Abstract
Persistent inflammation has been associated with severe disc degeneration (DD). This study investigated the effect of prolonged nuclear factor κB (NF-κB) activation in DD. Using an inducible mouse model, we genetically targeted cells expressing aggrecan, a primary component of the disc extra cellular matrix, for activation of the canonical NF-κB pathway. Prolonged NF-κB activation led to severe structural degeneration accompanied by increases in gene expression of inflammatory molecules (Il1b, Cox2, Il6, and Nos2), chemokines (Mcp1 and Mif), and catabolic enzymes (Mmp3, Mmp9, and Adamts4). Increased recruitment of proinflammatory (F4/80+,CD38+) and inflammatory resolving (F4/80+,CD206+) macrophages was observed within caudal discs. We found that the secretome of inflamed caudal disc cells increased macrophage migration and inflammatory activation. Lumbar discs did not exhibit phenotypic changes, suggestive of regional spinal differences in response to inflammatory genetic overactivation. Results suggest prolonged NF-κB activation can induce severe DD through increases in inflammatory cytokines, chemotactic proteins, catabolic enzymes, and the recruitment and activation of macrophage cell populations.
Persistent inflammatory activation of disc cells initiates macrophage recruitment and severe structural degradation.
INTRODUCTION
Chronic inflammation plays a critical role across musculoskeletal tissue diseases by contributing to degeneration and pain, which have a massive global impact on disability and wellbeing. Among musculoskeletal tissue diseases, low back pain (LBP) is the leading cause of disability globally, where a growing prevalence and limited therapeutic interventions drives an annual US economic encumbrance over $100 billion (1, 2). Although the etiology of LBP is multifactorial, intervertebral disc (IVD) degeneration is the most prevalent contributor to symptomatic LBP (3). A commonly proposed signaling driver of chronic musculoskeletal inflammation is the master transcription factor nuclear factor κB (NF-κB), which is known to mediate immune responses and has been observed to be elevated locally in connective tissues of patients with disc degeneration (DD) (4), and other musculoskeletal soft tissue diseases such as tendinopathy (5), knee osteoarthritis (OA) (6), and synovial rheumatoid arthritis (RA) (7).
Molecularly, canonical NF-κB activation is mediated by degradation of the inhibitor of NF-κB (IκBα) by the IκB kinase (IKK) complex, containing the regulating subunit, IKKγ, and catalytic subunits, IKKα and IKKβ. IκBα degradation allows NF-κB to freely translocate from the cytoplasm to the nucleus to regulate transcription. During canonical pathway activation, NF-κB subunits, most commonly p50 and p65, regulate transcription of downstream inflammatory signaling (8, 9). Within the human IVD and during degeneration, inflammatory cytokines (10, 11) and catabolic enzymes (12–14) have been found to be persistently elevated. Coinciding with persistent inflammation, increased innate immune cell presence, specifically macrophages, has been observed in human lumbar (15) and murine caudal (16) DD samples and is thought to engage in inflammatory driven cross-talk with IVD cells (15–17). The master transcription factor, NF-κB, is known to regulate a number of these inflammatory cytokines and catabolic enzymes, as well as mediate immune cell recruitment and activation (8, 18, 19).
Although chronic inflammation is thought to be a key driver of DD, there is a lack of in vivo models of inflammatory driven DD. Unexpectedly, prior studies evaluating global inflammatory mutations have yielded mixed outcomes on the effects to IVD integrity (20, 21). Specifically, investigations of global overexpression of human TNFα in Tg197 mice resulted in systemic inflammation with higher incidence of spontaneous herniation, increased immune cell presence, and degenerative changes in vertebral bone, but no overt evidence of DD was observed in the lumbar or caudal IVDs, suggesting that human TNFα-driven systemic inflammation does not produce severe DD (20). In a model of interleukin-1 (IL-1)–mediated inflammation, IL-1 receptor antagonist (IL-1ra) knockout mice were found to exhibit features of DD in the lumbar spine (structural degeneration and increased catabolic enzyme expression), suggesting that deletion of the natural inhibitor of IL-1 (IL-1ra) created a global IL-1 imbalance, which could serve as a possible driver of DD (21). Yet, another study that investigated IVD integrity in IL-1α/β knockout mice found little to no effect on lumbar or caudal IVD integrity, despite evidence of decreased systemic cytokine levels (22). Moreover, global deletion of IL-1ra had no appreciable effect on the response of caudal IVDs to puncture injury (23). Together, the unexpected and disparate effects observed using global overactivation or knockout models highlight the complexity of using regulation of systemic inflammatory signaling to study local effects on IVD integrity, in part because the IVD naturally exists in an avascular niche, which may limit the impact of systemic inflammation on the IVD. Therefore, there remains a gap in knowledge on whether persistent local IVD inflammation can produce severe DD in vivo.
To directly investigate the role of discal inflammation in the development of DD, we used an inducible aggrecan-Cre–driven genetic mouse model to target constitutive activation of NF-κB, throughout all compartments of the lumbar and caudal IVD, of which aggrecan is a major extra cellular matrix (ECM) component. We assessed tissue structural, compositional, and biological changes in the lumbar and caudal IVDs, with the hypothesis that prolonged canonical NF-κB pathway activation within IVD cells will induce a chronic proinflammatory microenvironment that mimics inflammatory features of clinical DD, which in turn will activate an innate immune response and produce advanced morphological DD in otherwise healthy adult mice. Findings indicate that inducing persistent inflammation in the IVD is sufficient to cause severe caudal, but not lumbar, DD mediated by increased proinflammatory cytokine, chemokine, and catabolic enzyme expression and macrophage recruitment. Furthermore, we demonstrate that the secretome of caudal IVD cells overexpressing IKKβ directly promote an inflammatory macrophage phenotype, and that this modulation could be mitigated by paracrine factors derived from inflammatory-resolving macrophages.
RESULTS
Aggrecan-linked Cre recombination targets IVD cells for sustained IKKβ–NF-κB overactivation
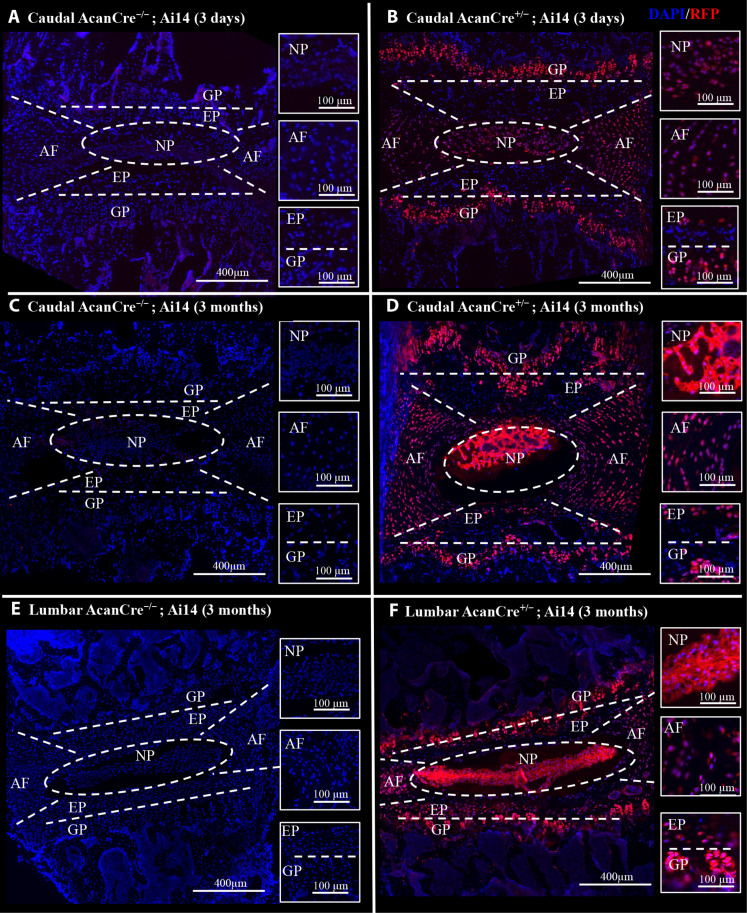
We used a mouse carrying the inducible Cre recombinase construct, CreERT2, in the aggrecan (Acan) gene to target signaling specifically in IVD Acan+ cells. To validate the presence of Cre-mediated genetic recombination within the adult IVD, an AcanCreERT2 mouse was crossed with a Ai14 fluorescent reporter mouse (AcanCre+/−;Ai14) and IVDs were evaluated following intraperitoneal (IP) tamoxifen injections. Red fluorescent protein (RFP) expression indicative of Cre activity was evaluated throughout all tissue compartments of the caudal and lumbar IVDs, including the nucleus pulposus (NP), annulus fibrosis (AF), cartilage endplate (EP), and vertebral growth plates (GPs). RFP expression was observed throughout all tissue compartments of the caudal IVD at 3 days (Fig. 1B) and 3 months (Fig. 1D) following IP tamoxifen injections (or after recombination). Similarly, RFP expression was observed throughout all tissue compartments of lumbar IVDs, including the NP, AF, EPs, and GPs 3 months after recombination (Fig. 1F). No RFP expression was observed in caudal or lumbar IVDs from AcanCre−/−;Ai14 mice (Fig. 1, A, C, and E). Following successful validation that AcanCreERT2 mice target IVD tissues for genetic recombination, we next crossed AcanCreERT2 mice with Ikk2cafl/fl mice to induce IKKβ–NF-κB overactivation in Acan+ IVD cells upon tamoxifen IP injection (AcanCreERT2/+;Ikk2cafl/fl, referred to from here on as IKKβCA mice). To validate NF-κB activation downstream of Cre activity, we first evaluated IKKB gene (Ikk2) expression 1 week after recombination in lumbar and caudal whole IVDs. A significant increase in Ikk2 gene expression (~8-fold) was detected in the caudal IVDs of IKKβCA mice, compared to AcanCre−/−;Ikk2cafl/fl, or control mice (P < 0.0001) (Fig. 2A). A significant increase in Ikk2 expression (~3-fold) was also detected in lumbar IVDs of IKKβCA mice compared to lumbar control IVDs (P = 0.0099) (Fig. 2A). However, comparing across caudal and lumbar spinal levels of IKKβCA mice, caudal IKKβCA IVDs had significantly higher increases in Ikk2 expression compared to lumbar IKKβCA IVDs (P = 0.0001), both normalized to their respective control IVDs (Fig. 2A). Next IKKβ protein expression was assessed in IKKβCA mice compared to the control mice (Acan+/+;Ikk2cafl/fl) at 1 month after recombination. Staining for cells positive for IKKβ were detected in all tissue compartments (NP, AF, EP, and GP) of caudal and lumbar IVDs from IKKβCA mice (Fig. 2B), in a pattern similar to the Cre activity observed in reporter mice (Fig. 1). Little to no detectable staining for IKKβ was seen in control caudal or lumbar IVDs (Fig. 2B). Validation results showed this in vivo model effectively targets all tissue compartments of the caudal and lumbar IVDs, producing an increased and sustained IKKβ expression at both gene and protein levels, although caudal IKKβCA IVDs exhibited a more robust increase in IKKβ at the gene expression level.
Fig. 1. The AcanCreERT2 mouse targets all compartments of the IVD and GP.
(A to D) Representative images of immunofluorescent (IF) staining for RFP within mid-sagittal sections of caudal AcanCre−/−;Ai14 and AcanCre+/−;Ai14 reporter mice 3 days and 3 months following tamoxifen IP injections. (E and F) Images of IF staining for RFP within mid-sagittal sections of lumbar AcanCre−/−;Ai14 and AcanCre+/−;Ai14 reporter mice 3 months following tamoxifen IP injections. NP, AF, EP, and GP compartments are delineated (white dashed lines).
