Abstract
Background
Lynch syndrome is a hereditary cancer predisposition syndrome caused by germline mutations in DNA mismatch repair genes, which lead to high microsatellite instability and frameshift mutations at coding mononucleotide repeats in the genome. Recurrent frameshift mutations in these regions are thought to play a central role in the increased risk of various cancers, but no biomarkers are currently available for the surveillance of high microsatellite instability-associated cancers.
Methods
A frameshift mutation-based biomarker panel was developed and validated by targeted next-generation sequencing of supernatant DNA from cultured high microsatellite instability colorectal cancer cells. This panel supported selection of 122 frameshift mutation targets as potential biomarkers. This biomarker panel was then tested using matched tumor, adjacent normal tissue, and buffy coat samples (53 samples) and blood-derived cell-free DNA (cfDNA) (38 samples) obtained from 45 high microsatellite instability and mismatch repair-deficient patients. We also sequenced cfDNA from 84 healthy participants to assess background noise.
Results
Recurrent frameshift mutations at coding mononucleotide repeats were detectable not only in tumors but also in cfDNA from high microsatellite instability and mismatch repair-deficient patients, including a Lynch syndrome carrier, with a varying range of target detection (up to 85.2%), whereas they were virtually undetectable in healthy participants. Receiver operating characteristic curve analysis showed high sensitivity and specificity (area under the curve = 0.94) of the investigated panel.
Conclusions
We demonstrated that frameshift mutations can be detected in cfDNA from high microsatellite instability and mismatch repair-deficient patients and asymptomatic carriers. The 122-target frameshift mutation panel described here has promise as a tool for improved surveillance of high microsatellite instability and mismatch repair-deficient patients, with the potential to reduce the frequency of invasive screening methods for this high-cancer-risk cohort.
Lynch syndrome, also known as hereditary nonpolyposis colorectal cancer (CRC), affects about 1 in 279 individuals (1). Lynch syndrome is caused by monoallelic germline mutations in DNA mismatch repair genes (eg, MLH1, MSH2, MSH6, and PMS2), which predispose Lynch syndrome carriers to frameshift mutations at coding mononucleotide repeats in the genome. Recurrent mutations in coding mononucleotide repeats are thought to play a central role in increased cancer risk, with up to 82% lifetime risk for CRC and 60% for endometrial cancer (2). Early detection is key to reducing cancer incidence in this high-risk population.
The National Comprehensive Cancer Network recommends a surveillance strategy for Lynch syndrome CRC prevention: colonoscopy every 1 to 2 years starting at 20 to 25 years of age for MLH1 and MSH2 carriers and at 30 to 35 years of age for MSH6 and PMS2 carriers (3), though a recent simulation study proposed a gene-specific strategy for optimal surveillance (4). All recommended strategies involve frequent colonoscopy (every 1-3 years), however, with the caveat that adherence remains low (38%) due to patient burden and costs (5,6). Epi proColon (Epigenomics, Inc, Berlin, Germany) and Cologuard (Exact Sciences Corporation, Madison, WI) are 2 US Food and Drug Administration-approved tests for sporadic CRC screening but not for Lynch syndrome. Thus, a noninvasive cancer surveillance strategy for this high-risk population is needed (7). Recurrent frameshift mutations at coding mononucleotide repeats may serve as sensitive biomarkers in the evolution of Lynch syndrome-associated tumors, and resulting neoantigens are the current basis for vaccine development (8).
Recent advances in liquid biopsy research, including that directed toward multicancer early detection, show promise for cancer detection and monitoring. Blood-derived cell-free DNA (cfDNA) and circulating tumor DNA have been shown to contain variant DNA that can serve as biomarkers, with implications for treatment (9), prognosis (10), and therapeutic response monitoring (11-14). We hypothesized that the frameshift mutations characteristic of mismatch repair deficiency observed in Lynch syndrome should be detectable in blood of tumor-bearing patients and potentially in asymptomatic carriers. To test this hypothesis, we assessed cfDNA from high microsatellite instability and mismatch repair-deficient patients with cancer, asymptomatic carriers, and healthy participants for the presence of frameshift mutations at commonly altered coding mononucleotide repeats.
Methods
Cell culture
Supernatant or cellular DNA from cultured high microsatellite instability and microsatellite-stable CRC cell lines was used for target panel development through polymerase chain reaction (PCR) fragment size analysis and next-generation sequencing (Supplementary Methods, available online).
Patient sample cohort
Deidentified samples from 29 matched patients (including 29 tumors, 23 cfDNA samples, 10 adjacent normal tissue samples, and 14 buffy coat samples) and 16 cfDNA-only patients, for a total of 45 Lynch syndrome and high microsatellite instability patients were obtained from multiple sources (Table 1 and Supplementary Figure 1, available online). For cfDNA (total 39 patients), each patient had only 1 cfDNA sample, except for 1 with 2 samples, the only Lynch syndrome carrier without active cancer detected at the time of 2 blood collections. Eighty-four plasma and 7 buffy coat samples from healthy participants (self-reported as cancer free at the time of blood collection; not screened for Lynch syndrome and microsatellite instability) were from the National Institutes of Health (NIH) Clinical Center (Bethesda, MD) and a commercial vendor (ZenBio, Research Triangle, NC). Buffy coat DNA, representing germline DNA, is widely used as a control or reference for somatic mutation calling. Thus, we included buffy coat from a subset of matched patients for frameshift mutation analysis. DNA sources used at various stages of panel development are shown diagrammatically in Supplementary Figure 2 (available online). Sample collection was approved by institutional review boards at each study site, and informed consent was obtained from all participants at their respective collection sites. The use of biospecimens was reviewed and determined exempt by the Frederick National Laboratory for Cancer Research, and the study was conducted in accordance with the US Common Rule.
Table 1.
