Abstract
The 5′ untranslated region, also called the leader, of oncoretroviruses and lentiviruses is long and is formed of several structured domains critically important in virus replication. The 5′ leader of murine leukemia virus (MLV) RNA contains an internal ribosomal entry segment (IRES) which promotes synthesis of Gag and glyco-Gag polyprotein precursors. In the present study we investigated the translational features of the 5′ leader of MLV subgenomic RNA (env RNA) encoding the Env polyprotein precursor. When the env leader was inserted between two genes, such as lacZ and the neomycin resistance cassette, in a dicistronic vector, it allowed IRES-dependent translation of the 3′ cistron in the rabbit reticulocyte lysate system and in murine cells. The drug rapamycin and the foot-and-mouth disease virus L protease, known to inhibit cap-dependent translation, caused an enhancement of the translation driven by the env leader sequence, consistent with an IRES activity promoting Env expression. Analysis of several deletion mutants led us to localize the minimal env IRES between the splice junction and the env AUG start codon.
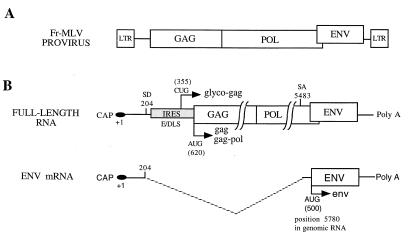
The murine leukemia viruses (MLV) belong to the simple retrovirus family bearing in common a genomic RNA with the gag, pol, and env genes. The structural proteins and enzymes of the virion core encoded by the gag and pol genes, respectively, are translated by full-length viral RNA, whereas the envelope proteins, encoded by env, are translated by a spliced RNA (env RNA) (19, 32) (Fig. 1). In the Moloney strain of MLV, the splice donor site is located at position 204 of the genomic RNA and the splice acceptor site is in pol at position 5483 (27). As a consequence, (i) the dimerization and encapsidation sequences present in the 5′ leader of the genomic RNA are absent in the spliced RNA, thus preventing its packaging into virions (16); (ii) the 5′ env leader derives from the pol coding region from position 205 to the env initiation codon at position 5780; and (iii) the genomic RNA and the spliced RNA share the same 5′ leader sequences from the 5′ cap to the splice donor site, probably with the same stable secondary structures (18, 29). The presence of secondary structures between the cap and the initiation codon has been proposed to strongly interfere with the scanning mechanism of translation initiation (11). In this model, well established for most eukaryotic mRNAs, translation is initiated after a linear scanning of the 40S ribosomal subunit from the 5′ end of the mRNA (the cap structure) to the initiation site (generally the first AUG codon) (12, 17). The features of a 5′ leader that optimize the efficiency of translation initiation include the presence of a 5′ cap, a short length, the absence of stable secondary structures as well as AUG codons upstream from the initiation site, and an initiation codon in a good context (A/GCCAUGG). A large number of eukaryotic mRNAs share these features, but a growing number of cellular and viral RNAs lack them. Studies of picornaviruses were the first to show that translation initiation can occur by a mechanism different from the canonical scanning. In these viruses, protein synthesis is initiated by internal ribosome binding to the RNA (8, 22). This implies that a segment of the viral 5′ untranslated region, now known as IRES (internal ribosomal entry segment), guides the ribosomes to the translation initiation site. IRESs are not restricted to viral RNAs but have also been identified in many cellular mRNAs (5, 7, 15, 20, 30). Recently, it has been shown that the 5′ leader of MLV genomic RNA contains an IRES that promotes synthesis of Gag and glyco-Gag polyprotein precursors (2). When inserted between two genes in a dicistronic vector, the 5′ leader of MLV genomic RNA can promote expression of a downstream cistron in vitro and in vivo. Interestingly, deletion of the 5′ first 280 nucleotides of the leader results in a fourfold decrease of IRES activity, suggesting that this region plays an important role in the ribosome entry mechanism (2).
FIG. 1.
Genetic organization and expression of Friend murine leukemia virus (Fr-MLV). (A) Fr-MLV provirus. LTR, long terminal repeat. (B) Viral RNAs: full-length viral RNA (used as genomic RNA, pre-mRNA, and mRNA) and splice RNA (used as messenger for env). SD, splice donor site; SA, splice acceptor site; E/DLS, dimerization and encapsidation sequence; Poly A, polyadenylate sequence. The numbering is with respect to the 5′ cap (+1).