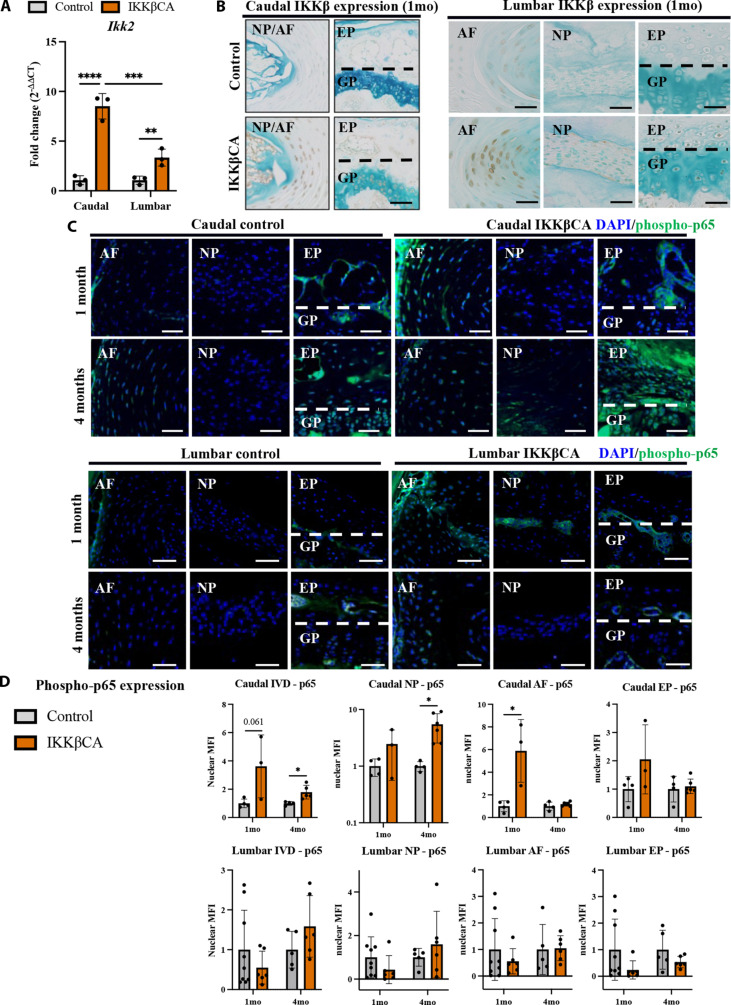
Fig. 2. IKKβ overexpression and NF-κB activation within IKKβCA mice.
(A) Gene expression of Ikk2 within caudal and lumbar IVDs expressed as fold change relative to control (AcanCre−/−;Ikk2cafl/fl) 1 week after recombination (n = 3 mice, one disc per mouse). (B) IHC staining for IKKβ within mid-sagittal sections of 1 month control and IKKβCA IVDs. Scale bar, 100 μm. (C) Representative IF staining for phosphorylated p65 (green) within mid-sagittal sections of control and IKKβCA IVDs. Scale bars, 50 μm. (D) Nuclear MFI quantification (normalized to control within time point) of phosphorylated p65 (n = 3 to 6 mice per group, one to two IVDs per mouse). *P < 0.05, **P < 0.01, and ***P < 0.001.
We then assessed activation of p65 subunit in the canonical IKKβ–NF-κB pathway, with staining for phosphorylated p65 (phospho-p65) (Fig. 2C) (8). In caudal IVDs, increased immunofluorescence staining of phospho-p65 was detected in all IVD tissues of IKKβCA mice compared to control mice at 4 (P = 0.032) and 6 months (P = 0.0073) after activation (Fig. 2, C and D, and fig. S1). Analysis of p65 activation within caudal tissue compartments revealed significant increases in phospho-p65 nuclear mean fluorescence intensity (MFI) in the AF at 1 month (P = 0.032) and in the NP at 4 months (P = 0.034) after recombination in IKKβCA mice compared to control mice (Fig. 2, C and D). In lumbar IVDs, phospho-p65–positive staining was observed throughout the AF and NP tissues at 1 month and within the AF at 4 months (Fig. 2C). However, when quantified, the magnitude of phosphor-p65 was similar in control and IKKβCA IVDs from the lumbar region (Fig. 2D). These results demonstrate that this model produces expression of constitutively active IKKβ within the caudal IVD, resulting in increased and prolonged activation of downstream canonical NF-κB signaling pathway activation. Results also reveal variability in phospho-p65 across compartments of the caudal IVD and in the lumbar IVD levels with time after recombination, suggesting an influence of differences in Cre activation, molecular signaling, and/or cellularity that individually or in combination may lead to the observed differences.
IKKβ overexpression up-regulates IVD inflammatory cytokine, chemokine, catabolic enzyme, and neurotrophic factor gene expression over time
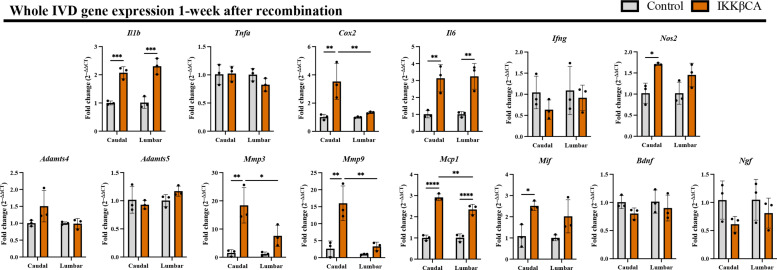
NF-κB mediates inflammatory responses via an up-regulation in inflammatory cytokine, chemokine, and catabolic enzyme expression (8, 18, 19). To assess this type of activation in IKKβCA IVDs, gene expression was assessed from RNA isolated from whole caudal and lumbar IVD tissue, containing the AF, NP, and EPs. To assess for early activation of NF-κB IVD tissue was harvested at 1 week after recombination. Gene expression of the inflammatory mediators, Il1b (P = 0.0005), Cox2 (P = 0.0042), Il6 (P = 0.0039), and Nos2 (P = 0.0174), was increased in IKKβCA caudal IVDs compared to control (Fig. 3). Significant increases in catabolic enzymes, Mmp3 (P = 0.0016) and Mmp9 (P = 0.0013) and proinflammatory chemokines, Mcp1 (P < 0.0001) and Mif (P = 0.0218), were also observed in IKKβCA caudal IVDs compared to control 1 week after recombination (Fig. 3). In lumbar analysis, we observed significant increases in the proinflammatory cytokines, Il1b (P = 0.0002) and Il6 (P = 0.0039) and the proinflammatory chemokine Mcp1 (P < 0.0001) (Fig. 3). Like the comparison of Ikk2 expression between caudal and lumbar IVDs of IKKβCA mice, we observed that expression of Cox2 (P = 0.0063), Mmp3 (P = 0.0147), Mmp9 (P = 0.0013), and Mcp1 (P = 0.0053) to be more robustly increased versus respective controls in caudal IKKβCA IVDs relative to lumbar IKKβCA IVDs (Fig. 3). Extending this analysis of gene expression changes to 2 months after recombination within caudal IVDs, Cox2 (P = 0.034), Nos2 (P = 0.011), Mmp3 (P = 0.042), Mcp1 (P = 0.0091), and Mif (P = 0.0057) remained significantly increased in caudal IKKβCA IVDs compared to control (fig. S2). The proinflammatory cytokine Ifng (P = 0.042), catabolic enzyme Adamts4 (P = 0.0068), and neurotrophic factors implicated in pathological nerve ingrowth in DD, Ngf (P = 0.017) and dnf (P = 0.020), exhibited a unique pattern of significant up-regulation at 2 months, but not at 1 week, in caudal IKKβCA IVDs compared to control (fig. S2). Sustained increased Ikk2 expression (~3-fold) was also observed at 2 months after recombination in caudal IKKβCA IVDs compared to control (P = 0.0011) (fig. S2). Further validating up-regulations in IVD inflammatory gene expression, a serum analysis from whole blood collected at 1 week and 6 months after recombination also revealed significant increases in circulating inflammatory cytokines and chemokines, including IL-1α, IL-1β, IL-6, interferon-β (IFN-β), tumor necrosis factor–α (TNFα), and monocyte chemoattractant protein-1 (MCP-1) in IKKβCA mice compared to control (fig. S3). Of note, increases in circulating cytokine levels may be a contribution of not only the targeted IVD compartments but also other ACAN-expressing cells throughout the body (e.g., chondrocytes in synovial joints). Together, gene expression results suggest NF-κB overactivation contributed early prodegenerative molecular changes, which included an up-regulation of proinflammatory markers, chemokines and immune cell activation mediators, catabolic enzymes, and neurotrophic factors within IVD cells, with several targets being more robustly increased in caudal IVDs compared to lumbar.
Fig. 3. IKKβ overexpression up-regulates inflammatory cytokine, chemokine, catabolic enzyme, and neurotrophic factor gene expression.
Gene expression changes (relative to control) from total RNA isolated from control (AcanCre−/−;Ikk2cafl/fl) and IKKβCA whole caudal and lumbar IVDs containing NP, AF, and EP, 1 week after recombination. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (n = 3 mice per genotype, one IVD per mouse).
NF-κB overactivation in IVD cells produces severe morphological caudal IVD degeneration
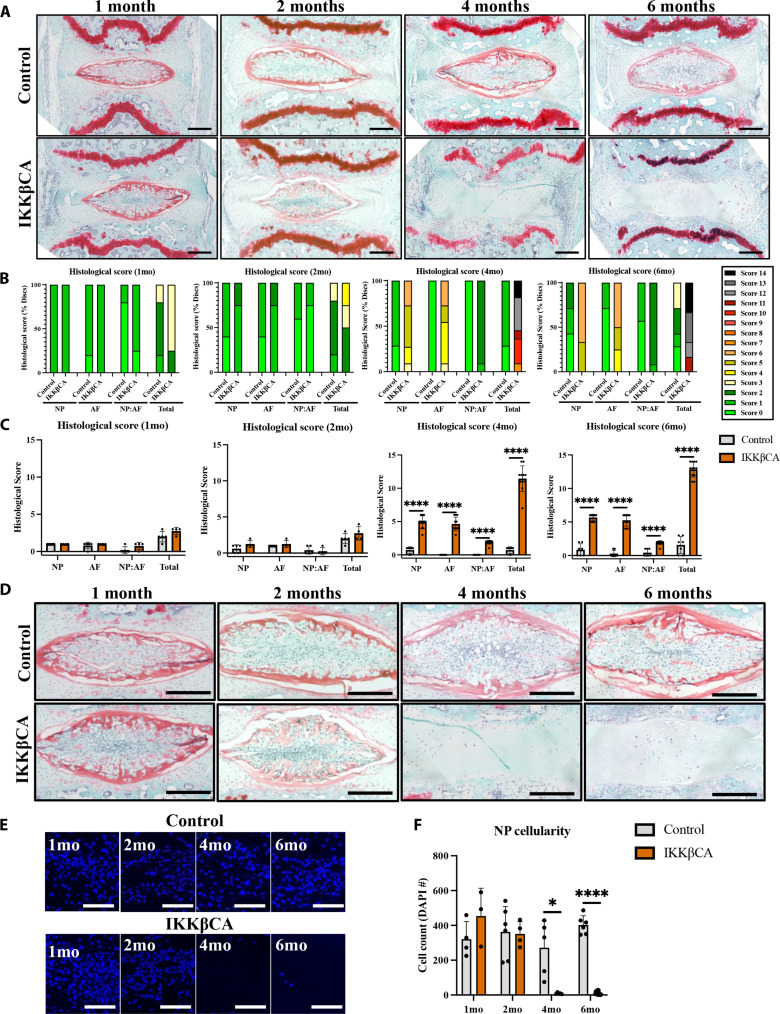
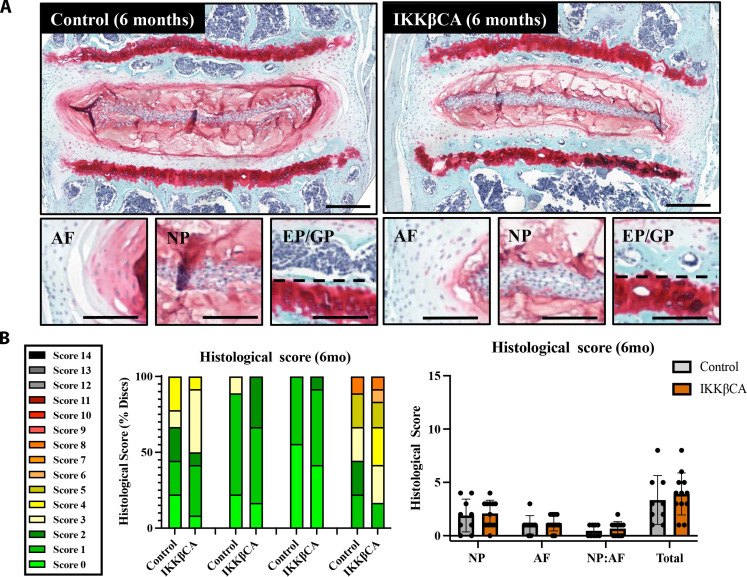
The IVD is a composite fibro-cartilaginous connective tissue structure consisting of distinct tissue types: The AF is a highly fibrous outer ring, which encompasses a gelatinous proteoglycan-rich NP, and the cartilage EPs anchor the IVD to the adjacent vertebrae. To determine the effect of NF-κB–mediated inflammation on IVD integrity, caudal IVD motion segments were assessed for degenerative histomorphological changes (24). At 1 and 2 months after recombination, mild degenerative changes within the NP (reduced cellularity and increased cell clustering) and AF (loss in concentric lamellae structure and cellularity) could be seen in IKKβCA mice but not control IVDs (Fig. 4A). However, these mild observable differences did not produce statistically significant changes in histological grading scores between groups at 1 or 2 months (Fig. 4A). IKKβCA mice at 4 and 6 months after recombination showed histological characteristics of severe DD in caudal IVDs. Compared to control mice, caudal IVDs from IKKβCA mice showed decreased cellularity, loss of Safranin-O staining, loss of NP tissue structure, and disruption of the border that differentiates the NP from the AF regions (i.e., NP:AF border) (Fig. 4A). AF tissue integrity in caudal IVDs was also altered, including the presence of rounded cells near the NP:AF border and inner half of the AF, loss of cellularity in the inner half of AF, and loss of lamellae structure or widened lamellae (Fig. 4A). Degenerative changes were not detected in caudal IVDs of control mice. This led to significant increases in histological scores in IKKβCA caudal IVDs across all scoring criteria at 4 (P < 0.0001) and 6 months (P < 0.0001) after recombination compared to control (Fig. 4, B and C).
