Abstract
Somatic mutations in T cells can cause cancer but also have implications for immunological diseases and cell therapies. The mutation spectrum in nonmalignant T cells is unclear. Here, we examined somatic mutations in CD4+ and CD8+ T cells from 90 patients with hematological and immunological disorders and used T cell receptor (TCR) and single-cell sequencing to link mutations with T cell expansions and phenotypes. CD8+ cells had a higher mutation burden than CD4+ cells. Notably, the biggest variant allele frequency (VAF) of non-synonymous variants was higher than synonymous variants in CD8+ T cells, indicating non-random occurrence. The non-synonymous VAF in CD8+ T cells strongly correlated with the TCR frequency, but not age. We identified mutations in pathways essential for T cell function and often affected lymphoid neoplasia. Single-cell sequencing revealed cytotoxic TEMRA phenotypes of mutated T cells. Our findings suggest that somatic mutations contribute to CD8+ T cell expansions without malignant transformation.
Profiling of T cells reveals their somatic mutation spectrums and identifies mutations related to CD8+ T cell expansions.
INTRODUCTION
While somatic mutations are essential to malignant transformation, their role in nonmalignant cells and phenotypes is increasingly recognized (1, 2). In addition to clock-like mutagenesis generating mutations in all cells during aging (3, 4), other endo- and exogenous mutagenic processes shape the somatic mutation landscape throughout our lifetimes (5). Different tissues and cell types have varying mutational burden and spectrum of mutated genes. One example of a tissue-specific mutation landscape is clonal hematopoiesis (CH), where myeloid driver gene mutations accumulate in aging hematopoietic stem cells (6–8).
Somatic mutations in T cells can be derived from hematopoietic stem or progenitor cells (9) (central CH) or occur in mature T cells after T cell receptor (TCR) rearrangement (peripheral CH) (10). Rapid proliferation of T cells follows their activation through antigen-specific TCR. In addition to enhanced mitotic rate (3, 4), inflammation can accelerate the generation and expansion of mutated cells, possibly explained by reactive oxygen species (11), and inflammatory cytokines (12–14). Somatic mutations can affect T cell function with clinical consequences: secondary genetic alterations in chimeric antigen receptor (CAR) T cells have been suggested to improve immune cell therapy outcomes (15–18). In addition, somatic mutations in T cells have been linked with aberrant immune responses in large granular lymphocyte (LGL) leukemia, which is characterized by somatic STAT3 mutations in expanded cytotoxic T cells and autoimmunity (19).
In other immune-mediated hematological diseases than LGL leukemia, the somatic mutation spectrum in T cells is not yet fully discovered. Somatic mutations in T cells have been reported in patients with immunodeficiency (20), chronic graft-versus-host disease (cGVHD) (21), and various autoimmune conditions (22–30). In some patients, somatic mutations occurred in expanded T cell clones that were shown to induce apoptosis of target cells or mirrored the clinical course of auto- or alloimmune disorder (21, 30).
To investigate the spectrum of somatic mutations in T cells and their association with T cell clonality, we designed a custom-made gene panel consisting of 2533 genes related to immunity, cell proliferation, and survival. With gene panel sequencing, we characterized somatic mutations in 197 sorted T cell samples from 90 patients with various hematological disorders and compared them with the clonal structure of T cells defined by TCRβ deep sequencing. In addition, we analyzed 42 T cell samples from 21 age-matched healthy donors with the same gene panel. Last, we performed paired single-cell transcriptome and TCRαβ sequencing to investigate phenotypes of T cell clones with somatic mutations. The study outline is presented in Fig. 1A.
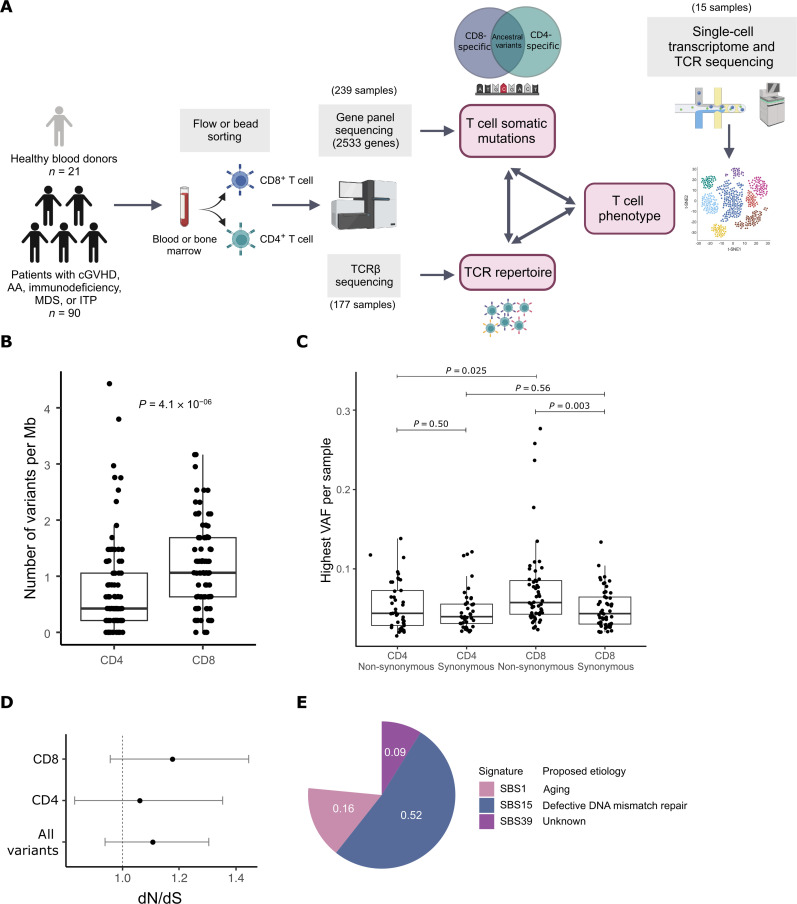
Fig. 1. Lineage-specific somatic mutations in CD4+ and CD8+ T cells.
(A) Study outline. We sequenced DNA from flow- or bead-sorted CD4+ or CD8+ T cells, with a gene panel consisting of 2533 genes related to cell survival or immunity (immunogene panel). From the same samples, we analyzed the clonal structure of T cells with TCRβ sequencing. Last, we combined the variant analysis of T cell phenotype with genotyping of single-cell transcriptome data. The schematic has been created with BioRender.com. (B) Lineage-specific mutation burden (number of variants per megabase) in CD4+ and CD8+ T cells of hematological patients. P value has been calculated with the Mann-Whitney test. (C) The highest non-synonymous and synonymous lineage–specific variant allele frequencies (VAFs) per sample in CD4+ and CD8+ T cells of hematological patients. P values have been calculated with the Mann-Whitney test. (D) Global estimates of dN/dS ratio for somatic mutations in T cells of hematological patients. dN/dS ratio > 1 (marked with a dashed line) indicates positive selection. Global dN/dS for all non-synonymous substitutions with their associated confidence intervals are shown separately for CD8+ T cells, CD4+ T cells, and both. (E) Mutational signature analysis of T cell mutations in hematological patients. The pie plot shows the weights for each identified signature.
RESULTS
CD8+ T cells from patients with hematological diseases harbor a higher number of somatic mutations than CD4+ T cells
We performed immunogene panel sequencing of sorted CD8+ and CD4+ populations from altogether 197 samples from patients with cGVHD (n = 38), aplastic anemia (AA; n = 18) (30), myelodysplastic syndrome (MDS; n = 15), immunodeficiency (n = 17) (20), and immune thrombocytopenia (ITP; n = 2) (table S1). The mean sequencing depth was 389× (fig. S1, B to E). Variants were filtered for variant allele frequency (VAF) and other parameters described in detail in Supplementary Materials and Methods, and only somatic variants with high confidence were included in the analysis. Together, we found 923 lineage-specific and 34 ancestral mutations, with mean VAF of 4.3% (1.0 to 28.9%) and 9.6% (2.3 to 28.7%), respectively (tables S2 and S3). CD8+-specific mutations were identified in 97% (87 of 90) and CD4+-specific mutations in 84% (76 of 90) of patients, with an average of 5.6 (CD8+) and 3.6 (CD4+) mutations per patient (fig. S2A). Mutational burden was significantly higher in CD8+ T cells, when compared to CD4+ T cells (Mann-Whitney test, P = 4.1 × 10−6; Fig. 1B). The same trend was detected in all cohorts when patients with different diagnoses were analyzed separately (fig. S2B). The sequencing depth did not explain the difference in mutational burden between CD8+ and CD4+ T cells (fig. S1, F and G). Between cohorts, there were no statistically significant differences in mutation burden, although it was slightly lower in the CD8+ T cells of patients with immunodeficiency (fig. S2, C and D). The mean VAF of all mutations was 4.3% and there was no significant difference between CD4+ and CD8+ VAFs if all variants were included in the analysis (Mann-Whitney test, P = 0.48; fig. S3A). However, when comparing the biggest non-synonymous or synonymous VAF per sample, CD8+ T cells had significantly higher non-synonymous VAF when compared to synonymous VAF in CD8+ T cells or non-synonymous VAF in CD4+ T cells (P = 0.003 and P = 0.025, respectively, Mann-Whitney test; Fig. 1C), suggesting that protein-coding mutations may drive clonal proliferation in CD8+ T cells. Furthermore, while the non-synonymous to synonymous ratio (dN/dS) was 1.11 for all T cell mutations indicating positive selection [confidence interval (CI) 0.94 to 1.30], it was higher in CD8+ T cells (dN/dS 1.18, CI 0.96 to 1.44) than CD4+ T cells (dN/dS 1.06, CI 0.83 to 1.24; Fig. 1D), although these results were not statistically significant due to low number of variants.
Most somatic mutations were missense variants (fig. S3B). Mutational signature analysis indicated that somatic mutations in T cells are mainly derived from endogenous mutational processes: clock-like signature SBS1 and defective DNA mismatch repair–associated signature SBS15 explained most lineage-specific mutations (Fig. 1E) and T cell mutations in different patient groups (fig. S3C). SBS1 results from spontaneous deamination of thymine, resulting in C > T transitions (31). In addition, we detected the signature SBS39, which has an unknown etiology. In CD8+ T cells alone, two additional defective DNA mismatch repair–associated signatures (SBS6 and SBS26) were detected (fig. S3D).
