Abstract
INTRODUCTION:
Lung adenocarcinomas harboring EGFR mutations do not respond to immune checkpoint blockade therapy as well as their EGFR wildtype counterpart. The mechanisms underlying this lack of clinical response have been investigated but remain incompletely understood.
METHODS:
We analyzed three cohorts of resected lung adenocarcinomas (PROSPECT, ICON and TCGA) and compared tumor immune microenvironment of EGFR-mutant tumors to EGFR-wildtype (WT) tumors, to identify actionable regulators to target and potentially enhance the treatment response.
RESULTS:
EGFR-mutant NSCLC exhibited low PD-L1, low tumor mutational burden, decreased number of cytotoxic T cells, and low T cell receptor clonality, consistent with an immune-inert phenotype, though T cell expansion ex vivo was preserved. In an analysis of 75 immune checkpoint genes, the top up-regulated genes in the EGFR-mutant tumors (NT5E and ADORA1) belonged to the CD73/adenosine pathway. Single-cell analysis demonstrated that the tumor cell population expressed CD73, both in the treatment-naïve and resistant tumors. Using co-culture systems with EGFR-mutant NSCLC cells, T regulatory cell proportion was decreased with CD73 knockdown. In an immune-competent mouse model of EGFR-mutant lung cancer, the CD73/adenosine pathway was markedly upregulated and CD73 blockade significantly inhibited tumor growth.
CONCLUSIONS:
Our work demonstrated that EGFR-mutant NSLCLC has an immune-inert phenotype. We identified the CD73/adenosine pathway as a potential therapeutic target for EGFR-mutant NSCLC.
INTRODUCTION:
Though immune checkpoint blockade has been successfully utilized in patients with NSCLC, the benefit of immunotherapy for patients with advanced EGFR-mutant NSCLC has been limited. Patients with EGFR-mutant lung cancers experience a response rate of 3–8% to single-agent anti-PD-1/PD-L1(1, 2), compared to 19–45% in EGFR wild-type (WT) tumors(3, 4). The presence of EGFR mutations is often associated with never or light smoking history in a relatively younger patient population(5, 6) with low tumor mutational burden (TMB)(7) and PD-L1 expression(8). The number of CD8+ tumor-infiltrating T cells (TIL) were also found to be low in EGFR-mutant tumors(8). However, it is unclear whether low TMB, PD-L1 and CD8+ T cells are the only mechanisms of resistance to anti-PD-1/PD-L1 therapy in EGFR-mutant NSCLC or whether other factors may contribute to the immune-inert phenotype of EGFR-mutant NSCLC. In a retrospective analysis focusing on high PD-L1 NSCLC (>50%) patients’ response to anti-PD1 immunotherapy, EGFR-mutant tumors responded very poorly (overall response rate [ORR] at 12.5%, 1 responder in 8 EGFR-mutant patients) compared to all never- to light-smoker patients (total cohort n=45, ORR 31.6%)(9), suggesting that smoking status and PD-L1 expression are not the only factors leading to poor responses to anti-PD1/PD-L1 therapy in EGFR-mutant patients.
CD73, encoded by gene ecto-5’-nucleotidase (NT5E), is a known regulator of the immune microenvironment through its effects on the generation of extracellular adenosine(10–13). ATP released from stressed or injured cells can be dephosphorylated from ATP to ADP and AMP by CD39(14), and CD73 then dephosphorylates and converts extracellular adenosine monophosphate (AMP) into adenosine and inorganic phosphate(15). Extracellular adenosine generated by this pathway binds to G-protein-coupled adenosine receptors and inhibits T cell function and promotes T cell apoptosis, leading to immune escape by the tumor(15–17). CD73 expression has been observed in EGFR-mutant NSCLC(10), although whether it represents a potential target for immune modulation in this context is unclear.
Here, we performed an integrated analysis of the tumor immune microenvironment in EGFR-mutant and EGFR-WT lung adenocarcinomas in three independent cohorts of resected NSCLC. We characterized tumor immune regulatory genes, immune cell composition, and the reactivity of T cells in EGFR-mutant tumors. We focused our analyses on identifying key targetable immunosuppressive pathways. This study sheds light on the mechanisms underlying tumor immune suppression and represents a step towards designing appropriate immunotherapies for EGFR-mutant lung cancer patients.
METHODS:
Clinical datasets
We queried three independent datasets. The first dataset is the Profiling of Resistance Patterns of Oncogenic Signaling Pathways in Evaluation of Cancer of Thorax, referred to as the PROSPECT cohort(18). There were 94 surgically resected stage I-III lung adenocarcinomas (LUADs) with adequate clinical and genetic information for analysis. The second dataset is the Immune Genomic Profiling of NSCLC, referred to as the ICON cohort (MDACC PA15–1112). There were 72 surgically resected stage I-III lung adenocarcinomas with adequate clinical and genetic information for analysis. The last dataset is The Cancer Genome Atlas (TCGA, LUAD n=511). There were 511 surgically resected lung adenocarcinomas with adequate clinical and genetic information for analysis.
Immunohistochemistry and immunofluorescent staining
For samples from the PROSPECT cohort, 10 immune markers were tested by immunohistochemistry(18), including PD-L1, PD-1, CD3, CD4, CD8, CD45RO, CD57, CD68, FoxP3 and Granzyme B. PD-L1 expression in malignant cells was analyzed using a cell membrane staining algorism and quantified using a 4-value intensity score and the percentage (0–100%) of the extent of reactivity. H-score (range, 0–300) is a typical approach for scoring these analyses and is obtained by multiplying the percentage of staining positive cells by the staining intensity ordinal (0–3). For ICON cohort, multiplex immunofluorescence (mIF) staining was performed for profiling analysis of PD-1, CD3, CD4, CD8, CD45ro, CD57, GZB, FOXP3, and CD68. The density of cells expressing these markers was evaluated by quantifying positive cells in five selected regions (1 mm2) that were randomly selected during the automated imaging analysis in both intra-tumoral and peritumoral compartments. Each area examined was identified and assessed in the sequential immunohistochemical slides in order to quantify each immune marker at similar spatial locations in the specimen slides. The average total number of cells positive for each marker across the five areas was expressed as cell density per mm2.
TCR sequencing
Immunosequencing of the CDR3 regions of human TCRβ chains was performed using the immunoSEQ Assay (Adaptive Biotechnologies). T cell clonality, density and richness were calculated as previously defined(19). To adjust for TMB between EGFR-mutant and WT tumors, only cases with total synonymous mutations less than 100 were used for comparison for the ICON cohort.
T cell expansion
Tumor-infiltrating lymphocytes (TIL) were cultured from tumor fragments. Briefly, resected lung cancer tumor samples were cut into 1–3 mm2 fragments and placed in complete TIL culture media (TIL-CM: RPMI 1640 with Glutamax supplemented with 2 mM L-Glutamine, 1 mM Pyruvate, 1x of HEPES, 50 μM 2-mercaptoethanol, 1X Pen-Strep (Invitrogen, Carlsbad, CA) and 10% heat-inactivated human AB Serum (Sigma-Aldrich)) with 6,000 IU/mL IL-2 (Prometheus; San Diego, CA) in culture treated 24-well plates for a period of 3 to 5 weeks, as previously described(20).
mRNA expression profiling and fusion detection
For PROSPECT cohort, the array-based expression profiling of PROSPECT tumors was performed using the Illumina Human WG-6 v3 BeadChip according to the manufacturer’s protocol and gene expression data have been previously deposited in the GEO repository (GSE42127). For ICON cohort, the RNA sample libraries were prepared using the SureSelectXT RNA Direct Protocol (version A0, July 2017) from Agilent Technologies and sequenced on the Illumina HiSeq2000 platform. The raw reads were aligned to the hg19 reference genome using STAR (v2.3.0e). Then raw read counts mapped to each gene were obtained using htseq-count (v0.6.1p1). Paired-end transcriptome sequencing reads were aligned to the human reference genome (GRCh37/hg19) using a RNA-seq spliced read mapper Tophat2 (Tophat 2.1.0)(21), with “–fusion-search” option turned on to detect potential gene fusion transcripts. Potential false-positive fusion candidates were filtered out using “Tophat-Fusion-Post” module. For TCGA cohort, we used level-3 TCGA pan-cancer data(22), including RNAseqV2, copy number, mutation, and clinical data. The fusion information was extracted from TCGA fusions dataset (23) and https://www.tumorfusions.org/.
For other oncogene driver annotations, we focused on ALK-, ROS1-, RET-, NTRK1/2/3-fusion, and BRAF V600E and MET exon 14 skipping, based on FDA-approved targeted therapies. They were analyzed as “other” oncogene driver cases. We also annotated KRAS mutations, but kept KRAS cases within the “WT” group. We did not annotate for ERBB2, PIK3CA, PTEN loss, or NRG-fusion. In the TCGA set, we identified the following: ALK-fusion (5 cases), ROS1-fusion (5 cases), RET-fusion (1 case), NTRK2-fusion (1 case), BRAF V600E (3 cases) and MET exon 14 skipping (6 cases). In addition, there were total of 152 cases of KRAS mutations G12C/V/D (143 cases) and G13C/D (9 cases) in the TCGA cohort. In the ICON cohort, there were one EML4-ALK, one NCOA4-RET case, one METex14 case (D1028Y), no BRAF V600E, and 20 KRAS mutations. In the PROSPECT cohort, the fusion detection was not performed, because the dataset was on an elder microarray platform. From the DNA-sequencing data where the EGFR mutation status was called, we identified 31 KRAS mutation cases.
Single-cell RNAseq
Tumor samples were obtained per MDACC PA14–0276 research protocol. The samples were dissociated and prepared for single-cell RNAseq using 10xGenomics platform (Chromium Single Cell 3’ Library & Gel Bead Kit v3). The libraries were sequenced at MDACC genomics core facility. A bioinformatics pipeline for single-cell RNAseq was applied for data analysis. Only EPCAM-positive cells were selected for subsequent analysis. Seraut(24) was used for unsupervised clustering of the single-cell data and UMAP(25) was applied for dimensional reduction.