We wanted to investigate the translational features of the 5′ leader of env RNA. Therefore, we analyzed the expression of dicistronic MLV-derived vectors in vitro and in murine NIH 3T3 cells under conditions that inhibit cap-dependent translation. In the present report we show that the env leader contains an IRES which most probably directs expression of the Env polyprotein precursor.
MATERIALS AND METHODS
General methods.
Standard procedures were used for restriction nuclease digestion and plasmid DNA construction (26). Escherichia coli HB101 strain 1035 (recA mutant) was used for plasmid DNA amplification. Details of plasmid construction are given below. The numbering is with respect to the env RNA cap site (position +1) unless otherwise stated.
Reverse transcription-PCR amplification and cloning of the 5′ leader of env RNA.
Reverse transcription of the 5′ leader of env RNA was performed as follows. First-strand cDNA synthesis was performed on 100 ng of purified mRNA with the You-Prime First-Strand Beads kit (Pharmacia Biotech). By using Oligotex suspension (Qiagen), template mRNAs for the reverse transcription were extracted from total RNA purified from MLV-infected Mus dunni cells (9). Subsequently, a DNA fragment corresponding to the entire 5′ leader of env RNA was generated by PCR with first-strand cDNA as a template and specific oligonucleotides with an NheI restriction site at their 5′ ends. This PCR fragment was ligated into pBluescript in order to generate pBenv.
DNA constructs for translation assays in the rabbit reticulocyte lysate (RRL) system.
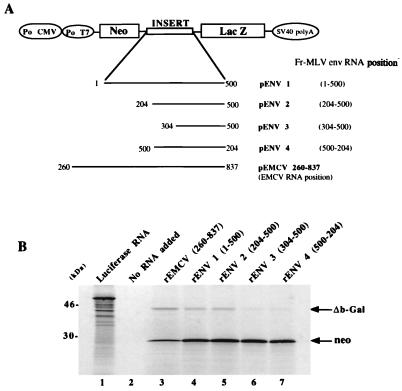
A DNA fragment corresponding to the 5′ leader of env RNA from position 1 to 500 was generated by digesting pBenv with NheI. DNA segments corresponding to the env leader from positions 204 to 500, 150 to 500, and 304 to 500 were generated by PCR with the pBenv plasmid as a template followed by digestion with NheI (PCR-added restriction site). To construct the dicistronic pENV 1 (1-500), pENV 2 (204-500), pENV 3 (304-500), and pENV 4 (500-204), each respective fragment was inserted between neo and lacZ of pEMCV-D 260-837 previously digested with NheI (2).
In vitro RNA synthesis.
Capped and uncapped RNAs were synthesized with T7 RNA polymerase (mMessage; Ambion) with the linearized DNA template according to the manufacturer's instructions. A SspI site in the lacZ gene (position 1240) was used to linearize template DNA. RNAs were precipitated by lithium chloride, washed with 70% ethanol, and then resuspended in 50 μl of water. RNA integrity was verified on a 1% agarose gel, and the concentration was determined spectrophotometrically.
Translation in the RRL system.
Capped and uncapped dicistronic RNAs were translated in nuclease-treated RRL (Flexy; Promega) at 50% of the original concentration with 60 mg of RNA/ml (20 mg/ml was used for encephalomyocarditis virus [EMCV] RNA) and 0.6 mCi of [35S]methionine (Amersham) per ml at 30°C for 1 h. All assay mixtures were supplemented with potassium chloride to a final concentration of 100 mM KCl and with 0.2 mM MgCl. The effect of L protease on the translation of capped RNAs was assayed in the Flexy lysate as described above, but the lysate was pretreated with 1.2 mg of purified recombinant L protease (kindly provided by S. J. Morley, Department of Biochemistry, The University of Sussex, Sussex, United Kingdom)/ml. After translation, the reaction mixtures were treated at 70°C for 5 min in 62.5 mM Tris-HCl (pH 6.8)–2% sodium dodecyl sulfate (SDS)–10% glycerol–5% β-mercaptoethanol–0.002% bromophenol blue, and the 35S-labeled proteins were analyzed by electrophoresis on 15% (wt/vol) polyacrylamide gels. Bands were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Cell culture, DNA transfection, and selection.