Clinical characteristics of patients and carriers
| Variable | Patients, No. (%) (n = 45) |
|---|---|
| Sample sources | |
| Colon Cancer Family Registry | 9 (20.0) |
| Tissue For Research Ltda | 13 (28.9) |
| Heidelberg | 18b (40.0) |
| Weill Cornell | 5b (11.1) |
| Samples | |
| Plasma or serum only | 16 (35.6) |
| Plasma or serum with matched tumor | 23c (51.1) |
| Tumors without matched plasma or serum | 6 (13.3) |
| Cancer type | |
| Colorectum and cecum | 34 (75.6) |
| Small intestine | 1 (2.2) |
| Prostate | 1 (2.2) |
| Ovarian | 1 (2.2) |
| Endometrial | 3 (6.7) |
| Glioblastoma | 1 (2.2) |
| Lynch syndrome carrier without active cancer | 1d (2.2) |
| Stomach | 1 (2.2) |
| Unknown | 2 (4.4) |
| Microsatellite instability status | |
| High microsatellite instability | 31 (68.9) |
| Mismatch repair-deficient without microsatellite instability status | 8 (17.8) |
| Unknown | 6 (13.3) |
| Tumor stage | |
| I | 2 (4.4) |
| II | 4 (8.9) |
| III | 14 (31.1) |
| IV | 8 (17.8) |
| Unknown | 17 (37.8) |
| Tumor grade | |
| 1 | 2 (4.4) |
| 2 | 12 (26.7) |
| 3 | 11 (24.4) |
| 4 | 1 (2.2) |
| Unknown | 19 (42.2) |
| Race | |
| Black or African American | 1 (2.2) |
| White | 12 (26.7) |
| Unknown | 32 (71.1) |
| Age, y | |
| ≤50 | 13 (28.9) |
| >50 | 19 (42.2) |
| Unknown | 13 (28.9) |
Only 4 serum samples were used in this study from this source. All the remaining blood samples were plasma.
One cell-free DNA sample from each of these 2 sites did not complete the Archer analysis pipeline because of low reads and thus was excluded from subsequent analysis.
Ten patients also had matched adjacent normal tissue.
The only Lynch syndrome carrier with 2 plasma samples available (no active cancer detected at either blood collection time).
All 45 patients included in the study were confirmed as mismatch repair-deficient or high microsatellite instability. All samples were obtained from treatment-naive patients and collected before surgery. Matched blood and tumor and tissue samples were collected on the same day. Each site provided demographic, diagnostic, and prognostic information for each patient and demographic information about healthy participants, if available.
Statistical analysis
All statistical analyses were 2-sided and performed using GraphPad Prism (GraphPad Software, Boston, MA). P less than .05 was considered statistically significant. For other methods and detailed data analyses, please refer to Supplementary Methods and Supplementary Tables 1 through 4 (available online).
Results
Frameshift mutation detection in culture supernatant of high microsatellite instability CRC cells using PCR fragment size analysis
DNA extracted from the supernatant of cultured high microsatellite instability tumor cell lines has previously been shown to contain several recurrent frameshift mutations (15). We first developed and validated a PCR-based detection assay by examining the frameshift mutations in supernatant DNA from high microsatellite instability (HCT116, KM12, LoVo, HCT15, and RKO) and microsatellite-stable (Colo205 and HT29) CRC cell lines. The size of DNA fragments varied mainly between 100 and 200 base pairs (Supplementary Figure 3, available online; lane 2-3). This finding is consistent with the notion that DNA in the culture supernatant represents fragmented pieces of genomic DNA (Supplementary Figure 3, available online; lane 4-5).
A panel of 12 genes with frameshift mutations at coding mononucleotide repeats was selected (16-19) and assessed by PCR fragment size analysis. The allele profiles of microsatellite-stable cells were used as controls. Frameshift mutations were detectable in all 5 high microsatellite instability cell lines with homozygous [eg, TGFBR2(‒1) in HCT116 cells] or heterozygous [eg, TGFBR2(‒1) in KM12] variations but not detected in microsatellite-stable cells (Table 2). Some frameshift mutations had more than 1 nucleotide deletion (eg, ‒2 or ‒3 variants [eg, TAF1B(‒3) in HCT116]) or rarely 1 nucleotide insertion (+1).
Table 2.
Summary of frameshift mutation detection in DNA from cell culture supernatants of high microsatellite instability cells by polymerase chain reaction fragment size analysisa
| Target | Coding mononucleotide repeat | HCT116 | LoVo | KM12 | HCT15 | RKO |
|---|---|---|---|---|---|---|
| ACVR2A K437 | A8 | m1 | m1 | m1 | m1 | m1 |
| AIM2 T343 | A10 | m1 | m1 | wt | wt | p1, m1 |
| ASTE1 R657 | A11 | m1, m2 | m1, wt | m1, m2 | p1, m1 | m2 |
| EXO1 D731T | A7 | wt | m1, wt | wt | wt | wt |
| LIG3 I157S | A8 | wt | wt | wt | wt | wt |
| MLH3 E586N | A9 | wt | wt | wt | wt | m2, wt |
| MLH3 N674I | A8 | wt | wt | m1, wt | wt | wt |
| MSH3 K383 | A8 | m1 | wt | wt | wt | m1 |
| PTEN K267R | A6 | wt | wt | m1, wt | wt | wt |
| PTEN N323M | A6 | wt | wt | wt | wt | wt |
| TAF1B N66 | A11 | m3, wt | m1, wt | m2, wt | wt | m2, wt |
| TGFBR2 K128 | A10 | m1 | m1, m2 | m1, wt | m1, wt | m2, wt |
Two microsatellite-stable cell lines (Colo205 and HT29) were also tested, and no frameshift mutations were detected for any targets. m = minus (deletion); p = plus (insertion); wt = wild type.
High microsatellite instability tumors typically harbor many frameshift mutations at coding mononucleotide repeat loci throughout the genome. We expanded from 12 to 30 targets by selecting those with high variant frequency in Lynch syndrome CRC (20,21) and assessed the frameshift mutations in the same supernatant DNA. As expected, frameshift mutations were highly detectable in high microsatellite instability cells, with a variable detection rate (eg, 17 of 30 for HCT116 [56.7%] and 7 of 24 tested targets for HCT15 [29.2%]) (Supplementary Table 5, available online) but not detected in microsatellite-stable cells (data not shown). Fragment size data were confirmed by Sanger sequencing (data not shown).