We also analyzed lineage-specific copy number variants (CNVs) in CD4+ and CD8+ T cells in regions targeted by the sequencing panel. After filtering spurious CNV calls (described in Supplementary Materials and Methods), we detected 13 lineage-specific CNVs in 9 of 90 (10%) of the patients (fig. S4A; list of CNVs in table S4). Most CNVs were detected in CD8+ samples, while four CNVs were detected in CD4+ samples. Chromosomal regions that were recurrently affected by CNVs included ATM and inflammasome genes (fig. S4B). CNVs were associated with higher lineage-specific somatic mutation burden in CD8+ T cells (P = 0.0061, Mann-Whitney test; fig. S4C), but not with age or TCRβ clonotype size (P = 0.54 and P = 0.079, Mann-Whitney test; fig. S4, D and E).
Comparison to healthy donor T cell mutations
In the original analysis, we used healthy donor T cells (n = 21, number of samples 42) in the somatic variant filtering [panel of normals (PON); see Materials and Methods], biasing the results toward alterations that are absent in healthy donor T cells. To enable comparison between patients with hematological diseases and age-matched healthy donors, we repeated the variant filtering with an alternative PON formed from all samples, including the patient samples (see Supplementary Materials and Methods for details; list of all refiltered somatic variants in table S5). As in patients, healthy donor CD8+ T cells had larger TCRβ clones than healthy CD4+ cells (P = 1.4 × 10−5, Mann-Whitney test; fig. S5, A and B). However, in healthy donors, the somatic variant burden was similar in CD4+ and CD8+ T cells (P = 0.52, Mann-Whitney test; fig. S5C). The mutation burden of healthy donor CD8+ T cells was significantly lower than in patients with GVHD, AA, MDS, or ITP, but not different from patients with immunodeficiency (P < 0.05 for GVHD, AA, MDS, and ITP, P = 0.64 for immunodeficiency, Mann Whitney test with Benjamin-Hochberg adjustment for multiple comparisons; fig. S5D). Only three ancestral variants were found in healthy donors. While the difference between the highest non-synonymous and synonymous VAF in CD8+ T cells was validated in patient samples with alternative PON (P = 0.0081, Mann-Whitney test), this was not observed in healthy CD8+ T cells (P = 0.56, Mann-Whitney test; fig. S5E). Five healthy donors with CD4+ mutations showed higher maximum non-synonymous VAF in comparison with synonymous VAF in CD4+ T cells (P = 0.016), which was not seen in patients (fig. S5E). Like in patient T cells, most of the T cell variants in healthy donors were explained by SBS1 and SBS15 in the mutational signature analysis (fig. S5F). Sequencing coverage was similar in healthy donor CD4+ and CD8+ T cells (median coverage 437× in CD4+ and 423× in CD8+ samples, P = 0.52, Mann-Whitney test; fig. S5G).
Variant allele frequency of non-synonymous mutations associates with clonal T cell expansion in CD8+ T cells
We performed TCRβ deep sequencing to integrate T cell clonal structures with somatic mutations in 135 patient samples (table S6). The size of the largest TCRβ expansion was associated with the highest VAF of non-synonymous lineage–specific mutations in CD8+ T cells (Pearson’s R = 0.73, P = 2 × 10−11) but not in CD4+ T cells (Pearson’s P = 0.1; Fig. 2A and fig. S6A).
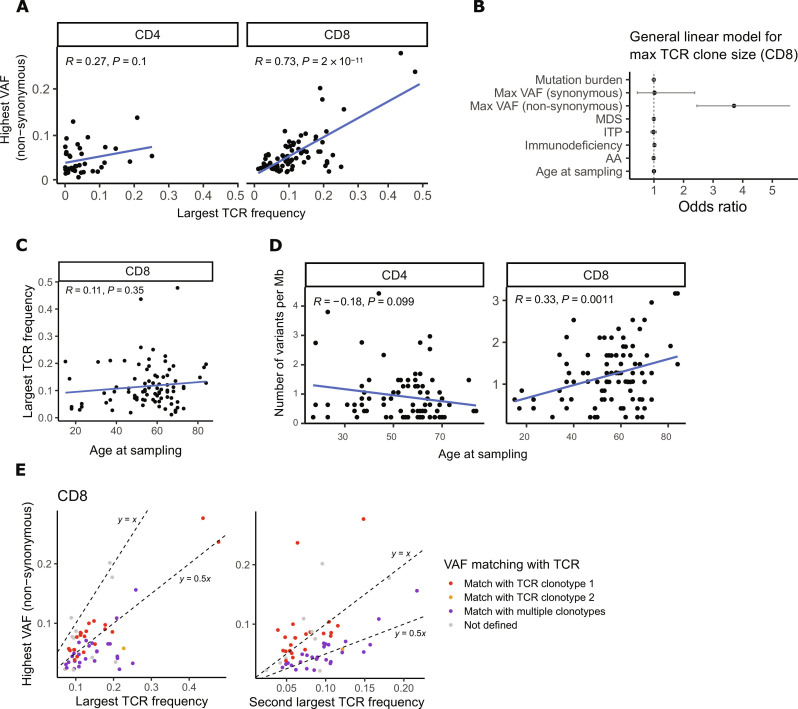
Fig. 2. Variant and TCR associations in hematological patient samples.
(A) The size of the largest TCRβ expansion (x axis) and the highest non-synonymous VAF (y axis) in CD4+ and CD8+ T cells. P value and correlation coefficient have been calculated with Pearson’s test. (B) A multivariate general linear model was used to study associations of age and mutations to the largest TCRβ expansion size. Odds ratios with their associated 95% confidence intervals are shown for each variable included in the model [mutation burden (number of variants per megabase), the highest non-synonymous and synonymous VAF, diagnosis (GVHD, AA, MDS, ITP, and immunodeficiency), and age at sampling]. (C) Age at sampling and the largest TCRβ frequency in CD8+ T cells. P value and correlation coefficient have been calculated with Pearson’s test. (D) Age at sampling and lineage-specific mutation burden (number of variants per megabase) in CD8+ and CD4+ T cells. P value and correlation coefficient have been calculated with Pearson’s test. (E) Comparison of the highest non-synonymous VAF with the top four largest TCRβ clonotype frequencies (top 2 clonotype frequencies are shown). We assumed the mutations to be heterozygous meaning that the size of the matching TCRβ clone would be double the VAF % (y ~ 0.5x). Variants matching with multiple TCRβ clonotypes have been marked with purple dots. Dotted lines representing y = x and y = 0.5 have been added to the plots for reference.
In a multivariate linear regression model, the highest non-synonymous VAF was the only significant factor associated with TCRβ expansion size in CD8+ T cells, when the highest synonymous VAF, mutation burden, age, and cohort were taken into account [odds ratio (OR) 3.71, 95% CI 2.45 to 5.61; Fig. 2B and fig. S6, B and C]. As indicated in the multivariate analysis, age was not associated with the largest TCRβ frequency in CD8+ T cells (Pearson’s R = 0.11, P = 0.35; Fig. 2C). However, age had a weak correlation with somatic mutation burden in CD8+ T cells but not in CD4+ T cells (Pearson’s R = 0.33, P = 0.0011; for CD4+ T cells, P = 0.1; Fig. 2D). When comparing the highest non-synonymous VAF with four largest TCRβ clonotype frequencies in CD8+ samples (see Materials and Methods), we could identify a matching TCRβ for 41% of patients (Fig. 2E). Twenty of 21 VAFs were matched with the largest TCRβ and one with the second largest TCRβ. When analyzing the antigen specificity of VAF-matching TCRs with antigen prediction tool TCRGP (32) as well as hard-matching CDR3s to the VDJdb database (33), two mutated TCRβ clones were found to be specific for CMV epitopes (table S7).
Mutations in pathways important for T cell function
We performed pathway enrichment analysis from all lineage-specific mutations with Oncodrive-fm (34). We identified several significantly mutated pathways related to T cell activation: T cell costimulation (q = 0.019), positive regulation of mitogen-activated protein (MAP) kinase activity (q = 0.019), and closely associated positive regulation of extracellular signal–regulated kinase 1 (ERK1) and ERK2 cascade (q = 0.030). In addition, interleukin-7 (IL-7)–mediated signaling pathway (q = 0.0014) and negative regulation of cell growth (q = 0.00074) were significantly mutated in T cells. Mutated genes linked to these five pathways are presented in Fig. 3A and fig. S7A.
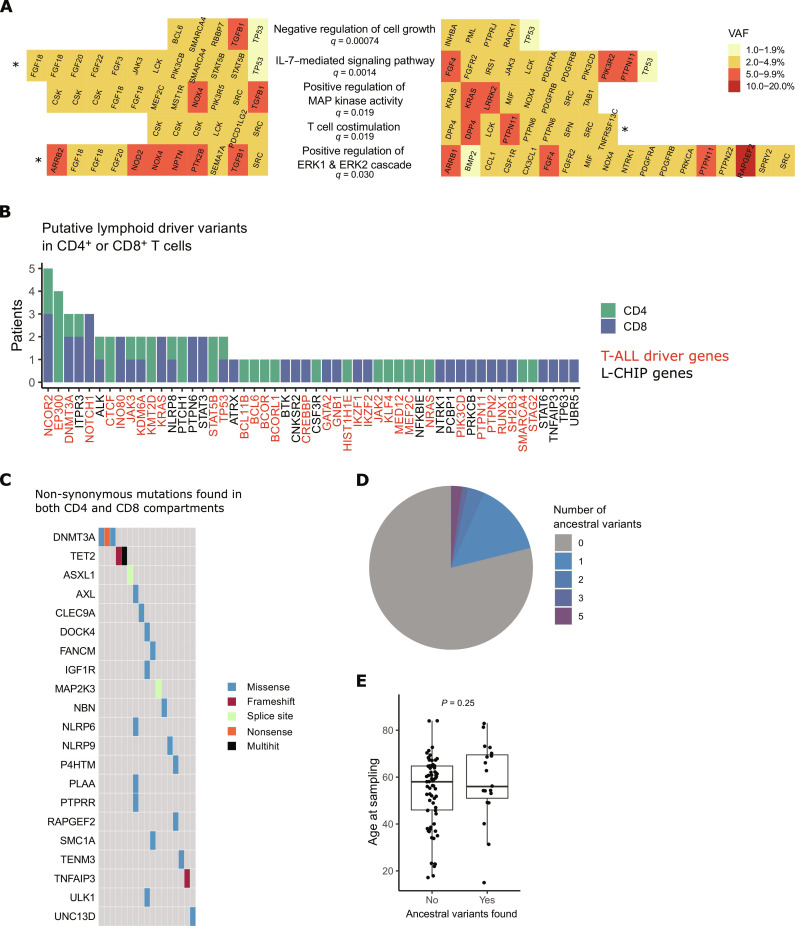
Fig. 3. Pathways and genes affected by somatic mutations in hematological patients.
