h of infection, the plaques were visualized by a black-plaque immunoreactivity assay with gB antiserum (1:200). The assay was performed essentially as described above except that the plaques were first treated with 2% paraformaldehyde for 10 min at room temperature to fix the cells prior to addition of the primary antibody (diluted in PBS containing 3% BSA and 0.5% saponin). The diameters of 20 random immunoreactive plaques were measured on an Olympus inverted microscope with an ocular reticule.
(vi) Western blot analysis.
Monolayers of PK15 cells were infected with PRV Becker, PRV 160, PRV 161, PRV 160R, or PRV 161R at a multiplicity of infection (MOI) of 10. At 15 h after infection, the cells were washed with PBS and harvested in lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.8], 1% Triton X-100, 1% sodium deoxycholate). Viral particles were isolated from the medium of infected cells by centrifugation through sucrose as described previously (5). Approximately 10 μg of total cellular extract or 1 μg of purified virions was separated on a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel and transferred to nitrocellulose (Amersham) by a semidry transfer apparatus. The Us9, gE, and gI proteins were visualized by incubation of the nitrocellulose with primary antibodies followed by enhanced chemiluminescence detection (SuperSignal; Pierce).
Animal infections. (i) Animals.
Adult male Sprague-Dawley rats weighing 210 to 290 g were used in this assay. The photoperiod was standardized to 14 h of light and 10 h of dark (light on at 0600 h). Food and water were freely available throughout the experiments. Experimental protocols were approved by the Animal Welfare Committees at Princeton University and the University of Pittsburgh and were consistent with the regulations of the American Association for Accreditation of Laboratory Animal Care and those in the Animal Welfare Act (Public Law 99-198). During the course of the experiment, all animals were confined to a biosafety level 2 laboratory and experiments were conducted with the specific safeguards noted by Enquist and Card (14). Animals were deeply anesthetized by intraperitoneal or intramuscular injection of ketamine (60 mg per kg of body weight) and xylazine (7 mg/kg) prior to all experimental manipulations.
(ii) Intravitreal injections.
Injection of PRV Becker into the vitreous humor of the rat eye leads to first-order infection of retinal ganglion cells and subsequent anterograde transynaptic infection of all regions of the brain that receive retinal input (10, 12). Infection of retinorecipient nuclei occurs in two separated waves that target the dorsal geniculate nucleus (DGN) and superior colliculus (SC), areas involved in visual perception and reflex movements of the eyes, followed by regions of the diencephalon (suprachiasmatic nuclei [SCN] and intergeniculate leaflets [IGL]) responsible for imparting temporal organization to behavior and physiological processes (10, 12). Identical injection of PRV Bartha and strains isogenic with PRV Becker but lacking gE or gI produces a restricted pattern of central infection characterized by replication of virus in the SCN and the IGL and a failure of virus to replicate in the DGN or SC (10–12, 45). Using this model, we examined the invasiveness of the Us9 mutants in 11 rats. Infection of the retina was performed as described previously (12). Briefly, 2.5 μl of virus (approximately 108 to 109 PFU/ml) was injected into the vitreous humor of the left eyes of anesthetized animals with a Hamilton microliter syringe equipped with a 26-gauge sharpened needle. The inoculum was injected slowly over a period of 10 min, and the needle was left in the eye for an additional 10 min in an effort to reduce leakage of virus into the orbit. Four additional animals received intravitreal injection of 4 μl of either PRV 160 or PRV 161 (two animals each) to determine if the viral concentration affected the pattern of anterograde infection. For the complementation experiments, a 1:1 (vol/vol) mixture of PRV 161 (4 × 108 PFU/ml) and either PRV 91 (gE null; 6 × 108 PFU/ml) or PRV 98 (gI null; 7 × 108 PFU/ml) was prepared just prior to inoculation. The animals were anesthetized and killed by transcardiac infusion of buffered aldehyde solutions when symptoms of infection were overt. Survival times for animals infected with wild-type virus or with Us9 null strains ranged from 60 to 86 h and 67 to 112 h, respectively. The brains were then removed and prepared for immunohistochemical localization of infected neurons by procedures detailed below.
(iii) PFC injections.
Anterograde transneuronal spread of viral mutants resulting from intracerebral injection of mutants into the PFC was characterized in 35 rats. This model takes advantage of the unidirectional projection between the PFC and the subjacent striatum and was used in a prior analysis to demonstrate differences in the levels of invasiveness of PRV Bartha and PRV Becker (9). The basic organization of the circuitry that it is based upon is described in that paper. Injections were conducted in the following manner according to the methods established by Card and colleagues (9). The heads of anesthetized animals were secured in a stereotaxic frame, and 100 to 200 nl of virus was injected into the medial PFC by using coordinates derived from the rat brain atlas of Paxinos and Watson (33). Virus was injected slowly over a period of 5 min with a 1-μl syringe equipped with a 26-gauge sharpened needle, and the needle was left in the brain for 10 min following completion of the injection to reduce reflux of the inoculum along the injection tract. After survival times of 47 to 66 h, animals were anesthetized and killed for immunohistochemical localization of infected neurons by the procedures described below.
(iv) Immunohistochemical procedures.
Following transcardiac perfusion fixation with buffered paraformaldehyde solutions the brain was postfixed in the primary fixative and cryoprotected in a 20% phosphate-buffered sucrose solution. Each brain was then sectioned serially in the coronal plane with a freezing microtome at 35 μm. Six bins of tissue were collected and transferred to a glycol-based cryopreservant (44) and stored at −20°C until they were processed for immunohistochemical localization of infected neurons. We have previously shown that this method of tissue storage preserves the antigenicity of viral antigens for a minimum of 6 years. One bin of sections (a section frequency of 210 μm) was processed for immunohistochemical localization of infected neurons with a rabbit polyclonal antiserum (Rb133) generated against acetone-inactivated PRV Becker. This procedure utilized the avidin-biotin immunoperoxidase procedure developed by Hsu and colleagues (19), species-specific affinity-purified secondary antibodies (Jackson ImmunoResearch), and Vectastain Elite reagents (Vector Laboratories). After immunoperoxidase processing, sections were mounted on gelatin-coated slides, dehydrated with a graded ethanol series, cleared with xylene, and coverslipped with Cytoseal60 (Stephens Laboratories). Specific details regarding this procedure have been published previously (7, 14).