Detection sensitivity assessment using the Archer next-generation sequencing platform and DNA spike-in assay
The enrichment of cfDNA is a determining factor for detection sensitivity. To develop a sensitive assay for frameshift mutation detection, cfDNA was enriched using Anchored Multiplex PCR (AMP) chemistry (Archer, now Integrated DNA Technologies, Inc, Coralville, IA) for custom primer design and the Illumina NextSeq platform (San Diego, CA) for targeted sequencing. To assess the sensitivity of the platform, different amounts of supernatant DNA from high microsatellite stability cells (variant DNA from HCT116, KM12, or LoVo) were serially spiked into supernatant DNA from microsatellite-stable cells (wild type DNA from HT29). Results were analyzed using the Archer Analysis pipeline, and data from microsatellite-stable cells were used for background error correction. Frameshift mutations across the panel were detectable in the range of 5% down to 0.05% variant spike-in samples (Supplementary Table 6, available online). Three frameshift mutations [CASP5(‒1), MARCKS(‒1), and MYH11(‒1)] were detected only in 5% of spike-in samples, while 7 targets were wild type in these high microsatellite stability cell lines (eg, BEND5, EXO1, LIG3, MLH3-E586, PPP3CA, PTEN-N323, and WNT11) (Supplementary Table 7, available online).
Given the disease heterogeneity in Lynch syndrome tumors, we opted to increase detection sensitivity in cfDNA by further expanding the target panel to include a total of 168 targets with reported high frameshift mutation frequency in Lynch syndrome (20). The detection sensitivity was assessed using the same assay and platform described earlier. Frameshift mutations were highly detectable in 20%, 10%, and 5% spike-in samples (up to 96.0%, 91.4%, and 83.4% of targets with frameshift mutations detected), respectively (Table 3), and up to 59.6% and 6.8% of targets were detected in 1% and 0.05% spike-in samples, respectively, suggesting that frameshift mutation detection was target locus and cell line-dependent. Therefore, we established a final panel of 122 targets by selecting those detected in less than 5% of spike-in samples, of which 88 targets are represented as frameshift peptides in the NOUS-209 vaccine (22).
Table 3.
Number of frameshift mutations detected and percentage detection rate using a 168-target panel and Illumina NextSeq platform in a DNA spike-in assay
| Variant DNA (No. of frameshift mutations detected in cellular DNA) | % Variant DNA spike-ina, No. (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| 20% | 10% | 5% | 1% | 0.50% | 0.25% | 0.10% | 0.05% | |
| HCT116 (151) | 145 (96.0) | 138 (91.4) | 126 (83.4) | 90 (59.6) | 29 (19.2) | 8 (5.3) | 8 (5.3) | 6 (4.0) |
| KM12 (140) | 134 (95.7) | 100 (71.4) | 74 (52.8) | 59 (42.1) | 49 (35.0) | 25 (17.9%) | 8 (5.7) | 4 (2.9) |
| LoVo (132) | 94 (71.2) | 84 (63.6) | 75 (56.8) | 62 (47.0) | 38 (28.8) | 22 (16.7) | 9 (6.8) | 9 (6.8) |
High microsatellite instability variant supernatant DNA was spiked into microsatellite-stable wild type supernatant DNA from HT29.
Panel validation in tumor, adjacent normal tissue, and buffy coat DNA from high microsatellite stability patients
To further assess the 122-target panel, DNA from 29 tumors and 10 matched adjacent normal tissue and 14 buffy coat samples from high microsatellite instability and mismatch repair-deficient patients (Table 1) were sequenced using the NextSeq platform. Seven buffy coat samples from healthy participants were used for background error correction during data analysis. All tumors had a high number of frameshift mutations detected (Table 4) (28.7%-91.8% of targets), except 1 glioblastoma patient with only 1 target detected. Most targets had detectable ‒1 variants, while some had ‒2 and +1 but rarely ‒3 variants (data not shown). As expected, adjacent normal tissue had lower detectable frameshift mutations than matched tumors (3.3%-26.2% of targets). Matched-buffy coat samples had no or low frameshift mutation detection (0%-0.8%, with 1 sample having 4% of targets).
Table 4.
Number of frameshift mutations detected in matched high microsatellite instability tumor, buffy coat, and adjacent normal tissue samples
| Sample ID | Tumor |
Buffy coat |
Adjacent normal |
|
|---|---|---|---|---|
| Variants, No. | Targets, No. (%) | Targets, No. (%) | Targets, No. (%) | |
| Colon Cancer Family Register (formalin fixed, paraffin embedded) | ||||
| P30 | 68 | 55 (45.1) | 1 (0.8) | — |
| P31 | 37 | 35 (28.7) | 0 (0) | — |
| P32 | 74 | 65 (53.3) | 0 (0) | — |
| P33 | 62 | 55 (45.1) | 0 (0) | — |
| P34 | 66 | 57 (46.7) | 0 (0) | — |
| P35 | 147 | 98 (80.3) | 1 (0.8) | — |
| P36 | 189 | 112 (91.8) | 0 (0) | — |
| P37 | 179 | 109 (89.3) | 5 (4.1) | — |
| P38 | 158 | 100 (82.0) | 0 (0) | — |
| Heidelberg (frozen) | ||||
| P1 | 158 | 100 (82.0) | — | 22 (18.0) |
| P2 | 78 | 63 (51.6) | — | 4 (3.3)a |
| P3 | 125 | 78 (63.9) | — | 19 (15.6) |
| P4 | 84 | 70 (57.4) | — | 6 (4.9)a |
| P5 | 132 | 89 (73.0) | — | 26 (21.3) |
| P6 | 67 | 53 (43.4) | — | 9 (7.4)a |
| P7 | 70 | 52 (42.6) | — | 17 (13.9) |
| P8 | 76 | 69 (56.6) | — | 32 (26.2) |
| Tissue For Research Ltd (frozen) | ||||
| P50 | 118 | 81 (66.4) | — | — |
| P51 | 115 | 74 (60.7) | — | — |
| P52 | 93 | 71 (58.2) | — | — |
| P53 | 41 | 35 (28.7) | — | — |
| P54 | 52 | 38 (31.1) | — | — |
| P55 | 72 | 63 (51.6) | — | 4 (3.3) |
| P56 | 79 | 54 (44.3) | 0 (0) | 7 (5.7) |
| P57 | 97 | 74 (60.7) | — | — |
| P58 | 80 | 53 (43.4) | 0 (0) | — |
| P59 | 56 | 42 (34.4) | 1 (0.8) | — |
| P60 | 70 | 57 (46.7) | 0 (0) | — |
| P61 | 1 | 1 (0.8) | 0 (0) | — |
Failed quality control due to low unique DNA start sites per gene-specific primer 2 (<50). These samples were not resequenced because of limited DNA availability. — = No matched samples available.