Murine NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (GIBCO-BRL) containing 10% newborn calf serum at 37°C in the presence of 5% CO2. Mus dunni cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum at 37°C in the presence of 5% CO2. Transfections were performed by the calcium phosphate method (2a, 2b) with 20 μg of plasmid DNA. Selection for neomycin resistance was performed with G418 at 0.8 mg/ml for 1 month.
Histochemical staining.
For placental alkaline phosphatase (plap) histochemical staining, cells were fixed in phosphate-buffered saline (PBS) containing 2% formaldehyde and 0.2% glutaraldehyde. After two washes in 1× PBS, the cells were incubated at 65°C for 30 min in 1× PBS. The cells were washed twice with AP buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, and 50 mM MgCl2 in 1× PBS) and incubated for 5 h in staining solution (0.1 mg of 5-bromo-4-chloro-3-indolyl phosphate [BCIP], 1 mg of nitroblue tetrazolium salt/ml, and 1 mM levamisole in 1× AP buffer).
Effect of rapamycin on dicistronic plasmid expression.
Stably transfected murine NIH 3T3 cells were grown to 70% confluency before serum-free medium was added for 48 h. Then, complete medium containing rapamycin to a final concentration of 100 ng/ml was added for 20 h at 37°C, after which cellular proteins were extracted and enzymatic activities were determined.
Protein extraction and enzymatic activities.
Cellular proteins were extracted with the β-galactosidase enzyme assay system (Promega). The protein concentration was determined with the Micro BSA kit (Pierce). Neomycin phosphotransferase (Neo) activity was measured by [γ32-P]ATP phosphate transfer to neomycin (25). In order to check for linearity of the assay and to quantify the neomycin activity in each cell extract, an internal standard curve was used by serial dilutions of the cell extract giving the strongest Neo activity. Alkaline phosphatase activity was determined spectrophotometrically (alkaline phosphatase substrate kit; Bio-Rad). Alkaline phosphatase (in units) was estimated by using commercial calf intestine alkaline phosphatase (Boehringer Mannheim) as an activity standard.
RESULTS
Characterization of an IRES in the 5′ leader of the env RNA.
Recent studies have shown that the 5′ leader of MLV genomic RNA contains an IRES directing synthesis of Gag and glyco-Gag polyproteins (Fig. 1) (2). This finding prompted us to search for an IRES in the 5′ leader of env RNA. Various fragments of the env leader were inserted between the neomycin phosphotransferase gene (neo) and lacZ sequences in dicistronic constructs (Fig. 2A). Subsequently, capped RNAs were generated in vitro and their translational activities were monitored in the RRL system. In this system, translation initiation of the 5′ cistron (neo) is cap dependent (2, 14) while that of the 3′ cistron (lacZ) can only occur via either (i) an internal ribosome binding to the intercistronic region (IRES-dependent translation) (22) or (ii) a cap-dependent mechanism, such as a reinitiation mechanism, and/or a shunt mechanism (10). As a positive control for IRES-dependent translation, we used pEMCV (260-837) containing the EMCV IRES from position 260 to 837 in the intercistronic region. In order to estimate the background level of β-galactosidase produced in the RRL by a non-IRES-dependent mechanism, we constructed a dicistronic plasmid, pENV 4 (500-204). In this plasmid, the intercistronic region contains the fragment of env leader from position 204 to 500 but inserted in the antisense orientation.
FIG. 2.
Translation of capped dicistronic RNAs in the RRL system. (A) Schematic representation of the plasmid constructs used for in vitro RNA synthesis: DNA containing genetic elements of the MLV env RNA leader located between the neomycin phosphotransferase and lacZ genes under the control of the T7 promoter (Po T7). The numbering is with respect to the subgenomic env RNA 5′ cap (+1). Po CMV, cytomegalovirus promoter. (B) Translation of cap dicistronic RNAs in the Flexy RRL system (Promega). After heat denaturation, 35S-labeled proteins were analyzed by SDS–15% PAGE. The positions of Neo (28 kDa) and of the carboxy-terminal-truncated β-Gal protein (46 kDa) are indicated. Luciferase RNA was used as the positive control (lane 1), and the negative control had no RNA added (lane 2).