(A) The most significantly mutated biologically relevant pathways in CD4+ and CD8+ T cells of hematological patients, based on Oncodrive-fm analysis. Lineage-specific mutations in CD4+ T cells are shown on the left and CD8+ mutations on the right. False discovery rate correction was used to adjust P values for multiple comparisons, and q values are shown for each pathway. Gene names are in alphabetical order. The significance level for pathways that were significantly mutated in CD4+ or CD8+ T cells when analyzed separately is marked with an asterisk (*). (B) Putative lymphoid driver mutations in CD4+ or CD8+ T cells of hematological patients were analyzed as described in Materials and Methods. The y axis shows the number of patients with a putative lymphoid driver mutation in each gene. T-ALL driver genes are marked with red color and other putative lymphoid driver mutations with black color. (C) Oncoplot of ancestral non-synonymous mutations (identified in CD4+ and CD8+ compartments in the same patient). (D) We found one to five ancestral mutations in 21% (19 of 91) of patients. Mutated genes are shown in panel (C). (E) Age at sampling in patients with or without ancestral mutations in T cells. P value has been calculated with the Mann-Whitney test. L-CHIP, lymphoid clonal hematopoiesis.
Mutations in genes related to T cell costimulation included missense variants in CSK, LCK, DPP4, SRC, PTPN6, PTPN11, SPN, and TNFRSF13C, as well as a splicing variant in PDCD1LG2 gene (Fig. 3A and fig. S7A). CSK, LCK, DPP4, and PTPN6 were each mutated in multiple patients. The same DPP4 missense mutation (p.V635I, COSM329327) was detected in the CD8+ T cells of two patients with cGVHD (VAFs 3.0 and 5.2%). PTPN11 p.V432M mutation had the highest VAF (7.8% in CD8+ T cells) and the same mutation has been found in various cancer types (COSMIC ID: COSM546362). T cell costimulation pathway was significantly mutated also in CD8+ T cells alone (Fig. 3A).
MAP kinase and ERK1/ERK2 pathways are among the major pathways transmitting TCR activation signals. Genes on positive regulation of MAP kinase activity pathway included 28 non-synonymous variants in 17 patients, with five recurrently mutated genes (genes mutated in multiple patients): KRAS, CSK, SRC, NOX4, and FGF18 (Fig. 3A and fig. S7A). The highest VAFs were detected in LRRK2 (10% in CD8), TFGB1 (9.4% in CD4), NOX4 (9.0% in CD4), and KRAS (VAFs 3.0 to 7.8% in CD8 in two different patients, COSMIC IDs: COSM87288 and COSM19905; table S2). Positive regulation of ERK1 and ERK2 cascade was partially overlapping with T cell costimulation and MAPK pathway, without additional recurrently mutated genes (fig. S7A). It was significantly mutated also in CD4+ T cells alone (Fig. 3A).
IL-7–mediated signaling pathway genes were mutated in 20 patients (Fig. 3A and fig. S7A). Among these pathway genes, STAT5B mutations included one SH2 domain mutation (p.T628S) as well as one missense mutation (p.418 K) in the signal transducers and activators of transcription (STAT) binding domain, both in the CD4+ T cells of patients with immunodeficiency with 3.1% VAF (table S2). Both patients had their highest TCRβ frequency in CD4+ T cells at <1% (table S6), suggesting that mutations have emerged before TCR rearrangement (central CH). Two JAK3 mutations were detected in our study: one in CD4+ (VAF 2.1%) and another one in CD8+ T cells (VAF 3.5%). Ten patients harbored mutations in the negative regulation of cell growth pathway genes (Fig. 3A and fig. S7A). TP53 was the only recurrently mutated gene and except for TGFB1, all VAFs were below 5% (table S2).
We performed the Bradley-Terry model to study the clonal dominance of selected significantly mutated pathways (fig. S7, B and C). The results were strongly affected by the mutation load of each pathway, with the ERK1/ERK2 pathway first and the negative regulation of the cell growth pathway last (fig. S7B). This was in line with the observation that in most samples, only one of the studied pathways was mutated. Focusing only on co-occurring mutations and their relative orders, T cell costimulation was the first pathway to mutate in CD8+ but not in CD4+ T cells (fig. S7C). However, this result was not statistically significant due to the low number of samples with co-occurring mutations.
Putative lymphoid driver mutations in CD4+ or CD8+ T cells
We identified 78 T cell lineage–specific mutations in previously described lymphoid driver genes (35, 36) in 43 of 90 (47.7%) patients (Fig. 3B). We identified several mutations in pathways involved in the pathogenesis of T cell neoplasia, including NOTCH1 signaling (NOTCH1), Ras signaling (KRAS, NRAS, and PTPN11), Janus kinase (JAK)–STAT signaling (STAT3, STAT5B, JAK3, JAK2, PTPN2, PTPN6, and SH2B3), epigenetic regulation (EP300, KDM6A, CREBBP, INO80, KMT2D, and SMARCA4), transcription factors (BCL11B, BCL6, MED12, and RUNX1), and PI3K pathway (PIK3CD).
None of the three identified NOTCH1 mutations were in the hotspot region while all mutations in KRAS, NRAS, and PTPN11 (Ras signaling pathway) have been previously reported in cancer (COSMIC IDs: COSM19905, COSM87288, COSM573, and COSM546362). NRAS mutation was detected in CD4+ T cells of patient AA-2 with 4.6% VAF.
Both STAT3 mutations were in the SH2 domain hotspot (p.Y640F) and found in CD8+ T cells of patients with AA with high VAFs (17.7 and 23.2%), as reported previously (29, 30). A JAK2 hotspot mutation V617F (VAF 1.8%) was identified in CD4+ T cells in patient ID-10 whose biggest CD4+ TCRβ clonotype was 0.2% (tables S2 and S6), not consistent with the VAF. With manual inspection, three variant reads were also detected in CD8+ T cells (VAF 0.5%), pointing toward central CH. Four distinct missense mutations in CD8+ T cells were identified in PTPN2 (VAF 5.8%), PTPN6 (VAFs 3.6 to 4.2%), and SH2B3 (VAF 4.0%) genes, which are regulators of JAK-STAT signaling.
From genes related to epigenetic regulation, EP300 was the most frequently mutated (Fig. 3B). Four distinct EP300 missense mutations were detected in the CD4+ T cells of patients with AA, MDS, and cGVHD, VAFs varying from 1.0 to 6.3%. We had TCRβ sequencing data available from two patients with cGVHD with EP300 mutations: in GVHD-37 (VAF 6.3%), the highest TCRβ expansion in CD4+ was 2.3%, and in cGVHD-30 (VAF 2.4%), there was a large (18.8%) TCRβ expansion in CD4+ T cells (tables S2 and S6). In any of the patients with EP300 mutations, no variant reads were detected in the CD8+ compartment even with manual inspection. One of the detected EP300 mutations (p.L415P) has been shown to be recurrently mutated in hematological malignancies, including follicular and diffuse large B cell lymphoma as well as B-ALL (COSM221269). In addition to EP300, epigenetic regulator genes INO80, KDM6A, and KMT2D were recurrently mutated.
Mutated transcription factors with putative lymphoid driver mutations included BCL11B, BCL6, MED12, and RUNX1. None of the genes were recurrently mutated in our cohort, and the largest VAF was in RUNX1 (VAF 2.4% in the CD8+ T cells of patients with MDS). In the patient with RUNX1 mutation (MDS-1), no variant reads were detected in CD4+ T cells, and the largest TCRβ expansion in CD8+ T cells was 5.8% (tables S2 and S6).
In summary, putative driver mutations were detected in T cells, including several genes and pathways that are recurrently mutated in lymphoid neoplasia. The somatic mutation profile in nonmalignant T cells was still different from T cell malignancies: We did not identify any loss-of-function CDKN2A/B or ATM mutations, which are among the most frequently mutated tumor suppressor genes in T-ALL (37) and T-prolymphocytic leukemia (T-PLL) (38).
Ancestral mutations
Because CH mutations have been shown to pass on to lymphocytes (39), we sought to identify central CH mutations in T cells. We initially deducted that mutations derived from HSCs should be detected in both CD4+ and CD8+ T cells (ancestral mutations) if emerged before lineage differentiation. We found altogether 34 ancestral mutations in T cells in 19 patients (Fig. 3, C and D, and table S3). Only two genes were shown to have recurrent mutations: DNMT3A (three mutations in three patients) and TET2 (three mutations in two patients) (Fig. 3D), which are the most mutated CH genes in a healthy population (40). Other CH-associated genes with ancestral mutations in T cells included ASXL1 (splice-site mutation) and SMC1A (p.H307Q; Fig. 3D and table S3). In addition, we detected a frameshift deletion in TNFAIP3, which is a known driver gene in lymphoid malignancies (41) (VAF 2.8 and 9.2% in CD8+ and CD4+ T cells, respectively). There was no difference in age between patients with or without ancestral mutations in T cells (P = 0.25, Mann-Whitney test; Fig. 3E).
With our analysis strategy, we also identified lineage-specific mutations in DNMT3A and TET2 (both in three patients’ CD4+ or CD8+ cells; table S2). Mutations included both known hotspots (p.R882H and Y735C in DNMT3A; p.H3180Y and a nonsense mutation in TET2) as well as previously unidentified missense mutations (table S2). VAFs varied from 1.3 to 4.2%. From two of three TET2 and one of three DNMT3A mutations, no variant reads were detected in other T cell compartments even with manual inspection. To rule out central CH, simultaneous ultra-deep sequencing of the myeloid compartment would be needed.
Somatic mutations enrich to cGVHD-associated TEMRA cell phenotypes
To understand the phenotype of mutated T cell clones, we analyzed available samples from nine patients with cGVHD with paired single-cell transcriptome and TCR sequencing (scRNA + TCRαβ-seq; Fig. 4 and figs. S8 and S9). We genotyped the found mutations with Vartrix (42) and imputed the mutated clonotypes based on TCRβ of cells with mutation (Fig. 4, B and F; detailed description of the analysis in Supplementary Materials and Methods). We also compared the phenotypes of cGVHD T cells to those of age-matched healthy controls (Fig. 4, C, D, G, and H, and figs. S8 and S9).