Background noise detection in cfDNA from healthy participants
Recent reports suggest that genomic DNA with coding mononucleotide repeat is present in the blood of healthy participants (23-25). To confirm this finding, cfDNA was isolated from 2 mL plasma from 23 healthy participants. Because of low yield, cfDNA from multiple donors was pooled to reach 50 ng as input for library construction using the same 122-target panel and NextSeq platform. An additional 7 pooled cfDNA samples were sequenced and used as a normal dataset for background error correction during the analysis. As expected, wild type coding mononucleotide repeat sequences in cfDNA from healthy participants were detectable, but a small number of frameshift mutations were also detected (Table 5) (an average of 1.5 frameshift mutations per donor). To determine the background noise, cfDNA from an additional 61 healthy participants was sequenced. Again, because of low DNA yield, 8 pools from 17 participants were made (at least 25-34 ng). Consistently, fewer than 2 frameshift mutations were detected in individual and pooled samples (1.7 and 1.3, respectively) (Table 5). Among 44 individual samples, frameshift mutations were not detected in 11 samples (25%), 1 frameshift mutation was detected in 13 samples (29.5%), and 2 frameshift mutations were detected in 8 samples (18.2%) (Supplementary Table 8, available online). Among 122 targets, ADD3(‒1), USP35(‒1), PRDM2(‒1), and FBXL3(‒1) were detected in more than 5 samples (Supplementary Table 9, available online), which contributed most prominently to background noise.
Table 5.
Number of frameshift mutations detected in cell-free DNA from healthy participants
| Sample set | No. of samples | No. of frameshift mutations detected | Average No. of frameshift mutations/sample (SEM) |
|---|---|---|---|
| First pooled sample set | 23 | 35 | 1.5 (1.7) |
| Individual samples | 44 | 74 | 1.7 (0.2) |
| Second pooled sample set | 17 | 22 | 1.3 (0.7) |
| All samplesa | 84 | 131 | 1.6 (0.4) |
Including pooled samples. SEM = Standard Error of the Mean.
Frameshift mutation detection in cfDNA from high microsatellite instability patients
With an average background noise of 1.6 frameshift mutations in healthy participants (Table 5), we postulated that blood-derived frameshift mutations in high microsatellite instability patients would be detected above the background noise. To this end, we assessed frameshift mutations in cfDNA from high microsatellite instability patients, including a Lynch syndrome carrier (Table 6). All extracted cfDNA (1-50 ng) was used for library construction. Up to 81 (66.4%) frameshift mutations were detected in 1 patient and as few as 2 (1.6%) in 2 patients (excluding 3 patients with failed quality control during the data analysis) (Table 6). The unique molecule sequencing depth covering the variant (depth) and the variant allele frequency from deep molecular bins were relatively high in some cfDNA samples compared with their matched tumors (Supplementary Table 10, available online), indicating that some variants may be enriched in cfDNA. This occurrence did not appear to be target or patient-dependent. Unfortunately, data from 10 cfDNA samples failed quality control during the analysis, possibly because of low starting plasma or serum volumes (0.5-2 mL) and low cfDNA yield. We also analyzed 2 cfDNA samples from an asymptomatic Lynch syndrome carrier (Table 6). Interestingly, higher frameshift mutations were detected in blood collected later than that collected early from the same patient (11 vs 2 frameshift mutations, respectively). Because samples were deidentified, we cannot follow up on this finding. As expected, the overall number of frameshift mutations detected in cfDNA was lower than that in the matched tumor (Tables 4 and 6 and Supplementary Table 11, available online). Among 23 matched patients, an average of 34% of frameshift mutations detected in cfDNA were also detected in tumors, and 9 of 23 (39%) patients with more than 40% frameshift mutations detection in cfDNA were also detected in the matched tumor (Supplementary Table 11, available online).
Table 6.
Number of frameshift mutations detected in cell-free DNA from high microsatellite instability patients
| Sample ID | Variants, No. | Targets, No. | Cell-free DNA yield, ng |
|---|---|---|---|
| Heidelberg | |||
| P1a | 4 | 4 | 6.10 |
| P2a | 2 | 2 | 3.67 |
| P3a | 1 | 1 | 9.16 |
| P4a | 23 | 21 | 12.07 |
| P5 | 105 | 81 | 10.84 |
| P6 | 9 | 9 | 5.91 |
| P7a | 3 | 3 | 4.90 |
| P8 | 2 | 2 | 6.16 |
| P9a | 1 | 1 | 4.76 |
| P10a | 2 | 2 | 4.96 |
| P11 | 3 | 3 | 2.22 |
| P12 | 7 | 6 | 1.74 |
| P13 | 7 | 7 | 2.29 |
| P14 | 8 | 8 | 1 |
| P15 | 47 | 45 | 1.65 |
| P16 | 104 | 79 | 10.25 |
| P17a | 3 | 3 | < 1 |
| Weill Cornell | |||
| P20-1b | 2 | 2 | 11.33 |
| P20-2b | 11 | 10 | 10.63 |
| P21 | 4 | 4 | 64.9 |
| P22 | 11 | 10 | 177.1 |
| P23 | 18 | 17 | 56.1 |
| Tissue For Research Ltd | |||
| P50 | 72 | 61 | 12.43 |
| P51 | 61 | 56 | 4.71 |
| P52 | 6 | 6 | 7.74 |
| P53a | 14 | 14 | 2.51 |
| P54a | 8 | 8 | 4.68 |
| P55c | 9 | 9 | 444 |
| P56c | 6 | 6 | 21.45 |
| P57c | 3 | 3 | 552 |
| P61 | 5 | 5 | 2.18 |
| P62c | 53 | 50 | 22.95 |
| Colon Cancer Family Registry | |||
| P30 | 10 | 9 | 1.49 |
| P31 | 10 | 10 | 3.67 |
| P34 | 9 | 9 | 4.4 |
| P35 | 2 | 2 | 1.87 |
| P36 | 4 | 4 | 21.89 |
| P37 | 4 | 4 | 36.41 |
Failed quality control due to low unique DNA start sites per gene-specific primer 2 (<50).
A Lynch syndrome carrier without active tumor detected at the blood collection time points.
Serum was used in the analysis. All the remaining cell-free DNA were from plasma.