Figure 2 shows the protein products generated by translating the dicistronic RNAs and subsequently separated by 15% polyacrylamide gel electrophoresis (PAGE). Translation of the 3′ cistron, ΔLacZ, with recombinant ENV1 (1-500) [rENV1 (1-500)] and rENV2 (204-500) RNAs was efficient in comparison with that of the positive control RNA containing the EMCV IRES (rEMCV) (lanes 4 and 5). In contrast, with the rENV3 (304-500) and rENV4 (500-204) RNAs, translation of the 3′ cistron was very low (lanes 6 and 7). Translation of the 5′ cistron, neo, was very efficient for all RNAs (Fig. 2). These results suggest that the 5′ leader of env RNA contains an IRES and that the minimal IRES sequence should be located between nucleotide 204 and the env initiation codon.
Effect of L protease and cap on activity of env IRES.
To confirm that translation of the 3′ cistron in rENV1 (1-500) and rENV2 (204-500) is cap independent, we examined the effect of foot-and-mouth disease virus (FMDV) L protease on translation driven by the putative env IRES in the RRL system. The FMDV L protease cleaves eukaryotic initiation factor 4G (eIF4G), resulting in a partial inhibition of cap-dependent translation (4). IRES-dependent translation is not inhibited by L protease but can even be enhanced (33). After translation in RRL and separation of the protein products by gel electrophoresis (Fig. 3), the relative intensities of the β-Gal and Neo proteins were quantified by scanning densitometry and the effect of FMDV L protease was thus estimated. The results are presented as the percentage of protein synthesized in untreated RRL (summarized in Table 1). In addition, as complementary experiments, translation assays were performed with uncapped RNAs.
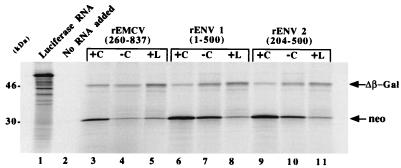
FIG. 3.
Effect of cap and FMDV L protease on RNA translation in the RRL system. Translation of dicistronic uncapped RNAs (−C) and capped RNAs in the Flexy RRL system (Promega) with (+L) or without (+C) L protease. After heat denaturation, 35S-labeled proteins were analyzed by SDS-12% PAGE. The positions of Neo (28 kDa) and the carboxy-terminal-truncated β-Gal protein (46 kDa) are indicated. Luciferase RNA was used as the positive control (lane 1), and the negative control had no RNA added (lane 2).
TABLE 1.
Relative variation of translation caused by FMDV L protease in the RRL systema
| RNA | Relative level of Neo (%) | Relative level of β-Gal (%) |
|---|---|---|
| rEMCV (837-260) | 44 | 175 |
| rENV1 (1-500) | 10 | 260 |
| rENV2 (204-500) | 8 | 220 |
The gel shown in Fig. 3 was scanned, and the data were used to calculate the effect of FMDV L protease as a percentage of variation relative to translation in untreated RRL.
As shown in Fig. 3, translation of the 5′ cistron, neo, was always severely reduced in the RRL system treated with L protease (compare lanes 3, 6, and 9 with 5, 8, and 11). In contrast, synthesis of β-galactosidase was enhanced with rEMCV (260-837), rENV 1 (1-500), and rENV 2 (204-500) RNAs (lanes 3, 5, 6, 8, and 11). Similar results were obtained with uncapped RNAs in that translation of the 5′ cistron was reduced while that of the 3′ cistron was unchanged or enhanced (lanes 4, 7, and 10). These results strongly suggest that termination-reinitiation or another cap-dependent mechanism cannot account for the translation of the 3′ cistron. In fact, in the RRL programmed with uncapped rather than capped RNA or in the presence of L protease, fewer ribosomes would be expected to reach the lacZ initiation codon and hence translation of the second cistron would be diminished. However translation of the 3′ cistron was either slightly increased (RNAs minus cap) or clearly increased (addition of L protease) (Table 1). These data show that the 3′ cistron is translated in a cap-independent fashion and thus indicate that the 5′ leader of env RNA contains an IRES. These data also indicate that the minimal env IRES is probably located between nucleotide 204 and the env start codon.
The env IRES directs gene expression in NIH 3T3 cells.