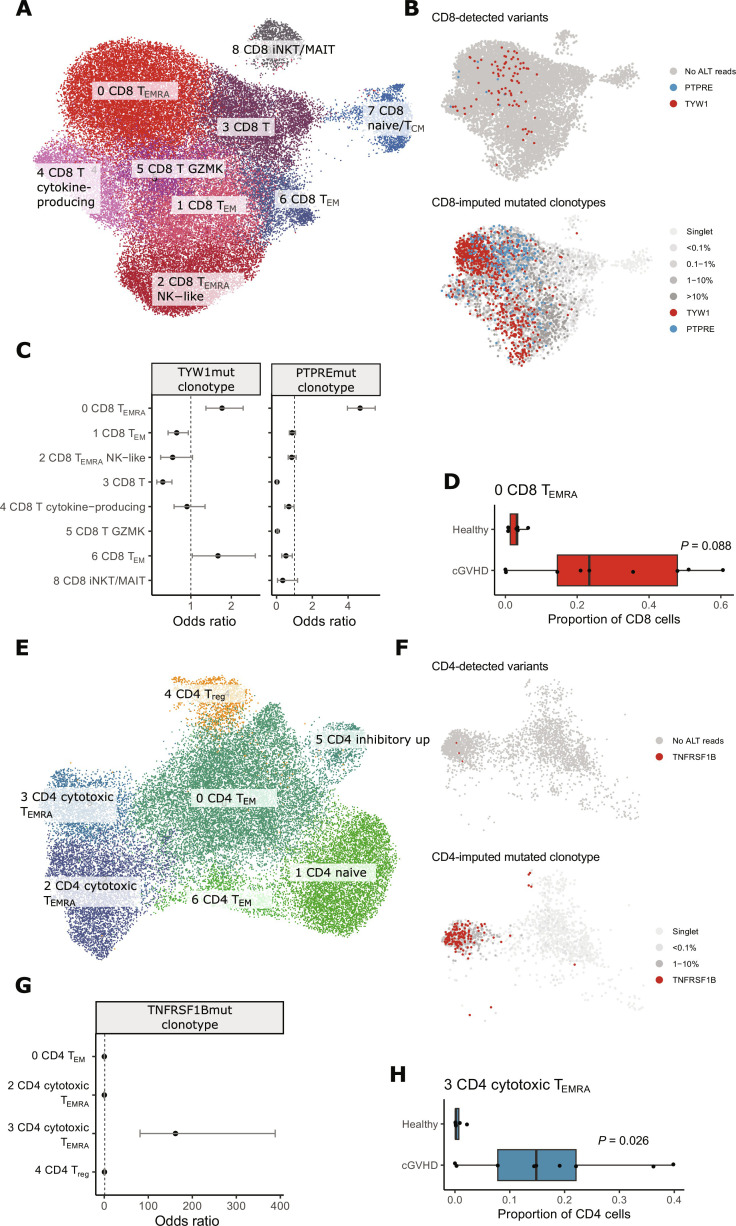
Fig. 4. Single-cell transcriptome analysis of nine patients with cGVHD and six age-matched healthy controls.
Analysis for CD8+ T cells is shown in (A) to (D) and for CD4+ T cells in (E) to (H). (A) Uniform Manifold Approximation and Projection (UMAP) and cluster annotation of CD8+ T cells. (B) We first identified cells with variants with genotyping of mutations found in immunogene panel sequencing with the Vartrix tool (top). CD8+ T cells with variant reads are marked with red (TYW1) or blue (PTPRE). Next, we imputed clonotypes of mutated cells based on TCRβ (shown on the bottom panel). (C) Phenotypes of mutated CD8+ clonotypes. Odds ratios and their associated 95% confidence intervals for mutated clonotypes against other CD8+ T cell clonotypes from the same patient. (D) Cluster 0 (CD8 TEMRA, i.e., terminally differentiated CD8+ T cells) was expanded in seven of nine patients with cGVHD but not in healthy controls. The x axis shows the proportion of cluster 0 phenotype in CD8+ T cells for each sample. P value has been calculated with the Mann-Whitney test. (E) UMAP and cluster annotation of CD4+ T cells. (F) CD4+ T cells with variant reads in TNFRSF19 (top) or imputed clonotype (bottom) are marked with red. (G) Phenotype of mutated CD4+ clonotype. Odds ratio and their associated 95% confidence intervals for mutated clonotype against other CD8+ T cell clonotypes from the same patient. (H) Cluster 3 (CD4 cytotoxic TEMRA) was expanded in seven of nine patients with cGVHD but not in healthy controls. The x axis shows the proportion of cluster 3 phenotype in CD4+ T cells for each sample. P value has been calculated with the Mann-Whitney test. TEM, effector memory T cell; Treg, regulatory T cell; TCM, central memory T cell; NK, natural killer, iNKT, invariant natural killer T cell; MAIT, mucosal-associated invariant T cell; GZMK, granzyme K.
Because of the fact that droplet-based single-cell transcriptome sequencing covers just short parts of the target genes (43), only three somatic mutations in three different individuals could be reliably confirmed from the transcriptome data: mutated genes included TYW1 (in CD8+), PTPRE (in CD8+), and TNFRSF1B (in CD4+ T cells) (see Supplementary Materials and Methods and fig. S10A). No CNVs were detected in the transcriptome data with Numbat (v1.3.0) (fig. S10B). Mutated clonotypes in CD8+ T cells enriched in the CD8+ cluster 0 (CD8 TEMRA) in both patients [OR 4.67 (CI 3.96 to 5.51) and 1.7 (CI 1.37 to 2.29) for TYW1 and PTPRE mutated clonotypes respectively; Fig. 4C]. Cluster 0 (CD8 TEMRA) was expanded in seven of nine patients with cGVHD but in none of the healthy controls (Mann-Whitney test, P = 0.088; Fig. 4D), while healthy controls’ expanded CD8+ T cell clones enriched to cluster 2 (CD8 TEMRA natural killer–like, Mann-Whitney test, P = 0.09; fig. S8, A and E). TNFRSF1B-mutated clonotype enriched in the CD4+ cluster 3 (CD4 cytotoxic TEMRA, OR 161.76 [CI 81.15–388.27]; Fig. 4G), which was significantly expanded in patients with cGVHD when compared to healthy controls (Mann-Whitney test, P = 0.026; Fig. 4H). When comparing the mutated clonotypes to other cells within the same (dominating) phenotypic cluster, the strongest signal in differentially expressed (DE) gene analysis was from TCR-related genes in all cases (fig. S11), and predicted cell-cell interactions with other cell types were comparable with other TEMRA cell clonotypes (fig. S12). None of the mutated TCRβ clonotypes were found in VDJdb (33) or predicted to be specific to common viral epitopes with TCRGP (32).
DISCUSSION
With the most comprehensive published dataset of the mutation spectrum of nonmalignant T cells to date (number of individuals = 111, number of samples = 239, 2533 genes), we showed that somatic mutations in T cells are ubiquitous in patients with oligoclonal T cell expansions. All patients in our study had a hematological disease and few patients fulfilled the criteria for LGL lymphocytosis, but notably, none of them developed a lymphoid malignancy during follow-up. In addition, samples from healthy volunteers were analyzed. Our observation complements previous findings from other somatic tissues (4, 44) and highlights the complex link between somatic mutations and clonal evolution.
Differentiated cells have less proliferative capacity, and hence, mutation timing is critical for leukemic transformation potential. We used main T cell lineage markers (CD4 and CD8) to distinguish between mutations acquired by HSCs or lymphoid progenitor cells versus mature T cells. For clonal structure analysis within T cells, we also used their somatically rearranged TCR profiles. Only 3.5% of somatic variants were detected in both CD4+ and CD8+ T cells of the same patient, suggesting that most mutations were acquired or selected after lineage differentiation. Our results indicating peripheral mutagenesis are in line with a recent study by Machado and colleagues (45), where they elegantly showed in six healthy individuals that T cells have a distinct mutational profile and that they acquire a manifold of mutations when compared to HSCs. In our study, the highest non-synonymous VAF was positively correlated with the size of the largest clonal TCRβ expansion in CD8+ T cells. Our results support the hypothesis that most mutations in CD8+ T cells expand after TCR rearrangement and T cell lineage segregation, resulting in higher mutation accumulation in highly differentiated T cells than in lymphoid precursors or HSCs (45). However, for individual mutations, we cannot exclude the possibility of central CH without mutation screening of the myeloid compartment (10). In addition, we detected lineage-specific CNVs in nine patients, although this analysis is limited by the usage of a targeted gene panel instead of whole-genome sequencing.
Our study confirms previous results suggesting that CD8+ T cells have increased mutation burden when compared to CD4+ T cells, independently of the tested hematological disease (23, 24, 30). As CD8+ T cells have larger clonal expansions than CD4+ T cells, this can lead to higher VAFs and more sensitive detection of CD8+ T cell mutations. However, in healthy donors, the mutation burden of CD8+ T cells was not higher than that of CD4+ T cells, despite larger TCRβ expansions in donors’ CD8+ T cells in comparison to CD4+ T cells. Although the healthy donor T cell cohort included fewer individuals (n = 21) than the patient cohort (n = 90), our results suggest that the acquisition or selection of somatic mutations in expanded CD8+ T cell clones is characteristic of patients with immunological hematological disease. In addition, only the highest non-synonymous mutation VAF was increased in CD8+ T cells of patients, while the overall VAFs and the highest synonymous VAFs were similar in both lineages, and the difference between the highest synonymous and non-synonymous VAFs was not detected in healthy donor CD8+ T cells. Furthermore, the largest TCRβ expansion size was not correlated with variant burden nor the highest synonymous VAF in patients’ CD8+ T cells, indicating that the mutation accumulation and expansion are not random. In CD4+ T cells, we did not observe an association between the largest TCRβ clone size and VAF, which implies that CD8+ T cells may be more prone to accumulate coding somatic mutations preceding or during peripheral expansion of a clone. In contrast to CH, our observations also suggest that age is not the primary factor driving mutated CD8+ T cell expansions.
The clinical relevance of somatic mutations in lymphocytes is increasingly recognized (10). Apart from predisposing to lymphoid malignancy (35), somatic mutations may alter lymphocyte function toward hyper- or hypo-reactiveness or limit T cell exhaustion. Hyper-reactiveness can contribute to the break of immune tolerance and auto- or alloimmunity. Somatic STAT3 mutations have been found to be prevalent in human autoimmune diseases mediated by cytotoxic CD8+ T cells (19, 28–30) and STAT3 gain-of-function mutations lead to oligoclonal accumulation of CD8+ T cells driving autoimmunity in mice (46). In addition to somatic STAT3 mutations, somatic FAS (47, 48) and KRAS (49) mutations are found in syndromes with autoimmunity and somatic mTOR mutation with alloimmunity (21). In our study, we identified STAT3 and KRAS mutations in CD8+ T cells of patients with autoimmunity but not in healthy donor T cells. Because of the cohort size and low number of recurrently mutated genes, we had limited power to evaluate associations with lymphoid malignancy or other health outcomes.