RPPA analysis
Protein lysate was prepared from pellets from tumor tissues as previously described(26). Briefly, lysis buffer was added to the samples, followed by microcentrifugation. Clear supernatants were collected, followed by protein quantitation. The cell lysate was mixed with SDS sample buffer without bromophenol blue [three parts cell lysate plus one part 4× SDS sample buffer]. Before using the 4X SDS sample buffer, 10% β-mercaptoethanol was added. The samples were boiled for 5 min. Then, the samples were serially diluted with dilution buffer [three parts lysis buffer : one part 4X SDS sample buffer + β-mercaptoethanol]. To each of the diluted samples, an equal amount of 60% glycerol/2× PBS solution was added, after which the diluted samples were transferred to 384-well plates. Reverse-phase protein arrays (RPPA) were produced and analyzed. Protein arrays were printed on nitrocellulose-coated glass slides (GRACE BIO-LABS, Bend, Oregon 97702) using an Aushon Biosystems 2470 arrayer (Aushon Biosystem, Billerica, MA01821). Antibody staining of each array was done using a Dako AutostainerPlusLink (Dako North America Inc. Carpinteia, CA93013) as previously described. Antibody CD73 was purchased from Cell Signaling Technology. Phosphorylated EGFR Y1173 and CD38 antibodies were purchased from abcam. RPPA data were quantified using a SuperCurve method which detects changes in protein level by Microvigene software (VigeneTech) and an R package developed at MDACC.
In vitro experiments:
Cell culture, small interfering RNA and recombinant CD7:
NSCLC cell lines (NCI-H1975 and HCC4006) were grown in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum. Small interfering RNAs targeting CD73 were purchased from Thermo Fisher and transfected by Lipofectamine 2000 (Invitrogen). Recombinant human 5’-nucleotidase/CD73 enzyme (R&D systems) was used at 100 ng/mL.
Conditioned-media PBMC assay and flow cytometry:
Serum-free conditioned media from CD73 siRNA transfected cell lines (H1975 and HCC4006) and from the respectively siRNA control was collected after 48 hours of transfection. Healthy donor peripheral blood mononuclear cells (PBMC) were co-incubated with conditioned media for 12 hours. The PBMCs were Fc-blocked with purified rhesus anti-human (Catalog 564219, BD Pharmingen) for 30 minutes at 4°C. Cells were then stained using the Zombie NIR Fixable Viability kit (Biolegend 423106) and samples were stained for markers using directly labeled primary antibodies against CD3 (Biolegend 200326), CD4 (Biolegend 300506), CD8 (Biolegend 300920), FoxP3 (Biolegend 320126), and Ki-67 (Biolegend 15210). The experiment was performed in triplicates. Cell surface CD73 expression was evaluated using an anti-CD73 antibody (Biolegend 344006). Staining was visualized by fluorescence-activated cell sorting (FACS) analysis using a BD Fortessa X-20 Analyzer (BD Biosciences) and analyzed using FlowJo v10 software (FlowJo LLC).
Immune cell cytotoxicity assay and IFN-gamma ELISA:
Tumor cells (H1975) were seeded in a 96-well plate at 2000 cells/well. Healthy donor PBMCs (Stem Cell Techology, Catalog # 70025.1) were co-cultured with tumor cells at the ratio of PBMCs/Tumor=10:1 for 24 hours. Human anti-CD3/28 antibodies (biolegend, Catalog # 300402, 302902) were added to the culture at the final concentration of 1μg/ml. Human anti-PD1 and anti-CD73 antibodies (Biolegend, Catalog # 621602, 344002) were added to the culture at the final concentration of 5μg/ml. 50μl of cell culture medium was harvested for LDH cytotoxicity assay. Cytotoxicity was evaluated by CyQUANT™ LDH Cytotoxicity Assay Kit (Thermofisher, Catalog # 20300) following the manufacturer’s instructions. The final percentage of lysis was calculated by the formula %lysis=100*(Release-Tumor spontaneous release-PBMC spontaneous release)/(Tumor maximum release- Tumor spontaneous release). 15μl of cell culture medium was harvested for IFN-gamma ELISA. IFN-gamma concentration was evaluated by IFN ELISA kit (Biolegend human IFNg ELISA Max).
Mouse studies
In the EGFR model used in the study is EGFR T790M L858R is under the control of a tet-inducible promoter. Mice were crossed CC10-RTTA restricting the expression of transactivator to club cells. Double mutant mice or CC10RTTA littermates (WT/controls) were fed doxycycline diet to induce EGFR expression and tumor formation as before (27). Once tumors were confirmed by MRI, mice were euthanized and tumors were collected. For IHC: Tissues were fixed in 10% formalin, embedded and sectioned. Immunohistochemistry was performed using Cell signaling Rabbit monoclonal antibody against CD73 (NT5E) # 13160, 1/200, following the manufacturer’s instructions. For microarray expression analysis: lung tumors (n=10) or healthy lungs (n=12) were snap-frozen in liquid nitrogen and subjected to microarray analysis in the previous study (28). Metabolite profiling data was also extracted from the same study(28). Briefly, metabolites were extracted from collected lung tumors and healthy lung tissue and Mass spectrometry was performed. Treatment studies: Once the tumors were confirmed by MRI mice were either treated with vehicle (n=6) or CD73 blocking antibody (n=4) (BioXcell, in vivo monoclonal anti-mouse CD73 antibody clone: TY/23) twice a week, 100ug/dose per mouse. Mice were imaged with Bruker 7T MRI at Dana-Farber Cancer Institute Lurie Family Imaging Center. MRI quantification was done using 3D slicer software https://www.slicer.org. Tumors were highlighted in 8 slices in the raw files by the observer for the software to determine and calculate a volume for each mouse(29). All mouse work was approved by DFCI institutional animal care and use committee.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism software (version8) or the R system. The unpaired t-test was used for comparisons between two group means, where the data could be assumed to be from a normal or near-normal distribution. Mann-Whitney U test was used to compare the mean ranks between two groups. All p values are two-tailed, and for all analyses, p<0.05 is considered statistically significant, unless otherwise specified.
RESULTS:
EGFR-mutant tumors exhibited lower PD-L1 expression and tumor mutational burden
We queried three independent datasets of surgically resected stage I-III lung adenocarcinomas (LUADs): MD Anderson Cancer Center (MDACC) PROSPECT (18), MDACC ICON and TCGA. We identified tumors with EGFR activating mutations (EGFR-mutant) in each dataset, 14 in PROSPECT, 15 in ICON and 70 in TCGA and tumors without EGFR driver mutations (WT, see Methods). For tumors without EGFR driver mutations, further annotation was performed for TCGA and ICON datasets to identify other oncogenic driver alterations (see Methods and Table 1). We applied various approaches to profile the tumor immune characteristics, immune cell compositions, T cell reactivities and expansion of the EGFR-mutant tumors. Due to various success rates for each assay, we used all available cases for analyses. The number of cases in each cohort, for each assay, by genotype was detailed in Table. 1.
Table 1.
PROSPECT, ICON and TCGA datasets and analyses performed.
| Cohorts | PROSPECT | ICON | TCGA | |||||
|---|---|---|---|---|---|---|---|---|
| Immune markers | yes | yes | no | |||||
| EGFR 14 | WT 80 | EGFR 15 | others 3 | WT 35 | ||||
| TMB | no | yes | yes | |||||
| EGFR 15 | others 3 | WT 35 | EGFR 70 | others 21 | WT 420 | |||
| TCR analysis | yes | yes | no | |||||
| EGFR 18 | WT 168 | EGFR 15 | others 3 | WT 54 | ||||
| Flow cytometry | yes | no | ||||||
| EGFR 8 | others 1 | WT 22 | ||||||
| T cell expansion | yes | no | ||||||
| EGFR 13 | others 3 | WT 32 | ||||||
| RPPA analysis | yes | no | ||||||
| EGFR 14 | others 2 | WT 29 | ||||||
| Expression data | yes | yes | yes | |||||
| EGFR 14 | WT 80 | EGFR 15 | others 3 | WT 35 | EGFR 70 | others 21 | WT 420 | |
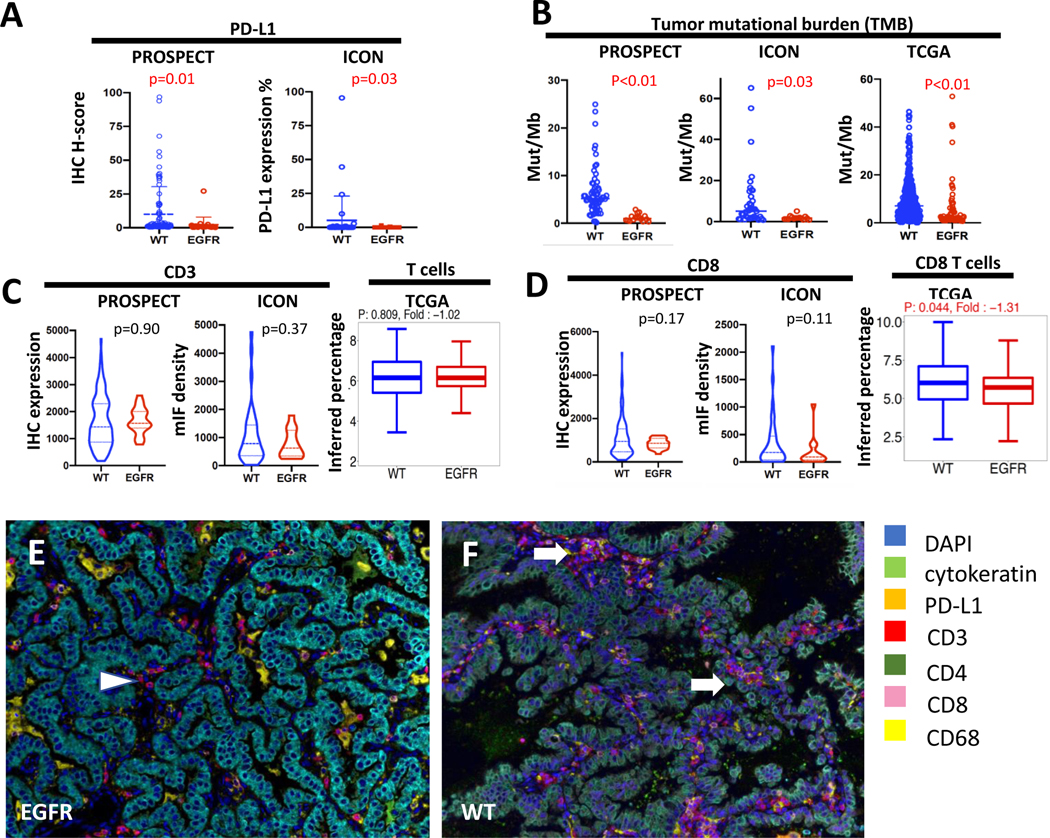
We first evaluated tumor features known to affect immunotherapy response such as PD-L1 expression and tumor mutational burden (TMB). Consistent with prior studies(8), PD-L1 expression on the malignant cells was significantly lower in EGFR-mutant tumors compared to EGFR WT tumors in both datasets (0 vs 7.28, p=0.05 by IHC H-score in PROSPECT cohort; 0.17 vs. 5.19%, p=0.03, in the ICON cohort by density by multiplex immunofluorescence [mIF](30), Fig. 1A). TMB was also significantly lower in EGFR-mutant tumors (1.19 vs. 6.51 mut/Mb, p<0.01 in PROSPECT cohort; 1.82 vs. 9.99 mut/Mb, p=0.03 in ICON cohort; 6.13 vs. 9.70 mut/Mb, p<0.01 in TCGA cohort, Fig. 1B), consistent with the previous studies(7). When other driver-oncogene tumors were removed from the analysis, the TMB remains to be relatively low in the EGFR-mutant tumors compared to WT (1.82 vs. 10.67 mut/Mb, p=0.03 in ICON cohort; 6.13 vs. 9.04 mut/Mb, p=0.086 in TCGA cohort, Supplementary Figure 1A).