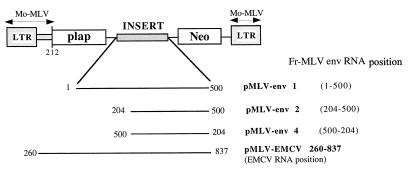
In order to examine the functionality of the env IRES in murine cells, dicistronic vectors were transfected in NIH 3T3 cells. The transfected constructs contained all or part of the env IRES inserted between the plap gene and neo (Fig. 4). Expression of the 5′ cistron (plap) is cap dependent (2, 14), while that of the 3′ cistron (neo) was IRES dependent (or cap dependent if a translation reinitiation mechanism could occur). We used pMLV-EMCV (260-837) with the EMCV IRES from position 260 to 837 between plap and neo as a positive IRES control. After 1 month of G418 selection, Neor foci were histochemically stained for alkaline phosphatase and cellular proteins were extracted to determine the relative level of alkaline phosphatase and neomycin phosphotransferase expression in each cell population (the results are summarized in Table 2).
FIG. 4.
Schematic representation of the DNA constructs used in cell culture experiments. The plasmids contain different genetic elements of the 5′ env RNA leader located between the placental alkaline phosphatase gene (plap) and the neomycin gene (Neo). LTR, long terminal repeat. The numbering is with respect to the env mRNA 5′ cap.
TABLE 2.
Expression of dicistronic plasmids in cell culturea
| Cell line or plasmid | Alkaline phosphatase sp act (U/μg of total protein) | Neomycin phosphotransferase sp act (arbitrary units) | neo/plap (%) |
|---|---|---|---|
| NIH 3T3 | 0 | 0 | 0 |
| pMLV-EMCV (260-837) | (1.0 ± 0.4) × 10−6 | 100 ± 35 | 100 ± 35.0 |
| pMLV-env 1 (1-500) | (7.2 ± 0.5) × 10−6 | 86 ± 19 | 12 ± 2.6 |
| pMLV-env 2 (204-500) | (5.7 ± 0.7) × 10−6 | 38 ± 14 | 7 ± 2.5 |
| pMLV-env 4 (500-204) | (2.6 ± 0.4) × 10−5 | 28 ± 10 | 1 ± 0.4 |
Murine NIH 3T3 cells were transfected with the dicistronic plasmids described previously (Fig. 4A). After 3 weeks of selection with G418, total proteins were extracted and enzymatic activities were determined as described in Materials and Methods. The data shown correspond to the averages (± standard deviations) of three independent experiments. The ratio of neo to plap expression is shown as percentages in the third column. In the absence of a functional IRES between the two cistrons, there is clearly a large increase in plap (first cistron) expression (see pMLV-env 4).
Vectors pMLV-EMCV (260-837), pMLV-env 1 (1-500), pMLV-env 2 (204-500), and pMLV-env 4 (500-240; antisense orientation) were able to confer G418 resistance. Based on histochemical staining, we determined that all Neor foci were expressing plap. Enzymatic activities were measured on cellular extracts and showed that the level of neo expression was high in NIH 3T3 stably transfected with pMLV-EMCV (260-837) and pMLV env 1 (1-500), medium with pMLV-env 2 (204-500), and low with pMLV-env 4 (500-204). These data indicate that the env IRES is functional in NIH 3T3 cells and confirm results obtained in the RRL system. The negative control, pMLV-env 4 (500-240; antisense), was also able to confer G418 resistance, but the number of Neor foci was at least 10 times less than with pMLV-env 2 (data not shown), thus indicating that neo expression was an inefficient process with pMLV-env 4 and probably results from a translation reinitiation of the ribosomes coming from the 5′ cistron (compare rapamycin [Table 3]).
TABLE 3.
Variation of dicistronic plasmid expression caused by rapamycin in cell culturea
| Cell line (plasmid) | Variation (%) in alkaline phosphatase sp act | Variation (%) in neomycin phosphotransferase sp act |
|---|---|---|
| pMLV-EMCV (260-837) | −20 | +826 |
| pMLV env 1 (1-500) | −26 | +316 |
| pMLV env 2 (204-500) | −30 | +800 |
| pMLV env 4 (500-204) | −15 | −16 |
Murine NIH 3T3 cells were transfected with the dicistronic plasmids described previously (Fig. 4A). After 3 weeks of selection with G418, the cells were serum starved for 48 h and then serum was added as well as rapamycin to a final concentration of 50 ng/ml. Total proteins were extracted 20 h later from both rapamycin-treated and untreated cells, and enzymatic activities were determined. The data shown correspond to the means of three independent experiments. The level of the reporter gene, measured as enzymatic activity, in the presence or absence of rapamycin was used to calculate the effect of the drug as a percentage increase or decrease relative to the level in untreated cells.