Further analysis was performed to assess whether the number of frameshift mutations detected in cfDNA and tumors had any correlation with the germline mismatch repair gene status. Perhaps because of the limitations of the cohort (eg, low volume of plasma or serum, different cancer type and disease stage and grade, and small sample size), no statistically significant differences were observed between MSH2-, MLH1-, and PMS2-deficient groups (Supplementary Figure 4, available online).
Sensitivity and specificity analysis
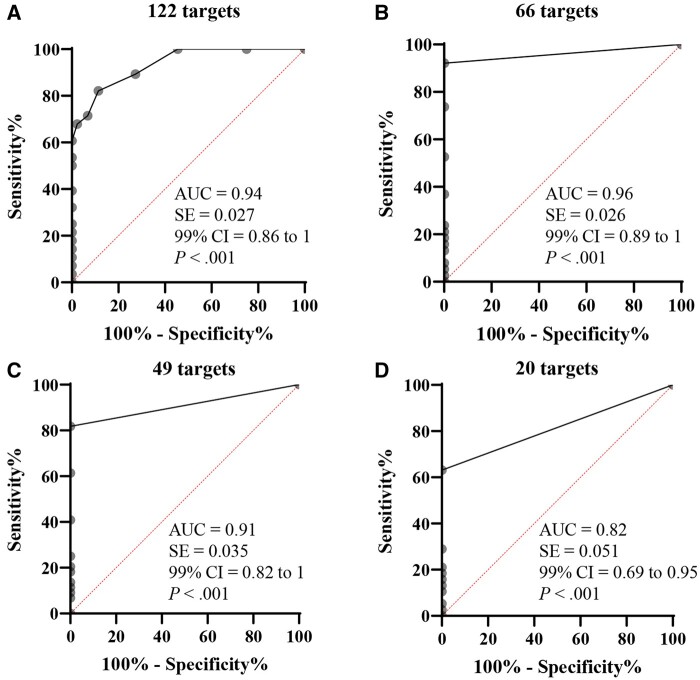
Receiver operating characteristic curve analysis was performed to evaluate the performance of the panel in distinguishing high microsatellite instability and mismatch repair-deficient patients from healthy participants. The area under the curve (AUC) was 0.94 (99% confidence interval = 0.86 to 1.00; P < .001), demonstrating high ability of our panel to distinguish between high microsatellite instability and mismatch repair-deficient patients and healthy participants (Figure 1, A). To determine whether the performance can be improved, we modeled the receiver operating characteristic curve analysis by removing those targets detected in healthy participants. The AUC was 0.96 (99% confidence interval = 0.89 to 1.00; P < .001) for 66 targets (Figure 1, B). Reducing the targets further resulted in decreased AUC values (eg, AUC = 0.91 for 49 targets [Figure 1, C] and AUC = 0.82 for 20 targets [Figure 1, D]), indicating that a panel with too few targets may not have an appropriate coverage of the frameshift mutation spectrum for high microsatellite instability and mismatch repair-deficient patients or carriers.
Figure 1.
Receiver operating characteristic curve analysis of frameshift mutation comparison between high microsatellite instability and mismatch repair-deficient patients and healthy participants. The receiver operating characteristic curve was generated by comparing the number of frameshift mutations detected between high microsatellite instability and Lynch syndrome patients and healthy participants. Pooled samples were excluded in this analysis. A) All 122 targets in the panel were included in the analysis. B, C, D) Receiver operating characteristic curve modeling analysis: By removing those targets, frameshift mutations were also detected in healthy participants (B, 66 targets), and by further removing those targets frameshift mutations were detected in ≤2 patients (C, 49 targets) or in ≤5 patients (D, 20 targets). AUC = area under the curve; CI = confidence interval.
Discussion
Liquid biopsy-based biomarkers have been evaluated for detection of different types of cancer (26-30). Two mutation-based test panels [Guardant360 CDx (Guardant Health, Redwood City, CA) (31) and FoundationOne Liquid CDx (Foundation Medicine, Inc, Cambridge, MA) (32)] have been approved by the Food and Drug Administration. Combinatorial approaches with other types of biomarkers have been explored to increase the sensitivity and specificity of these blood tests. CancerSEEK, combining circulating tumor DNA and protein biomarkers, has a median sensitivity of 73% for stage II and 43% for stage I cancers across 8 solid cancer types and 65% for all-stage CRC (26). A prospective interventional study (Detecting cancers Earlier Through Elective mutation-based blood Collection and Testing; DETECT-A) demonstrated a 28.3% positive predictive value and a 99.6% specificity when the blood test was combined with imaging (33). Other types of liquid biopsy-based biomarkers (eg, methylomics and fragmentomics) alone or in combination with approved screening tools have also been actively pursued for noninvasive cancer detection (28,34-38). A methylation-based study (PATHFINDER) revealed a 38% positive predictive value and 99.1% specificity for a population multicancer screening (39,40). The low sensitivity highlights the challenges for pan-cancer early detection using liquid biopsies. None of these tests are tailored for or have been tested in high-risk high-microsatellite instability and mismatch repair-deficient patients with cancer. One study used methylated DNA markers in tissue DNA to discriminate Lynch syndrome CRC and endometrial cancer from healthy participants (AUC = 0.92 for both Lynch syndrome cancers) (41), but it remains to be determined whether this methylation panel can be applied to cfDNA for Lynch syndrome cancer detection. Here, we took advantage of recurrent frameshift mutations in high microsatellite instability and mismatch repair-deficient tumors and assessed the feasibility of using targeted frameshift mutations in blood for disease monitoring. In this proof-of-principle study, we showed that frameshift mutations were detectable in cfDNA with high sensitivity and specificity. They were also detectable from an asymptomatic carrier, which is consistent with the progressive emergence of frameshift mutations in the blood of Lynch syndrome carriers during disease progression. It remains to be determined whether the number of frameshift mutations or variant allele frequency is correlated with disease progression in Lynch syndrome carriers.
Frameshift mutations included in the panel were selected mainly based on reported variant frequencies in high microsatellite instability tumors (20,21). It is not known, however, whether the variant frequency is mismatch repair gene-specific or cancer type-specific. With advances in sequencing technology and data analysis pipelines, it is vital to analyze the frameshift mutations in high microsatellite instability tumors by mismatch repair genes and cancer types so that specific panels can be developed in the event that frameshift mutations are different among mismatch repair gene variations and cancer types.