In addition to putative driver mutations, we identified several pathways that were significantly mutated in T cells based on Oncodrive-fm analysis (50). Functionally interesting pathways were related to T cell activation and survival (cell costimulation, positive regulation of MAP kinase activity, and positive regulation of ERK1 and ERK2 cascade), cell proliferation (negative regulation of cell growth), and self-renewal (IL-7–mediated signaling pathway). Notably, IL-7 signaling is one of the key kinase signaling pathways altered in acute T cell lymphoblastic leukemia (37, 51) and its germline variation is associated with immune-related toxicity during checkpoint blockade therapy (52, 53).
Recent studies have shown that STAT3 mutations are found at negligible VAFs (0.007 to 0.07%) also in healthy individuals (25, 26), which indicates that a mutated T cell clone alone is not enough to drive abnormal immune reactions. Considering this and the fact that clonal expansion does not alone make a T cell clone pathogenic, we studied antigen specificities and phenotypes of expanded T cell clones with somatic mutations. We found that mutated T cells were mainly of cytotoxic TEMRA phenotypes that were expanded in patients with cGVHD but not in healthy controls and were not predicted to recognize common viral epitopes. In our previous study, we noted a similar cytotoxic CD4+ TEMRA phenotype in a patient with chronic skin GVHD and an alloreactive CD4+ T cell clone with somatic mTOR mutation (21).
Somatic mutations can also benefit health if they occur in appropriate T cell clones. Donor CH in HSCT has been associated with lower relapse rates and increased cGVHD risk, indicative of immunological mechanisms (54). In CAR T cell therapies, secondary genetic alterations at the lentiviral vector insertion site can improve CAR T cell function and lead to clonal expansion in TET2 (15), CBL (16), and other genes (17) in human, and in DNMT3A in animal models (18). Several genes associated with CAR T clonality (TET2, DNMT3A, CREBBP, KMT2D, and KDM6A) were also identified in our study, indicating that the same genetic mechanisms can contribute to clonal expansion in not genetically modified T cells and mutations can already occur in cells in which CAR T construct is inserted.
In conclusion, our study shows that somatic mutations occur frequently in T cells. A subset of mutations may promote abnormal immune reactions and clonal expansion and alter T cell survival and function, but this can also be affected by the antigen specificity of mutated T cells, hematologic disease itself, and changes in the microenvironment. In the era of cellular immunotherapies and the increasing prevalence of autoimmune diseases, the evaluation of central and peripheral CH in T cells has substantial clinical relevance and should be an object of future studies developing more targeted and efficient therapies for patients.
MATERIALS AND METHODS
Samples used for immunogene panel and TCRβ sequencing
The cohorts included patients with cGVHD (number of patients: 38), MDS (n = 14), ITP (n = 2), AA [n = 19; previously published (30)], immunodeficiency [n = 17; previously published (20)], and healthy donor T cells [n = 21; previously published (20, 30)]. Patient characteristics are presented in table S1. Age was similar in all cohorts (fig. S1A). Patients were enrolled in the study in the Department of Hematology at the Helsinki University Hospital (Finland), Turku University Hospital (Finland), and Karolinska Institute (Sweden). Blood or bone marrow samples were collected from patients during routine laboratory follow-ups. Healthy blood donor buffy coats (n = 21) were provided by the Finnish Red Cross Blood Service. Blood donation criteria exclude patients with hematological diseases, acute viral and bacterial infections, coronary heart disease, progressive neurological disease, insulin-treated diabetes, organ transplantation, rheumatoid diseases in the symptomatic phase, or treated with other than NSAID or hydroxychloroquine.
Samples were processed as previously described (20, 30). When frozen, mononuclear cells (MNCs) were suspended in prechilled phosphate-buffered saline with 10% fetal bovine serum. From either fresh or frozen MNCs, we isolated CD4+ and CD8+ cells with either flow-assisted cell sorting (FACSAria II, Beckton Dickinson) or magnetic bead separation (Miltenyi Biotechnologies, catalog no. 130-045-101 or 130-045-201) according to the manufacturer’s instructions. Purities of bead-sorted fractions were confirmed by flow cytometry, after staining samples with BD Multitest (anti–CD45-PerCP, anti–CD3-FITC, anti–CD4-APC, and anti–CD8-PE-Cy7; catalog no. 340499) according to the manufacturer’s instructions. For flow-sorted fractions, we did not perform additional purity analysis because the purities of flow-sorted samples are nearly 100% based on previous experiments. The gating strategy for flow-assisted cell sorting and purity analysis of bead-sorted samples is presented in fig. S13 (A and B). Samples with low purity (less than 60% of live cells were CD4+ or CD8+ T cells) were excluded from the lineage-specific variant analysis. DNA was extracted with a DNA tissue kit (Macherey-Nagel, Cat. No. 740952) according to the manufacturer’s instructions.
Immunogene panel sequencing
A custom gene panel based on 2533 candidate genes was used, including genes in pathways important in innate and adaptive immunity (20, 30). Next-generation sequencing (NGS) was performed as previously described (20, 30) with the HiSeq 2500 instrument at the Institute for Molecular Medicine Finland, HiLIFE.
Short variant analysis
Single-nucleotide and indel variant discoveries were performed according to a previously described Genome Analysis Toolkit practice (30). All datasets, including aligned sequencing reads from previously published cohorts (20, 30), were processed similarly to allow integrated analysis. See Supplementary Materials and Methods for details regarding sequence data processing, alignment, variant calling, filtering, and quality control. Variants detected in both CD4+ and CD8+ compartments from the same patient were called ancestral variants, and the rest of the variants are called lineage-specific variants. With forced genotyping of detected variants, 92.2% of lineage-specific variants had no variant read support in the paired sample. The sequencing depths at (lineage-specific) mutated loci resembled each other in the mutated samples and their paired samples without mutation (fig. S1G).
dN/dS ratio
To estimate the rate of selection, we used the “dndscv” function of the dNdScv R package (55), restricting the analysis to immunogene panel target genes by the “gene_list” option. We performed the test for CD4+ T cell, CD8+ T cell, and all mutations separately. Global dN/dS estimates of all non-synonymous substitutions with their associated CIs were reported.
Mutational signatures
After filtering steps described in detail in Supplementary Materials and Methods, all synonymous and non-synonymous single-nucleotide variants were included in mutational signature analysis. Identification of mutational signatures was done using the deconstructSigs18 software (56) with default parameters and cancer profiles downloaded from the COSMIC website on 4 July 2020 (v91). COSMIC profiles were complemented with the SBSblood signature profile provided by Machado et al. (45). The mapSeqlevels function from the GenomeInfoDb package was used to convert EnsEMBL to UCSC chromosome nomenclature.
Pathway enrichment and putative lymphoid driver variant analysis
Putative lymphoid driver mutations were identified on the basis of literature (35, 36). Significantly mutated pathways were identified with the Oncodrive-fm tool (34). Pathways queried with Oncodrive-fm included all Gene Ontology pathways, but only biologically relevant pathways were reported. All target genes of the immunogene panel sequencing were used as the background gene set. P values were corrected with a false discovery rate (FDR) approach.
Lymphoid driver genes were identified on the basis of previously curated lists of pathogenic and putative lymphoid driver gene variants (35) supplemented with driver variants in acute lymphoblastic leukemia (36).
Bradley-Terry analysis
Bradley-Terry models were used to assess the clonal dominance of mutations. In the analysis, comparisons were made between matched sample pairs, and variant data were collapsed at the pathway level by choosing the variant calls with the highest VAF. Both fraction-specific and ancestral variants were used. VAFs of unaffected pathways were set to 0, indicating the absence of mutations. The analysis was also repeated using only co-occurring mutations. The Bradley-Terry estimates and 95% CIs were then calculated with the R package “BradleyTerry2.”
Copy number variant analysis
CNVs were investigated by reprocessing sequencing data from short variant analysis alignment files using DRAGEN Bio-IT Platform version 4.2 (Illumina) (57) with Illumina Multigenome Graph Reference hg38 for DRAGEN v4.2 (Illumina). Alterations were annotated with AnnotSV (58). Further details of the analysis are provided in Supplementary Materials and Methods.
TCRβ repertoire analysis
Sorted CD8+ or CD4+ cells were analyzed with multiplexed PCR assay targeting the variable CDR3 region of the rearranged TCRβ locus. NGS was performed and data were analyzed with the immunoSEQ (Adaptive Biotechnologies) and VDJtools (59) platforms, and non-productive TCRβs were filtered (FilterNonFunctional function in VDJtools) before calculating clonotype abundances. Viral predictions were performed with TCRGP (32): We interpreted a prediction to be positive if the TCRGP probability exceeded the FPR = 0 threshold for models in the publication by Jokinen et al. (32). In addition, we performed hard-matching of TCRs to epitope-specific TCRs from VDJdb database (confidence score ≥ 1 TCR downloaded on 14 Match 2023) (33). For seven patients without available TCRβ sequencing data, we analyzed their TCR Vβ families with the IOTest Beta Mark TCR Vβ Repertoire Kit, as described previously (30).
Comparison of VAF with TCRβ clonotype frequency
We compared the highest non-synonymous VAF with the four largest TCRβ clonotype frequencies in samples with at least one TCR clonotype with ≥7% frequency. We assumed the mutations to be heterozygous, and hence, the highest non-synonymous VAF was assigned as “matching” if the VAF was between 35 and 75% of a TCRβ clonotype frequency within the sample. The cutoffs were set on the basis of previous experience in validating variants in Vβ-sorted clonotypes (30). We reasoned that variants with VAFs above 75% of the largest TCRβ clonotype frequency were likely to represent central CH (appearing across distinct TCRβ clonotypes), whereas variants with VAF below 35% of a TCRβ clonotype frequency could either be present in less frequent TCRβ clonotype(s) or be a subclonal event, in other words, the mutation has not been acquired at the beginning of the clonal proliferation of the examined TCRβ clone. If the highest non-synonymous VAF was “matching” with more than one TCRβ clonotype, then it was marked as “multiple-matching.”