In order to confirm the above data, we analyzed the variation of plap and neo expression caused by rapamycin, an inhibitor of cap-dependent translation.
env IRES-dependent translation is enhanced in rapamycin-treated murine cells.
Rapamycin has been shown to partially inhibit cap-dependent translation. This inhibition results from the dephosphorylation and consequent activation of 4E-BP1, a repressor of the cap-binding protein eIF-4E (1, 6, 13). To further examine the translational properties of the env IRES in cells, we analyzed the effect of rapamycin on plap and neomycin expression in Neor populations (the results are summarized in Table 3). As predicted, in the presence of rapamycin the expression of the 5′ cistron (plap) was always decreased. This confirms the capacity of rapamycin to inhibit cap-dependent translation in murine cells. Similarly, in NIH 3T3 cells stably transfected with pMLV env 4 (500-204), the expression of the 3′ cistron (neo) was inhibited in the presence of rapamycin. These results indicate that the fragment of the 5′ leader env RNA from position 204 to 500 but in an inverted orientation does not possess an IRES activity. This supports the view that expression of the 3′ cistron (neo) in pMLV env 4 (500-204) occurs via a cap-dependent and reinitiation mechanism. In contrast, in rapamycin-treated cells stably transfected with pMLV-EMCV (260-837), pMLV env 1 (1-500), pMLV env 2 (150-500), and pMLV env 3 (204-500), we observed a drastic increase of neo activity (Table 3). These results show that the full-length env leader, as well as env leader sequences from position 204 to 500, allows cap-independent translation of a downstream cistron in vivo. These data are in agreement with those obtained in vitro, indicating that a minimal env IRES is present between the splice junction and the initiation codon of the env RNA.
DISCUSSION
The genomic RNA of MLV and MLV-like type C retroviruses has a long 5′ leader formed of structured domains involved in key functions of the viral life cycle, such as splicing, translation, dimerization and packaging, and reverse transcription (3, 31). Stable secondary structures in the 5′ leader of MLV are thought to prevent translation initiation of the Gag precursor by the classical ribosome scanning mechanism. The presence of an IRES upstream of the gag initiation codon allows the local recruitment of ribosomes and synthesis of the Gag precursor (2). This prompted us to search for an IRES in the 5′ leader of the env RNA, since the genomic and the subgenomic RNAs of MLV have the same sequence upstream of the splice junction. Therefore, we constructed dicistronic plasmids containing part or all of the env leader inserted between two reporter genes. The translational activity of these constructs was monitored in the RRL system and in NIH 3T3 cells. The results show that the 5′ leader of env RNA contains an IRES directing translation of a downstream cistron independently of the first cistron. Furthermore, either the drug rapamycin or FMDV L protease, which are known to specifically shut off cap-dependent translation, caused an increase in translation driven by the env leader, whereas they both reduced expression of the 5′ cap-dependent cistron present in the bicistronic RNA. Analysis of the expression of pENV 2 (204-500) and pENV 3 (304-500) constructs led us to identify a functional segment of the env IRES between the splice junction and the env initiation codon. Thus, the gag and env IRESes have common sequences at their 5′ ends, and in agreement with this, the region from position 1 to 260 increases translation initiation directed by the gag IRES (2). Similarly, our data show that in murine cells the full-length env leader possesses a stronger IRES activity than the region extending from position 204 to 500 (Table 2). Therefore, it is tempting to speculate that the activity of both the gag and env IRESes might be coregulated by cellular factors binding to the same 5′ sequence of the gag and env RNAs. Differential regulation of Gag and Env expression would then depend on 3′ sequences unique to each IRES. This is presently under investigation.