Figure 1.
EGFR-mutant tumors had lower PD-L1, TMB and CD8+ T cells. (A) PD-L1 by IHC H score in PROSPECT and density in ICON, (B) Tumor mutational burden by mutations per megabase (mut/Mb) in PROSPECT, ICON, TCGA, (C) CD3+ T cells by IHC in PROSPECT, density by multiplex immunofluorescent (mIF) in ICON, and the percent of inferred T cells using MCP Counter in TCGA, (D) CD8+ T cells by IHC in PROSPECT, density by immunofluorescent in ICON, and the percent of inferred CD8+ T cells using MCP Counter in TCGA, (E-F) representative mIF images from EGFR-mutant and WT tumor sections. Color keys are at the right of the images. White arrows and arrowhead indicate CD3+CD8+ T cells.
The number of CD8+ T cells was reduced in EGFR-mutant lung adenocarcinomas
We then characterized the immune cell composition in the tumor immune microenvironment. In the PROSPECT and ICON cohorts, CD3+ T cells were not significantly different in the EGFR-mutant vs. WT groups (1631 vs. 1657, p=0.90 in PROSPECT by IHC with EGFR n= 14 vs. WT n=80, 771 vs. 1068, p=0.37 in ICON by mIF with EGFR n= 15 vs. WT n=35, Fig. 1C). In both PROSPECT and ICON, EGFR-mutant tumors showed a trend towards lower CD8+ T cell infiltration in the tumor immune microenvironment compared to WT, although these results did not meet statistical significance (833 vs. 1123, p=0.17 in PROSPECT by IHC with EGFR n= 14 vs. WT n=80; 189 vs. 344, p=0.11 in ICON by mIF with EGFR n= 15 vs. WT n=35, Fig. 1D–1F). CD4+ T cells were found to be higher in EGFR-mutant tumors in the PROSPECT cohort (p=0.029), whereas FoxP3+ T regulator cells did not show any difference (Supplementary Fig 2A). Other immune cell populations, including CD20+ and CD68+ cells, were not significantly different between EGFR-mutant and WT tumors (Supplementary. Fig 2B). When other driver-oncogene tumors were removed from the analysis, the EGFR-mutant versus WT analyses results remained largely unchanged (Supplementary Fig. 2C). Because immune marker staining was not available for the TCGA dataset, we used the computational tool MCP Counter(31) to infer immune cell composition. Consistent with the direct immune marker staining in PROSPECT and ICON cohorts, the total CD3+ T cell population was no different between EGFR-mutant (n=70) and WT (n=421) tumors (6.10 vs. 6.13, p=0.81, Fig. 1C), whereas the fraction of CD8+ T cells was lower in EGFR-mutant tumors (5.62 vs. 6.02, p=0.044, Fig. 1D). The inferred immune cell population analysis also showed lower NK cells, cytolytic T cell populations, but higher myeloid dendritic cells, in EGFR-mutant tumors, whereas B cells and monocytes did not show any difference (Supplementary Fig. 3A and Supplementary Table 1). When other driver-oncogene tumors were removed from the analysis, the EGFR-mutant versus WT analyses results remained largely unchanged (Supplementary Fig. 1B and Supplementary Fig. 3B). It is worth noting that other oncogene driver tumors had similar immune cell composition to EGFR-mutant tumors with all comparisons having non-significant differences (Supplementary Fig. 1B).
Flow cytometry was performed in resected tumor samples from the ICON cohort when the immune cells were successfully isolated (EGFR-mutant, n=8 and WT n=23). Using total alive cells as the parental gate, CD45+, CD3+, CD4+, CD8+ populations were not different between the two groups (Table 2. top panel), overall consistent with IHC and mIF results.
Table 2.
Flow cytometry to compare T cell subpopulations. The top panel used alive cells as the parental gate. The bottom panel used CD45+ alive cells (total immune cells) as the parental gate. (Abbreviations: SSC-A: Side SCcatter parameter A; GRZB: granzyme B; PERF: perforin; Tregs: T regulatory cells, GITR: Glucocorticoid-Induced TNFR-Related protein; Tconv: T conventional cells, defined as CD4+FOXP3-CD25low; CD8+ naïve T cells: defined as CD8+CCR7+CD45RA+; Avg: Average; Diff: difference)
| Population | Avg, EGFR | Avg, WT | Diff. | p Value |
|---|---|---|---|---|
| CD45+ SSC-A subset | 32.47 | 32.59 | −0.12 | 0.991 |
| CD3+ | 20.05 | 17.39 | 2.66 | 0.730 |
| CD4+ | 12.79 | 9.42 | 3.37 | 0.457 |
| CD8+ | 6.09 | 6.67 | −0.58 | 0.840 |
| Population | Avg, EGFR | Avg, WT | Diff. | p Value |
| CD8+ | 25.58 | 33.53 | −7.94 | 0.079 |
| CD8+ PD1+ | 7.80 | 20.12 | −12.32 | 0.000 |
| CD8+ TIM3+ | 0.81 | 2.87 | −2.07 | 0.001 |
| CD8+ Ki67+ | 2.41 | 4.53 | −2.12 | 0.028 |
| CD8+ GRZB+ PD1+ | 0.35 | 1.25 | −0.90 | 0.016 |
| CD8+ GRZB+ PERF1+ | 0.62 | 2.18 | −1.56 | 0.026 |
| CD8+ CD103+ | 7.20 | 13.22 | −6.02 | 0.045 |
| CD8+ CD103- | 12.82 | 10.31 | 2.51 | 0.415 |
| CD8+ Naïve | 0.06 | 0.25 | −0.19 | 0.039 |
| CD4+ | 64.03 | 53.70 | 10.33 | 0.046 |
| CD4+ Tregs | 1.32 | 1.25 | 0.06 | 0.911 |
| CD4+ Tregs GITR+ | 1.66 | 0.40 | 1.26 | 0.001 |
| CD4+ Tconv | 39.56 | 30.66 | 8.90 | 0.072 |
Next, we used CD45+ cells to further dissect the relative composition of leukocyte subpopulations (Table 2, bottom panel). CD4+ T cells were higher (64% vs. 53%, p=0.043) while CD8+ T cells were lower (25% vs. 33%, p=0.079) in the EGFR-mutant tumors. CD8+PD1+ (activated T cells, 7.8% vs. 19.7%, p<0.01), CD8+Ki67+ (proliferating T cells, 2.4% vs. 4.5%, p=0.028), CD8+ naïve (0.06% vs. 0.25%, p=0.039) T cells were significantly lower in EGFR-mutant tumors. Among CD4+ T cells, Tregs were similar between EGFR-mutant and WT tumors (1.3% vs. 1.2%, p=0.91), but CD4+ conventional T cells showed a trend towards higher in the EGFR-mutant tumors (39% vs. 30%, p=0.07). When other driver-oncogene tumor were removed from the analysis, the EGFR-mutant (n=8) versus WT (n=22) analyses results remained largely unchanged (Supplementary Table 2).
Taken together, our immune cell population analyses from immune staining as well as flow cytometry demonstrated that the total T cell population was not significantly different in EGFR-mutant versus WT tumors, whereas CD8+ T cells were decreased but not depleted in EGFR-mutant lung adenocarcinomas. By flow cytometry, we dissected the proportion of each functional subgroups of T cells, and found activated and proliferating CD8+ T cells (CD8+PD1+ and CD8+Ki67+) were reduced in EGFR-mutant tumors. Interestingly, CD4+ conventional T cells were elevated in EGFR-mutant cancers. Lastly, the regulatory T cell population did not show a significant increase in EGFR-mutant tumors.
T cell receptor (TCR) clonality was lower but T cell ex vivo expansion was preserved in EGFR-mutant tumors.