The frameshift mutation-based biomarkers described here have the potential to reduce the frequency of colonoscopy at young ages as a personalized screening approach rather than using age (20-25 years) as a basis for colonoscopy initiation. Frameshift mutations could help identify Lynch syndrome carriers most likely to harbor CRC, which can be followed up with colonoscopy or other screening tools. In a Lynch syndrome mouse model with MSH2 deletion in the intestine, fewer frameshift mutations were detected in the mucosa of young mice (non-tumor bearing; < 6 months) than old mice (data not shown). Thus, frameshift mutation-based biomarkers may be useful for disease surveillance and reduce the frequency of colonoscopy.
There are several limitations to our study. First, the biomarker panel described here was tested in a retrospectively collected cohort from different sources with limited clinical data and processed without a standardized protocol. Thus, correlation cannot be made between the number of frameshift mutations detected and disease stage and grade, or mismatch repair gene status. Second, compared with an average of 5 to 8 mL plasma used in other liquid biopsy biomarker studies (26), a smaller volume (0.5-2 mL) was used in the current study, which may have contributed to the variability of the number of frameshift mutations detected and limited concordance in frameshift mutations detected between matched cfDNA and tumor (42). In addition, different germline mismatch repair mutations, cancer types, and disease stage and grade may also contribute to the variability observed. The variability was also present in tumors, for which different representations of tumor cells in the tissue used for DNA extraction may be partially accountable. Possibly because of these limitations, there was no statistically significant difference in the number of frameshift mutations detected among different germline mismatch repair gene variation groups. Third, cfDNA from healthy participants was used as a control. To determine whether this panel is specific to Lynch syndrome-related cancers, it is necessary to test cfDNA from microsatellite-stable patients. We are currently in the process of requesting samples for a follow-up study. Finally, only 1 Lynch syndrome carrier was included in the study. It is not known how early and what range of frameshift mutations can be detected in carriers. To overcome these limitations, a validation study using a large prospective cohort of patients with or carriers of Lynch syndrome is needed to better understand the variability of frameshift mutations detected using this assay. This study may enable us to set a cutoff value for the number of frameshift mutations detected for monitoring disease development and treatment.
In summary, we demonstrated that frameshift mutations at coding mononucleotide repeats can be detected in cfDNA from high microsatellite instability and mismatch repair-deficient patients with high sensitivity and specificity using a 122-target panel and the NextSeq platform. Longitudinal tracking of frameshift mutations in blood may serve as a surveillance strategy for Lynch syndrome carriers, which may result in reduced frequency of colonoscopy at young ages. Frameshift neoantigen-based vaccines are showing promising activity clinically and preclinically for treatment and prevention of high microsatellite instability and mismatch repair-deficient cancers (ClinicalTrials.gov identifier NCT05078866) (22,43-47). This frameshift mutation-based biomarker panel could be used in parallel with Lynch syndrome monitoring in treatment and prevention settings.
Supplementary Material
Acknowledgements
The authors would like to thank the National Institute of Health Clinical Center staff for providing blood samples from healthy participants; Dr. Felice H. Schnoll-Sussman from Weill Cornell Medicine for collecting patient samples; Kristen M. Pike, Drs Daniel R. Soppet and Stephen Hewitt, and the late Dr Gordon R. Whiteley at the Molecular Diagnostics Laboratory for project support; Drs Yuriko Mori and Rachana Agarwal at Biospecimen Research Group for procuring patient samples; and Cancer Immunoprevention Laboratory members and scientists at the Chemopreventive Agent Development Research Group at the National Cancer Institute (NCI) Division of Cancer Prevention, for data discussion. Finally, we would like to thank the patients for participating in this research.
The Colon Cancer Family Registry graciously acknowledges the generous contributions of its study participants, dedication of study staff, and financial support from NCI, without which this important registry would not exist.
The content of this publication is solely the responsibility of the authors and does not necessarily reflect the views or policies of the US Department of Health and Human Services or any of the collaborating centers in the Colon Cancer Family Registry, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government, any cancer registry, or the Colon Cancer Family Registry. The funders did not play a role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Ms Plona could not be reached by the Journal to verify her authorship and author contributions at the time of acceptance, but her direct supervisor, Dr Daniel Soppet, director of the Laboratory of Molecular Technology at Frederick National Laboratory for Cancer Research, and the co-authors verified her authorship and author contributions on her behalf.
Contributor Information
Yurong Song, Vaccine, Immunity and Cancer Directorate, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Holli Loomans-Kropp, Division of Cancer Prevention, National Cancer Institute, Bethesda, MD, USA; Now at Department of Internal Medicine, The Ohio State University, Columbus, OH, USA.
Ryan N Baugher, Molecular Diagnostics Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Brandon Somerville, Vaccine, Immunity and Cancer Directorate, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Shaneen S Baxter, Vaccine, Immunity and Cancer Directorate, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Travis D Kerr, Vaccine, Immunity and Cancer Directorate, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Teri M Plona, Molecular Diagnostics Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Stephanie D Mellott, Molecular Diagnostics Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Todd B Young, Molecular Diagnostics Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Heidi E Lawhorn, Molecular Diagnostics Laboratory, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
Lei Wei, Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA.
Qiang Hu, Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA.
Song Liu, Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA.
Alan Hutson, Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA.
Ligia Pinto, Vaccine, Immunity and Cancer Directorate, Frederick National Laboratory for Cancer Research, Frederick, MD, USA.
John D Potter, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA; Research Centre for Hauora and Health, Massey University, Wellington, New Zealand; School of Public Health, University of Washington, Seattle, WA, USA.
Shizuko Sei, Division of Cancer Prevention, National Cancer Institute, Bethesda, MD, USA.
Ozkan Gelincik, Department of Medicine, Weill Cornell Medicine, New York, NY, USA.
Steven M Lipkin, Department of Medicine, Weill Cornell Medicine, New York, NY, USA.
Johannes Gebert, Department of Applied Tumor Biology, Institute of Pathology, University Hospital Heidelberg, Heidelberg, Germany.
Matthias Kloor, Department of Applied Tumor Biology, Institute of Pathology, University Hospital Heidelberg, Heidelberg, Germany.
Robert H Shoemaker, Division of Cancer Prevention, National Cancer Institute, Bethesda, MD, USA.
Data availability
The data presented in this study are available in the National Center for Biotechnology Information Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/PRJNA1033850 and can be accessed with BioProject accession No. PRJNA1033850.