Fig. 4. IKKβ overexpression produces severe caudal DD.
(A) Representative images of safranin-O–stained mid-sagittal sections of control (AcanCre−/−;Ikk2cafl/fl) and IKKβCA caudal IVDs 1, 2, 4, and 6 months after recombination. Scale bars, 250 μm. Histological scoring legend, ranging from 0 (healthy) to 14 (most severe). (B) Distribution of histological scores. (C) Histological scoring within NP, AF, and NP:AF border compartments, and total score. (D) Representative images of safranin-O–stained mid-sagittal sections of the NP region of control and IKKβCA caudal IVDs at 1, 2, 4, and 6 months after recombination (n = 4 to 6 mice per genotype and time point, one to three caudal IVDs per mouse). Scale bars, 250 μm. (E) Representative images of 4′,6-diamidino-2-phenylindole (DAPI; nuclear)–stained mid-sagittal caudal IVD sections. Scale bars, 100 μm. (F) Quantification of caudal NP cellularity within hand-drawn ROIs of DAPI nuclear-stained mid-sagittal sections. *P < 0.05 and ****P < 0.0001. (n = 4 to 6 mice per genotype and time point, one to three caudal IVDs per mouse).
Among the defined histomorphological characteristics of the degenerative IKKβCA caudal IVD, a notable change following prolonged NF-κB pathway activation was the loss of NP cellularity compared to IVDs of control mice (Fig. 4D). While no significant changes in NP cell count were detected between IKKβCA and control caudal IVDs at 1 and 2 months, a significant decrease in NP cell count was detected in IKKβCA caudal IVDs at 4 (P = 0.018) and 6 months after recombination (P < 0.0001) compared to control mice (Fig. 4, E and F).
Degenerative EP changes in cellularity and organization, clefts and microfractures, and Schmorl’s node presence were also observed based on a murine EP–specific grading criteria (25) that was not part of the IVD histologic scoring criteria used for evaluating the NP and AF. Degeneration within the EP was observed in IKKβCA caudal IVDs at 4 and 6 months after recombination, including clear evidence of EP fissures and microfractures and EP cellular disorganization (Fig. 4A). Furthermore, potential infiltrating cell populations were detected within the EP of IKKβCA caudal IVDs at 4 and 6 months, as indicated by nuclear stain, which was not observed in control IVDs (Fig. 4A). Last, degeneration within the adjacent GPs of IKKβCA caudal IVDs was also observed at 4 and 6 months and included GP erosion and discontinuity (Fig. 4A). These results suggest that all tissue compartments of the IVD and the closely associated GP are severely affected by prolonged NF-κB activation.
NF-κB overactivation in lumbar IVD cells did not induce morphological lumbar IVD degeneration
Unexpectedly, and contrary to the caudal spine, no degenerative phenotype was observed within lumbar IVDs of IKKβCA mice by 6 months after recombination (Fig. 5A). Both IKKβCA and control lumbar IVDs exhibited a healthy IVD structure, with very similar histological grading scores across NP, AF, NP:AF border, and total IVD categories (Fig. 5B). Although validation revealed Cre activity throughout the lumbar IVD (Fig. 1) and increased Ikk2 and several other downstream proinflammatory gene expression (Fig. 3), the healthy histopathology observed in the lumbar spine highlights a regional difference in the response of the IVD to proinflammatory signaling within this model.
Fig. 5. IKKβ overexpression produces no significant changes to lumbar disc health/structure at 6 months.
(A) Representative images of safranin-O–stained mid-coronal sections of control (AcanCre−/−;Ikk2cafl/fl) and IKKβCA lumbar IVDs 6 months after recombination. Total IVD scale bars = 400 μm. Compartment image scale bars = 200 μm. (B) Distribution of histological grades within NP, AF, and NP:AF border compartments and total score. Histological scoring severity legend ranging from 0 (healthy) to 14 (most severe). No significant differences observed (n = 3 mice per genotype, three to four lumbar IVDs per mouse).
NF-κB overactivation in IVD cells disrupts functional mechanical properties in the caudal IVD degeneration
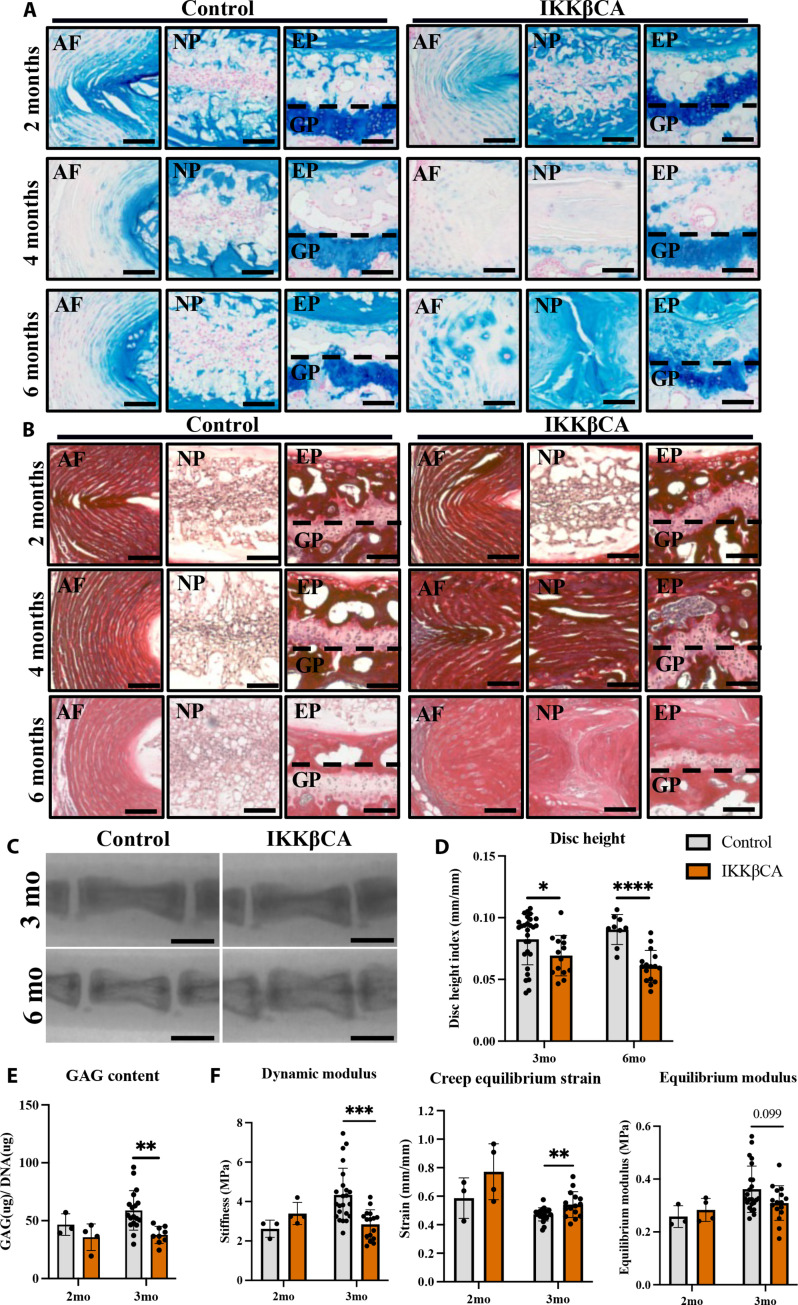
Given the robust phenotypic morphological changes observed in caudal IVDs, we further characterized changes in the mechanical and structural properties of caudal IVDs from IKKβCA mice. To determine whether the observed degeneration in caudal IVDs was associated with ECM changes, specific histological staining was used to evaluate changes in glycosaminoglycans (GAGs; Alcian blue) and collagen (Picrosirius red) content. Compared to control mice, a mild decrease in GAG staining intensity was observed in the inner AF of IKKβCA mice at 2 months, while a more notable loss was observed throughout the IVD of IKKβCA mice at 4 months after recombination (Fig. 6A). However, at 6 months after recombination IKKβCA mice exhibited an NP that lacked the notochordal cell pattern and was replaced by an acellular disorganized matrix that stained positively for GAGs (Fig. 6A). In the AF, an increase in GAG staining in the pericellular matrix of large, rounded AF cells was observed in 6-month IKKβCA IVDs (Fig. 6A). Evaluation of collagen staining revealed no major differences between IKKβCA and control IVDs at 2 months after recombination (Fig. 6B). Whereas at 4 and 6 months after recombination, more pronounced collagen staining was detected within the NP and irregular lamellar AF structures of IKKβCA IVDs compared to control IVDs (Fig. 6B). The differences in GAG and collagen staining suggest that prolonged activation of NF-κB contributes to ECM remodeling and displacement of AF tissue into the NP region, which transitioned into a fibrotic-like NP structure. Overall, histological assessment demonstrated that IVD IKKβ–NF-κB overactivation leads to severe DD, consistent with features of human DD.
Fig. 6. IKKβ overexpression mediates a loss of caudal ECM, disc height, and weakened mechanical properties.
(A) Representative Alcian blue (GAG) and (B) Picrosirius red (collagen)–stained images of control (AcanCre−/−;Ikk2cafl/fl) and IKKβCA caudal IVDs mid-sagittal sections at 2, 4, and 6 months after recombination. Scale bars, 100 μm. (C) Representative fluoroscopic images of control and IKKβCA C6-C8 IVDs 3 and 6 months after recombination. Scale bars, 1 mm. (D) IVD height quantified via DHI of control and IKKβCA caudal IVDs 3 and 6 months after recombination (n = 3 to 6 mice per genotype and time point, three to five IVDs per mouse). (E) GAG content (μg) normalized to total DNA content (μg) within control and IKKβCA caudal IVD digests 2 and 3 months after recombination (n = 3 to 6 mice per genotype, one to three discs per mouse). (F) Dynamic modulus (MPa), creep equilibrium strain (mm/mm), and equilibrium modulus (MPa) of control and IKKβCA caudal IVDs 2 and 3 months after recombination (n = 3 to 6 mice per genotype, one to three discs per mouse). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Having observed structural and compositional changes, changes in ECM biochemistry, functional biomechanics, and IVD height with prolonged inflammation were investigated at time points before (2 months after recombination) and during the onset of morphological degeneration (3 months after recombination). For this, whole IVDs were digested for a quantification of GAG content (normalized to total DNA content). The GAG content was found to be similar in IKKβCA and control IVDs (Fig. 6E) at 2 months, consistent with the mild degenerative structural changes observed. However, at the 3-month time point, a significant loss in GAG content was observed in IKKβCA IVDs compared to control (P = 0.0033; Fig. 6E). Quantification of GAG loss within IKKβCA IVDs is consistent with differences in GAG staining, and further demonstrates that localized NF-κB overactivation mediates ECM degradation within the IVD.