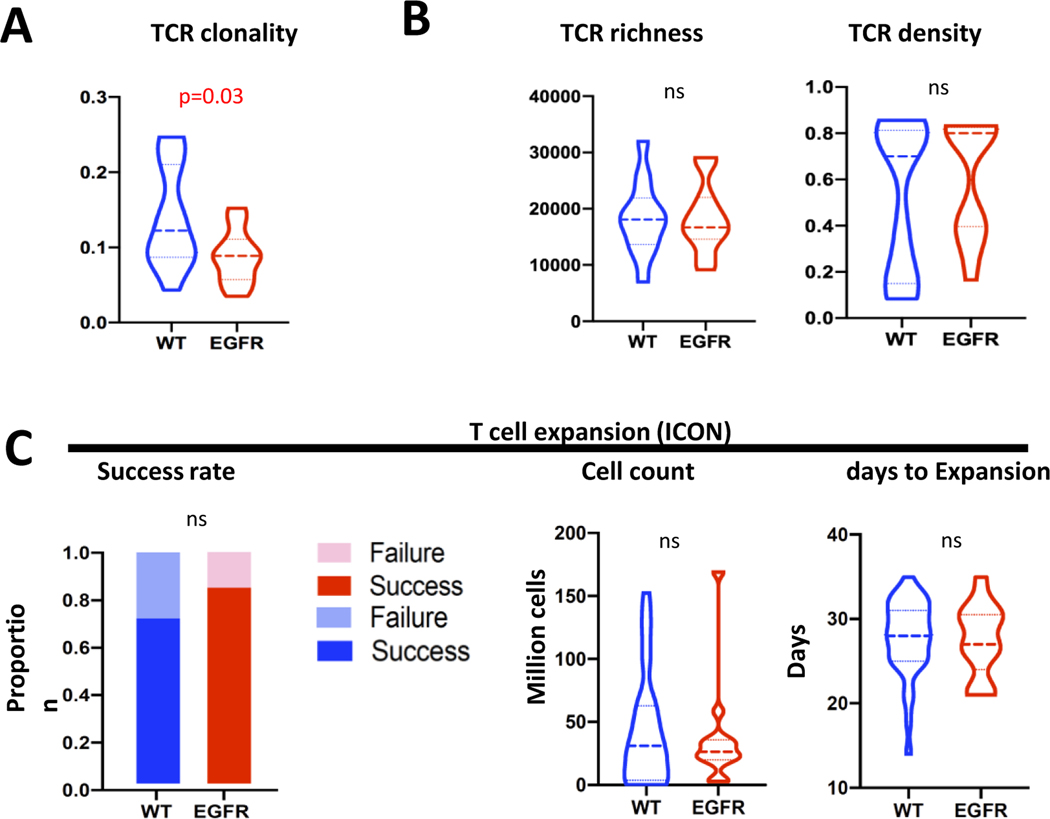
We next sequenced the T cell repertoire(19), to compare TCR density, diversity (richness) and reactivity (clonality) of T cells in EGFR-mutant and WT tumors, in ICON (EGFR n=15 vs. WT n=57) and PROSPECT (EGFR n=12 vs. WT n=92) cohorts. TCR clonality trended lower in EGFR-mutant tumors (0.098 vs. 0.140, p=0.09 in ICON cohort, Supplementary Fig. 4A and 0.117 vs. 0.155, p=0.001 in PROSPECT cohort(32), Supplementary Fig. 4D). Because EGFR-mutant tumors had lower TMB, Reuben et al has reported that most of the EGFR-mutant tumors were found to exhibit a TMB within the bottom tertile of the cohort(32), which was around 100 synonymous mutations per tumor. In order to adjust for this difference in TMB, we focused exclusively on tumors having TMB <100 synonymous mutations to remove TMB as the confounding factor. After this adjustment, TCR clonality was found to be significantly lower in the EGFR-mutant tumors (EGFR n= 10 vs. WT n= 11, 0.088 vs. 0.140, p=0.03, ICON cohort, Fig. 2A). Richness or density was not different between EGFR-mutant and WT tumors with or without adjusting for TMB (Fig. 2B and Supplementary Fig. 4A). Similar results were found in the PROSPECT cohort(32). When other driver-oncogene tumors were removed from the ICON cohort TCR analysis, the EGFR-mutant (n=15) versus WT (n=54) analysis results remained largely unchanged (Supplementary Fig. 4B–C). These data suggest that EGFR-mutant tumors have a less reactive T cell repertoire independent of TMB, consistent with prior studies(32).
Figure 2.
EGFR-mutant tumors had decreased TCR clonality but intact ex vivo T cell expansion capacity. (A) TCR analysis from ICON cohort for clonality, after adjusting for TMB difference between EGFR-mutant and WT tumors (only cases with total non-synonymous mutations less than 100 were used for comparison). (B) TCR analysis from ICON cohort for richness and density, after adjusting for TMB difference between EGFR-mutant and WT tumors. (C) T cell ex vivo expansion capacity, including success rate, final cell count after successful expansion, and days to successful expansion.
In the ICON cohort, T cells collected from tumor samples were subjected to ex vivo expansion(20, 33) and their ability to expand successfully was evaluated. Success rates (able to expand to more than 12 million T cells) were not significantly different between EGFR-mutant and WT tumors (11/13, 85% in EGFR-mutant vs. 25/35, 74% in WT tumors, Fisher exact test, p=0.35, Fig. 2C) as was the case with expanded T cell numbers (42 million cells vs. 37 million cells, p=0.69) and the number of days to successful expansion (27.5 days vs. 27.8 days, p= 0.86, Fig. 2C). When other driver-oncogene tumors were removed from the ICON cohort T cell ex vivo expansion analysis, the EGFR-mutant (n=13) versus WT (n=32) analysis results remained unchanged (Supplementary Fig. 5). This suggests that T cells in EGFR-mutant tumors were less activated with a decreased clonality, but preserved their ability to expand ex vivo. These data support the hypothesis that decreased activation of T cells in EGFR-mutant tumors may be associated with a suppressive tumor immune microenvironment.
EGFR-mutant tumors were less inflamed than their WT counterparts
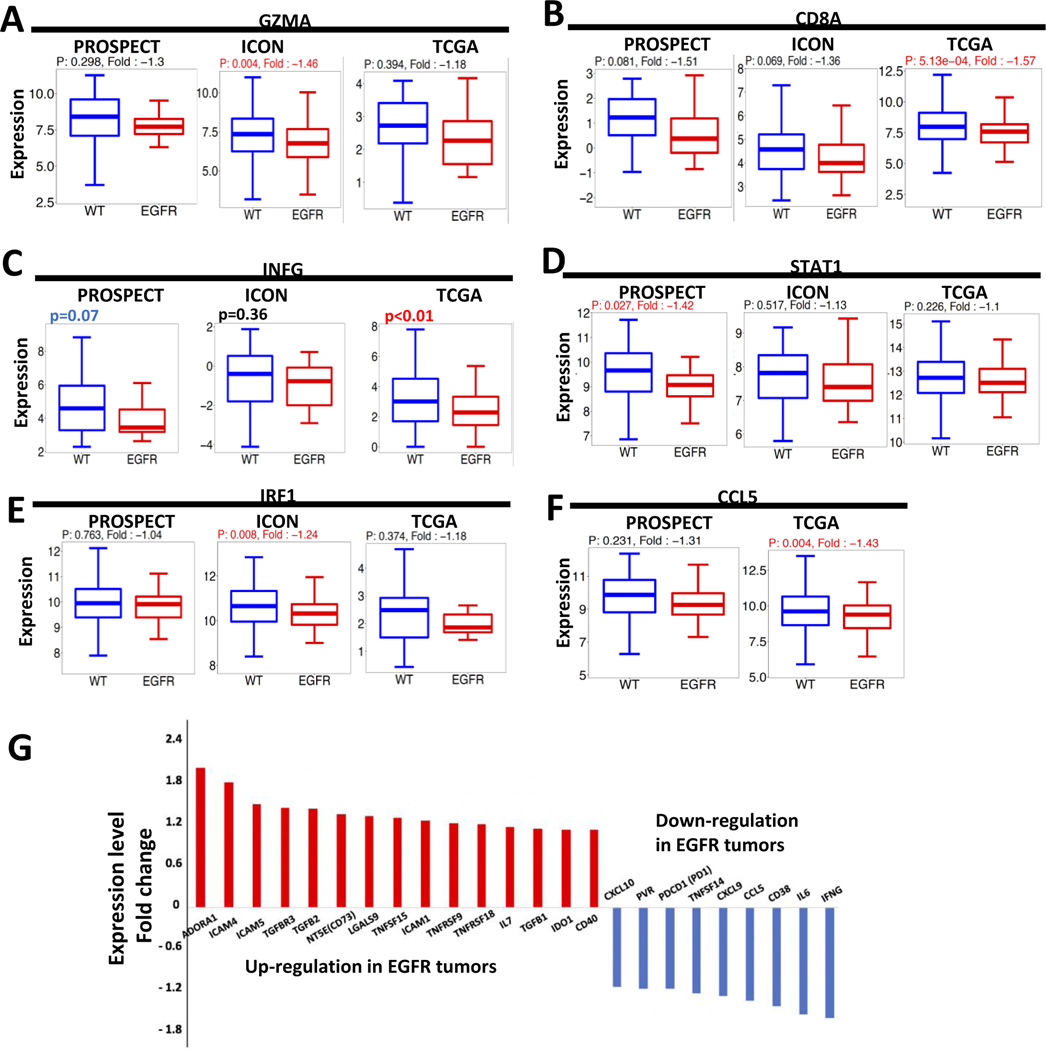
Next, we analyzed transcriptional data from the three cohorts to understand the key features of EGFR-mutant tumors’ immune microenvironment. Consistent with decreased cytolytic CD8+ T cells (Fig. 1D), CD8A and GMZA mRNA expression levels were lower in EGFR-mutant tumors (Fig. 3A–B and Supplementary Fig. 6A–B). Interferon-gamma (IFNG) expression also demonstrated a trend towards lower in the EGFR-mutant tumors across all datasets (Fig. 3C and Supplementary Fig. 6C) while expression levels of its downstream targets, including STAT1, IRF-1, CCL5 also trended towards decreased in EGFR-mutant tumors (Fig. 3D–F and Supplementary Fig. 6D–F), consistent with a less inflamed tumor immune microenvironment.
Figure 3.
EGFR-mutant tumors had decreased INF-gamma signaling. (A-F) GZMA, CD8A, INFG, STAT1, IRF1 and CCL5 RNA expression levels were compared between EGFR-mutant and WT tumors in PROSPECT, ICON and TCGA. Fold-change was calculated comparing expression of EGFR-mutant tumors to WT, using the lower expressed as the denominator. A positive fold-change value indicates overexpression in EGFR-mutant tumors and a negative value indicates decreased expression in EGFR-mutant tumors. The univariant analysis was used for p-value. (G) gene expression fold change in the TCGA dataset. The genes with a fold change (EGFR/WT) >1.2 or < −1.2 were included.