Author contributions
Yurong Song, PhD (Conceptualization; Data curation; Formal analysis; Investigation; Project administration; Resources; Supervision; Visualization; Writing—original draft; Writing—review & editing), Holli Loomans-Kropp, PhD, MPH (Investigation; Methodology; Writing—review & editing), Ryan N. Baugher, MSc (Data curation; Formal analysis; Investigation; Methodology; Validation), Brandon Somerville, MSc (Investigation; Methodology), Shaneen S. Baxter, PhD (Investigation; Methodology), Travis D. Kerr, MSc (Investigation; Methodology), Teri M. Plona, BA (Investigation; Methodology; Validation), Stephanie D. Mellott, BSc (Investigation; Methodology), Todd B. Young, BSc (Investigation; Methodology), Heidi E. Lawhorn, BSc (Investigation; Methodology), Lei Wei, PhD (Formal analysis), Qiang Hu, PhD (Formal analysis), Song Liu, PhD (Formal analysis), Alan Hutson, PhD (Funding acquisition; Resources; Supervision), Ligia Pinto, PhD (Funding acquisition; Resources; Supervision), John D. Potter, MBBS, PhD (Writing—review & editing), Shizuko Sei, MD (Supervision; Writing—review & editing), Ozkan Gelincik, PhD (Investigation; Methodology), Steven M. Lipkin, MD, PhD (Funding acquisition; Resources; Supervision; Writing—review & editing), Johannes Gebert, PhD (Investigation; Methodology; Writing—review & editing), Matthias Kloor, MD, PhD (Funding acquisition; Resources; Supervision; Writing—review & editing), Robert H. Shoemaker, PhD (Conceptualization; Resources; Supervision; Writing—review & editing).
Funding
This research was supported in part with federal funds from the National Cancer Institute (NCI), National Institute of Health (NIH), under contract No. HHSN261201500003I; IOTN Moonshot grant U24CA232979-01, and NCI ARTNet grant U24CA274159 (L.W., Q.H., S.L., and A.H.); U54 CA272688 (O.G. and S.M.L.); and the NCI PREVENT program (HHSN2612015000391) (O.G., S.M.L., J.G., and M.K.). Dr Holli Loomans-Kropp was supported by a postdoctoral fellowship from the Division of Cancer Prevention at NCI. The Colon Cancer Family Registry (www.coloncfr.org) is supported in part by funding from NCI, NIH (award No. U01 CA167551). Support for patient ascertainment was provided in part from the Surveillance, Epidemiology, and End Results program and the following US state cancer registries (Arizona, Colorado, Minnesota, North Carolina, and New Hampshire) and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada).
Conflicts of interests
The authors declared no conflict of interest.
References
- 1. Win AK, Jenkins MA, Dowty JG, et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2017;26(3):404-412. doi: 10.1158/1055-9965.EPI-16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yurgelun MB, Hampel H. Recent advances in lynch syndrome: diagnosis, treatment, and cancer prevention. Am Soc Clin Oncol Educ Book. 2018;38:101-109. doi: 10.1200/EDBK_208341. [DOI] [PubMed] [Google Scholar]
- 3. Provenzale D, Gupta S, Ahnen DJ, et al. Genetic/familial high-risk assessment: colorectal version 1.2016, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2016;14(8):1010-1030. doi: 10.6004/jnccn.2016.0108. [DOI] [PubMed] [Google Scholar]
- 4. Kastrinos F, Ingram MA, Silver ER, et al. Gene-specific variation in colorectal cancer surveillance strategies for lynch syndrome. Gastroenterology. 2021;161(2):453-462.e15. doi: 10.1053/j.gastro.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singal AG, Gupta S, Skinner CS, et al. Effect of colonoscopy outreach vs fecal immunochemical test outreach on colorectal cancer screening completion: a randomized clinical trial. JAMA. 2017;318(9):806-815. doi: 10.1001/jama.2017.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. D’Andrea E, Ahnen DJ, Sussman DA, Najafzadeh M. Quantifying the impact of adherence to screening strategies on colorectal cancer incidence and mortality. Cancer Med. 2020;9(2):824-836. doi: 10.1002/cam4.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ Precis Oncol. 2018;2:23. doi: 10.1038/s41698-018-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hernandez-Sanchez A, Grossman M, Yeung K, et al. Vaccines for immunoprevention of DNA mismatch repair deficient cancers. J Immunother Cancer. 2022;10(6):e004416. doi: 10.1136/jitc-2021-004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castellanos-Rizaldos E, Grimm DG, Tadigotla V, et al. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin Cancer Res. 2018;24(12):2944-2950. doi: 10.1158/1078-0432.CCR-17-3369. [DOI] [PubMed] [Google Scholar]
- 10. Mohrmann L, Huang HJ, Hong DS, et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin Cancer Res. 2018;24(1):181-188. doi: 10.1158/1078-0432.CCR-17-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hellmann MD, Nabet BY, Rizvi H, et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L)1 blockade in NSCLC. Clin Cancer Res. 2020;26(12):2849-2858. doi: 10.1158/1078-0432.CCR-19-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71-88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 13. Kilgour E, Rothwell DG, Brady G, Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37(4):485-495. doi: 10.1016/j.ccell.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 14. Garcia-Pardo M, Makarem M, Li JJN, et al. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: opportunities and challenges. Br J Cancer. 2022;127(4):592-602. doi: 10.1038/s41416-022-01776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fricke F, Lee J, Michalak M, et al. TGFBR2-dependent alterations of exosomal cargo and functions in DNA mismatch repair-deficient HCT116 colorectal cancer cells. Cell Commun Signal. 2017;15(1):14. doi: 10.1186/s12964-017-0169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwitalle Y, Kloor M, Eiermann S, et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134(4):988-997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 17. Pinheiro M, Pinto C, Peixoto A, et al. Target gene mutational pattern in Lynch syndrome colorectal carcinomas according to tumour location and germline mutation. Br J Cancer. 2015;113(4):686-692. doi: 10.1038/bjc.2015.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woerner SM, Kloor M, Mueller A, et al. ; German HNPCC Consortium. Microsatellite instability of selective target genes in HNPCC-associated colon adenomas. Oncogene. 2005;24(15):2525-2535. doi: 10.1038/sj.onc.1208456. [DOI] [PubMed] [Google Scholar]