RESULTS
Construction of Us9 null viruses.
We constructed isogenic strains of PRV Becker containing two different null mutations in the Us9 gene. As the transcription of this region is complex and not well understood, deletion mutations may alter local transcripts qualitatively and quantitatively. Accordingly, we constructed a Us9 nonsense mutant as well as a deletion mutant to increase confidence that the phenotypes we determined reflected the loss of Us9 expression. PRV 160 contains a nonsense mutation at position 4 of the Us9 open reading frame. PRV 161 contains a 258-bp deletion removing the majority of the Us9 coding sequences. The construction of these viruses is described in Materials and Methods (Fig. 1).
Us9 null viruses express gE and gI proteins.
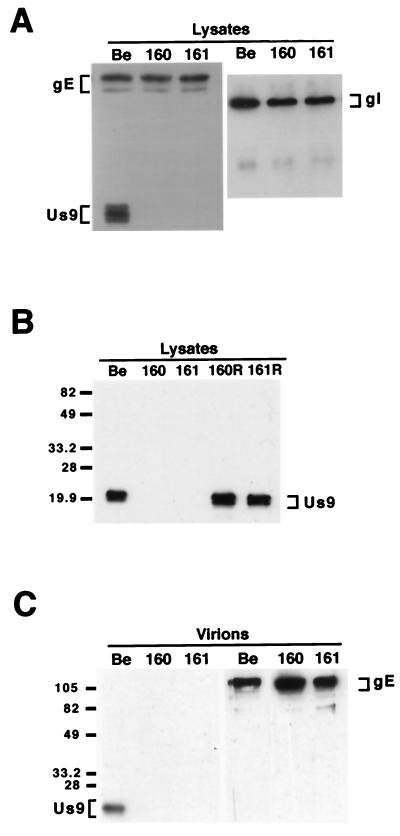
To ensure that PRV 160 and PRV 161 no longer expressed the Us9 protein yet maintained wild-type expression levels of the gE and gI proteins, we examined the steady-state levels of Us9, gE, and gI in PRV Becker-, PRV 160-, and PRV 161-infected cells. Cell lysates were prepared from monolayers of PK15 cells infected with PRV Becker, PRV 160, or PRV 161 at an MOI of 10 and were Western blotted with Us9, gE, or gI antiserum. As anticipated, Us9-immunoreactive polypeptides were detected in PRV Becker-infected PK15 cells but were absent from PRV 160- and PRV 161-infected cell lysates (Fig. 2A). The Us9 protein is present in PRV Becker-infected cell lysates as three polypeptides ranging from approximately 17 to 20 kDa. Western blot analysis with gE and gI antisera revealed that PRV Becker, PRV 160, and PRV 161 all produced identical species of both the gE and gI proteins (Fig. 2A). The gE protein is present in infected cell lysates as both an immature precursor from migrating at approximately 93 kDa and a mature glycosylated form migrating at approximately 110 kDa. The gI protein is expressed as a 65-kDa precursor that is glycosylated to a mature form, which is not well recognized by the gI antiserum used in this experiment. As the steady-state levels of the gE and gI proteins in the PRV 160- and 161-infected cells are indistinguishable from those in PRV Becker-infected cells, we are confident that the engineered mutations in the Us9 gene did not interfere with expression of either gE or gI proteins. The expression and processing of the glycoproteins gB and gC were also unaffected by the loss of Us9 (data not shown). Figure 2B shows the result of Western blot analysis with Us9-specific antiserum of PK15 cells infected with PRV Becker, PRV 160, PRV 161, and the revertant viruses PRV 160R and PRV 161R. The wild-type steady-state level of the Us9 protein in both PRV 160R and PRV 161R clearly demonstrates that the full-length Us9 gene has been restored to these viruses.
FIG. 2.
Western blot analysis of Us9 null viruses and revertants. Monolayers of PK15 cells were infected with PRV Becker, PRV 160, and PRV 161 (A and B) and the revertant viruses PRV 160R and PRV 161R (B) at an MOI of 10. Cellular extracts were prepared after 15 h of infection, and approximately 10 μg of total cell lysate was fractionated on an SDS–12.5% polyacrylamide gel, transferred to nitrocellulose, and analyzed by Western blotting with gE, gI, or Us9 antiserum. (C) Viral particles were isolated from the medium of PK15 cells infected for 15 h with either PRV Becker, PRV 160, or PRV 161 (MOI = 10) by centrifugation through sucrose. Virions were analyzed by Western blotting with either gE or Us9 antiserum. The migration of molecular mass markers is indicated on the left in kilodaltons.
We also examined purified viral particles by Western blot analysis to ensure that the absence of the Us9 protein did not interfere with the incorporation of other glycoproteins into the virion envelope. Specifically, we isolated virions from the medium of PK15 cells infected with PRV Becker, PRV 160 and PRV 161 (MOI = 10) by centrifugation through sucrose. The purified virions were separated on an SDS–12.5% polyacrylamide gel and Western blotted with antisera against Us9 and gE. As anticipated, only PRV Becker virions incorporated Us9 into the viral envelope as PRV 160 and PRV 161 do not express the Us9 protein (Fig. 2C). Western blot analysis with gE-specific antiserum revealed that gE was efficiently incorporated into both PRV 160 and PRV 161 viral particles despite the absence of Us9 (Fig. 2C). Further examination of these viral particles showed that the absence of Us9 also did not affect the incorporation of either the gI or gC membrane proteins into the virion envelope (data not shown).
Us9 null mutants have no observable defects in growth or cell-to-cell spread.
The ability of a virus to form plaques on monolayers of cultured epithelial cells is believed to reflect cell-to-cell spread of a virus in vivo. PRV gE and gI null viruses replicate as well as wild-type virus in single-step growth experiments but form small plaques on MDBK cells (20, 21, 47). We performed single-step growth analysis with PRV 160 on PK15 cells. PRV 160 produced and released equivalent amounts of virus with the same kinetics as those observed for both the wild-type strain, PRV Becker, and the revertant strain PRV 160R (data not shown). To test if the Us9 null viruses have a defect in cell-to-cell spread as measured by plaque size in tissue culture, confluent monolayers of MDBK cells were infected with approximately 200 PFU of PRV Becker, PRV 160, or PRV 161. The sizes of PRV Bartha and PRV 91 plaques on MDBK cells were also measured for comparison. The results of this experiment are shown in Table 1. Unlike PRV Bartha and PRV 91, which form significantly smaller plaques on MDBK cells than PRV Becker (36 and 74% of wild type, respectively), both PRV 160 and PRV 161 formed plaques that were indistinguishable from those of PRV Becker in both shape and size.