To identify potentially targetable immune regulators, we curated a list of 75 key immune regulatory genes, including immune checkpoint molecules(34), cytokines, and extra-cellular molecules and evaluated differences in their expression levels between EGFR-mutant and WT tumors. Using a fold-change >1.2, corresponding to most of the gene having a significant difference between EGFR and WT group, 15 genes were upregulated in EGFR-mutant tumors in the TCGA cohort (Supplementary Table 3). The top upregulated gene was ADORA1 (upregulation fold change 2.16, p<0.001, Fig. 3G and Fig. 4C), encoding the adenosine receptor 1, and the sixth upregulated gene was NT5E (upregulation fold change 1.44, p=0.02, Fig. 3G and Fig. 4A and Supplementary Fig. 7A), encoding CD73, both of which are key molecules in the CD73/adenosine pathway. Three TGF-beta and receptor molecules, TGFBR3, TGFB1, and TGFB2, were also upregulated (Fig. 3G and Supplementary Fig. 8), as were IDO1 and ICAM1/4/5. IFNG and IL-6 were the two genes most significantly down-regulated in EGFR-mutant cancers. Other immune regulatory genes such as CTLA4, B2M, LAG3, IL-2, IL-10, and VEGF-A showed no difference between groups (Fig. 3G and Supplementary Table 3).
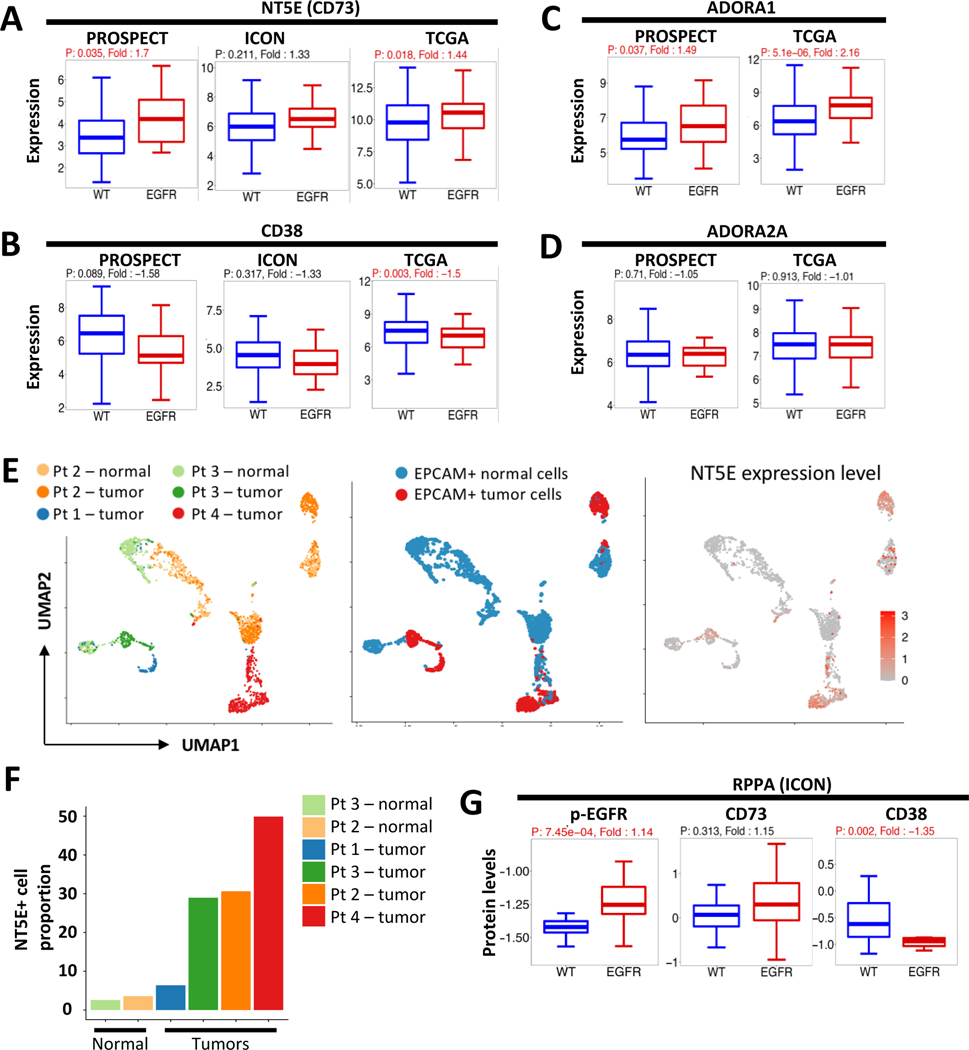
Figure 4.
CD73/adenosine pathway was upregulated in EGFR-mutant tumors. (A-D) NT5E (CD73), CD38, ADORA1 and ADORA2A RNA expression levels were compared between EGFR-mutant and WT tumors in PROSPECT, ICON and TCGA. Fold-change was calculated comparing expression of EGFR-mutant tumors to WT, using the lower expressed as the denominator. A positive fold-change value indicates overexpression in EGFR-mutant tumors and a negative value indicates decreased expression in EGFR-mutant tumors. The univariant analysis was used for p-value. (E) Feature plots showing EPCAM+ cells from the 3 single-cell RNAseq samples (right panel: cells by sample; middle panel: cells by malignant versus normal; left panel, the expression levels of NT5E in each cell) (F). Porportion of NT5E expression cells in each sample. (G) phosphorylated EGFR, CD73 and CD38 protein levels were compared between EGFR-mutant and WT tumors in the ICON cohort, from the reverse-phase protein array (RPPA).
CD73/Adenosine pathway was upregulated in treatment-naïve EGFR-mutant tumors
CD73 dephosphorylates and converts extracellular adenosine monophosphate (AMP) into adenosine, a known immune suppressant. Given the prominent upregulation of two key molecules in the CD73/adenosine pathway, we sought to evaluate other genes in the pathway. CD39, which dephosphorylates ATP to ADP and AMP, was not differently expressed in EGFR-mutant tumors, though CD38(35, 36), another key regulator of the adenosine pathway, was suppressed in EGFR-mutant tumors (Fig. 4B and Supplementary Fig.7B). At the adenosine receptor level, ADORA1 was upregulated in EGFR-mutant tumors (Fig. 4C and Supplementary Fig. 7C), though ADORA2a and ADORA2b were not (Fig. 4D and Supplementary Fig. 7D).
Next, we evaluated CD73 expression at the single-cell level in epithelial cells of EGFR-mutant NSCLC using 3’ single-cell RNA-sequencing approach. Six samples from four patients whose tumor harboring EGFR activating mutations were analyzed, with patient 1 and 3 being treatment-naïve and patient 2 and 4 having an EGFR tyrosine kinase inhibitor (TKI)-resistant tumor. We compared NT5E (CD73) expression in the EPCAM+ cell from the 6 samples: the tumor sample from all four patients and the uninvolved lung sample from patient 2 and 3. We found most of the malignant cells having a moderate to high NT5E expression, whereas most of the normal epithelial cells expressing low to none (Fig. 4E, middle panel and right panel). Tumor samples had a high proportion of cells expressing NT5E (range from 6.3–49.9%) compared to the normal lung epithelial cells (2.6–3.6%). Both treatment-naïve and EGFR TKI-resistant tumor cells had positive expression of NT5E, with the TKI-resistant sample having the highest proportion of cells expressing NT5E (resistant tumors at 30.6. and 49.9% vs. treatment-naïve 6.3 and 28.9%, Fig. 4F).
To confirm the differential expression at the protein level, we performed reverse-phase protein arrays (RPPA) in the ICON cohort, and found that phospho-EGFR levels were significantly higher in EGFR-mutant tumors, as expected. CD38 levels were lower (fold change −1.35, p<0.01) and CD73 slightly higher (fold change 1.15, p=0.3) in the EGFR-mutant tumors (n=14) compared to WT (n=31, Fig. 4G and Supplementary Fig. 7E). Taken together, these data suggested the CD73-mediated, but not CD38-mediated, adenosine accumulation may contribute to a suppressed tumor immune microenvironment in EGFR-mutant tumors.
CD73 blockade modulated T cell composition and function in EGFR-mutant non-small cell lung cancer cells.