Furthering the functional evaluation, unconfined compression testing was performed using two impermeable platens, where dynamic modulus (MPa), equilibrium strain (mm/mm), and equilibrium modulus (MPa) were calculated during 20 cycles of dynamic loading to 0.25 N (1× body weight) followed by an applied static load of 0.25 N. The dynamic modulus of IKKβCA IVDs at 3 months after recombination was lower than that of control (P = 0.00058; Fig. 6F). Creep testing revealed an increase in creep equilibrium strain in 3 months after recombination IKKβCA IVDs compared to control (P = 0.0097; Fig. 6F). This was associated with a trend change in the equilibrium modulus within IKKβCA IVDs compared to control (P = 0.099) (Fig. 6F). Before structural changes, at 2 months after recombination, mechanical testing revealed no significant differences in dynamic modulus, creep equilibrium strain, or equilibrium modulus between groups (Fig. 6F). Ultimately, changes in compressive properties of IKKβCA IVDs revealed a loss of dynamic compression functionality and resistance to compressive loading.
In another functional output, fluoroscopy imaging was used to assess changes in the disc height index (DHI), commonly used as a clinical indicator of DD (26). DHI analysis of caudal spines from IKKβCA and control mice showed discernable qualitative decreases in IVD height between caudal vertebrae of IKKβCA mice compared to control mice (Fig. 6C). Quantitative analysis revealed a significant decrease in caudal DHI in IKKβCA mice compared to control mice at 3 (P = 0.044) and 6 months (P < 0.0001) after recombination (Fig. 6D). The loss of IVD height in IKKβCA caudal spine reveals another functional consequence of prolonged NF-κB activation possibly associated with the loss of GAG content.
NF-κB overactivation in caudal IVD cells promotes macrophage recruitment and activation
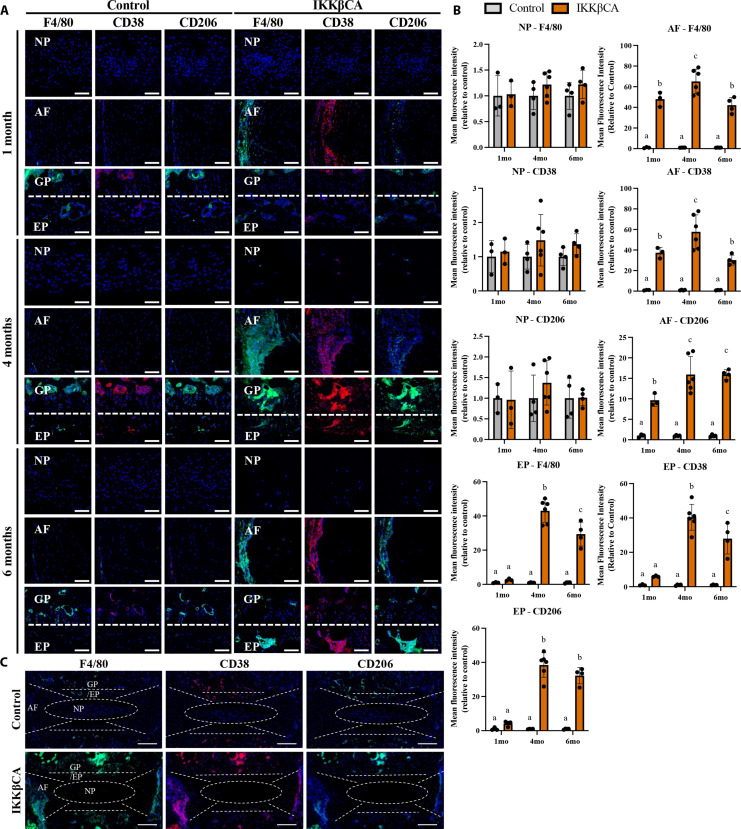
In addition to IVD tissue degeneration, hematoxylin and eosin staining revealed increased cellularity in the outer AF regions of IKKβCA caudal IVDs at all time points (Fig. 4A and fig. S4). In quantitative analysis of cell nuclei in the outer AF regions, compared to control mice, caudal IVDs from IKKβCA mice had significantly increased cellularity at 1 (P = 0.028), 2 (P = 0.00020), 4 (P = 0.028), and 6 months (P = 0.046) after recombination (fig. S4). To further investigate the identity of these cell populations, we performed immunostaining for the pan-macrophage marker F4/80. Additional phenotyping of these cells using CD38 as a marker of proinflammatory (M1) macrophages and CD206 as a marker of the inflammatory resolving (M2) macrophage were performed. Increased expression level of F4/80+ cells was observed within IKKβCA caudal AF compartments across all time points (1, 4, and 6 months after recombination, P < 0.0001), and within IKKβCA caudal EP compartments at 4 and 6 months (P < 0.0001) after recombination when compared to control caudal IVDs (Fig. 7, A and B). Cells expressing the M1 marker, CD38, increased within IKKβCA AF compartments at 1 (P = 0.0013), 4 (P < 0.0001), and 6 months (P = 0.0029) after recombination and within IKKβCA caudal EP compartments at 4 (P < 0.0001) and 6 months (P < 0.0001) after recombination when compared to control caudal IVDs (Fig. 7, A and B). CD206+ cells also significantly increased in IKKβCA caudal AF compartments at 1 (P = 0.0039), 4 (P < 0.0001), and 6 months (P < 0.0001) after recombination and within IKKβCA caudal EP compartments at 4 (P < 0.0001) and 6 months (P < 0.0001) after recombination when compared to control caudal IVDs (Fig. 7, A and B). The temporal pattern of expression of CD206+ cells differed from CD38+ cells. While expression of both F4/80+ and CD38+ cells peaked within AF and EP compartments of IKKβCA caudal IVDs at 4 months before slightly but significantly decreasing at 6 months after recombination, levels of CD206+ cells remained elevated at peak levels at 6 months after recombination (Fig. 7, A and B). Notably, there were no significant differences in any macrophage marker expression observed within the NP compartments between IKKβCA and control caudal IVDs (Fig. 7, A and B). In the lumbar spine, where no degenerative phenotype was observed, little to no positive staining for F4/80+ macrophages was observed at 1 or 4 months after recombination, yielding no significant differences between IKKβCA and control lumbar IVDs (fig. S5). Together, these results suggest that NF-κB overactivation within IVD cells, and the associated molecular changes, initiates an immune response by recruiting and maintaining a population of macrophages within the AF and EP regions of the caudal IVD. The IKKβCA microenvironment promoted a greater abundance of proinflammatory (CD38+) over inflammatory-resolving (CD206+) macrophage subsets in the caudal IVD, based on immunofluorescence imaging, suggesting that the IKKβCA caudal IVD promoted a more proinflammatory microenvironment.
Fig. 7. IKKβ overexpression increases macrophage presence within the caudal IVD.
(A) Representative images of IF staining for F4/80, CD38, and CD206 in mid-sagittal sections of control (AcanCre−/−;Ikk2cafl/fl) and IKKβCA caudal IVDs 1, 4, and 6 months after recombination. DAPI nuclear stain (blue). Scale bars, 100 μm. (B) MFI quantification of F4/80, CD38, and CD206 expression within individual NP, AF, and EP compartments. Letters (a), (b), and (c) indicate statistically significant (P < 0.05) different groupings. (C) Representative images of whole disc sagittal sections of control and IKKβCA IVDs 4 months after recombination with delineation of tissue compartments. DAPI nuclear stain (blue). Scale bars, 200 μm. (n = 3 to 6 mice per group, one to two discs per mouse).
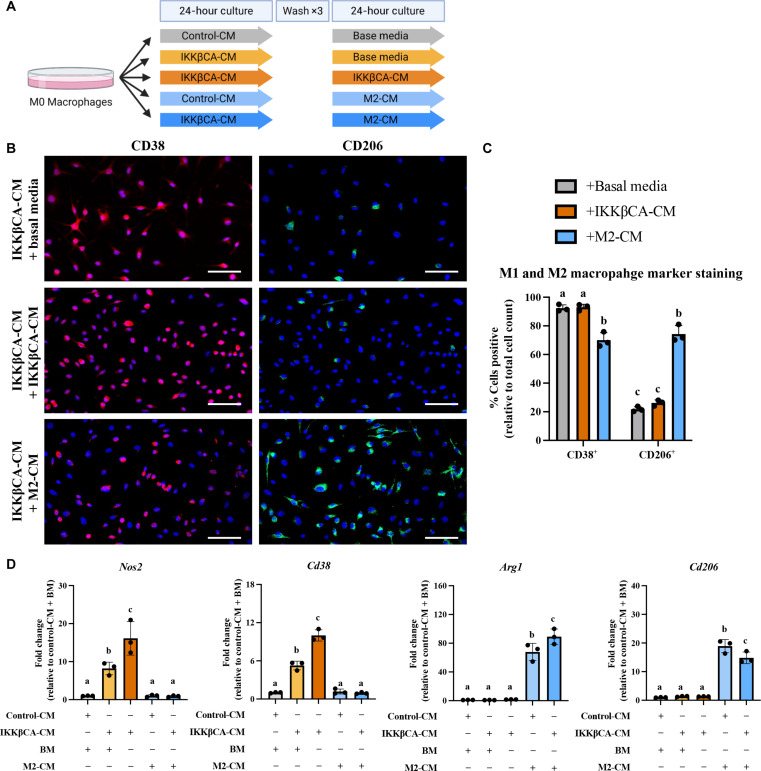
The proinflammatory effects of caudal IVD cell secretome are mitigated by the secretome of inflammatory-resolving macrophages
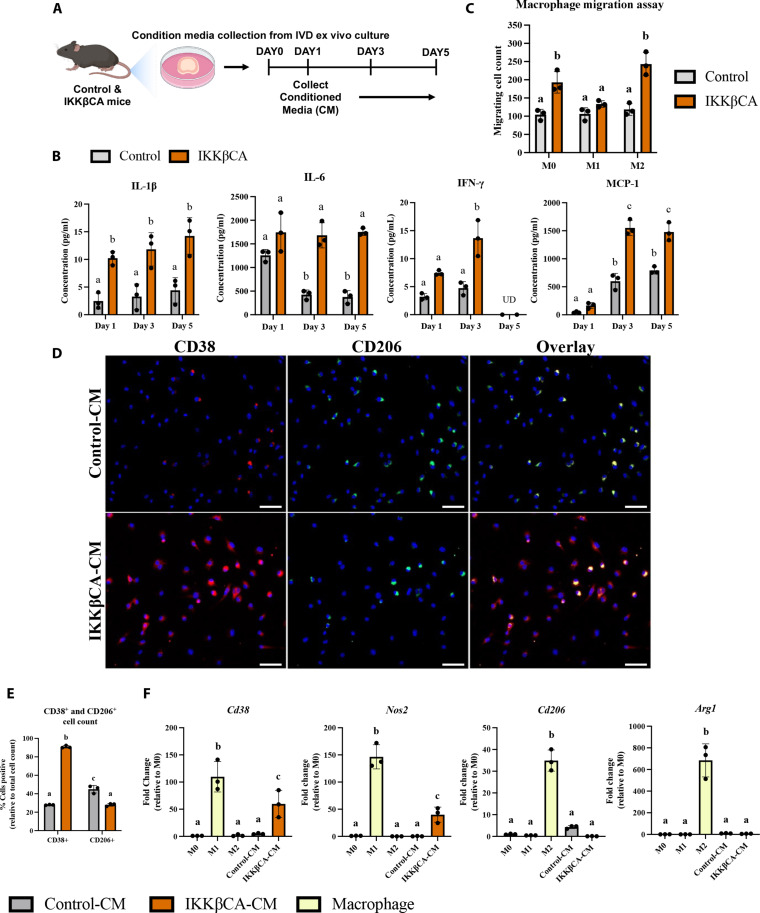
To identify possible mechanisms responsible for the early responses of macrophages to the IVD microenvironment, we analyzed the secretome released by IKKβCA caudal IVD cells 1 week following recombination using whole IVD organ culture (Fig. 8A). Conditioned media (CM) from IKKβCA caudal IVD organ cultures had significantly greater levels of inflammatory cytokines, IL-1β, IL-6, and IFN-γ and the chemokine, MCP-1, compared to CM from control caudal IVDs (Fig. 8, A and B). Next, in evaluating functional and phenotypic changes of macrophages exposed to IKKβCA caudal IVDs, we investigated macrophage migration behavior toward control or IKKβCA-CM. Macrophage migration across a transwell membrane (8 μm) was quantified following culture with each CM. We saw an increase in naïve (M0) (P = 0.0028) and M2-like (P = 0.0001) cell migration to IKKβCA-CM when compared to control-CM (Fig. 8C and fig. S6). No difference was observed in M1-like macrophage migration (P = 0.64) (Fig. 8C). To further investigate the effect of IKKβCA microenvironment on macrophage phenotype, phenotypic markers for M1 (CD38 and NOS2) and M2 macrophages (CD206 and ARG1) were evaluated following in vitro stimulation of naïve macrophages with IKKβCA or control IVD-CM. In IKKβCA-CM, cells positive for CD38 were significantly higher (P < 0.0001), while cells positive for CD206 (P < 0.0001) were significantly lower compared to control-CM conditions (Fig. 8, D and E). At the gene expression level, M1 macrophage markers Cd38 (P = 0.0123) and Nos2 (P = 0.0117) were significantly increased in M0 macrophages stimulated with IKKβCA-CM compared to control-CM, while no significant changes were observed for the M2 macrophage markers Cd206 and Arg1 (Fig. 8F). In vitro results support the in vivo findings where NF-κB overactivation initiated inflammatory molecular changes in IVD cells, which directly increased macrophage migration and polarization toward an M1 phenotype while suppressing M2 polarization, possibly through secreted inflammatory mediators, IL-1β, IL-6, and IFN-γ.