TABLE 1.
Plaque size on MDBK cells
| Virus | Plaque size (mm)a | SD (mm) | % Wild type | Pb |
|---|---|---|---|---|
| PRV Be | 0.95 | 0.11 | 100 | |
| PRV 91 | 0.70 | 0.09 | 74 | <0.005 |
| PRV Ba | 0.34 | 0.06 | 36 | <0.005 |
| PRV 160 | 0.95 | 0.10 | 100 | 0.924 |
| PRV 161 | 0.96 | 0.09 | 101 | 0.898 |
An average of 20 plaques measured 48 h after infection.
P with respect to PRV Becker (Be) was determined by Student's t test.
Anterograde transneuronal spread of PRV 160 and PRV 161 through visual projections.
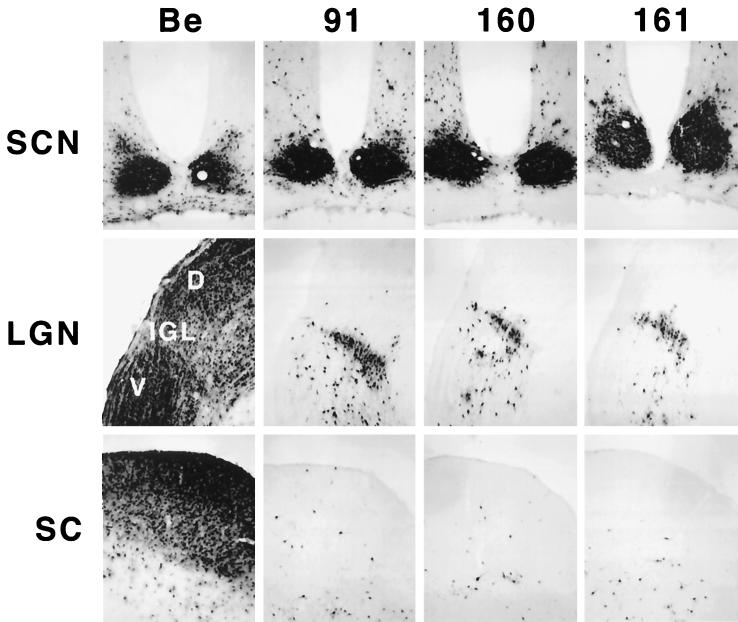
To determine if the Us9 protein plays a role in anterograde spread in the visual system, we injected the Us9 null viruses, PRV 160 and PRV 161, into the vitreous humor and examined the resulting pattern of infection in the brain as described in Materials and Methods. Specifically, we injected 2.5 × 105 to 106 PFU of PRV 160 or PRV 161 (Table 2) into the vitreous body and characterized the central patterns of infection at survival times extending from 67 to 112 h postinoculation. PRV Becker (1.2 × 106 PFU) and PRV 91 (5 × 105 PFU) were injected into separate groups of animals as controls (Table 2). At the time of imminent death, the animals were sacrificed and the brains were removed, fixed, and prepared for immunohistochemical localization of infected neurons. PRV Becker and PRV 91 produced the expected patterns of central infection. Viral antigen was detected in all retinorecipient areas in PRV Becker-infected animals, whereas virus was restricted to the SCN and IGL of animals infected with PRV 91 (Fig. 3). PRV 160 and PRV 161 produced a restricted pattern of infection essentially identical to that produced by PRV 91 and other mutants lacking gE or gI (10–12, 45). Both of the Us9 null mutants produced a robust infection of the SCN and IGL but failed to produce productive replication of virus in the DGN and SC, even though the postinoculation survival intervals substantially exceeded those shown to be adequate for infection of these areas with PRV Becker. Injection of a larger amount of virus (2 × 106 to 5 × 106 PFU in 4 μl) produced the same restricted phenotype, demonstrating that the failure to infect neurons in the DGN and SC was not related to the concentration of the inoculum.
TABLE 2.
Intravitreal injections
| Virus | Genotype | na | PFU/injection (105) | Mean time to death (h) |
|---|---|---|---|---|
| PRV Beb | Wild type | 2 | 12 | 66.9 |
| PRV 91 | gE null | 3 | 5.0–20 | 88.3 ± 16.1 |
| PRV 160 | Us9 null | 6 | 2.5–10 | 96.8 ± 12.9 |
| PRV 161 | Us9 null | 5 | 2.5 | 79.2 ± 8.5 |
| PRV 160R | Revertant | 4 | 5.0–30 | 72.4 ± 7.9 |
| PRV 161R | Revertant | 4 | 5.0–18 | 69.6 ± 11.6 |
Number of animals.
PRV Be, PRV Becker.
FIG. 3.
Anterograde transport of Us9 null viruses in the rodent visual system. Approximately 2.5 × 105 to 2 × 106 PFU of PRV Becker (Be), PRV 91, PRV 160, or PRV 161 was injected into the vitreous humor of Sprague-Dawley male rats. At the time of imminent death, the animals were sacrificed, and the brains were fixed and sliced into 35-μm-thick coronal sections with a freezing microtome. Viral antigen was detected with rabbit polyvalent antiserum Rb133. Representative sections are shown for each virus. LGN, lateral geniculate nuclei; D, dorsal; V, ventral.
The restricted-spread phenotype reflects absence of Us9.