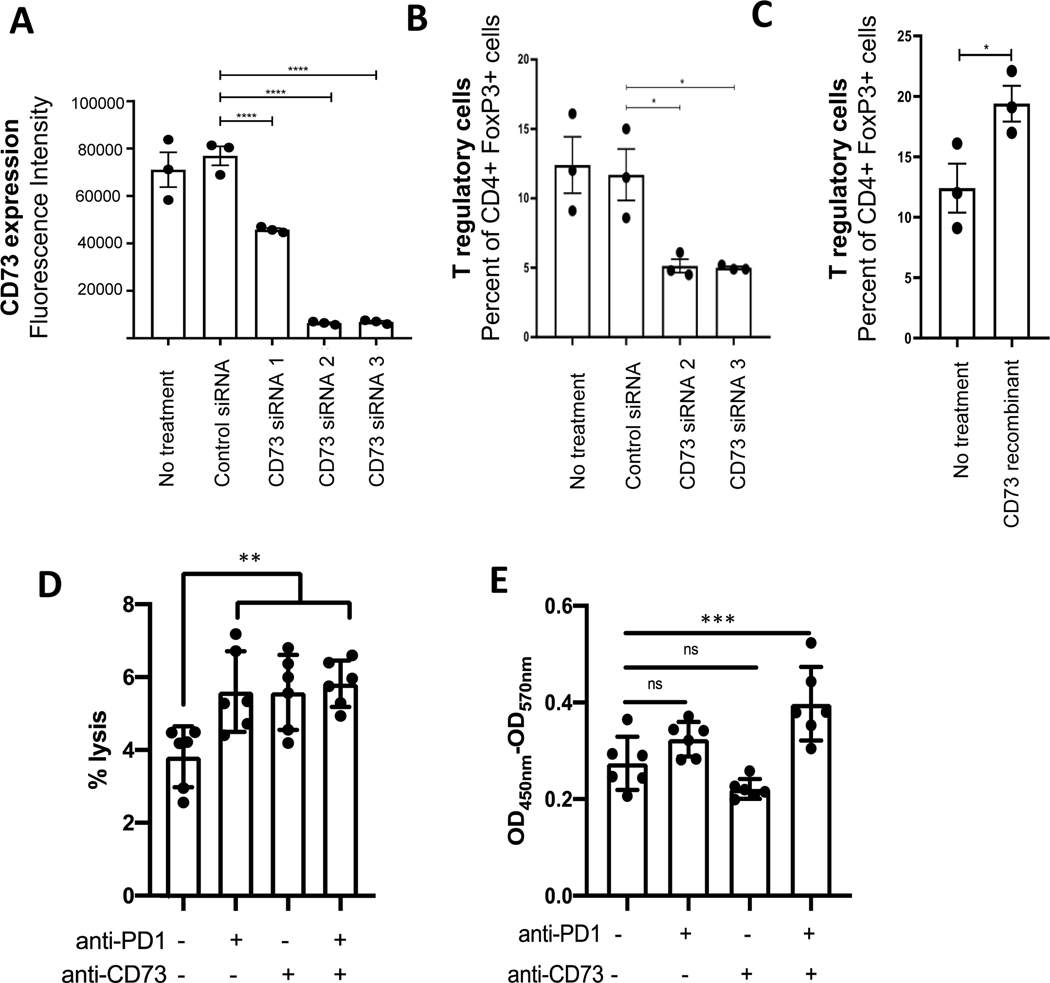
Multiple lines of evidence have established that CD73/adenosine blockade can favorably impact solid tumor immune microenvironment, especially on T cells. In an in vitro system, T cell proliferation and division was suppressed by the addition of AMP, but restored with anti-CD73 antibody (oleclumab/MEDI9447)(37). In the in vivo setting, tumor growth was inhibited in CD73 and A2AR knockout mouse models(38). Furthermore, NK cells, CD8+ T cells, and interferon-gamma were required for the anti-tumor activity(38). We hypothesized that in the EGFR-mutant NSCLC context, CD73 blockade could also modulate T cell composition and function. To test the hypothesis, we first evaluated the cell surface expression of CD73 in NSCLC cell lines by flow cytometry. Two EGFR-mutant NSCLC cell lines, H1975 and HCC4006, had high CD73 surface expression (Supplementary Fig. 9A). Two siRNAs (2 and 3) targeting CD73 had high knockdown efficiency and were selected for subsequent use (Fig. 5A and Supplementary Fig. 9 B). Next, PBMCs collected from the healthy donors were co-incubated with the conditioned media collected from cells with CD73 siRNA and control siRNA, and different T cell populations were evaluated. By flow cytometry, the percent of proliferating CD4+ and CD8+ T cells was not changed (Supplementary Fig. 9C), but T regulatory (CD4+FoxP3+) cells were significantly decreased with conditioned media from cells with CD73 siRNA treatment (5.133% (control siRNA) vs. 11.70% (CD73 siRNA2) and 11.70% (CD73 siRNA3), p=0.027 and 0.022 respectively in H1975 cells, Fig. 5C). Consistent with this finding, the addition of recombinant CD73 resulted in an increase of T regulatory cell proportion (12.40% vs. 19.39%, p=0.04 in H1975 cells, Fig. 5C). Similar results were observed in HCC4006 cells (Supplementary Fig. 9D).
Figure 5.
CD73 blockade modulates T cell composition and function in EGFR-mutant non-small cell lung cancer cells. (A) CD73 cell surface expression on H1975 cells was assessed by flow cytometry with or without control and CD73 siRNAs. (B) Healthy donor PBMCs were incubated with conditioned median collected from H1975 cells after siRNA treatment. T regulatory cell population was assessed in each condition by flow cytometry using CD4+ and FoxP3+ as markers. (C) Healthy donor PBMCs were incubated with conditioned median collected from H1975 cells or H1975 after recombinant CD73 overexpression. T regulatory cell population was assessed by flow cytometry using CD4+ and FoxP3+ as markers. (D) Healthy donor PBMCs were co-cultured with H1975 cells with anti-CD73 and anti-PD-1 treatment, alone or in combination. Percent of tumor cell lysis was assessed using cytotoxicity assay. (E) Healthy donor PBMCs were co-cultured with H1975 cells with anti-CD73 and anti-PD-1 treatment, alone or in combination. The cell medium was collected and assessed for interferon release by ELISA INF-gamma assay. Student t-test was used, statistical significance: * indicates p value less than 0.05, ** indicates p value less than 0.005, *** indicates p value less than 0.0005, and **** indicates p value less than 0.0001.
Next, we evaluated the immune cell cytotoxicity using an established PBMC co-culture system (39) with anti-CD73 blockade. In EGFR-mutant H1975 cells, treatments with anti-CD73 antibody alone or in combination with anti-PD-1 antibody led to increased tumor cell lysis with human peripheral blood monocytes (PBMC) (control 12% vs. anti-CD73 18% vs. combination 19%, p=0.0012 between control and anti-CD73, Fig. 5D). Furthermore, the interferon release was measured in the same four conditions. There was an increase in interferon-gamma release in anti-CD73 and anti-PD-1 combination treatment conditions (control OD450nm-570nm=0.27 vs. vs. combination 0.40, p=0.0005, Fig. 5E). These data provided evidence that blockade of CD73 in EGFR-mutant NSCLC cells may lead to a reversal of T cell suppression. When combined with anti-PD-1 treatment, anti-CD73 leads to increased interferon-gamma mediated T cell anti-tumor effect.
CD73 as a therapeutic target for EGFR-mutant lung adenocarcinomas
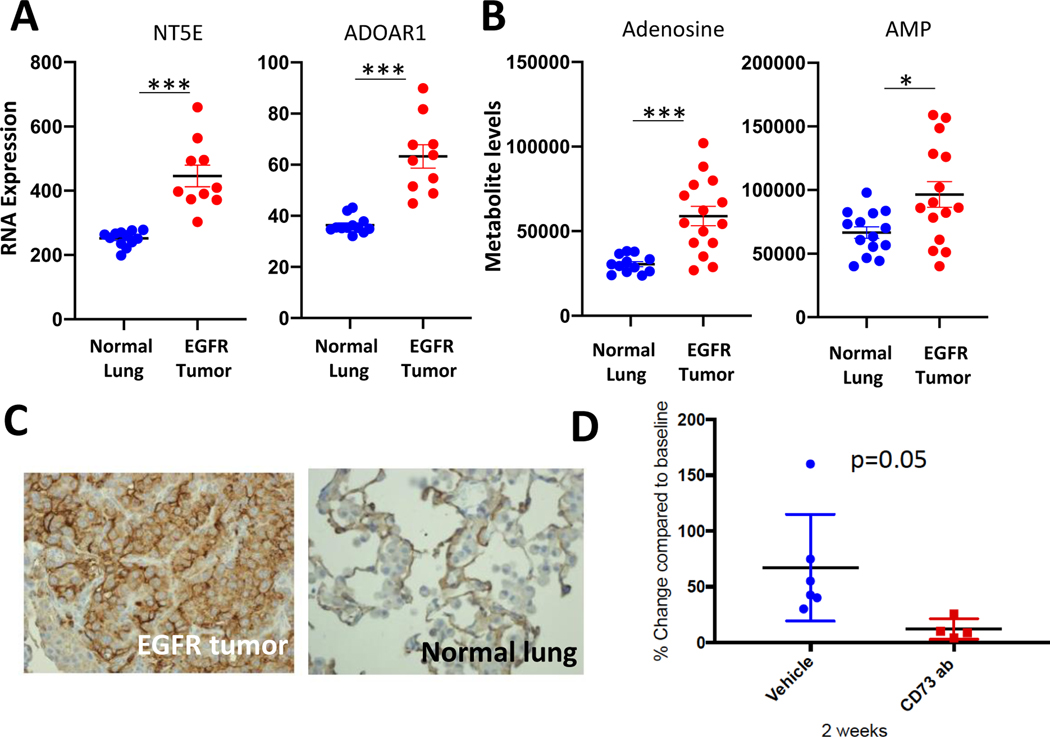
Next, we used an immune-competent EGFR-mutant genetically-engineered mouse model (GEMM) to evaluate the therapeutic efficacy of CD73 targeting,. RNA expression of several genes in the adenosine pathway in tumors from EGFR-L858R-T790M; CCSP-rtTA mice(27) were compared to lung tissues from mice carrying the CCSP-rtTA driver. Significantly elevated CD73 and adenosine receptor 1A RNA levels were detected in the EGFR-L858R-T790M murine tumors (Fig. 6A). Immunohistochemistry also confirmed CD73 overexpression in the tumor (Fig. 6C), indicating that this model recapitulates human findings. However, in contrast to patient samples, CD39 expression was elevated in murine tumors along with several adenosine receptors (Supplementary Fig. 10A). Metabolite levels were analyzed on these tumors to determine the functional role of these expression signatures. Interestingly, both AMP and adenosine levels were significantly elevated in EGFR-L858R-T790M tumors (Fig. 6B), confirming that gene expression correlates with metabolic signatures and CD73/adenosine pathway activation in this EGFR-mutant mouse model.
Figure 6.
Anti-CD73 therapy-induced tumor reduction in EGFR-mutant murine lung cancers. (A) NT5E (CD73) and ADORA1 RNA expression levels in EGFR-mutant tumors and normal lung. (B) adenosine and AMP metabolite levels in EGFR-mutant tumors and normal lung. (C) CD73 immunohistochemistry for EGFR-mutant tumors and adjacent normal lung. (D) The change of tumor sizes with 2 weeks of treatment was documented and compared between vehicle-treated and anti-CD73 treated groups.
We then evaluated the therapeutic potential of CD73 blockade in this immune-competent model. Murine lung cancers were induced with doxycycline and tumor formation was confirmed by small animal MRI. Anti-CD73 antibody (100ug/dose per mouse intraperitoneal twice week) versus vehicle control was given to tumor-bearing mice for 2 weeks and mouse tumors were monitored by MRI. Tumor reduction was observed in the anti-CD73 antibody group, whereas continued tumor growth was seen in the vehicle group (Fig. 6D and Supplementary Fig. 10B). These results suggest that targeting CD73/adenosine pathway for EGFR-mutant lung cancers may represent a viable therapeutic strategy.
DISCUSSION:
In this study, we performed comprehensive multi-cohort analyses to characterize the immune landscape of EGFR-mutant versus EGFR-WT NSCLC with the goal of identifying factors that potentially contribute to the lack of response to anti-PD1/PD-L1 therapy in EGFR-mutant lung cancers. While the overall number of T cells was not lower in EGFR-mutant tumors, they exhibited lower levels of PD-L1 protein, TMB, and CD8+ T cell density, consistent with prior studies(7, 8). T cell clonality was also low, suggestive of a less reactive T cell repertoire. These features have been linked to the lack of response to immune checkpoint blockade therapy(40–43). Furthermore, single-cell analysis hinted that the tumor cell population expressed CD73 in EGFR-mutant NSCLC, both in the treatment-naïve and TKI-resistant tumors.