Fig. 8. Caudal IKKβCA-CM increases macrophage migration and polarization toward an inflammatory phenotype.
(A) Study design schematic of whole-organ in vitro culture and CM collection from caudal control (AcanCre−/−;Ikk2cafl/fl) and IKKβCA IVDs harvested 1 week after recombination. (B) Protein concentrations (pg/ml) within CM analyzed after 1, 3, and 5 days in culture. UD, undetectable reading. Letters (a), (b), and (c) indicate statistically significant (P < 0.05) different groupings (n = 3 per genotype). (C) Quantification of M0, M1, and M2 macrophage migration through a transwell membrane via DAPI nuclear count (n = 3). (D) Representative images of IF staining for CD38 and CD206 within BMDMs cultured in 2D monolayer and stimulated with control or IKKβCA CM. DAPI nuclear stain (blue). Scale bars, 100 μm. (E) Quantification of % positivity for CD38 and CD206 within bone marrow–derived macrophages treated with CM (n = 3). (F) Gene expression of M1 and M2 phenotypic markers within M0, M1, or M2 macrophages in basal media, or M0 macrophages with or without IVD CM stimulation (n = 3). Letters (a), (b), and (c) indicate statistically significant (P < 0.05) different groupings.
To identify whether the inflammatory macrophage phenotype induced by IKKβCA caudal IVDs could be reversed, we evaluated the effect of subsequent treatment with an inflammatory-resolving M2-like secretome on macrophages initially stimulated by the IKKβCA-CM or control-CM (Fig. 9A). Macrophages treated with basal media following IKKβCA-CM stimulation exhibited the same level of positivity for CD38 and CD206 as macrophages maintained in IKKβCA-CM for the duration of the experiment (Fig. 9, B and C). This suggests that washout of IKKβCA-CM is not sufficient to eliminate or reverse the proinflammatory phenotypic state. However, when cells from IKKβCA-CM group are subsequently treated with M2-like CM, a significant decrease in CD38+ cells and significant increase in CD206+ cells was observed (P < 0.0001) (Fig. 9, B and C). Gene expression analysis results further support the findings, where M2-like CM significantly decreased expression of Nos2 (P < 0.0001) and Cd38 (P < 0.0001), and increased expression of Arg1 (P < 0.0001) and Cd206 (P < 0.0001), when compared to groups treated with IKKβCA-CM for duration of the study (Fig. 9D). Basal media treatment following IKKβCA-CM also decreased Nos2 (P < 0.01) and Cd38 (P < 0.0001) expression compared to IKKβCA-CM treatment for duration of the study, although expression levels remained significantly higher than cells treated with M2-like CM following IKKβCA-CM (P < 0.05) (Fig. 9D). A greater increase in Arg1 was achieved in M2-CM treatment following IKKβCA-CM compared to M2-like CM treatment of control cells (P < 0.0001) (Fig. 9D). Ultimately, these results suggest that M2-like CM can reverse inflammatory polarization of macrophages activated by IVD cells overexpressing IKKβ, highlighting a therapeutic potential of M2 macrophages in initiating this resolution.
Fig. 9. Caudal IKKβCA-CM mediated macrophage inflammatory responses are mitigated by costimulation with an M2 secretome.
(A) Study design schematic for CM stimulation of M0 macrophages. (B) Representative images of IF staining for CD38 and CD206 within BMDMs cultured in 2D monolayer and stimulated with IKKβCA-CM followed by basal media or M2 macrophage CM. Scale bars, 100 μm. (C) Quantification of % positivity for CD38 and CD206 within BMDMs following CM stimulation (n = 3). (D) Gene expression of M1 and M2 phenotypic markers within M0 macrophages following CM stimulation (n = 3). Letters (a), (b), and (c) indicate statistically significant (P < 0.05) different groupings.
DISCUSSION
Using an AcanCreERT2/+;Ikkβca mouse model, our findings indicate that prolonged NF-κB activation in IVD cells leads to severe structural caudal degeneration with a complete loss of NP cellularity, loss of GAG content, shift of notochordal NP to a cartilaginous NP, and eventual fibrosis of the NP. These changes were associated with functional loss of caudal IVD height and compressive mechanical properties. IVD structural changes were accompanied by increased macrophage recruitment and increases in gene expression of inflammatory cytokines, chemokines, catabolic enzymes, and neurotrophic factors within IVD tissue. Within the lumbar spine, no evidence of degeneration was observed in addition to a less robust increase of inflammatory gene expression and no evidence of increased macrophage presence. Together, these findings fill in a gap in evidence on the effects of prolonged inflammatory activation and macrophage infiltration on degenerated IVD integrity and support the utility of the AcanCreERT2/+;Ikkβca mouse to address key questions and evaluate therapeutics for treatment of persistent IVD inflammation and degeneration.
Within initial characterization of this model, we observed Cre activation and subsequent IKKβ overexpression at gene and protein levels within both lumbar and caudal IVDs. While Ikk2 gene expression, a primary indicator of successful IKKβ overactivation, was up-regulated in both lumbar and caudal IKKβCA IVDs relative to control IVDs, a greater magnitude was observed in the caudal IKKβCA IVDs (~8-fold increase versus control) compared to lumbar IKKβCA IVDs (~3-fold increase versus control). Results also revealed variability in phospho-p65 between the caudal and the lumbar IKKβCA IVDs with time after recombination, suggesting an influence of differences in activation, molecular signaling, and/or cellularity that individually or in combination may contribute to the observed differences by spinal levels. Resulting differences in the Ikk2 gene expression, and/or NF-κB pathway activation via other canonical NF-κB family members (p50 and c-Rel), which were not evaluated, may have varying effects on the resulting phosphor-p65 (or RelA) in IKKβCA IVDs. A study of knee OA using this IKKβCA mouse model found that p50 limited cartilage degeneration in settings of joint inflammation, including advanced age (27). In the current study, proinflammatory gene expression changes in whole caudal and lumbar IVDs isolated at 1 week after recombination were similar, with significant increases of comparable magnitudes (two to fourfold increase versus controls) in Il1β and Il6 genes, which are downstream of NF-κB pathway. However, catabolic gene expression (Mmp3 and Mmp9) was found to be higher in magnitude in caudal IKKβCA IVDs compared to lumbar, which could contribute to the severe degenerative phenotype in caudal but not lumbar IVDs. In addition, the potent chemotactic factor, Mcp1, was significantly up-regulated in caudal IKKβCA IVDs compared to lumbar. Macrophages (F4/80+) were also identified throughout the AF of caudal IKKβCA IVDs as early as 1 month and up to 6 months after recombination; however, little positive staining for F4/80+ cells could be observed at either 1 or 4 months after recombination in lumbar IVDs, with no clear differences from lumbar control IVDs. These findings provide possible explanations for the differences in phenotypes across lumbar and caudal IVDs. Given that the degenerative phenotype in the caudal spine appears to be driven in part by increased recruitment of macrophages to the IVD, the lack of degenerative phenotype in the lumbar spine may be the result of a less robust increase in catabolic gene expression, and specifically the macrophage recruiting chemokine Mcp1, leading to subsequent macrophage recruitment to the IVD.
While macrophages are the most prominent immune cell present in degenerated human discs (15, 28, 29), there are conflicting reports about macrophage phenotype found in human DD. One study found that both M1 (CCR7+) and remodeling M2c (CD163+) cells increased with human DD severity, in particular in regions with structural irregularities and defects (15). However, other human analyses found M2 cells to decrease significantly with degeneration, while the proportion of M1 polarized cells increased (30). In examining mechanisms contributing to severe DD within IKKβCA IVDs, we observed increased presence of macrophages (F4/80+) within the outer AF of IKKβCA mice, which consisted of M1 and M2 cell subsets, with an overall higher prevalence of M1 (CD38+) over M2 (CD206+) cells. The ratio of M1-to-M2 cells in IKKβCA IVDs varied over time, with the highest levels occurring at 1 and 4 months after activation. However, by 6 months, the levels of M1 cells decreased significantly from peak levels, while the M2 cell levels remained elevated, resulting in an overall decrease in the M1-to-M2 ratio. These findings provide longitudinal evidence for the existence of temporal regulation of macrophage polarization in DD in vivo. Since signals encountered within the microenvironment drive the polarization of macrophages into distinct subsets (31), the findings on temporal changes in macrophage subsets suggests that macrophages may play a role in resolving the inflammatory cascade and promoting fibrosis in the IVD, which was present in IKKβCA IVDs. The shift toward increased proresolution occurred after the onset of substantial NP cell loss and matrix degradation; however, the microenvironment was not devoid of M1 cells. Our results of dynamic macrophage populations during DD are consistent with prior studies identifying the presence of both M1 and M2 cells in a degenerative human IVD (15) and within a puncture injury model of murine caudal DD, where early recruitment of M1 macrophages followed by a delayed but sustained recruitment of M2 macrophages was observed using flow cytometry (16). The results of the current study, therefore, extend findings in the literature regarding macrophage phenotype dynamics and localization across different tissue compartments in IVDs undergoing degeneration, in a more clinically relevant model than puncture wounding of the IVD. Whether this is the result of the heterogeneity of the macrophage population driven by phenotypic shift versus recruitment of varying cell subsets remains unknown.