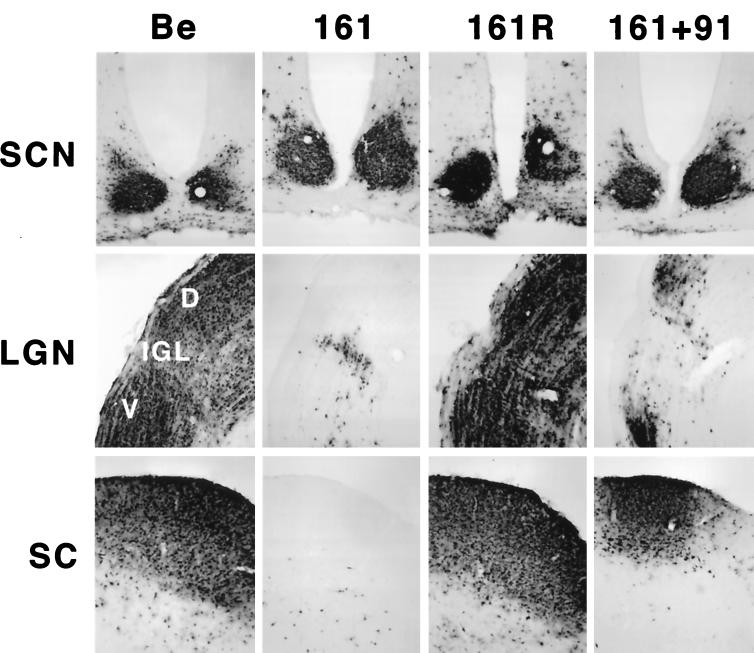
Several measures were taken to ensure that the phenotype observed for the Us9 null viruses was due to the absence of Us9 rather than other mutations. We were particularly concerned that unexpected mutations may have been introduced into the adjacent gE gene during construction of the viruses. First, we sequenced the plasmids containing the Us9 mutations that were used for construction of the recombinant viruses in their entirety in both directions. This analysis demonstrated that there were no additional mutations in regions adjacent to the Us9 mutations in PRV 160 and PRV 161. Second, we constructed revertants of both Us9 mutants by reintroducing the wild-type Us9 gene by homologous recombination. To construct the revertants, PRV 160 or PRV 161 viral DNA was cotransfected with a 1,004-bp fragment of PRV Becker DNA containing the complete Us9 gene sequence as well as 373 bp of upstream flanking gE gene and intergenic sequences and 334 bp of downstream flanking intergenic and Us2 gene sequences. Revertants PRV 160R (data not shown) and PRV 161R (Fig. 4) both produced the wild-type pattern of brain infection after injection into the vitreous body.
FIG. 4.
Reversion and complementation analysis of the Us9 null spread defect. The spread patterns of virus following intraocular injection of animals with PRV Becker (Be), PRV 161, PRV 161R, and a 1:1 (vol/vol) mixed population of PRV 161 and PRV 91 are shown. The infected tissue was processed and analyzed as described in the legend for Fig. 3. Abbreviations are as used for Fig. 3.
Pair-wise, mixed infection of individual mutants restores wild-type spread.
To further test that the spread defect observed for the two Us9 null mutants was due to the absence of Us9 and not because the mutations affected the adjacent gE and gI genes, we used mixed-infection experiments with pairs of gE, gI, and Us9 null mutants as described by Enquist et al. (15). The mixed-infection experiments also tested the hypothesis that the restricted spread of PRV 160 and PRV 161 is due to the lack of Us9 in their viral envelopes such that they no longer can enter into the retinal ganglion cells that project to the DGN and the SC. As demonstrated in Fig. 3, viruses, examined individually in the rat visual system, that lack either gE (PRV 91) or Us9 (PRV 160 and PRV 161) are incapable of infecting the DGN or SC. In the experiment shown in Fig. 4, a 1:1 mixture of a Us9 null virus (PRV 161) and a gE null virus (PRV 91) was injected into the vitreous body. When the animals were exhibiting terminal symptoms, they were euthanized, and their brains were examined for extent of virus spread. We observed marked restoration of anterograde spread of infection to both the DGN and the SC in animals that were injected with both mutant viruses. The smaller numbers of PRV-positive neurons in the DGN and SC of animals coinfected with PRV 91 and PRV 161 compared to those of animals infected with the wild-type strain, PRV Becker, most likely reflects the inefficiency of complementation and the architecture of the circuitry. Whereas both the SC and DGN are infected by anterograde transneuronal passage of virus through retinal ganglion cells, the DGN also receives projections from the SC. Therefore, infection of animals with the wild type strain, PRV Becker, can produce a direct anterograde infection of the DGN through retinal ganglion cells and an indirect infection through the SC. However, in a complementation experiment, both Us9 null and gE null viruses need to enter the same retinal ganglion cell to produce complemented virus that can spread in the anterograde direction to the SC and DGN, and the same process has to occur for indirect infection of the DGN through the SC. We also performed a similar mixed infection with PRV 161 (Us9 null) and PRV 98 (gI null) and observed full restoration of spread to all retinorecipient regions (data not shown). Taken together, these experiments strongly suggest that Us9 is not needed to enter retinal ganglion cells projecting to the visual centers and therefore that the anterograde spread defect associated with the loss of Us9 must occur after viral entry. Moreover, these results further confirm that the gE and gI genes in the Us9 null viruses are fully functional and that the restricted anterograde spread observed for PRV 160 and PRV 161 is due to the loss of the Us9 protein.
Directional spread of Us9 mutants injected into the PFC.