Single-cell gene expression and V(D)J transcript profiling and data analysis
Viably frozen blood MNCs from nine patients with cGVHD and six age-matched healthy controls were sorted with BD Influx cell sorter, to enrich for CD45+ cells. To ensure a fair comparison of T cell phenotypes with patients with cGVHD, originally, 12 healthy controls from the Finnish Red Cross Blood Service were screened with cytomegalovirus pp65–specific pentamer or Vβ antibody analysis, and from those, six were confirmed to have oligoclonal T cell expansions and were subjected to single-cell sequencing. Transcript profiles were studied with 10x Genomics Chromium Single Cell V(D)J and 5′ Gene Expression platform (v1.1) and analyzed with Seurat package (60) complemented with Vartrix (42) to detect mutations and Numbat (v1.3.0) (61) to detect CNVs in the transcriptome data. Batch correction was done with scVI (62) and cell-cell interactions were analyzed with CellChat (v1.6.1) (63). See Supplementary Materials and Methods for further details regarding single-cell transcriptome sequencing and data analysis.
Statistical analysis
Statistical tests were performed with R version 4.2.1. The Mann-Whitney test was used for pairwise comparisons and the Kruskal-Wallis test for multiple comparisons. If the Kruskal-Wallis test result was significant, then post hoc analysis was performed with Dunn’s test. Pearson’s correlation was used to test the correlation between two variables and the general linear model (“glm” function in R) was used for multivariable analysis. In the case of multiple comparisons, P values were adjusted with Benjamini-Hochberg, FDR (OncoDrive-fm analysis), or Bonferroni (scRNA-seq DE analysis) method.
Ethical approval
This study was approved by the local university hospitals’ ethics committees, and the principles of the Helsinki Declaration were followed. This study was conducted under ethical approval number 303/13/03/01/11. All patients had given their written informed consent before sample collection.
Acknowledgments
This research was funded with research grants from the European Research Council (M-IMM and STRATIFY projects), Academy of Finland, Sigrid Juselius Foundation, Cancer Foundation Finland, Blood Disease Research Foundation, and Instrumentarium Science Foundation. S.L. was supported by Biomedicum Helsinki Foundation and Finnish Medical Foundation. We acknowledge P. Ellonen from the Institute of Molecular Medicine Finland (FIMM) for advice on gene panel design. We thank H. Lähteenmäki, T. Kasanen, and J. Klievink from the Hematology Research Unit Helsinki for excellent technical assistance. We acknowledge J. Huuhtanen for expertise and advice with TCR data analysis. Single-cell sequencing and immunogene panel sequencing were performed at FIMM Single-cell Analytics and Sequencing units supported by HiLIFE and Biocenter Finland. We acknowledge the Finnish Red Cross Blood Service for providing healthy control samples. The authors thank all the patients for their generous participation. We acknowledge the IT Center for Science Ltd. for data storage and computational resources and Aalto Science-IT project for computational resources.
Author contributions: Conceptualization: S.L., M.A.I.K., P.S., T.K., and S.M. Methology: S.L., M.A.I.K., T.K., P.S., and J.S. Validation: S.L., P.S., and M.K. Formal analysis: S.L., T.J., J.T., M.K., and J.S. Investigation: S.L., M.A.I.K., P.S., D.K., H.R., and J.S. Resources: M.A.I.K., U.S., G.W., P.S., H.R., E.H.-L., M.I.-R., and S.M. Data curation: S.L., M.A.I.K., U.S., G.W., P.S., H.R., and M.I.-R. Visualization: S.L. Supervision: S.M., M.M., and M.K. Writing—original draft: S.L. Writing—review and editing: All authors. Funding acquisition: S.M.
Competing interests: S.M. has received honoraria and research funding from BMS and Novartis, research funding from Pfizer, and honoraria from Dren-Bio (none of these are related to this project). The other authors declare that they have no competing interests.
Data and materials availability: The scRNA + TCRαβ-seq raw data and immunogene panel sequencing raw data have been deposited at the European Genome-phenome Archive (EGA; https://ega-archive.org), which are hosted by the EBI and the CRG, under accession numbers EGAD00001012121 and EGAD50000000352. The access is restricted and can be requested on the EGA website through the Data Access Committee, due to data privacy concerns. The processed scRNA + TCRαβ-seq data are available in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-13419. TCRβ sequencing data are available at Zenodo under 10.5281/zenodo.11106370 and 10.5281/zenodo.11106372. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S13
Legends for tables S1 to S11
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S11
REFERENCES AND NOTES
- 1.Mustjoki S., Young N. S., Somatic mutations in “benign” disease. N. Engl. J. Med. 384, 2039–2052 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Kakiuchi N., Ogawa S., Clonal expansion in non-cancer tissues. Nat. Rev. Cancer 21, 239–256 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Alexandrov L. B., Jones P. H., Wedge D. C., Sale J. E., Campbell P. J., Nik-Zainal S., Stratton M. R., Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blokzijl F., de Ligt J., Jager M., Sasselli V., Roerink S., Sasaki N., Huch M., Boymans S., Kuijk E., Prins P., Nijman I. J., Martincorena I., Mokry M., Wiegerinck C. L., Middendorp S., Sato T., Schwank G., Nieuwenhuis E. E. S., Verstegen M. M. A., van der Laan L. J. W., de Jonge J., IJzermans J. N. M., Vries R. G., van de Wetering M., Stratton M. R., Clevers H., Cuppen E., van Boxtel R., Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore L., Cagan A., Coorens T. H. H., Neville M. D. C., Sanghvi R., Sanders M. A., Oliver T. R. W., Leongamornlert D., Ellis P., Noorani A., Mitchell T. J., Butler T. M., Hooks Y., Warren A. Y., Jorgensen M., Dawson K. J., Menzies A., O’Neill L., Latimer C., Teng M., van Boxtel R., Iacobuzio-Donahue C. A., Martincorena I., Heer R., Campbell P. J., Fitzgerald R. C., Stratton M. R., Rahbari R., The mutational landscape of human somatic and germline cells. Nature 597, 381–386 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Genovese G., Kähler A. K., Handsaker R. E., Lindberg J., Rose S. A., Bakhoum S. F., Chambert K., Mick E., Neale B. M., Fromer M., Purcell S. M., Svantesson O., Landén M., Höglund M., Lehmann S., Gabriel S. B., Moran J. L., Lander E. S., Sullivan P. F., Sklar P., Grönberg H., Hultman C. M., McCarroll S. A., Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P. V., Mar B. G., Lindsley R. C., Mermel C. H., Burtt N., Chavez A., Higgins J. M., Moltchanov V., Kuo F. C., Kluk M. J., Henderson B., Kinnunen L., Koistinen H. A., Ladenvall C., Getz G., Correa A., Banahan B. F., Gabriel S., Kathiresan S., Stringham H. M., McCarthy M. I., Boehnke M., Tuomilehto J., Haiman C., Groop L., Atzmon G., Wilson J. G., Neuberg D., Altshuler D., Ebert B. L., Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie M., Lu C., Wang J., McLellan M. D., Johnson K. J., Wendl M. C., McMichael J. F., Schmidt H. K., Yellapantula V., Miller C. A., Ozenberger B. A., Welch J. S., Link D. C., Walter M. J., Mardis E. R., Dipersio J. F., Chen F., Wilson R. K., Ley T. J., Ding L., Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shlush L. I., Zandi S., Mitchell A., Chen W. C., Brandwein J. M., Gupta V., Kennedy J. A., Schimmer A. D., Schuh A. C., Yee K. W., McLeod J. L., Doedens M., Medeiros J. J. F., Marke R., Kim H. J., Lee K., McPherson J. D., Hudson T. J., HALT Pan-Leukemia Gene Panel Consortium, Brown A. M. K., Yousif F., Trinh Q. M., Stein L. D., Minden M. D., Wang J. C. Y., Dick J. E., Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Beck K., von Beck T., Ferrell P. B. Jr., Bick A. G., Kishtagari A., Lymphoid clonal hematopoiesis: Implications for malignancy, immunity, and treatment. Blood Cancer J. 13, 5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Kenawi A., Ruffell B., Inflammation, ROS, and Mutagenesis. Cancer Cell 32, 727–729 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hormaechea-Agulla D., Matatall K. A., Le D. T., Kain B., Long X., Kus P., Jaksik R., Challen G. A., Kimmel M., King K. Y., Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell 28, 1428–1442.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avagyan S., Henninger J. E., Mannherz W. P., Mistry M., Yoon J., Yang S., Weber M. C., Moore J. L., Zon L. I., Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science 374, 768–772 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Caiado F., Kovtonyuk L. V., Gonullu N. G., Fullin J., Boettcher S., Manz M. G., Aging drives Tet2+/- clonal hematopoiesis via IL-1 signaling. Blood 141, 886–903 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraietta J. A., Nobles C. L., Sammons M. A., Lundh S., Carty S. A., Reich T. J., Cogdill A. P., Morrissette J. J. D., DeNizio J. E., Reddy S., Hwang Y., Gohil M., Kulikovskaya I., Nazimuddin F., Gupta M., Chen F., Everett J. K., Alexander K. A., Lin-Shiao E., Gee M. H., Liu X., Young R. M., Ambrose D., Wang Y., Xu J., Jordan M. S., Marcucci K. T., Levine B. L., Garcia K. C., Zhao Y., Kalos M., Porter D. L., Kohli R. M., Lacey S. F., Berger S. L., Bushman F. D., June C. H., Melenhorst J. J., Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah N. N., Qin H., Yates B., Su L., Shalabi H., Raffeld M., Ahlman M. A., Stetler-Stevenson M., Yuan C., Guo S., Liu S., Hughes S. H., Fry T. J., Wu X., Clonal expansion of CAR T cells harboring lentivector integration in the CBL gene following anti-CD22 CAR T-cell therapy. Blood Adv. 3, 2317–2322 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobles C. L., Sherrill-Mix S., Everett J. K., Reddy S., Fraietta J. A., Porter D. L., Frey N., Gill S. I., Grupp S. A., Maude S. L., Siegel D. L., Levine B. L., June C. H., Lacey S. F., Melenhorst J. J., Bushman F. D., CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J. Clin. Invest. 