Secretome analysis from IKKβCA IVDs provided evidence that soluble mediators are participating in the recruitment and activation of macrophages. We observed an increase in secreted levels of the inflammatory mediators (IL-1β, IL-6, and IFN-γ) and chemokines (MCP-1) from IVD into CM, which mimicked changes observed in IVD gene expression. Despite the naturally occurring low vascularity of the IVD, IKKβCA also led to early increases in circulating levels of MCP-1, IL-1α, IL-1β, and IL-6 and prolonged increases in circulating levels of TNFα and IFN-γ. The IKKβCA IVD secretome also caused activation of macrophages via an increase in migration and increased inflammatory phenotypic gene (Arg1 and Cd38) and cell surface markers (CD38), while decreasing anti-inflammatory phenotypic surface markers (CD206). Similar inflammatory-inducing IVD-macrophage cross-talk has been observed within multiple in vitro studies where degenerated bovine (32) and human (28) IVD tissue were seen to polarize human macrophages toward a proinflammatory phenotype via increases in various inflammatory markers. Circulating cytokine and/or secretome analyses reveal the chemokines, MCP-1 or MIF, which were found to be released by inflamed IVD cells, as potential targets for inhibition to ultimately mitigate degenerative changes mediated by infiltrating macrophages.
Coinciding with the histological degenerative changes in this model, we observed increases in Mmp3, Mmp9, and Adamts4 gene expression, losses in GAG staining and content, and loss of IVD height within IKKβCA caudal IVDs, which demonstrate a highly catabolic and degenerative tissue microenvironment. Further, Mmp3 and Mmp9 were observed to be more robustly in caudal IKKβCA IVDs compared to lumbar, where again, no degenerative phenotype was observed. In a possible mechanistic role, macrophages have been seen to exacerbate in vitro inflammatory driven catabolic responses of rat lumbar IVD cells through an up-regulation of inflammatory cytokine (Cox2) and catabolic enzyme (Mmp3 and Adamts4) gene expression (33). Further, in another murine model examining IL-1β–mediated inflammation and DD in the lumbar spine, global IL-1ra deficiency resulted in increases in catabolic enzyme expression and degenerative ECM changes, although the presence of infiltrating immune cells was not evaluated (21). Together, the transcriptional changes and inflammatory macrophage activation observed within this model suggest a macrophage-mediated mechanism driving the catabolic environment leading to severe DD.
Because of their plastic nature, macrophages provide a dynamic therapeutic opportunity for harnessing the associated inflammatory-resolving and regenerative functions. This has been explored in the context of the IVD, where stimulation with human M2 macrophage CM mitigated inflammatory responses initiated by TNFα in human NP cells (34). Similarly, our results found that stimulation with an M2-like macrophage secretome reversed the inflammatory macrophage responses induced by IVD cells overexpressing IKKβ. This ultimately adds support for harnessing the potential of an M2 macrophage to treating a chronically inflamed IVD tissue.
The results presented here are consistent with prior studies using NF-κB activation to study the role of persistent inflammation in musculoskeletal disease pathology, specifically tendinopathy and knee OA, and extend the understanding on immune cell cross-talk in musculoskeletal degeneration. Arra et al. (35) found that overexpression of IKKβ in aggrecan-rich articular cartilage in the knee, using the same cross of mice in the current study but otherwise referred to as IKK2caacan, increased gene expression of inflammatory factors such as IL-6 and MCP-1 and catabolic enzymes such as MMP13 and ADAMTS4 in chondrocytes, leading to articular cartilage degradation at 2 months after activation in vivo. Further, using the same model, Catheline et al. (27) found IKKβ-mediated using AcanCreERT2/+;Ikk2cafl/fl mice to accelerate a degenerative OA phenotype with evidence of degeneration and inflammation within articular cartilage, menisci, and synovium. Abraham et al. (5) also observed that IKKβ-mediated NF-κB overactivation within tendon fibroblasts contributed to rotator cuff tendon degeneration and impaired healing driven by an up-regulation in proinflammatory cytokines. Studies targeting the inhibition of NF-κB activation via genetic deletion or small interfering RNA knockdown of p65 within murine models observed a protection from OA progression (36, 37). Although not all studies evaluated immune cell infiltration, increased immune cell presence was observed within the tendon and synovium of NF-κB overexpression models; however, cross-talk between the inflamed tissue and immune cells was not evaluated (5, 27). Ultimately, the current study provides additional support for the importance of persistent inflammation as a primary cause of musculoskeletal disease progression while also adding insight into the role inflamed IVD cells play in recruiting and activating macrophages and DD. Moreover, the evidence that local activation of NF-κB signaling in aggrecan-rich tissues, which includes IVDs and other joint tissues, leads to elevations in circulating cytokine levels warrants further consideration of the role of cross-talk between the local and systemic microenvironments in mediating disease in these animal models.
Limitations to this study include the well-known differences between murine and human IVDs (38). Nonetheless, findings from in vivo animal models inform relevant biological processes that may contribute to the better understanding of human DD. A second limitation is the varied phenotype across spinal regions (lumbar versus caudal). While the exact underlying basis for regional differences in spinal phenotypes is not known, some hypotheses for the existence of regional variation include difference in Cre activation, molecular signaling, cellularity, biomechanical loading, vascularity or neural innervation of the respective regions, all of which may individually or in combination lead to phenotypic differences (39, 40). It is important to note other relevant studies evaluating inflammatory-driven murine models of DD, which have identified phenotypic differences across lumbar and caudal spinal regions, including a subtle but significant varied histopathology between lumbar and caudal IVDs within a murine model of human TNFα overexpression (20), and varied vertebral bone changes across lumbar and caudal IVDs within an IL-1KO murine model (22). These findings and those presented here demonstrate the importance of evaluating and reporting phenotypic differences within such models. Nevertheless, the phenotypic DD evidence in the current study appears to model hallmarks and mechanisms of human DD based on analyses of excised tissues, thus making the mechanistic and temporal observations made herein to be relevant to human DD broadly. The results presented here ultimately support our primary conclusions of prolonged inflammatory activation and subsequent macrophage recruitment driving severe DD. Last, although less robust than the caudal IVD, increased inflammatory activation was still observed in the lumbar spine. With this, the time point chosen for this study might have been insufficient to capture the changes in lumbar IVDs due to NF-κB overactivation, and thus, DD in lumbar IVDs may require longer time points after recombination. Future studies using this model will evaluate histopathology changes at longer time points throughout all spinal levels (cervical, thoracic, lumbar, sacral, and caudal).
In summary the findings of this study provide evidence that prolonged canonical IKKβ–NF-κB signaling pathway leads to accelerated DD by 4 months. Model characterization showed that IKKβ overexpression within the IVD led to a decrease in IVD height, loss of IVD structure, composition, and cellularity, and loss in compressive mechanical properties. Moreover, IKKβ overexpression led to the recruitment of an activated macrophage population to the IVD. The increased production of inflammatory cytokine, chemokine, catabolic enzyme, and neurotrophic factor genes and proteins downstream of canonical NF-κB activation is believed to mediate these degenerative changes and to directly recruit and activate innate immune cells, such as macrophages. Last, we identified that stimulation with an M2 macrophage secretome can mitigate the IVD cell driven inflammatory changes. Together, these results provide support for characterizing the NF-κB–mediated chronic inflammatory environment within the IVD and provide a model for which therapeutic targets, including downstream targets of NF-κB and the utility of an inflammatory-resolving M2 macrophage, may be investigated in future studies for their potential in mediating chronic inflammation and subsequent severe DD.
MATERIALS AND METHODS
Genetic mouse models
Procedures involving the use of animals in this study were performed after attaining approval from the Institutional Animal Care and Use Committee (IACUC) at Columbia University (IACUC protocol AC-AAAZ2452 and AC-AABQ8555). Conditional IKKβ “gain-of-function” R26StopFLikk2ca mice (JAX stock no. 008242) were used to induce canonical NF-κB pathway activation (41). Homozygous Ikk2cafl/fl mice were bred to mice heterozygous for aggrecan (Acan) knock-in allele carrying tamoxifen-inducible form of Cre recombinase (AcanCreERT2/+; JAX stock no. 019148) (42). Mice without CreERT2 recombinase were used as controls (Acan+/+;Ikk2cafl/fl, control) for comparison to CreERT2-positive mice (AcanCreERT2/+;Ikk2cafl/fl, IKKβCA). For initial in vivo histopathology analysis, Cre-mediated recombination was induced in skeletally mature (3 to 4 months of age) mice via IP tamoxifen injections (0.1 or 0.3 mg/g of body weight dissolved in sunflower seed oil; Sigma-Aldrich, catalog no. T5648) for five or three consecutive days, respectively. CreERT2-negative littermate control mice received the same tamoxifen injection. All studies used male and female mice. IVDs were isolated from either the caudal (C5-C11) or lumbar (L1-L6) spinal region between 1 to 6 months after IP recombination.
To evaluate Cre activity within the IVD, heterozygous AcanCreERT2/+ mice were crossed with conditional Ai14 reporter mice (JAX stock no. 007914) to generate AcanCre;Ai14 mice (43). Recombination was induced as described above, and IVDs were isolated from the caudal (C5-C10) and lumbar spines (L1-L6) either 3 days or 3 months following IP injection (both 0.3 mg/g and 0.1 mg/g of body weight doses) for assessment of localized Cre activity (Fig. 1).
Whole IVD gene expression analysis
Whole IVD tissues containing were isolated, snap frozen, and homogenized using a bead tissue homogenizer (Mikro-Dismembrator U, Sartorius) (n = 3mice per genotype per time point, one to two discs per mouse). Total RNA was extracted using TRIzol and chloroform phase separation followed by RNA cleanup using spin columns (Qiagen) according to the manufacturer’s protocol. Relative gene expression was quantified normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) using the ΔΔCT method (gene abbreviations and primer sequences are listed in table S1).
Blood serum inflammatory cytokine multiplex assay
Following euthanasia, control and IKKβCA mice at 1 week and 6 months after injection (n = 3 per genotype per time point) were subjected to serum analysis. For each animal, 500 μl of whole blood was collected through cardiac puncture using 25-gauge (G) needle and incubated at room temperature to coagulate. Serum separation was performed by centrifugation at 1500g for 15 min at 4°C and the serum (top phase) was collected. Serum cytokine analysis was performed using immunoassay against nine cytokines of LEGENDPlex Mouse Inflammation Panel (BioLegend): granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-β, IFN-γ, IL-1α, IL-1β, IL-6, IL-10, MCP-1 (CCL2), and TNF-α. Samples were prepared following the manufacturers protocol. Cytokine quantification was performed in ZE5 Cell Analyzer system (BioRad).
IVD histological analysis
Caudal bone-disc-bone spine segments were fixed (4% paraformaldehyde, 24 hours), decalcified (14% EDTA, 10 days), and either processed for paraffin embedding or soaked in sucrose and embedded in optimal cutting temperature for cryosectioning. Tissue structure was analyzed using paraffin-embedded sagittal sections (7 μm) stained with either Safranin-O (cartilage/mucin), Alcian blue (GAGs), or Picrosirius red (types I and III collagen). Stained slides were imaged using an Axio Observer (Axiocam 503 color camera, Zeiss). Histomorphological analysis was performed using a previously described mouse specific histological grading systems (24, 25), with higher scores indicating increased tissue degeneration (n = 4 to 6 mice per genotype and time point, one to four IVDs per mouse). Stained sections were scored blinded to experimental groups. Differences in histological scores between genotype groups was performed by comparing scores of IVDs collected from multiple levels in each animal.
IVD compartment specific cellularity measurements
Using the ImageJ software [National Institutes of Health (NIH)], hematoxylin (nuclear)– or 4′,6-diamidino-2-phenylindole (DAPI)–stained histological images were converted to 8-bit, autothresholded, converted to binary, and the cell number was quantified using analyze particles function within a custom-defined region of interests (ROIs) containing the outer AF or NP (44). Cell number measurements and ROI delineation were performed blinded to experimental group and analyzed comparing multilevel pooled IVDs between groups. (n = 4 to 6 mice per genotype and time point, one to four IVDs per mouse).
Fluoroscopy analysis of disc height
IVD height was determined for analysis of digital fluoroscopy images (Glenbrook Technologies) taken following euthanasia and before IVD isolation. The DHI was calculated by averaging the IVD height and normalizing to adjacent vertebral body length (n = 3 to 6 mice per genotype and time point, three to five IVDs per mouse) (45). Measurements were taken blinded to experimental group. DHI values were analyzed comparing multilevel pooled IVDs between groups.