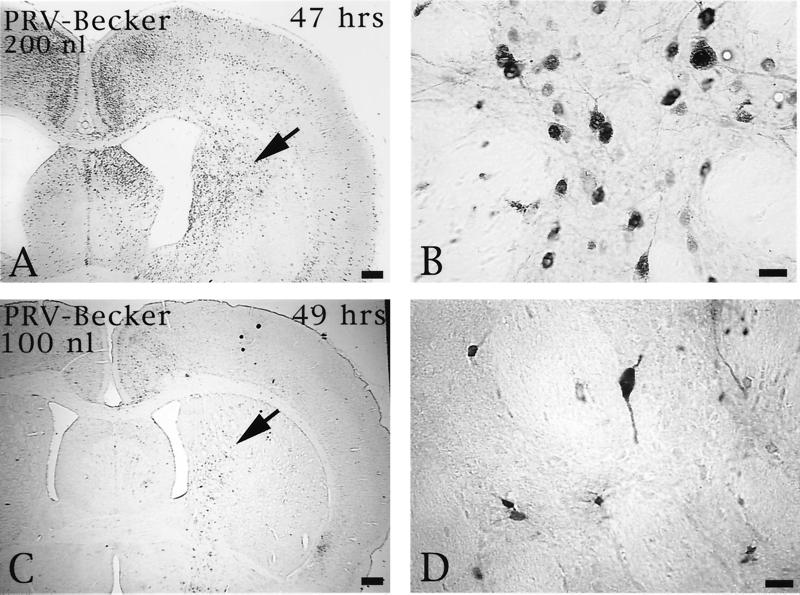
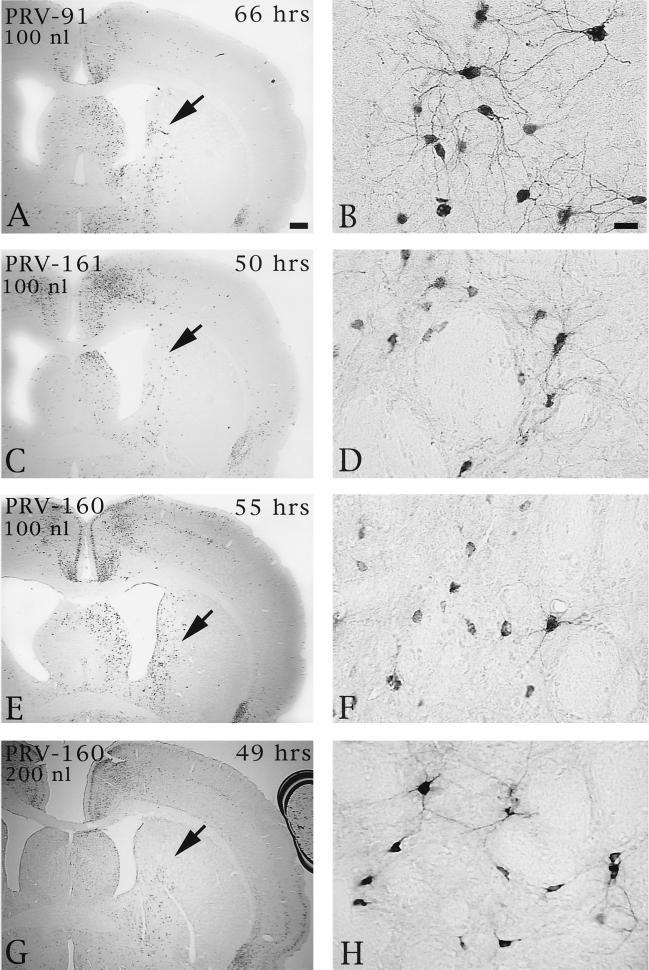
Injection of Us9 null mutants into the PFC permitted an assessment of the role of this envelope protein in both anterograde and retrograde transport through cortical circuits. As demonstrated previously, anterograde transynaptic passage of virus can be determined by the extent of infection in the striatum, an area that receives a projection from the prefrontal cortex but that does not reciprocate the projection (9). Retrograde transynaptic passage of this virus through projections to the PFC from other regions of the cortex and the thalamus were also accurately measured. Two days following injection of 100 or 200 nl of PRV Becker into the PFC, infected neurons were observed in the striatum in a pattern overlapping the known distribution of axons that project to this region from the PFC. Importantly, the magnitude of infection was concentration dependent, with a larger number of infected neurons present in animals infected with 200 nl of PRV Becker (Fig. 5). Deletion of either Us9 or gE reduced the magnitude of anterograde transneuronal infection of striatal neurons and also delayed the temporal sequence of infection (Fig. 6). A similar reduction in anterograde spread to striatal neurons was also observed for a gI null virus (PRV 98) (data not shown). In all cases, the striatum was free of infected neurons at 24 h and contained only a moderate number of infected striatal neurons in the longest-surviving animals. For example, 66 h after injection of PRV 91, there were only scattered infected striatal neurons, which were confined to the region that receives a projection from the injected region of PFC. A similar reduction in anterograde infection of this area was noted 50 and 55 h after injection of 100 nl of PRV 161 and PRV 160 (Fig. 6C and D and E and F, respectively). Examination of longer postinoculation intervals with these viruses was not possible given the increased virulence of PRV 160 and PRV 161 compared to that of PRV 91 or PRV 98 (see below). Importantly, injecting larger concentrations of the Us9 null mutants did not increase the magnitude of anterograde transynaptic infection of the striatum as it did with PRV Becker. This is readily apparent by comparing the restricted pattern of striatal infection 49 h after the injection of 200 nl of PRV 160 (Fig. 6G and H) with the extensive anterograde striatal infection 47 h after injection of an equivalent amount of PRV Becker (Fig. 5A and B).
FIG. 5.
The extent of anterograde transneuronal infection of the striatum approximately 48 h after injection of 100 (C and D) or 200 nl (A and B) of PRV Becker into the PFC. The areas of the striatum indicated by the arrows in panels A and C are shown at higher magnification in panels B and D, respectively. Note that the extent of anterograde transneuronal infection of the striatum was dramatically increased by injection of the higher concentration of virus. Bars: A and C, 550 μm; B and D, 20 μm.
FIG. 6.
The extent of anterograde transneuronal infection of striatal neurons following injection of PRV 91 (A and B), PRV 161 (C and D), or PRV 160 (E and F) into the medial PFC. The areas of the striatum indicated by the arrows in panels A, C, E, and G are shown at higher magnification in panels B, D, F, and H, respectively. Note that the relative magnitudes of striatal infection produced by injection of 100 nl of PRV 160 or PRV 161 into PFC are similar at 50 to 55 h following injection (compare panels C and D with E and F). In contrast, 66 h was required to achieve a similar magnitude of anterograde infection after injection of PRV 91. It is also important to note that injection of 100 (E and F) or 200 nl (G and H) of PRV 160 into the PFC produced similar magnitudes of anterograde transneuronal infection in the striatum. This contrasted with the concentration-dependent differences in the extent of infection produced by PRV Becker (Fig. 5). Bar in A, 500 μm (same scale for panels C, E, and G); bar in B, 20 μm (same scale for panels D, F, and H).