130, 673–685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prinzing B., Zebley C. C., Petersen C. T., Fan Y., Anido A. A., Yi Z., Nguyen P., Houke H., Bell M., Haydar D., Brown C., Boi S. K., Alli S., Crawford J. C., Riberdy J. M., Park J. J., Zhou S., Velasquez M. P., DeRenzo C., Lazzarotto C. R., Tsai S. Q., Vogel P., Pruett-Miller S. M., Langfitt D. M., Gottschalk S., Youngblood B., Krenciute G., Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci. Transl. Med. 13, eabh0272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koskela H. L. M., Eldfors S., Ellonen P., van Adrichem A. J., Kuusanmäki H., Andersson E. I., Lagström S., Clemente M. J., Olson T., Jalkanen S. E., Majumder M. M., Almusa H., Edgren H., Lepistö M., Mattila P., Guinta K., Koistinen P., Kuittinen T., Penttinen K., Parsons A., Knowles J., Saarela J., Wennerberg K., Kallioniemi O., Porkka K., Loughran T. P. Jr., Heckman C. A., Maciejewski J. P., Mustjoki S., Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 366, 1905–1913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savola P., Martelius T., Kankainen M., Huuhtanen J., Lundgren S., Koski Y., Eldfors S., Kelkka T., Keränen M. A. I., Ellonen P., Kovanen P. E., Kytölä S., Saarela J., Lähdesmäki H., Seppänen M. R. J., Mustjoki S., Somatic mutations and T-cell clonality in patients with immunodeficiency. Haematologica 105, 2757–2768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Park G., Huuhtanen J., Lundgren S., Khajuria R. K., Hurtado A. M., Muñoz-Calleja C., Cardeñoso L., Gómez-García de Soria V., Chen-Liang T. H., Eldfors S., Ellonen P., Hannula S., Kankainen M., Brück O., Kreutzman A., Salmenniemi U., Lönnberg T., Jerez A., Itälä-Remes M., Myllymäki M., Keränen M. A. I., Mustjoki S., Somatic mTOR mutation in clonally expanded T lymphocytes associated with chronic graft versus host disease. Nat. Commun. 11, 2246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savola P., Brück O., Olson T., Kelkka T., Kauppi M. J., Kovanen P. E., Kytölä S., Sokka-Isler T., Loughran T. P., Leirisalo-Repo M., Mustjoki S., Somatic STAT3 mutations in Felty syndrome: An implication for a common pathogenesis with large granular lymphocyte leukemia. Haematologica 103, 304–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savola P., Kelkka T., Rajala H. L., Kuuliala A., Kuuliala K., Eldfors S., Ellonen P., Lagström S., Lepistö M., Hannunen T., Andersson E. I., Khajuria R. K., Jaatinen T., Koivuniemi R., Repo H., Saarela J., Porkka K., Leirisalo-Repo M., Mustjoki S., Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nat. Commun. 8, 15869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valori M., Jansson L., Kiviharju A., Ellonen P., Rajala H., Awad S. A., Mustjoki S., Tienari P. J., A novel class of somatic mutations in blood detected preferentially in CD8+ cells. Clin. Immunol. 175, 75–81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valori M., Lehikoinen J., Jansson L., Clancy J., Lundgren S. A., Mustjoki S., Tienari P., High prevalence of low-allele-fraction somatic mutations in STAT3 in peripheral blood CD8+ cells in multiple sclerosis patients and controls. PLOS ONE 17, e0278245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Horebeek L., Dedoncker N., Dubois B., Goris A., Frequent somatic mosaicism in T lymphocyte subsets in individuals with and without multiple sclerosis. Front. Immunol. 13, 993178 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Z.-Y., Fan L., Wang L., Qiao C., Wu Y.-J., Zhou J.-F., Xu W., Li J.-Y., STAT3 mutations are frequent in T-cell large granular lymphocytic leukemia with pure red cell aplasia. J. Hematol. Oncol. 6, 82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawakami T., Sekiguchi N., Kobayashi J., Imi T., Matsuda K., Yamane T., Nishina S., Senoo Y., Sakai H., Ito T., Koizumi T., Hirokawa M., Nakao S., Nakazawa H., Ishida F., Frequent STAT3 mutations in CD8+ T cells from patients with pure red cell aplasia. Blood Adv. 2, 2704–2712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jerez A., Clemente M. J., Makishima H., Rajala H., Gómez-Seguí I., Olson T., McGraw K., Przychodzen B., Kulasekararaj A., Afable M., Husseinzadeh H. D., Hosono N., LeBlanc F., Lagström S., Zhang D., Ellonen P., Tichelli A., Nissen C., Lichtin A. E., Wodnar-Filipowicz A., Mufti G. J., List A. F., Mustjoki S., Loughran T. P. Jr., Maciejewski J. P., STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood 122, 2453–2459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundgren S., Keränen M. A. I., Kankainen M., Huuhtanen J., Walldin G., Kerr C. M., Clemente M., Ebeling F., Rajala H., Brück O., Lähdesmäki H., Hannula S., Hannunen T., Ellonen P., Young N. S., Ogawa S., Maciejewski J. P., Hellström-Lindberg E., Mustjoki S., Somatic mutations in lymphocytes in patients with immune-mediated aplastic anemia. Leukemia 35, 1365–1379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nik-Zainal S., Alexandrov L. B., Wedge D. C., Van Loo P., Greenman C. D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L. A., Menzies A., Martin S., Leung K., Chen L., Leroy C., Ramakrishna M., Rance R., Lau K. W., Mudie L. J., Varela I., McBride D. J., Bignell G. R., Cooke S. L., Shlien A., Gamble J., Whitmore I., Maddison M., Tarpey P. S., Davies H. R., Papaemmanuil E., Stephens P. J., McLaren S., Butler A. P., Teague J. W., Jönsson G., Garber J. E., Silver D., Miron P., Fatima A., Boyault S., Langerød A., Tutt A., Martens J. W. M., Aparicio S. A. J. R., Borg Å., Salomon A. V., Thomas G., Børresen-Dale A.-L., Richardson A. L., Neuberger M. S., Futreal P. A., Campbell P. J., Stratton M. R., Breast Cancer Working Group of the International Cancer Genome Consortium , Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jokinen E., Huuhtanen J., Mustjoki S., Heinonen M., Lähdesmäki H., Predicting recognition between T cell receptors and epitopes with TCRGP. PLoS Comput. Biol. 17, e1008814 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goncharov M., Bagaev D., Shcherbinin D., Zvyagin I., Bolotin D., Thomas P. G., Minervina A. A., Pogorelyy M. V., Ladell K., McLaren J. E., Price D. A., Nguyen T. H. O., Rowntree L. C., Clemens E. B., Kedzierska K., Dolton G., Rius C. R., Sewell A., Samir J., Luciani F., Zornikova K. V., Khmelevskaya A. A., Sheetikov S. A., Efimov G. A., Chudakov D., Shugay M., VDJdb in the pandemic era: A compendium of T cell receptors specific for SARS-CoV-2. Nat. Methods 19, 1017–1019 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Perez A., López-Bigas N., Functional impact bias reveals cancer drivers. Nucleic Acids Res. 40, e169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niroula A., Sekar A., Murakami M. A., Trinder M., Agrawal M., Wong W. J., Bick A. G., Uddin M. M., Gibson C. J., Griffin G. K., Honigberg M. C., Zekavat S. M., Paruchuri K., Natarajan P., Ebert B. L., Distinction of lymphoid and myeloid clonal hematopoiesis. Nat. Med. 27, 1921–1927 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady S. W., Roberts K. G., Gu Z., Shi L., Pounds S., Pei D., Cheng C., Dai Y., Devidas M., Qu C., Hill A. N., Payne-Turner D., Ma X., Iacobucci I., Baviskar P., Wei L., Arunachalam S., Hagiwara K., Liu Y., Flasch D. A., Liu Y., Parker M., Chen X., Elsayed A. H., Pathak O., Li Y., Fan Y., Michael J. R., Rusch M., Wilkinson M. R., Foy S., Hedges D. J., Newman S., Zhou X., Wang J., Reilly C., Sioson E., Rice S. V., Loyola V. P., Wu G., Rampersaud E., Reshmi S. C., Gastier-Foster J., Auvil J. M. G., Gesuwan P., Smith M. A., Winick N., Carroll A. J., Heerema N. A., Harvey R. C., Willman C. L., Larsen E., Raetz E. A., Borowitz M. J., Wood B. L., Carroll W. L., Zweidler-McKay P. A., Rabin K. R., Mattano L. A., Maloney K. W., Winter S. S., Burke M. J., Salzer W., Dunsmore K. P., Angiolillo A. L., Crews K. R., Downing J. R., Jeha S., Pui C.-H., Evans W. E., Yang J. J., Relling M. V., Gerhard D. S., Loh M. L., Hunger S. P., Zhang J., Mullighan C. G., The genomic landscape of pediatric acute lymphoblastic leukemia. Nat. Genet. 54, 1376–1389 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girardi T., Vicente C., Cools J., De Keersmaecker K., The genetics and molecular biology of T-ALL. Blood 129, 1113–1123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stengel A., Kern W., Zenger M., Perglerová K., Schnittger S., Haferlach T., Haferlach C., Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker. Gene. Chromosomes Canc. 55, 82–94 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Arends C. M., Galan-Sousa J., Hoyer K., Chan W., Jäger M., Yoshida K., Seemann R., Noerenberg D., Waldhueter N., Fleischer-Notter H., Christen F., Schmitt C. A., Dörken B., Pelzer U., Sinn M., Zemojtel T., Ogawa S., Märdian S., Schreiber A., Kunitz A., Krüger U., Bullinger L., Mylonas E., Frick M., Damm F., Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia 32, 1908–1919 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Bick A. G., Weinstock J. S., Nandakumar S. K., Fulco C. P., Bao E. L., Zekavat S. M., Szeto M. D., Liao X., Leventhal M. J., Nasser J., Chang K., Laurie C., Burugula B. B., Gibson C. J., Lin A. E., Taub M. A., Aguet F., Ardlie K., Mitchell B. D., Barnes K. C., Moscati A., Fornage M., Redline S., Psaty B. M., Silverman E. K., Weiss S. T., Palmer N. D., Vasan R. S., Burchard E. G., Kardia S. L. R., He J., Kaplan R. C., Smith N. L., Arnett D. K., Schwartz D. A., Correa A., de Andrade M., Guo X., Konkle B. A., Custer B., Peralta J. M., Gui H., Meyers D. A., McGarvey S. T., Chen I. Y.-D., Shoemaker M. B., Peyser P. A., Broome J. G., Gogarten S. M., Wang F. F., Wong Q., Montasser M. E., Daya M., Kenny E. E., North K. E., Launer L. J., Cade B. E., Bis J. C., Cho M. H., Lasky-Su J., Bowden D. W., Cupples L. A., Mak A. C. Y., Becker L. C., Smith J. A., Kelly T. N., Aslibekyan S., Heckbert S. R., Tiwari H. K., Yang I. V., Heit J. A., Lubitz S. A., Johnsen J. M., Curran J. E., Wenzel S. E., Weeks D. E., Rao D. C., Darbar D., Moon J.