- 19. Tougeron D, Fauquembergue E, Rouquette A, et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod Pathol. 2009;22(9):1186-1195. doi: 10.1038/modpathol.2009.80. [DOI] [PubMed] [Google Scholar]
- 20. Leoni G, D’Alise AM, Cotugno G, et al. A genetic vaccine encoding shared cancer neoantigens to treat tumors with microsatellite instability. Cancer Res. 2020;80(18):3972-3982. doi: 10.1158/0008-5472.CAN-20-1072. [DOI] [PubMed] [Google Scholar]
- 21. Roudko V, Bozkus CC, Orfanelli T, et al. Shared immunogenic poly-epitope frameshift mutations in microsatellite unstable tumors. Cell. 2020;183(6):1634-1649 e17. doi: 10.1016/j.cell.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Overman M, Fakih M, Le D, et al. Phase I interim study results of nous-209, an off-the-shelf immunotherapy, with pembrolizumab, for the treatment of tumors with a deficiency in mismatch repair/microsatellite instability (Dmmr/Msi). Journal for Immunotherapy of Cancer. 2021;9(Suppl 2):A441-A441. doi: 10.1136/jitc-2021-SITC2021.410. [DOI] [Google Scholar]
- 23. Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One. 2017;12(8):e0183915. doi: 10.1371/journal.pone.0183915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soung YH, Ford S, Zhang V, Chung J. Exosomes in cancer diagnostics. Cancers (Basel). 2017;9(1):8. doi: 10.3390/cancers9010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sauter ER. Exosomes in blood and cancer. Translational Cancer Research. 2017;6(S8):S1316-S1320. doi: 10.21037/tcr.2017.08.13. [DOI] [Google Scholar]
- 26. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926-930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Medina JE, Dracopoli NC, Bach PB, et al. Cell-free DNA approaches for cancer early detection and interception. J Immunother Cancer. 2023;11(9):e006013. doi: 10.1136/jitc-2022-006013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rolfo C, Russo A. Brave new world of cfDNA-omics for early cancer detection. J Immunother Cancer. 2023;11(9):e006309. doi: 10.1136/jitc-2022-006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dao J, Conway PJ, Subramani B, et al. Using cfDNA and ctDNA as oncologic markers: a path to clinical validation. Int J Mol Sci. 2023;24(17):13219. doi:10.1321910.3390/ijms241713219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Philipson TJ, Durie T, Cong Z, Fendrick AM. The aggregate value of cancer screenings in the United States: full potential value and value considering adherence. BMC Health Serv Res. 2023;23(1):829. doi: 10.1186/s12913-023-09738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015;10(10):e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woodhouse R, Li MJ, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15(9):e0237802., doi: 10.1371/journal.pone.0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lennon AM, Buchanan AH, Kinde I, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369(6499):49. doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142(2):248-256. quiz e25-26. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kriz D, Ansari D, Andersson R. Potential biomarkers for early detection of pancreatic ductal adenocarcinoma. Clin Transl Oncol. 2020;22(12):2170-2174. doi: 10.1007/s12094-020-02372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Foda ZH, Annapragada AV, Boyapati K, et al. Detecting liver cancer using cell-free DNA fragmentomes. Cancer Discov. 2023;13(3):616-631. doi: 10.1158/2159-8290.CD-22-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leal AIC, Mathios D, Jakubowski D, et al. Cell-free DNA fragmentomes in the diagnostic evaluation of patients with symptoms suggestive of lung cancer. Chest. 2023;164(4):1019-1027. 10.1016/j.chest.2023.04.033. doi: 10.1016/j.chest.2023.04.033. [DOI] [PubMed] [Google Scholar]
- 38. Bruhm DC, Mathios D, Foda ZH, et al. Single-molecule genome-wide mutation profiles of cell-free DNA for non-invasive detection of cancer. Nat Genet. 2023;55(8):1301-1310. doi: 10.1038/s41588-023-01446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schrag D, McDonnell CH, Nadauld L, et al. A prospective study of a multi-cancer early detection blood test. Annals of Oncology. 2022;33(7):S961. doi: 10.1016/j.annonc.2022.07.1029. [DOI] [Google Scholar]
- 40. Schrag D, Beer TM, McDonnell CH, et al. Blood-based tests for multicancer early detection (PATHFINDER): a prospective cohort study. Lancet. 2023;402(10409):1251-1260. doi: 10.1016/S0140-6736(23)01700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bramblet RM, Bakkum-Gamez JN, Slettedahl SW, et al. Methylated DNA markers for sporadic colorectal and endometrial cancer are strongly associated with lynch syndrome cancers. Cancer Prev Res (Phila). 2023;16(11):611-620. doi: 10.1158/1940-6207.CAPR-23-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jahangiri L, Hurst T. Assessing the concordance of genomic alterations between circulating-free DNA and tumour tissue in cancer patients. Cancers. 2019;11(12):1938. doi: 10.3390/cancers11121938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gebert J, Gelincik O, Oezcan-Wahlbrink M, et al. Recurrent frameshift neoantigen vaccine elicits protective immunity with reduced tumor burden and improved overall survival in a lynch syndrome mouse model. Gastroenterology. 2021;161(4):1288-1302.e13. doi: 10.1053/j.gastro.2021.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kloor M, Reuschenbach M, Pauligk C, et al. A frameshift peptide neoantigen-based vaccine for mismatch repair-deficient cancers: a phase I/IIa clinical trial. Clin Cancer Res. 2020;26(17):4503-4510. doi: 10.1158/1078-0432.CCR-19-3517. [DOI] [PubMed] [Google Scholar]
- 45. Roudko V, Cimen Bozkus C, Greenbaum B, et al. Lynch syndrome and MSI-H cancers: from mechanisms to "Off-The-Shelf" cancer vaccines. Front Immunol. 2021;12:757804. doi: 10.3389/fimmu.2021.757804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sellars MC, Wu CJ, Fritsch EF. Cancer vaccines: Building a bridge over troubled waters. Cell. 2022;185(15):2770-2788. doi: 10.1016/j.cell.2022.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sei S, Ahadova A, Keskin DB, et al. Lynch syndrome cancer vaccines: a roadmap for the development of precision immunoprevention strategies. Front Oncol. 2023;13:1147590. doi: 10.3389/fonc.2023.1147590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the National Center for Biotechnology Information Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/PRJNA1033850 and can be accessed with BioProject accession No. PRJNA1033850.