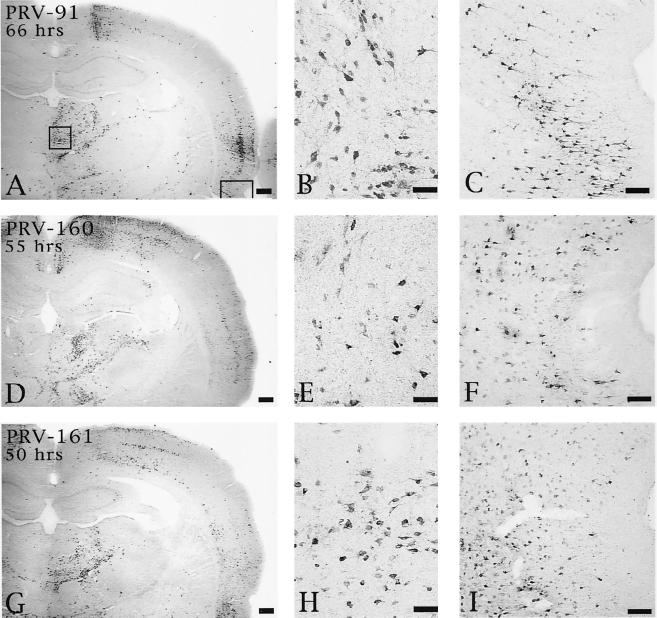
Although deletion of Us9, gE, or gI produced a comparable disruption in anterograde transynaptic infection of striatal neurons, the effects of these deletions upon retrograde spread of virus differed. Rats injected with PRV 91 exhibited delayed retrograde transynaptic transport of virus relative to animals infected with PRV Becker. Whereas PRV Becker exhibited extensive retrograde spread of virus by 48 h (data not shown, but see reference 9), the gE null mutant produced a much more restricted infection of the same circuitry at this postinoculation interval and only achieved a magnitude of retrograde infection equivalent to that of wild-type virus at 66 h (Fig. 7A to C). Similarly, 66 h was required for PRV 91 to produce a retrograde spread of virus that was equivalent to that produced by PRV 160 or PRV 161 at 55 and 50 h, respectively (Fig. 7D to F and G to I).
FIG. 7.
Extent of retrograde infection of thalamus and perirhinal cortex following injection of PRV 91 (A to C), PRV 160 (D to F), or PRV 161 (G to I) into PFC. The boxed areas shown in panel A designate the regions of the thalamus and perirhinal cortex shown at higher magnification in the photomicrographs to the right in each row (thalamus in panels B, E, and H; perirhinal cortex in panels C, F, and I). Note that the levels of retrograde infection produced by all three viruses are similar even though the animal infected with PRV 91 survived 66 h and those injected with PRV 160 and PRV 161 survived 55 and 50 h, respectively. Bars for A, D, and G, 500 μm; bars for B, E, and H, 50 μm; bars for C, F, and I, 100 μm.
Virulence of PRV 160 and PRV 161 in the rat eye model.
Viruses with gE and gI deleted not only have a defect in anterograde spread to the DGN and the SC but also are markedly less virulent in rats than the wild-type virus, PRV Becker, as measured by reduced symptoms of infection and delayed mean time to death (11, 45). Typical symptoms of PRV disease in the rat include ruffling of the fur, nasal discharge, hunched posture, labored breathing, uncoordinated movement, and scratching and rubbing of the face, particularly in the area surrounding the injected eye. These symptoms appear more rapidly after wild-type virus infection and are much more severe by comparison. In contrast, animals infected with mutant viruses lacking the gE or gI genes live longer and exhibit milder symptoms, which are delayed in onset. We found that animals infected with Us9 mutants also lived significantly longer (Table 2) and exhibited delayed, milder symptoms. After infection with PRV Becker, the mean time to death was 66.9 h (n = 2) and severe symptoms appeared on average as early as early as 62 h. For animals infected with PRV 91 (gE null) the mean time to death was 88.3 ± 16.1 h and symptoms appeared on average as early as 82 h but were very mild. For animals infected with PRV 160, the mean time to death was 96.8 ± 12.9 h and mild symptoms appeared on average as early as 93 h. For animals infected with PRV 161, the mean time to death was approximately 79.2 ± 8.5 h and mild symptoms appeared on average as early as 77 h. Animals infected with the revertant viruses PRV 160R or PRV 161R had mean times to death of 72.4 and 69.5 h, respectively. These animals also experienced the rapid and severe symptoms typical for animals infected with wild-type virus. Wild-type virulence was also restored in coinfection experiments of PRV 161 and either PRV 91 or PRV 98 (data not shown).
Virulence of PRV 160 and PRV 161 in the PFC injection model.
When wild-type PRV is injected directly into the PFC, animals rapidly develop severe symptoms and rarely survive longer than 48 h (9). In contrast, when PRV Bartha is injected similarly, animals survive to at least 65 h postinfection with only mild symptoms at time of death. PRV gE and gI null mutants produce somewhat extended times to death compared to wild-type virus, but both PRV Us9 null mutants were as virulent as wild-type virus in this model (Fig. 6). Animals injected with either of the Us9 null mutants exhibited a rapid onset of symptoms as they approached 48 h of survival and died shortly thereafter. However, in both instances the progression to death following the appearance of symptoms was very rapid and animals rarely lived beyond 55 h.
DISCUSSION
The lipid envelope of PRV virions contains at least 16 virally encoded membrane proteins (gB, gC, gD, gE, gI, gH, gK, gL, gM, gN, Us9, UL3, UL11, UL20, UL34, and UL43) (28). Among these envelope proteins, at least six have been shown to be nonessential for growth in tissue culture (28) and are therefore believed to play important roles in the pathogenesis and spread of the virus in vivo. We have previously identified the protein product of one of these non-essential PRV genes, the Us9 gene, to be a 98-amino-acid, phosphorylated membrane protein which is inserted into the viral envelope in a unique type II tail-anchored topology (5). In such a topology, PRV Us9 is predicted to have a 68-amino-acid cytoplasmic tail and a 3-amino-acid ectodomain. PRV Us9 localizes to the trans-Golgi network region in both infected and transfected cells and is maintained in this compartment by endocytosis from the plasma membrane (6). In this study, we constructed two Us9 null viruses and examined their growth properties in vitro and in vivo.
Neither PRV 160 nor PRV 161 has obvious defects associated with the loss of Us9 in vitro. The Us9 null viruses are indistinguishable from wild-type PRV Becker in single-step growth release and plaque size. Moreover, there is no defect with Us9 null viruses in the expression, processing, or intracellular localization (data not shown) of the gE, gI, gB, or gC envelope proteins. Nor does the removal of Us9 from the viral particle affect the incorporation of other viral glycoproteins into the virion envelope.