-Y., Tracy R. P., Buth E. J., Rafaels N., Loos R. J. F., Durda P., Liu Y., Hou L., Lee J., Kachroo P., Freedman B. I., Levy D., Bielak L. F., Hixson J. E., Floyd J. S., Whitsel E. A., Ellinor P. T., Irvin M. R., Fingerlin T. E., Raffield L. M., Armasu S. M., Wheeler M. M., Sabino E. C., Blangero J., Williams L. K., Levy B. D., Sheu W. H.-H., Roden D. M., Boerwinkle E., Manson J. E., Mathias R. A., Desai P., Taylor K. D., Johnson A. D., NHLBI Trans-Omics for Precision Medicine Consortium, Auer P. L., Kooperberg C., Laurie C. C., Blackwell T. W., Smith A. V., Zhao H., Lange E., Lange L., Rich S. S., Rotter J. I., Wilson J. G., Scheet P., Kitzman J. O., Lander E. S., Engreitz J. M., Ebert B. L., Reiner A. P., Jaiswal S., Abecasis G., Sankaran V. G., Kathiresan S., Natarajan P., Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato M., Sanada M., Kato I., Sato Y., Takita J., Takeuchi K., Niwa A., Chen Y., Nakazaki K., Nomoto J., Asakura Y., Muto S., Tamura A., Iio M., Akatsuka Y., Hayashi Y., Mori H., Igarashi T., Kurokawa M., Chiba S., Mori S., Ishikawa Y., Okamoto K., Tobinai K., Nakagama H., Nakahata T., Yoshino T., Kobayashi Y., Ogawa S., Frequent inactivation of A20 in B-cell lymphomas. Nature 459, 712–716 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Petti A. A., Williams S. R., Miller C. A., Fiddes I. T., Srivatsan S. N., Chen D. Y., Fronick C. C., Fulton R. S., Church D. M., Ley T. J., A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat. Commun. 10, 3660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng G. X. Y., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., McFarland C., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martincorena I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D. C., Fullam A., Alexandrov L. B., Tubio J. M., Stebbings L., Menzies A., Widaa S., Stratton M. R., Jones P. H., Campbell P. J., Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machado H. E., Mitchell E., Øbro N. F., Kübler K., Davies M., Leongamornlert D., Cull A., Maura F., Sanders M. A., Cagan A. T. J., McDonald C., Belmonte M., Shepherd M. S., Vieira Braga F. A., Osborne R. J., Mahbubani K., Martincorena I., Laurenti E., Green A. R., Getz G., Polak P., Saeb-Parsy K., Hodson D. J., Kent D. G., Campbell P. J., Diverse mutational landscapes in human lymphocytes. Nature 608, 724–732 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masle-Farquhar E., Jackson K. J. L., Peters T. J., Al-Eryani G., Singh M., Payne K. J., Rao G., Avery D. T., Apps G., Kingham J., Jara C. J., Skvortsova K., Swarbrick A., Ma C. S., Suan D., Uzel G., Chua I., Leiding J. W., Heiskanen K., Preece K., Kainulainen L., O’Sullivan M., Cooper M. A., Seppänen M. R. J., Mustjoki S., Brothers S., Vogel T. P., Brink R., Tangye S. G., Reed J. H., Goodnow C. C., STAT3 gain-of-function mutations connect leukemia with autoimmune disease by pathological NKG2Dhi CD8+ T cell dysregulation and accumulation. Immunity 55, 2386–2404.e8 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Holzelova E., Vonarbourg C., Stolzenberg M.-C., Arkwright P. D., Selz F., Prieur A.-M., Blanche S., Bartunkova J., Vilmer E., Fischer A., Le Deist F., Rieux-Laucat F., Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med. 351, 1409–1418 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Dowdell K. C., Niemela J. E., Price S., Davis J., Hornung R. L., Oliveira J. B., Puck J. M., Jaffe E. S., Pittaluga S., Cohen J. I., Fleisher T. A., Rao V. K., Somatic FAS mutations are common in patients with genetically undefined autoimmune lymphoproliferative syndrome. Blood 115, 5164–5169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niemela J. E., Lu L., Fleisher T. A., Davis J., Caminha I., Natter M., Beer L. A., Dowdell K. C., Pittaluga S., Raffeld M., Rao V. K., Oliveira J. B., Somatic KRAS mutations associated with a human nonmalignant syndrome of autoimmunity and abnormal leukocyte homeostasis. Blood 117, 2883–2886 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mularoni L., Sabarinathan R., Deu-Pons J., Gonzalez-Perez A., López-Bigas N., OncodriveFML: A general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 17, 128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fattizzo B., Rosa J., Giannotta J. A., Baldini L., Fracchiolla N. S., The physiopathology of T- cell acute lymphoblastic leukemia: Focus on molecular aspects. Front. Oncol. 10, 273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor C. A., Watson R. A., Tong O., Ye W., Nassiri I., Gilchrist J. J., de los Aires A. V., Sharma P. K., Koturan S., Cooper R. A., Woodcock V. K., Jungkurth E., Shine B., Coupe N., Payne M. J., Church D. N., Naranbhai V., Groha S., Emery P., Mankia K., Freedman M. L., Choueiri T. K., Middleton M. R., Gusev A., Fairfax B. P., IL7 genetic variation and toxicity to immune checkpoint blockade in patients with melanoma. Nat. Med. 28, 2592–2600 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Groha S., Alaiwi S. A., Xu W., Naranbhai V., Nassar A. H., Bakouny Z., El Zarif T., Saliby R. M., Wan G., Rajeh A., Adib E., Nuzzo P. V., Schmidt A. L., Labaki C., Ricciuti B., Alessi J. V., Braun D. A., Shukla S. A., Keenan T. E., Van Allen E., Awad M. M., Manos M., Rahma O., Zubiri L., Villani A.-C., Fairfax B., Hammer C., Khan Z., Reynolds K., Semenov Y., Schrag D., Kehl K. L., Freedman M. L., Choueiri T. K., Gusev A., Germline variants associated with toxicity to immune checkpoint blockade. Nat. Med. 28, 2584–2591 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frick M., Chan W., Arends C. M., Hablesreiter R., Halik A., Heuser M., Michonneau D., Blau O., Hoyer K., Christen F., Galan-Sousa J., Noerenberg D., Wais V., Stadler M., Yoshida K., Schetelig J., Schuler E., Thol F., Clappier E., Christopeit M., Ayuk F., Bornhäuser M., Blau I. W., Ogawa S., Zemojtel T., Gerbitz A., Wagner E. M., Spriewald B. M., Schrezenmeier H., Kuchenbauer F., Kobbe G., Wiesneth M., Koldehoff M., Socié G., Kroeger N., Bullinger L., Thiede C., Damm F., Role of donor clonal hematopoiesis in allogeneic hematopoietic stem-cell transplantation. J. Clin. Oncol. 37, 375–385 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Martincorena I., Raine K. M., Gerstung M., Dawson K. J., Haase K., Van Loo P., Davies H., Stratton M. R., Campbell P. J., Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenthal R., McGranahan N., Herrero J., Taylor B. S., Swanton C., deconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller N. A., Farrow E. G., Gibson M., Willig L. K., Twist G., Yoo B., Marrs T., Corder S., Krivohlavek L., Walter A., Petrikin J. E., Saunders C. J., Thiffault I., Soden S. E., Smith L. D., Dinwiddie D. L., Herd S., Cakici J. A., Catreux S., Ruehle M., Kingsmore S. F., A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 7, 100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.AnnotSV: An integrated tool for structural variations annotation, Bioinformatics | Oxford Academic; https://academic.oup.com/bioinformatics/article/34/20/3572/4970516. [DOI] [PubMed]
- 59.Shugay M., Bagaev D. V., Turchaninova M. A., Bolotin D. A., Britanova O. V., Putintseva E. V., Pogorelyy M. V., Nazarov V. I., Zvyagin I. V., Kirgizova V. I., Kirgizov K. I., Skorobogatova E. V., Chudakov D. M., VDJtools: Unifying post-analysis of T cell receptor repertoires. PLoS Comput. Biol. 11, e1004503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao T., Soldatov R., Sarkar H., Kurkiewicz A., Biederstedt E., Loh P.-R., Kharchenko P. V., Haplotype-aware analysis of somatic copy number variations from single-cell transcriptomes. Nat. Biotechnol. 41, 417–426 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez R., Regier J., Cole M. B., Jordan M. I., Yosef N., Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin S., Guerrero-Juarez C. F., Zhang L., Chang I., Ramos R., Kuan C.-H., Myung P., Plikus M. V., Nie Q., Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 (2013).
- 66.Picard Tools - By Broad Institute; https://broadinstitute.github.io/picard/.
- 67.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tate J. G., Bamford S., Jubb H. C., Sondka Z., Beare D. M., Bindal N., Boutselakis H., Cole C. G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S. C., Kok C. Y., Noble K., Ponting L., Ramshaw C. C., Rye C. E., Speedy H. E., Stefancsik R., Thompson S. L., Wang S., Ward S., Campbell P. J., Forbes S. A., COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adnan Awad S., Kankainen M., Ojala T., Koskenvesa P., Eldfors S., Ghimire B., Kumar A., Kytölä S., Kamel M. M., Heckman C. A., Porkka K., Mustjoki S., Mutation accumulation in cancer genes relates to nonoptimal outcome in chronic myeloid leukemia. Blood Adv. 4, 546–559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dufva O., Pölönen P., Brück O., Keränen M. A. I., Klievink J., Mehtonen J., Huuhtanen J., Kumar A., Malani D., Siitonen S., Kankainen M., Ghimire B., Lahtela J., Mattila P., Vähä-Koskela M., Wennerberg K., Granberg K., Leivonen S.-K., Meriranta L., Heckman C., Leppä S., Nykter M., Lohi O., Heinäniemi M., Mustjoki S., Immunogenomic landscape of hematological malignancies. Cancer Cell 38, 380–399.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Huuhtanen J., Bhattacharya D., Lönnberg T., Kankainen M., Kerr C., Theodoropoulos J., Rajala H., Gurnari C., Kasanen T., Braun T., Teramo A., Zambello R., Herling M., Ishida F., Kawakami T., Salmi M., Loughran T., Maciejewski J. P., Lähdesmäki H., Kelkka T., Mustjoki S., Single-cell characterization of leukemic and non-leukemic immune repertoires in CD8+ T-cell large granular lymphocytic leukemia. Nat. Commun. 13, 1981 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S13
Legends for tables S1 to S11
References
Tables S1 to S11