IRES-dependent translation does not use the cap-binding initiation factor eIF4E required for cap-dependent translation. This protein is a key target in the regulation of protein expression (21, 23, 24). It has been proposed that internal entry, which avoids eIF4E regulation, may represent a viral strategy to favor viral protein synthesis at the expense of that of cellular proteins. Such an advantage would, for example, enable virus production during mitosis, when cap-dependent translation is significantly decreased (28). For such a strategy to be effective it is logical that, like Gag, Env polyprotein should possess a cap-independent translation mechanism.
Results obtained with dicistronic constructs containing portions of the env leader indicate that a functional part of the env IRES encompasses the splice junction and env initiator codon. This situation raises the possibility that some Env polyprotein precursors could be translated after internal binding on genomic RNAs. This hypothesis does not agree with early oocyte injection experiments which showed that genomic RNAs could only produce Gag-Pol proteins in oocyte cytoplasm (19, 32). One can argue that these experimental procedures do not recreate optimal cellular conditions for IRES-dependent translation. A more likely hypothesis is that the env IRES is not functional when present as part of the pol coding region. This would also explain the splicing of the genomic RNA. This is presently under investigation in vitro and in cell culture.
ACKNOWLEDGMENTS
Thanks are due to S. J. Morley, Department of Biochemistry, The University of Sussex, United Kingdom, for kindly providing purified recombinant FMDV L protease.
This work was supported by grants from ANRS and MGEN. C. Deffaud is supported by a fellowship of the Fondation Mérieux.
REFERENCES
- 1.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 2.Berlioz C, Darlix J L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b.Chen C, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transformation system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–637. [PubMed] [Google Scholar]
- 3.Corbin A, Darlix J L. Functions of the 5′ leader of murine leukemia virus genomic RNA in virion structure, viral replication and pathogenesis, and MLV-derived vectors. Biochimie. 1996;78:632–638. doi: 10.1016/s0300-9084(96)80009-5. [DOI] [PubMed] [Google Scholar]
- 4.Devaney M A, Vakharia V N, Lloyd R E, Ehrenfeld E, Grubman M J. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol. 1988;62:4407–4409. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gan W, Celle M L, Rhoads R E. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 6.Graves L M, Bornfeldt K E, Argast G M, Krebs E G, Kong X, Lin T A, Lawrence J C., Jr cAMP- and rapamycin-sensitive regulation of the association of eukaryotic initiation factor 4E and the translational regulator PHAS-I in aortic smooth muscle cells. Proc Natl Acad Sci USA. 1995;92:7222–7226. doi: 10.1073/pnas.92.16.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 9.Khandjian E W, Meric C. A procedure for Northern blot analysis of native RNA. Anal Biochem. 1986;159:227–232. doi: 10.1016/0003-2697(86)90332-5. [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin T A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C., Jr Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Lastra M, Gabus C, Darlix J L. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum Gene Ther. 1997;8:1855–1865. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- 15.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 16.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 17.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mougel M, Tounekti N, Darlix J L, Paoletti J, Ehresmann B, Ehresmann C. Conformational analysis of the 5′ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res. 1993;21:4677–4684. doi: 10.1093/nar/21.20.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy E C, Jr, Campos III D, Arlinghaus R B. Cell-free synthesis of Rauscher murine leukemia virus “gag” and “env” gene products from separate cellular mRNA species. Virology. 1979;93:293–302. doi: 10.1016/0042-6822(79)90234-4. [DOI] [PubMed] [Google Scholar]
- 20.Oh S K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 21.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 22.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 23.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramesh N, Osborne W R. Assay of neomycin phosphotransferase activity in cell extracts. Anal Biochem. 1991;193:316–318. doi: 10.1016/0003-2697(91)90028-r. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook I, Maniatis T, Fritsch E. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 28.Sonenberg N. Remarks on the mechanism of ribosome binding to eukaryotic mRNAs. Gene Expr. 1993;3:317–323. [PMC free article] [PubMed] [Google Scholar]
- 29.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J L, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J Mol Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 30.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss R T N, Varmus H, Coffin J. RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. [Google Scholar]
- 32.Zaane D V, Gielkens A L, Hesselink W G, Bloemers H P. Identification of Rauscher murine leukemia virus-specific mRNAs for the synthesis of gag- and env-gene products. Proc Natl Acad Sci USA. 1977;74:1855–1859. doi: 10.1073/pnas.74.5.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziegler E, Borman A M, Kirchweger R, Skern T, Kean K M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]