To our knowledge, our study is one of the first studies to analyze immune cells composition and T cell subpopulations and functionality in EGFR-mutant NSCLC. Using MCPcounter to infer immune cell composition from TCGA cohort, we found that T cell population and monocytes macrophages were in a reverse relationship, with NK cells were also coordinately decreased with T cells in EGFR-mutant tumors compared to WT group. This observation is consistent with another study leveraging single-cell transcriptomics analysis focusing on the tumor immune microenvironment of EGFR-mutant and other oncogene-drivers NSCLC(44). By flow cytometry, we were able to evaluate T cells with a variety of functional markers, and found both activated T cells and proliferating T cells, as well as naïve CD8+ T cells were lower in EGFR-mutant tumors, suggesting a suppressed immune microenvironment, again consistent with single-cell level observations. Furthermore, in our bulk RNAseq analysis, IDO was found to be upregulated in EGFR-mutant tumors compared to the WT group. Maynard et al found that tumor cells having high IDO was associated with an immunosuppressive phenotype(44), consistent with the concept that EGFR-mutant tumors having a more immune inert phenotype. Additional evidence supporting the suppressive effect of the tumor microenvironment on T cells came from the lack of difference in T cell ex vivo expansion, when removed from the tumor microenvironment. This argues against intrinsic T cells dysfunction, but supports the importance of the tumor microenvironment-mediated immune suppression in EGFR-mutant tumors.
When we evaluated the tumor immune microenvironment with a focus on targetable immune checkpoint proteins, most known immune checkpoint genes were either not differentially regulated or down-regulated, such as CTLA-4, LAG3, TIM3, TIGIT, and IL-2. We found both CD73 and ADORA1, key factors in the CD73/adenosine pathway, to be significantly upregulated in EGFR-mutant tumors, consistent with prior studies(10, 45). Our observation was consistent across all cohorts at both the mRNA and protein levels. Upregulation of CD73/adenosine was also confirmed in murine models at mRNA, protein and adenosine metabolite product levels. Furthermore, analysis of the three main ectoenzymes CD73, CD39, and CD38 in the adenosine pathways suggested EGFR-mutant tumors preferentially engage CD73. Interestingly, CD38 was down-regulated in EGFR-mutant tumors, suggesting EGFR-WT tumors may engage this mechanism to mediate immune escape. This is consistent with the work of Chen et al showing that CD38 upregulation is associated with resistance to anti-PD1 therapy in EGFR-WT and KRAS-mutant lung cancer models(36). Our data highlight the importance of detailed dissection of tumor immune microenvironment to identify appropriate targets for each oncogene-driven lung cancer subgroup.
Our study has several limitations. First, although we were able to leverage three independent cohorts to compare key immune features between EGFR-mutant versus WT tumors using transcriptome profiling and key immune marker analyses, some other analyses, such as T cell subgroup flow cytometry analysis, T cell ex vivo expansion and RPPA analysis, came from only ICON cohort of about 60 cases (EGFR=8–15 vs. WT=23–45, depending on the assay success rates). Additional experiments, including the single-cell analysis for CD73 levels and mouse model experiments had small sample sizes, therefore, only provided preliminary evidence. The results were provocative but not conclusive. Second, more detailed mechanistic studies, especially in EGFR-mutant NSCLC in vivo models, are needed to illustrate the molecular detail on how blocking of CD73/adenosine pathway favorably modulate the tumor immune microenvironment. Furthermore, whether anti-CD73 adds efficacy to anti-PD-1 is a clinically relevant question requiring future investigation. We provided preliminary evidence that anti-CD73 and anti-PD1–1, alone or in combination, enhanced PBMC’s EGFR-mutant tumor cell killing. Our results are consistent with prior preclinical studies(37, 38) as well as a recent biomarker analysis report from a clinical trial in patients with renal cell carcinoma (RCC) treated with adenosine receptor inhibitor (ciforadenant/CPI-444) with or without atezolizumab(46). The ciforadenant’s clinical efficacy (by prolonged disease control) was directly associated with increased CD8+ T cell tumor infiltrates and TCR repertoire diversity in patient samples(46). Although we observed preliminary anti-tumor efficacy of CD73 blockade in co-culture systems and in a mouse model, further investigations in various immune-competent animal models with comprehensive survival analysis as well as tumor immune microenvironment analysis after anti-CD73 blockade, with and without anti-PD1 therapy, is warranted. It is also possible that additional anti-CD73’s benefit could be related to NK cell function and other components in the tumor immune microenvironment (47, 48). Third, our analyses were based on three cohorts of stage I-III EGFR-mutant tumors that were TKI-naïve. Although the basic tumor immune features, such as low PD-L1, low TMB and low CD8+ T cells are likely to remain similar in the TKI-naïve and TKI-resistant EGFR-mutant tumors, the tumor immune microenvironment is likely to be different due to TKI related changes(49). Future studies are necessary to characterize the tumor immune microenvironment in TKI-resistant EGFR-mutant tumors, to best design strategies for immunotherapy combinations for metastatic EGFR-mutant NSCLC.
Inhibiting CD73 and/or A2AR restores antitumor immunity in many preclinical studies, thus far, the clinical benefit from CD73/A2AR inhibitor immunotherapy clinical trials have been limited. Several anti-CD73 agents have entered clinical trials and demonstrated a good overall safety and tolerability profile(37, 38, 50), as single agents or in combinations. In a phase I/Ib trial of CPI-006 (anti-CD73)+/− ciforadenant (CPI-444, anti-A2AR) in solid tumors, T and B cells were successfully modulated with the treatment, but no RECIST-defined response was seen in the first 20 patients(50). When used in combination with anti-PD1/L1 therapy, partial responses were seen in 1 of 33 (3%) RCC patients treated with ciforadenant monotherapy and 4 of 35 (11%) RCC patients with ciforadenant atezolizumab combination, with an additional 24% (15 of 63 evaluable) of patients had tumor regression(46). Similar efficacy was observed in metastatic castration-resistant prostate cancers(51). Oleclumab (MEDI9447, anti-CD73) in combination with durvalumab has produced partial response in 1 of 21 colorectal cancer patients and 2 of 20 pancreatic cancer patients, with additional stable disease achieved(52). Some ongoing clinical trials involving newer CD73/A2AR inhibitors, including TJ004309 (anti-CD73 plus atezolizumab, Tarcon), LY3475070 (anti-CD73, plus pembrolizumab, Eli Lilly), NZV930 (Novartis), AK119 (Akeso), and AB680 (Arcus Biosciences) are under development. Currently, one trial of combination of oleclumab with osimertinib or oleclumab with AZD4635 (A2AR inhibitor) is ongoing (NCT03381274), evaluating CD73/adenosine pathway blockade in EGFR-mutant NSCLC. The results will shed light on the potential clinical benefit of anti-CD73 therapy in the EGFR-mutant NSCLC context.
One potential explanation for the underwhelming efficacy with CD73/adenosine blockade could be the lack of biomarker selection. In our study, we identified EGFR-mutant lung cancers had higher CD73 expression than other lung adenocarcinomas, which might represent a disease subtype that could derive differential benefit from the CD73/A2AR inhibition. Fong et al identified an adenosine expression signature, which was associated with response to adenosine receptor inhibitor response(46). Furthermore, the relationship between CD73 levels and response to checkpoint inhibitors remains to be determined. CD73/adenosine upregulated is generally associated with the inhibition of tumor-infiltrating lymphocytes and upregulation of T regulatory cells and cancer immune evasion, also shown in our study. But in one retrospective analysis, high CD73 EGFR-mutant patients (n=6) had longer progression-free survival from immunotherapy than low CD73 (n=11) EGFR-mutant patients. In non-EGFR patients, CD73 levels were not associated with the differential outcomes with immunotherapy(53). Future clinical studies of CD73 blockade in combination with other immunotherapy will help to address these questions.
Our study represents one of the first comprehensive tumor immune landscape characterizations dedicated to EGFR-mutant NSCLC. We leveraged three independent datasets using various methods to characterize the tumor cell immune features, immune cell composition, and tumor immune microenvironment features as well as single-cell RNAseq data from human EGFR-mutant tumors. We showed that EGFR-mutant NSCLC has low-PDL1, low TMB, low CD8+ cytotoxic T cells, consistent with immune-inert phenotype. Even after correction for low TMB, the TCR clonality remains low. Interestingly, though T cells appeared suppressed within the tumor microenvironment, they maintained their expansion capabilities ex vivo, suggesting modulation of immune microenvironment could enhance the T cell response. Though most immune checkpoint genes were not upregulated, the CD73/adenosine pathway was, possibly contributing to decreased T cell function. Furthermore, we provided preliminary evidence showing in immune-competent murine models that blockade of CD73 leads to EGFR-mutant lung cancer shrinkage, indicating the therapeutic potential of the CD73/adenosine pathway blockade. Taken together, our data demonstrate that the CD73/adenosine pathway may be a critical therapeutic target for enhancing antitumor immunity in EGFR-mutant NSCLC.
Supplementary Material
Funding Information:
The authors acknowledge funding support from University of Texas MD Anderson Lung Cancer Moon Shot Program and the MD Anderson Cancer Center Support Grant P30 CA01667, NIH R01 CA190628, UT Lung SPORE P50 CA70907, Rexanna’s Foundation for Fighting Lung Cancer, Bruton Endowed Chair in Tumor Biology, the Hallman fund, The Gil and Dody Weaver Foundation (to J.V.H.). X.L is supported by the Paul Calabresi Clinical Oncology Award (K-12) at MD Anderson Cancer Center, the Khalifa Scholars Award and Conquer Cancer Foundation ASCO Career Development Award. E.A.A is supported by the Cancer Prevention and Research Institute of Texas (CPRIT) Scholar Award RR160080, a Career Enhancement Award through National Institutes of Health 5P50CA070907, and Welch Foundation research grant (1975-20190330).