We next assessed the contribution of Us9 to transneuronal infection following either intravitreal or intracranial infection. Upon direct infection of the retina, PRV 160 and PRV 161 both exhibited an anterograde neuronal spread defect. In this model, both Us9 null viruses were defective in anterograde spread to visual centers involved in visual perception and reflex movement of the eyes, but replicated efficiently in retinal ganglion cells and their target neurons that are involved in biological timing. Interestingly, the pattern of central infection that is produced by PRV 160 or PRV 161 is indistinguishable from that observed in animals infected with either a gE or gI null virus. The restoration of wild-type spread to all retinorecipient areas following infection with the revertant viruses PRV 160R and PRV 161R indicated that the neuronal spread defect of PRV 160 and PRV 161 is due to the loss of Us9 and not to additional mutations. This was also supported by sequencing analysis and mixed-infection experiments with a Us9 null virus and either a gE or gI null virus. A defect in anterograde transynaptic infection was also observed after injection of gE, gI, or Us9 null mutants into the PFC. In prior work we had demonstrated that PRV Becker had the capacity to produce an anterograde transynaptic infection of striatal neurons and further showed that this ability was lacking in rats injected with the attenuated PRV Bartha strain. The data presented in this report strongly suggests that the inability of PRV Bartha to produce an anterograde transynaptic infection through cortical projections to the striatum is related, at least in part, to the absence to the gE, gI, and Us9 genes from the unique short region of the genome. Interestingly, all of the null mutations reduced the magnitude of anterograde infection of the striatum rather than causing a total block, suggesting that the products of these genes function together to increase the efficiency of anterograde infection. Further study is necessary to determine the validity of this hypothesis.
We also examined the contribution of Us9 to virulence in both the rat eye and PFC models. As observed for gE and gI null viruses, the mean time to death of animals infected with either PRV 160 or PRV 161 was extended following intravitreal injection compared to that due to the wild-type controls. Not only did animals injected with the Us9 null viruses live longer than PRV Becker-infected animals, they also displayed less-severe symptoms. In contrast, Us9 null mutants were as virulent as wild-type virus after cortical injection. Further work, however, is necessary to understand these results, as gE and gI null viruses are markedly attenuated in this model. It is noteworthy that a similar differential expression of virulence was observed with an HSV-1 mutant lacking Us9, Us10, Us11, and Us12 following peripheral or central infection (32).
In this report, we show that the Us9 protein, in addition to the gE and gI glycoproteins, is necessary but not sufficient for anterograde spread between some, but not all, synaptically connected neurons in two models of PRV invasiveness. Although mixed-infection experiments in the rat eye model suggest that all three PRV envelope proteins function at a step after entry of the virus into the primary retinal ganglion cell, several observations indicate that Us9 may play a role different from that of gE or gI in promoting anterograde spread. The first observation is that both gE and gI have been shown to form a heterodimer shortly after synthesis in the endoplasmic reticulum (45, 47, 48), and it is often believed that this complex is the functional unit involved in transneuronal spread. However, Us9 does not appear to form a complex with either gE or gI. By standard coimmunoprecipitation experiments, we cannot detect any physical interaction between Us9 and the gE-gI complex in either infected cells or in the viral particle (unpublished data). Further experiments to confirm this observation are in progress, as it is possible that Us9 may interact with gE or gI either transiently or only under certain conditions. The second observation is that both the gE and gI proteins in PRV, as well as in other alphaherpesviruses, are required for efficient cell-to-cell spread in a variety of tissue culture cell lines (2, 20, 21, 23, 24, 47). In contrast, there is no dependence of Us9 on cell-to-cell spread as determined by plaque formation in tissue culture cells. Us9 null viruses form wild-type-sized plaques in all cell lines examined. To account for these observations, we propose a model in which the gE-gI complex is part of the machinery promoting the spread of virus from the presynaptic axon terminal to the postsynaptic cell. These proteins are likely to act at the site of egress and need not be in virus envelopes to promote spread between synaptically connected neurons, as shown by Tirabassi et al. (42). Accordingly, we suggest that the gE-gI complex mediates spread through the interaction of the gE-gI ectodomains with a cellular receptor on the postsynaptic cell. Interaction of the gE-gI complex with this putative receptor would be necessary for entry into secondary neurons in the DGN or SC as spread to the retinorecipient circadian areas is gE-gI independent. A similar model involving the interaction of gE-gI with a cellular ligand has been recently proposed by Dingwell and Johnson for the spread of HSV-1 between epithelial cells (13). Yet the question remains as to how Us9 promotes anterograde spread in this model. It does not seem likely that Us9 has the same role as gE and gI because a Us9 null virus forms wild-type-sized plaques in tissue culture. Nor does it seem probable that the three amino acids that constitute the Us9 ectodomain interact with a receptor on an adjacent cell. Rather, we propose that Us9 serves a transport role in the infected neuron, perhaps delivering newly enveloped viral particles and/or vesicles containing envelope proteins, such as gE and gI, to the axon terminal. In the absence of Us9, neither the virions nor the glycoprotein vesicles would be efficiently targeted to the site of egress, thereby leading to a defect in transynaptic spread. In support of this idea, Us9 has a topology similar to that of proteins involved in vesicular transport, and we have identified several motifs in the Us9 cytoplasmic tail that are important for intracellular trafficking (5, 6). We are currently examining viral mutants defective in Us9 intracellular trafficking to see what effects these motifs have on Us9's ability to promote anterograde spread in the rodent central nervous system.
ACKNOWLEDGMENTS
We gratefully acknowledge the expert technical assistance of J. Goodhouse and Jen-Shew Yen. Plasmid pPH2 was kindly provided by P. Husak. A.D.B. thanks R. Tirabassi and P. Husak for advice and guidance with the rat eye model as well as the members of the Enquist laboratory for careful reading of the manuscript and helpful comments.
A.D.B. was supported by NIH training grant 5T32GM07388. This work was supported by NINDS grant 1RO133506 to L.W.E. and a grant from The John D. and Catherine T. MacArthur Foundation Research Network on Early Experience and Brain Development to J.P.C.