Immunofluorescence microscopy and image analysis
Paraffin-embedded tissue sections were baked (60°C, 35 min), deparaffinized with xylene, and rehydrated using a graded series of ethanol washes. Antigen retrieval was performed with 0.1% Triton X-100 (10 min). Tissue sections were blocked (45 min) for nonspecific binding using background buster (Innovex Biosciences). Sections were then incubated overnight at 4°C with primary antibodies. The next day, sections were incubated for 1 hour with secondary fluorescent antibodies. Primary and secondary antibodies and dilutions are listed in table S2. Sections were mounted with DAPI antifade mounting medium (Vector, H-1200) before imaging with Axio Observer (Axiocam 702 mono camera, Zeiss). Exposure settings were fixed across all tissue sections during imaging.
For protein expression quantification, fluorescence images were converted to 8-bit and autothresholded, and a MFI was calculated using the measurement of mean gray value function within ImageJ (NIH) software. MFI measurements were taken within custom defined ROIs (NP, AF, and EP) drawn blind to experimental groups. For nuclear MFI measurements, nuclear ROIs were created by converting DAPI-stained sections to nuclear masks and measuring MFI within (n = 3 to 6 mice per group, one to two discs per mouse).
Cryo-embedded tissues were sectioned (7 μm) and signal recovery of the fluorescent reporter tdTomato was carried out. Sections were re-hydrated, antigen retrieval and peptide blocking was performed as detailed above. To enhance the tdTomato signal, cryosectioned tissues were incubated overnight at 4°C with primary antibodies followed by a 1-hour incubation with secondary fluorescent antibodies at room temperature (table S2). Sections were mounted (DAPI) and imaged as described above.
Immunohistochemistry analysis of NF-κB activation and macrophage presence within IVDs
Paraffin-embedded tissue sections were deparaffinized as described prior. Antigen retrieval was performed using hyaluronidase solution (Sigma-Aldrich, H3506; 100 μg/ml) at 37°C for 12 min. Endogenous peroxidase and protein blocking steps were performed using reagents provided in the ABC detection kit (Abcam, ab64261). Sections were incubated overnight with primary antibodies at 4°C (table S2). The next day, sections were incubated with secondary antibodies and DAB staining using reagents provided in the Abcam kit, according to the manufacturer’s protocol. Sections were dehydrated, counterstained with 0.5% methyl green (Sigma-Aldrich, 198080), mounted with Permount mounting medium (Thermo Fisher Scientific, SP15), and imaged (Axiocam 503 color camera, Zeiss) (n = 3 to 6 mice per group, one to two discs per mouse).
IVD mechanical testing
IVDs were mechanically tested on a TA Electroforce DMA 3200 Mechanical Tester. Before mechanical testing IVDs were thawed in phosphate-buffered saline (PBS; 37°C, 1 hour). IVD height (mm) and cross-sectional area (area of an ellipse, mm2) of the IVDs were approximated using fluoroscopy imaging with ImageJ software. For IVD height and cross-sectional area measurements IVD samples containing the intact NP, AF, and CEPs were microdissected and imaged. Unconfined compression testing was performed between two impermeable platens (WinTest). A 0.02-N preload was applied to each sample followed by 20 cycles of sinusoidal loading at 0.1 Hz to a maximum load of 0.25 N (1× body weight). This was followed by equilibrium creep testing, where a load ramp of 0.25 N was applied over 5 s and held for 1200 s. Dynamic modulus (MPa) was calculated from the ratio of the applied stress and measured strain during the 20th cycle of dynamic loading to allow for repeatable sample displacement hysteresis. The resulting equilibrium strain (mm/mm) and equilibrium modulus (MPa, applied stress/strain) at end of the hold was measured. Measurements were analyzed using multilevel pooled IVDs between groups (n = 3 to 6 mice per genotype, one to three discs per mouse).
IVD GAG analysis
Whole IVDs were digested overnight in papain (0.3 mg/ml) in 100 mM sodium acetate, 10 mM cysteine HCl, and 50 mM EDTA. A dimethymethylene blue assay was used to quantify GAG content within IVD tissue digests (46). GAG content was normalized to total DNA measured within IVD tissue digests using pico green assay (n = 3 to 6 mice per genotype, one to three discs per mouse). Measurements were analyzed using multilevel pooled IVDs between groups.
IVD ex vivo culture and generation of CM
To isolate the effects of IVD inflammation, we established IVD ex vivo culture to generate CM. IKKβCA and control mice at 1 week after recombination was euthanized and caudal IVDs were microdissected (n = 3 mice per genotype, five IVDs per mouse). After serial washes in PBS and Hank’s balanced salt solution, IVDs were cultured in DMEM/F12 with 5% fetal bovine serum (FBS; Crystalgen, catalog no. FBS-500HI) and 1% penicillin-streptomycin. Media was changed every 2 days and CM from days 1, 3, and 5 in culture were collected.
Inflammatory cytokine multiplex immunoassay of IVD CM
Secreted cytokine levels into the collected IKKβCA- and control-CMs (n = 5 per genotype) were measured using 9-Plex LEGENDPlex mouse inflammation panel (BioLegend, catalog no. 740446) according to manufacturer’s protocol. The predefined panel enabled simultaneous quantification of nine cytokines: CCL2 (MCP-1), GM-CSF, IFN-β, IFN-γ, IL-1α, IL-1β, IL-6, IL-10, and TNF-α.
Bone marrow–derived macrophage isolation and culture
Femur and tibia isolated from wild type C57BL/6 mice (n = 3) at 3 months of age were washed in ice-cold PBS and stored in ice-cold RPMI media (Thermo Fisher Scientific, catalog no. 11875093). Following dissection, long bones were transferred to the sterile biosafety cabinet and their bone marrows were flushed using 1 ml of sterile RPMI media using 27G needle. The collected flow through was resuspended in 30 ml of complete macrophage media, containing RPMI, 10% FBS (GeminiBio, catalog no. 100-106), 30% L929 CM (LCM), and 1% penicillin-streptomycin. The cell suspension was split into three 6-cm petri dish and cultured until 80% confluency, with media change every 2 days.
Migration of polarized macrophages
Upon desired confluency, macrophages were chemically stimulated for 24 hours to take on either M1-like or M2-like phenotypes. For M1-like polarization, macrophages were treated with lipopolysaccharide (100 ng/ml; Sigma-Aldrich, catalog no. L2630-10MG) and IFN-γ (20 ng/ml; Shenandoah Biotech, catalog no. 200-16) in complete macrophage media. For M2-like polarization, macrophages were treated with IL-4 (20 ng/ml; Shenandoah Biotech, catalog no. 200-18) and 20 ng/ml IL-13 (Shenandoah Biotech, catalog no. 200-22). Cells cultured with complete macrophage media was used as M0 group.
After polarization, 1.0 × 105 cells were seeded at the top of the transwell insert (8-μm pore size, Corning, catalog no. 3422) positioned into the well with 500 μl of either IKKβCA-CM or control-CM collected at day 3 in culture. Cells were incubated in normoxia for 24 hours, fixed in 4% paraformaldehyde, and cells at the top of the transwell membrane were removed using cotton swab. Migrated cells located at the bottom side of the transwell membrane were permeabilized with 0.1% Triton X-100 in PBS and the transwell membrane was coverslipped with mounting medium containing DAPI (Vector Labs, catalog no. H-1800-2). For each transwell, four different fields of view were imaged, and the number of migrated cells was analyzed by counting the number of DAPI-stained nuclei using ImageJ. Cell count of each transwell is an average count of four different fields of view (n = 3 per experimental group).
Efficacy of macrophage polarization using CM
To investigate the effects of CM on macrophage polarization, M0 macrophages were exposed to either IKKβCA-CM or control-CM at 1:1 solution with complete macrophage media for 24 hours. Following exposure to the CMs, cells were washed with Dulbecco’s PBS (DPBS) and were subjected to either RNA isolation or fixed in 4% PFA for 10 min for immunocytochemistry (ICC) analysis (n = 3 per experimental group).
For sequential polarization study, M0 macrophages exposed to either IKKβCA-CM or control-CM for 24 hours were then washed three times with DPBS and cultured in M2-like CM for 24 hours. After sequential polarization, cells were harvested for RNA isolation and ICC analysis. M2-like CM was generated using chemically stimulated M2-like macrophages, washed three times in DPBS, cultured in complete macrophage media for 24 hours, after which the media was collected (Fig. 9A).
Immunocytochemistry analysis of macrophage polarization markers following CM treatment
Following fixation, cells were washed with PBS, permeabilized with 0.1% Triton X-100 for 15 min, and blocked using 1% bovine serum albumin in PBS for 45 min. Samples were double stained with primary antibodies against overnight at 4°C. After incubation, cells were washed with PBS three times, and cells were incubated with secondary antibodies for 1 hour at room temperature. Following staining, cells were washed three times with PBS and mounted onto a glass slide with VectaShield DAPI mounting solution (Vector Labs, catalog no. H-1800-10). For each sample, four different fields of view were imaged, and the total number of cells and cells positive for CD38 or CD206 were manually counted. Percent cell positivity for each surface marker was calculated by dividing the number of cells positive for each surface marker by total number of cells in the field of view. Each data point of percent cell positivity is an average count of four different fields of view (n = 3 per experimental group).
Statistical analysis
Study sample sizes were determined using power analysis (power = 0.8 and α = 0.05), yielding an n = 10 for mechanical testing/biochemistry, n = 3 for gene expression and in vitro biological evaluations, and n = 4 for histopathology evaluations. On average, 2 to 3 litters were pooled and used for each time point and experimental outcomes. All differences between genotype groups were analyzed with Student’s t test with multiple comparison correction using Holm-Šídák in Prism (V8.3.1). Outliers were identified and removed using ROUT outlier identification with Q = 1% in Prism (V8.3.1). Differences across in vitro CM stimulation groups were analyzed using analysis of variance (ANOVA) with multiple comparison correction using Holm-Šídák in Prism (V8.3.1). P < 0.05 is considered significant, and P < 0.1 is considered a trend.
Acknowledgments
Funding: This work was supported by NIH grants R01AR069668, R01AR077760, and R21AR080516 (to N.O.C.).