Disclosure:
X. L. receives a consultant fee from Eli Lilly AstraZeneca, EMD Serono, and Boehringer Ingelheim. B.S. receives a consultant fee from BMS. J.R. has sponsored research agreement from Genprex, Inc, has ownership interest (including stock, patents, etc.) in Genprex, Inc., and is a consultant/advisory board member for Genprex, Inc. J. Z. reports research funding and personal fees from BMS, AstraZeneca, Geneplus, OrigMed, Innovent, grant from Merck, Johnson & Johnson, outside the current work. C.B. and I.W. reports outside the submitted work grants and personal fees from Genentech/Roche, Bayer, Bristol-Myers Squibb, Astra Zeneca/Medimmune, Pfizer, HTG Molecular, Asuragen, Guardant Health, and Merck; personal fees from GlaxoSmithKline, Oncocyte, and MSD; and grants from Oncoplex, DepArray, Adaptive, Adaptimmune, EMD Serono, Takeda, Amgen, Karus, Johnson & Johnson, Iovance, 4D, Novartis, Akoya. C.H. serves on the scientific advisory board for Briacell. K.K.W. is a founder and equity holder of G1 Therapeutics. K.K.W. has sponsored Research Agreements with Medimmune, Takeda, TargImmune and BMS. K.K.W. has consulting & sponsored research agreements with AstraZeneca, Janssen, Pfizer, Novartis, Merck, Ono and Array. D.L.G. has served on scientific advisory committees for AstraZeneca, GlaxoSmithKline, Takeda, Sanofi and Janssen and has received research support from Janssen Research and Development, Takeda, Ribon Therapeutics, Mitobridge and AstraZeneca. J.V.H has served on scientific advisory boards for AstraZeneca, Biotree, BMS, Boehringer Ingelheim, EMD Serono, Genentech, GSK, Guardant Health, Hengrui, Lilly, Novartis, Seattle Genetics, Spectrum, and Synta. He receives research support from AstraZeneca, Bayer, GlaxoSmithKline, Spectrum, and Takeda, and royalties and licensing fees from Spectrum.
Footnotes
Other authors do not report any relevant conflict of interest.
REFERENCES:
- 1.Lee CK, Man J, Lord S, Links M, Gebski V, Mok T, et al. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol. 2017;12(2):403–7. [DOI] [PubMed] [Google Scholar]
- 2.Lisberg A, Cummings A, Goldman JW, Bornazyan K, Reese N, Wang T, et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naive Patients With Advanced NSCLC. J Thorac Oncol. 2018;13(8):1138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–33. [DOI] [PubMed] [Google Scholar]
- 4.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, and Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Offin M, Rizvi H, Tenet M, Ni A, Sanchez-Vega F, Li BT, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res. 2019;25(3):1063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res. 2016;22(18):4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gainor JF, Rizvi H, Aguilar EJ, Mooradian M, Lydon CA, Anderson D, et al. Response and durability of anti-PD-(L)1 therapy in never- or light-smokers with non-small cell lung cancer (NSCLC) and high PD-L1 expression. Journal of Clinical Oncology. 2018;36(15_suppl):9011-. [Google Scholar]
- 10.Inoue Y, Yoshimura K, Kurabe N, Kahyo T, Kawase A, Tanahashi M, et al. Prognostic impact of CD73 and A2A adenosine receptor expression in non-small-cell lung cancer. Oncotarget. 2017;8(5):8738–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii H, Azuma K, Kawahara A, Kinoshita T, Matsuo N, Naito Y, et al. Predictive value of CD73 expression for the efficacy of immune checkpoint inhibitors in NSCLC. Thorac Cancer. 2020;11(4):950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misumi Y, Ogata S, Ohkubo K, Hirose S, and Ikehara Y. Primary structure of human placental 5’-nucleotidase and identification of the glycolipid anchor in the mature form. Eur J Biochem. 1990;191(3):563–9. [DOI] [PubMed] [Google Scholar]
- 13.Yegutkin GG. Nucleotide- and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim Biophys Acta. 2008;1783(5):673–94. [DOI] [PubMed] [Google Scholar]
- 14.Kaczmarek E, Koziak K, Sevigny J, Siegel JB, Anrather J, Beaudoin AR, et al. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J Biol Chem. 1996;271(51):33116–22. [DOI] [PubMed] [Google Scholar]
- 15.Allard B, Longhi MS, Robson SC, and Stagg J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol Rev. 2017;276(1):121–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mediavilla-Varela M, Luddy K, Noyes D, Khalil FK, Neuger AM, Soliman H, et al. Antagonism of adenosine A2A receptor expressed by lung adenocarcinoma tumor cells and cancer associated fibroblasts inhibits their growth. Cancer Biol Ther. 2013;14(9):860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin D, Fan J, Wang L, Thompson LF, Liu A, Daniel BJ, et al. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. 2010;70(6):2245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadara H, Choi M, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol. 2017;28(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuben A, Gittelman R, Gao J, Zhang J, Yusko EC, Wu CJ, et al. TCR Repertoire Intratumor Heterogeneity in Localized Lung Adenocarcinomas: An Association with Predicted Neoantigen Heterogeneity and Postsurgical Recurrence. Cancer Discov. 2017;7(10):1088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forget MA, Tavera RJ, Haymaker C, Ramachandran R, Malu S, Zhang M, et al. A Novel Method to Generate and Expand Clinical-Grade, Genetically Modified, Tumor-Infiltrating Lymphocytes. Front Immunol. 2017;8:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, and Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018;23(1):227–38 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018. [DOI] [PubMed] [Google Scholar]
- 26.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19(1):279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12(1):81–93. [DOI] [PubMed] [Google Scholar]
- 28.Weaver Z, Difilippantonio S, Carretero J, Martin PL, El Meskini R, Iacovelli AJ, et al. Temporal molecular and biological assessment of an erlotinib-resistant lung adenocarcinoma model reveals markers of tumor progression and treatment response. Cancer Res. 2012;72(22):5921–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483(7391):613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget MA, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep. 2017;7(1):13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuben A, Zhang J, Chiou SH, Gittelman RM, Li J, Lee WC, et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat Commun. 2020;11(1):603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavera RJ, Forget MA, Kim YU, Sakellariou-Thompson D, Creasy CA, Bhatta A, et al. Utilizing T-cell Activation Signals 1, 2, and 3 for Tumor-infiltrating Lymphocytes (TIL) Expansion: The Advantage Over the Sole Use of Interleukin-2 in Cutaneous and Uveal Melanoma. J Immunother. 2018;41(9):399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson DG, and Bell JI. Isolation of a cDNA encoding the human CD38 (T10) molecule, a cell surface glycoprotein with an unusual discontinuous pattern of expression during lymphocyte differentiation. J Immunol. 1990;144(7):2811–5. [PubMed] [Google Scholar]
- 36.Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, et al. CD38-Mediated Immunosuppression as a Mechanism of Tumor Cell Escape from PD-1/PD-L1 Blockade. Cancer Discov. 2018;8(9):1156–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay CM, Sult E, Huang Q, Mulgrew K, Fuhrmann SR, McGlinchey KA, et al. Targeting CD73 in the tumor microenvironment with MEDI9447. Oncoimmunology. 2016;5(8):e1208875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young A, Ngiow SF, Barkauskas DS, Sult E, Hay C, Blake SJ, et al. Co-inhibition of CD73 and A2AR Adenosine Signaling Improves Anti-tumor Immune Responses. Cancer Cell. 2016;30(3):391–403. [DOI] [PubMed] [Google Scholar]
- 39.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J Thorac Oncol. 2015;10(6):910–23. [DOI] [PubMed] [Google Scholar]
- 40.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016;6(8):827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, et al. Therapy-Induced Evolution of Human Lung Cancer Revealed by Single-Cell RNA Sequencing. Cell. 2020;182(5):1232–51 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streicher K, Higgs BW, Wu S, Coffman K, Damera G, Durham N, et al. Increased CD73 and reduced IFNG signature expression in relation to response rates to anti-PD-1(L1) therapies in EGFR-mutant NSCLC. Journal of Clinical Oncology. 2017;35(15_suppl):11505-. [Google Scholar]
- 46.Fong L, Hotson A, Powderly JD, Sznol M, Heist RS, Choueiri TK, et al. Adenosine 2A Receptor Blockade as an Immunotherapy for Treatment-Refractory Renal Cell Cancer. Cancer Discov. 2020;10(1):40–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neo SY, Yang Y, Record J, Ma R, Chen X, Chen Z, et al. CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J Clin Invest. 2020;130(3):1185–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Lupo KB, Chambers AM, and Matosevic S. Purinergic targeting enhances immunotherapy of CD73(+) solid tumors with piggyBac-engineered chimeric antigen receptor natural killer cells. J Immunother Cancer. 2018;6(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isomoto K, Haratani K, Hayashi H, Shimizu S, Tomida S, Niwa T, et al. Impact of EGFR-TKI Treatment on the Tumor Immune Microenvironment in EGFR Mutation-Positive Non-Small Cell Lung Cancer. Clin Cancer Res. 2020;26(8):2037–46. [DOI] [PubMed] [Google Scholar]
- 50.Luke JJ, Powderly JD, Merchan JR, Barve MA, Hotson AN, Mobasher M, et al. Immunobiology, preliminary safety, and efficacy of CPI-006, an anti-CD73 antibody with immune modulating activity, in a phase 1 trial in advanced cancers. Journal of Clinical Oncology. 2019;37(15_suppl):2505-. [Google Scholar]
- 51.Harshman LC, Chu M, George S, Hughes BGM, Carthon BC, Fong L, et al. Adenosine receptor blockade with ciforadenant +/− atezolizumab in advanced metastatic castration-resistant prostate cancer (mCRPC). Journal of Clinical Oncology. 2020;38(6_suppl):129-. [Google Scholar]
- 52.Overman MJ, LoRusso P, Strickler JH, Patel SP, Clarke SJ, Noonan AM, et al. Safety, efficacy and pharmacodynamics (PD) of MEDI9447 (oleclumab) alone or in combination with durvalumab in advanced colorectal cancer (CRC) or pancreatic cancer (panc). Journal of Clinical Oncology. 2018;36(15_suppl):4123-. [Google Scholar]
- 53.Ishii H, Azuma K, Kinoshita T, Matsuo N, Naito Y, Tokito T, et al. Predictive value of CD73 expression in EGFR-mutation positive non-small-cell lung cancer patients received immune checkpoint inhibitors. Journal of Clinical Oncology. 2018;36(15_suppl):9065-. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.