REFERENCES
- 1.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 2.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 3.Banfield B W, Yap G S, Knapp A C, Enquist L W. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J Virol. 1998;72:4580–4588. doi: 10.1128/jvi.72.6.4580-4588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Porat T, Kaplan A S. Molecular biology of pseudorabies virus. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Publishing Corp.; 1985. pp. 105–173. [Google Scholar]
- 5.Brideau A D, Banfield B W, Enquist L W. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau A D, del Rio T, Wolffe E J, Enquist L W. Intracellular trafficking and localization of the pseudorabies virus Us9 type II envelope protein to host and viral membranes. J Virol. 1999;73:4372–4384. doi: 10.1128/jvi.73.5.4372-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Card J P, Enquist L W. The use of pseudorabies virus for definition of synaptically linked populations of neurons. In: Adolph K W, editor. Methods in molecular genetics. Orlando, Fla: Academic Press, Inc.; 1994. pp. 363–382. [Google Scholar]
- 8.Card J P, Enquist L W, Miller A D, Yates B J. Differential tropism of pseudorabies virus for sensory neurons in the cat. J Neurovirol. 1997;3:49–61. doi: 10.3109/13550289709015792. [DOI] [PubMed] [Google Scholar]
- 9.Card J P, Levitt P, Enquist L W. Different patterns of neuronal infection after intracerebral injection of two strains of pseudorabies virus. J Virol. 1998;72:4434–4441. doi: 10.1128/jvi.72.5.4434-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card J P, Rinaman L, Schwaber J S, Miselis R R, Whealy M E, Robbins A K, Enquist L W. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Two alpha-herpesvirus strains are transported differentially in the rodent visual system. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 13.Dingwell K S, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enquist L W, Card J P. Pseudorabies virus: a tool for tracing neuronal connections. In: Lowenstein P R, Enquist L W, editors. Protocols for gene transfer in neuroscience: towards gene therapy of neurological disorders. New York, N.Y: John Wiley & Sons Ltd.; 1996. pp. 333–348. [Google Scholar]
- 15.Enquist L W, Dubin J, Whealy M E, Card J P. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neutropism. J Virol. 1994;68:5275–5279. doi: 10.1128/jvi.68.8.5275-5279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 17.Garner J A, LaVail J H. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J Neurovirol. 1999;5:140–150. doi: 10.3109/13550289909021996. [DOI] [PubMed] [Google Scholar]
- 18.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 19.Hsu S M, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs L, Rziha H J, Kimman T G, Gielkens A L J, van Oirschot J T. Deleting valine-125 and cysteine-126 in glycoprotein gI of pseudorabies virus strain NIA-3 decreases plaque size and reduces virulence in mice. Arch Virol. 1993;131:251–264. doi: 10.1007/BF01378630. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs L. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch Virol. 1994;137:209–228. doi: 10.1007/BF01309470. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs L, Mulder W A, Priem J, Pol J M, Kimman T G. Glycoprotein I of pseudorabies virus (Aujeszky's disease virus) determines virulence and facilitates penetration of the virus into the central nervous system in pigs. Acta Vet Hung. 1994;42:289–300. [PubMed] [Google Scholar]
- 23.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimman T G, de Wind N, Oei-Lie N, Pol J M A, Berns A J M, Gielkens A L J. Contribution of single genes within the unique short region of Aujeszky's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 26.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type and gC-, gE- and gI-deleted mutants of Aujeszky's disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 27.Lomniczi B, Blankenship M L, Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the gene. J Virol. 1984;49:970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettenleiter T. Pseudorabies (Aujeszky's disease) virus: state of the art. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 29.Mettenleiter T C, Lukacs N, Rziha H J. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J Virol. 1985;56:307–311. doi: 10.1128/jvi.56.1.307-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mettenleiter T C, Lomniczi B, Sugg N, Schreurs C, Ben-Porat T. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J Virol. 1988;62:12–19. doi: 10.1128/jvi.62.1.12-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mettenleiter T C, Zsak L, Kaplan A S, Ben-Porat T, Lomniczi B. Role of structural glycoprotein of pseudorabies in virus virulence. J Virol. 1987;61:4030–4032. doi: 10.1128/jvi.61.12.4030-4032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama Y, Kurachi R, Daikoku T, Umene K. The Us 9, 10, 11, and 12 genes of herpes simplex virus type I are of no importance for its neurovirulence and latency in mice. Virology. 1993;194:419–423. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- 33.Paxinos G, Watson C. The rat brain in sterotaxic coordinates. New York, N.Y: Academic Press; 1997. [Google Scholar]
- 34.Petrovskis E A, Timmins J, Gierman T, Post L. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pol J M A, Wagenaar F, Gielkens A. Morphogenesis of three pseudorabies virus strains in porcine nasal mucosa. Intervirology. 1991;32:327–337. doi: 10.1159/000150217. [DOI] [PubMed] [Google Scholar]
- 36.Pol J M A, Quint W G V, Kok G L, Broekhuysen-Davies J M. Pseudorabies virus infections in explants of porcine nasal mucosa. Res Vet Sci. 1991;50:45–53. doi: 10.1016/0034-5288(91)90052-p. [DOI] [PubMed] [Google Scholar]
- 37.Pol J M A, Gielkens A L J, van Oirschot J T. Comparative pathogenesis of three strains of pseudorabies virus in pigs. Microb Pathog. 1989;7:361–371. doi: 10.1016/0882-4010(89)90039-9. [DOI] [PubMed] [Google Scholar]
- 38.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J C, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roizman B. Herpesviridae: a brief introduction. In: Fields B N, Knipe D M, editors. Fundamental virology. 2nd ed. New York, N.Y: Raven Press; 1991. pp. 841–847. [Google Scholar]
- 40.Rotto-Percelay D M, Wheeler J G, Osorio F A, Platt K B, Loewy A D. Transneuronal labeling of spinal interneurons and sympathetic preganglionic neurons after pseudorabies virus injections in the rat medial gastrocnemius muscle. Brain Res. 1992;574:291–306. doi: 10.1016/0006-8993(92)90829-x. [DOI] [PubMed] [Google Scholar]
- 41.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagenaar F, Pol J M A, Peeters B, Gielkens A L J, de Wind N, Kimman T G. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 44.Watson R E, Jr, Wiegand S J, Clough R W, Hoffman G E. Use of cryoprotectant to maintain long-term peptide immunoreactivity and tissue morphology. Peptides. 1986;7:155–159. doi: 10.1016/0196-9781(86)90076-8. [DOI] [PubMed] [Google Scholar]
- 45.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zemanick M C, Strick P L, Dix R D. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci USA. 1991;88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zsak L, Zuckermann F, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuckermann F A, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]