Author contributions: Conceptualization: K.G.B., M.K.M.K., A.C.A., and N.O.C. Investigation: K.G.B., M.K.M.K., and D.C.V. Formal analysis: K.G.B., M.K.M.K., and N.O.C. Resources: K.G.B., M.K.M.K., D.C.V., A.C.A., and N.O.C. Writing–original draft: K.G.B., M.K.M.K., and N.O.C. Writing–review and editing: K.G.B., M.K.M.K., A.C.A., and N.O.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
REFERENCES AND NOTES
- 1.Murray C. J., Atkinson C., Bhalla K., Birbeck G., Burstein R., Chou D., Dellavalle R., Danaei G., Ezzati M., Fahimi A., Flaxman D., Foreman, Gabriel S., Gakidou E., Kassebaum N., Khatibzadeh S., Lim S., Lipshultz S. E., London S., Lopez, MacIntyre M., Mokdad A. H., Moran A., Moran A. E., Mozaffarian D., Murphy T., Naghavi M., Pope C., Roberts T., Salomon J., Schwebel D. C., Shahraz S., Sleet D. A., Murray, Abraham J., Ali M. K., Atkinson C., Bartels D. H., Bhalla K., Birbeck G., Burstein R., Chen H., Criqui M. H., Dahodwala, Jarlais, Ding E. L., Dorsey E. R., Ebel B. E., Ezzati M., Fahami, Flaxman S., Flaxman A. D., Gonzalez-Medina D., Grant B., Hagan H., Hoffman H., Kassebaum N., Khatibzadeh S., Leasher J. L., Lin J., Lipshultz S. E., Lozano R., Lu Y., Mallinger L., McDermott M., Micha R., Miller T. R., Mokdad A. A., Mokdad A. H., Mozaffarian D., Naghavi M., Narayan K. M., Omer S. B., Pelizzari P. M., Phillips D., Ranganathan D., Rivara F. P., Roberts T., Sampson U., Sanman E., Sapkota A., Schwebel D. C., Sharaz S., Shivakoti R., Singh G. M., Singh D., Tavakkoli M., Towbin J. A., Wilkinson J. D., Zabetian A., Murray, Abraham J., Ali M. K., Alvardo M., Atkinson C., Baddour L. M., Benjamin E. J., Bhalla K., Birbeck G., Bolliger I., Burstein R., Carnahan E., Chou D., Chugh S. S., Cohen A., Colson K. E., Cooper L. T., Couser W., Criqui M. H., Dabhadkar K. C., Dellavalle R. P., Jarlais, Dicker D., Dorsey E. R., Duber H., Ebel B. E., Engell R. E., Ezzati M., Felson D. T., Finucane M. M., Flaxman S., Flaxman A. D., Fleming T., Foreman, Forouzanfar M. H., Freedman G., Freeman M. K., Gakidou E., Gillum R. F., Gonzalez-Medina D., Gosselin R., Gutierrez H. R., Hagan H., Havmoeller R., Hoffman H., Jacobsen K. H., James S. L., Jasrasaria R., Jayarman S., Johns N., Kassebaum N., Khatibzadeh S., Lan Q., Leasher J. L., Lim S., Lipshultz S. E., London S., Lopez, Lozano R., Lu Y., Mallinger L., Meltzer M., Mensah G. A., Michaud C., Miller T. R., Mock C., Moffitt T. E., Mokdad A. A., Mokdad A. H., Moran A., Naghavi M., Narayan K. M., Nelson R. G., Olives C., Omer S. B., Ortblad K., Ostro B., Pelizzari P. M., Phillips D., Raju M., Razavi H., Ritz B., Roberts T., Sacco R. L., Salomon J., Sampson U., Schwebel D. C., Shahraz S., Shibuya K., Silberberg D., Singh J. A., Steenland K., Taylor J. A., Thurston G. D., Vavilala M. S., Vos T., Wagner G. R., Weinstock M. A., Weisskopf M. G., Wulf S., Murray, U.S. Burden of Disease Collaborators , The state of US health, 1990-2010: Burden of diseases, injuries, and risk factors. JAMA 310, 591–608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katz J. N., Lumbar disc disorders and low-back pain: Socioeconomic factors and consequences. J. Bone Joint Surg. Am. 88, 21–24 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Luoma K., Riihimäki H., Luukkonen R., Raininko R., Viikari-Juntura E., Lamminen A., Low back pain in relation to lumbar disc degeneration. Spine 25, 487–492 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Nerlich A. G., Bachmeier B. E., Schleicher E., Rohrbach H., Paesold G., Boos N., Immunomorphological analysis of rage receptor expression and nf-κb activation in tissue samples from normal and degenerated intervertebral discs of various ages. Ann. N. Y. Acad. Sci. 1096, 239–248 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Abraham A. C., Shah S. A., Golman M., Song L., Li X., Kurtaliaj I., Akbar M., Millar N. L., Abu-Amer Y., Galatz L. M., Thomopoulos S., Targeting the NF-κB signaling pathway in chronic tendon disease. Sci. Transl. Med. 11, eaav4319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu R., Liu N., Luo S., Huang W., Zha Z., Yang J., MicroRNA-9 regulates the development of knee osteoarthritis through the NF-kappaB1 pathway in chondrocytes. Medicine 95, e4315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aupperle K. R., Bennett B. L., Boyle D. L., Tak P. P., Manning A. M., Firestein G. S., NF-κB regulation by IκB kinase in primary fibroblast-like synoviocytes. J. Immunol. 163, 427–433 (1999). [PubMed] [Google Scholar]
- 8.Liu T., Zhang L., Joo D., Sun S.-C., NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 17023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita T., Nolan G. P., Ghosh S., Baltimore D., Independent modes of transcriptional activation by the p50 and p65 subunits of NF-kappa B. Genes Dev. 6, 775–787 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Burke J., Watson R. W. G., McCormack D., Dowling F. E., Walsh M. G., Fitzpatrick J. M., Intervertebral discs which cause low back pain secrete high levels of proinflammatory mediators. J. Bone Joint Surg. Am. 84, 196–201 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Le Maitre C. L., Hoyland J. A., Freemont A. J., Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1β and TNFα expression profile. Arthritis Res. Ther. 9, R77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pockert A. J., Richardson S. M., le Maitre C. L., Lyon M., Deakin J. A., Buttle D. J., Freemont A. J., Hoyland J. A., Modified expression of the ADAMTS enzymes and tissue inhibitor of metalloproteinases 3 during human intervertebral disc degeneration. Arthritis Rheum 60, 482–491 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Le Maitre C. L., Freemont A. J., Hoyland J. A., Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J. Pathol. 204, 47–54 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Liacini A., Sylvester J., Li W. Q., Huang W., Dehnade F., Ahmad M., Zafarullah M., Induction of matrix metalloproteinase-13 gene expression by TNF-α is mediated by MAP kinases, AP-1, and NF-κB transcription factors in articular chondrocytes. Exp. Cell Res. 288, 208–217 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Nakazawa K. R., Walter B. A., Laudier D. M., Krishnamoorthy D., Mosley G. E., Spiller K. L., Iatridis J. C., Accumulation and localization of macrophage phenotypes with human intervertebral disc degeneration. Spine J. 18, 343–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakawaki M., Uchida K., Miyagi M., Inoue G., Kawakubo A., Satoh M., Takaso M., Changes in nerve growth factor expression and macrophage phenotype following intervertebral disc injury in mice. J. Orthop. Res. 37, 1798–1804 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kawakubo A., Uchida K., Miyagi M., Nakawaki M., Satoh M., Sekiguchi H., Yokozeki Y., Inoue G., Takaso M., Investigation of resident and recruited macrophages following disc injury in mice. J. Orthop. Res. 38, 1703–1709 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Tilstra J. S., Clauson C. L., Niedernhofer L. J., Robbins P. D., NF-κB in aging and disease. Aging Dis. 2, 449–465 (2011). [PMC free article] [PubMed] [Google Scholar]
- 19.Hayden M., West A., Ghosh S., NF-κB and the immune response. Oncogene 25, 6758–6780 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Gorth D. J., Shapiro I. M., Risbud M. V., Transgenic mice overexpressing human TNF-α experience early onset spontaneous intervertebral disc herniation in the absence of overt degeneration. Cell Death Dis. 10, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips K. L. E., Jordan-Mahy N., Nicklin M. J. H., le Maitre C. L., Interleukin-1 receptor antagonist deficient mice provide insights into pathogenesis of human intervertebral disc degeneration. Ann. Rheum. Dis. 72, 1860–1867 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Gorth D. J., Shapiro I. M., Risbud M. V., A new understanding of the role of IL-1 in age-related intervertebral disc degeneration in a murine model. J. Bone Miner. Res. 34, 1531–1542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swamy G., Salo P., Duncan N., Jirik F., Matyas J., IL-1Ra deficiency accelerates intervertebral disc degeneration in C57BL6J mice. JOR spine 5, e1201 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam V., Chan W. C. W., Leung V. Y. L., Cheah K. S. E., Cheung K. M. C., Sakai D., Cann M. R. M., Bedore J., Séguin C. A., Chan D., Histological and reference system for the analysis of mouse intervertebral disc. J. Orthop. Res. 36, 233–243 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Melgoza I. P., Chenna S. S., Tessier S., Zhang Y., Tang S. Y., Ohnishi T., Novais E. J., Kerr G. J., Mohanty S., Tam V., Chan W. C. W., Zhou C.-M., Zhang Y., Leung V. Y., Brice A. K., Séguin C. A., Chan D., Vo N., Risbud M. V., Dahia C. L., Development of a standardized histopathology scoring system using machine learning algorithms for intervertebral disc degeneration in the mouse model—An ORS spine section initiative. JOR spine 4, e1164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benneker L. M., Heini P. F., Anderson S. E., Alini M., Ito K., Correlation of radiographic and MRI parameters to morphological and biochemical assessment of intervertebral disc degeneration. Eur. Spine J. 14, 27–35 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catheline S. E., Bell R. D., Oluoch L. S., James M. N., Escalera-Rivera K., Maynard R. D., Chang M. E., Dean C., Botto E., Ketz J. P., Boyce B. F., Zuscik M. J., Jonason J. H., IKKβ–NF-κB signaling in adult chondrocytes promotes the onset of age-related osteoarthritis in mice. Sci. Signal. 14, eabf3535 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C., Cao P., Gao Y., Wu M., Lin Y., Tian Y., Yuan W., Differential expression of p38 MAPK α, β, γ, δ isoforms in nucleus pulposus modulates macrophage polarization in intervertebral disc degeneration. Sci. Rep. 6, 22182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nerlich A. G., Weiler C., Zipperer J., Narozny M., Boos N., Immunolocalization of phagocytic cells in normal and degenerated intervertebral discs. Spine 27, 2484–2490 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Ling Z., Liu Y., Wang Z., Zhang Z., Chen B., Yang J., Zeng B., Gao Y., Jiang C., Huang Y., Zou X., Wang X., Wei F., Single-cell RNA-seq analysis reveals macrophage involved in the progression of human intervertebral disc degeneration. Front. Cell Dev. Biol. 9, 833420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S.-A., Mardani F., Seifi B., Mohammadi A., Afshari J. T., Sahebkar A., Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Silva A. J., Ferreira J. R., Cunha C., Corte-Real J. V., Bessa-Gonçalves M., Barbosa M. A., Santos S. G., Gonçalves R. M., Macrophages down-regulate gene expression of intervertebral disc degenerative markers under a pro-inflammatory microenvironment. Front. Immunol. 10, 1508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H., Liu B., Liu Y., He D., Xing Y., An Y., Tian W., Secreted factors from intervertebral disc cells and infiltrating macrophages promote degenerated intervertebral disc catabolism. Spine 44, E520–E529 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Li X. C., Luo S. J., Fan W., Zhou T. L., Huang C. M., Wang M. S., M2 macrophage-conditioned medium inhibits intervertebral disc degeneration in a tumor necrosis factor-α-rich environment. J. Orthop. Res. 40, 2488–2501 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Arra M., Swarnkar G., Ke K., Otero J. E., Ying J., Duan X., Maruyama T., Rai M. F., O’Keefe R. J., Mbalaviele G., Shen J., Abu-Amer Y., LDHA-mediated ROS generation in chondrocytes is a potential therapeutic target for osteoarthritis. Nat. Commun. 11, 3427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi H., Chang S. H., Mori D., Itoh S., Hirata M., Hosaka Y., Taniguchi Y., Okada K., Mori Y., Yano F., Chung U.-I., Akiyama H., Kawaguchi H., Tanaka S., Saito T., Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat. Commun. 7, 13336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan H., Duan X., Pan H., Holguin N., Rai M. F., Akk A., Springer L. E., Wickline S. A., Sandell L. J., Pham C. T. N., Suppression of NF-κB activity via nanoparticle-based siRNA delivery alters early cartilage responses to injury. Proc. Natl. Acad. Sci. U.S.A. 113, E6199–E6208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alini M., Eisenstein S. M., Ito K., Little C., Kettler A. A., Masuda K., Melrose J., Ralphs J., Stokes I., Wilke H. J., Are animal models useful for studying human disc disorders/degeneration? Eur. Spine J. 17, 2–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brendler J., Winter K., Lochhead P., Schulz A., Ricken A. M., Histological differences between lumbar and tail intervertebral discs in mice. J. Anat. 240, 84–93 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarver J. J., Elliott D. M., Mechanical differences between lumbar and tail discs in the mouse. J. Orthop. Res. 23, 150–155 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Sasaki Y., Derudder E., Hobeika E., Pelanda R., Reth M., Rajewsky K., Schmidt-Supprian M., Canonical NF-κB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity 24, 729–739 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Henry S. P., Jang C. W., Deng J. M., Zhang Z., Behringer R. R., Crombrugghe B., Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis 47, 805–814 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda K., Aota Y., Muehleman C., Imai Y., Okuma M., Thonar E. J., Andersson G. B., An H. S., A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: Correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine 30, 5–14 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Farndale R. W., Sayers C. A., Barrett A. J., A direct spectrophotometric microassay for sulfated glycosaminoglycans in cartilage cultures. Connect. Tissue Res. 9, 247–248 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2