Abstract
Hepatitis C virus (HCV) NS5B protein possesses an RNA-dependent RNA polymerase (RdRp) activity, a major function responsible for replication of the viral RNA genome. To further characterize the RdRp activity, NS5B proteins were expressed from recombinant baculoviruses, purified to near homogeneity, and examined for their ability to synthesize RNA in vitro. As a result, a highly active NS5B RdRp (1b-42), which contains an 18-amino acid C-terminal truncation resulting from a newly created stop codon, was identified among a number of independent isolates. The RdRp activity of the truncated NS5B is comparable to the activity of the full-length protein and is 20 times higher in the presence of Mn2+ than in the presence of Mg2+. When a 384-nucleotide RNA was used as the template, two major RNA products were synthesized by 1b-42. One is a complementary RNA identical in size to the input RNA template (monomer), while the other is a hairpin dimer RNA synthesized by a “copy-back” mechanism. Substantial evidence derived from several experiments demonstrated that the RNA monomer was synthesized through de novo initiation by NS5B rather than by a terminal transferase activity. Synthesis of the RNA monomer requires all four ribonucleotides. The RNA monomer product was verified to be the result of de novo RNA synthesis, as two expected RNA products were generated from monomer RNA by RNase H digestion. In addition, modification of the RNA template by the addition of the chain terminator cordycepin at the 3′ end did not affect synthesis of the RNA monomer but eliminated synthesis of the self-priming hairpin dimer RNA. Moreover, synthesis of RNA on poly(C) and poly(U) homopolymer templates by 1b-42 NS5B did not require the oligonucleotide primer at high concentrations (≥50 μM) of GTP and ATP, further supporting a de novo initiation mechanism. These findings suggest that HCV NS5B is able to initiate RNA synthesis de novo.
Hepatitis C virus (HCV) is the major causative agent of non-A, non-B viral hepatitis (23). Although acute HCV infection is often asymptomatic, nearly 80% of cases resolve to chronic hepatitis, which may lead to progressive liver disease, such as cirrhosis, and liver failure. Chronic HCV infection is also associated with the development of hepatocellular carcinoma (23). It is estimated that 170 million people worldwide and more than 4 million people in the United States are currently infected with HCV (2, 65). Obviously, HCV infection remains a major threat to the public health all over the world.
HCV is an enveloped RNA virus containing a single-stranded positive-sense RNA genome approximately 9.5 kb in length (14, 31, 56). The RNA genome consists of a 5′-untranslated region (5′ UTR) of 341 nucleotides (12, 13), a large open reading frame (ORF) encoding a single polypeptide of 3,010 to 3,040 amino acids (14, 31, 56), and a 3′-untranslated region (3′ UTR) of variable length (33, 57, 66). HCV is similar in amino acid sequence and genome organization to flaviviruses and pestiviruses (41), and therefore HCV was classified as a third genus of the family Flaviviridae (18, 50).
Studies of HCV replication and the search for specific anti-HCV agents have been hampered by the lack of an efficient tissue culture system for HCV propagation, the absence of a suitable small-animal model for HCV infection, the low level of viral replication, and the considerable genetic heterogeneity (quasispecies) associated with the virus (6, 50, 51). Our current understanding of the structures and functions of the HCV genome and encoded proteins is primarily derived from in vitro studies using various recombinant systems (6, 50). The 5′ UTR is one of the most conserved regions of the viral genome and plays a pivotal role in the initiation of translation of the viral polyprotein (6, 12, 13, 50). A single long ORF encodes a polyprotein which is co- or posttranslationally processed into structural (core, E1, and E2) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins by either cellular or viral proteinases (6, 50). The 3′ UTR consists of three distinct regions, the variable region, a polyuridine tract of variable length with interspersed substitutions of cytidines, and a highly conserved 98 nucleotides (nt) at the very 3′ end (33, 57, 66). It has been speculated that the 3′ UTR contains cis elements for replication of the viral RNA genome (33, 57, 66). Recent studies have revealed that two cellular proteins, p58 (PTB) and p35, specifically interact with the 3′ UTR (26, 39, 60) and may be involved in regulating the translation of the viral polyprotein (27) or viral replication. Unlike what is found for other RNA viruses, to which reverse genetics is applicable (1, 47, 64), functions of the cis signal sequences and viral proteins of HCV in the context of virus replication have not been amenable to examination in tissue culture (6, 50). Recent advances in establishing an HCV infection from cDNA-derived genomic RNA in chimpanzees have opened up new avenues for genetic analysis of the viral genome and viral proteins (34, 68). However, limited resources and cost will likely constrain the scope of application for such a system.
In spite of the lack of an efficient reverse-genetics system for HCV study, a great deal of progress on the structures and functions of the viral proteins has been made through utilization of recombinant systems involving the expression of individual viral gene products (6, 50). Biochemical studies of the recombinant viral proteins have identified four different virus-specific enzymatic activities. Two distinct viral proteases are responsible for specific cleavage of the viral polyprotein. The NS2-NS3 protease is a metalloprotease that catalyzes the cleavage at the NS2-NS3 junction (19, 22), while NS3 is a serine protease responsible for cleavages at the NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions (4, 20, 22, 40, 59). The NS4A protein acts as a cofactor of NS3 protease to promote proteolytic cleavage (5, 16). RNA helicase and NTPase activities have also been identified in the NS3 protein (32, 54). The N-terminal one-third of the NS3 protein functions as a protease, and the remaining two-thirds of the molecule acts as the helicase/ATPase that was speculated to be involved in HCV replication (6, 32, 54). The fourth viral enzyme, NS5B, is an RNA-dependent RNA polymerase (RdRp), a key component responsible for replication of the viral RNA genome (7, 15, 36). The RdRp activity of NS5B was first experimentally demonstrated by Behrens et al. with NS5B protein expressed from a recombinant baculovirus and a synthetic nonviral RNA as a substrate (7). It was subsequently confirmed and further characterized through the use of the HCV RNA genome as a substrate (36, 37). Recent studies have shown that NS5B with a C-terminal 21-amino-acid truncation expressed in Escherichia coli is also active for in vitro RNA synthesis (17, 36, 67).
In an effort to further evaluate the intrinsic properties of the RdRp activity of HCV NS5B, a highly active NS5B protein was cloned, expressed, purified, and characterized in an in vitro RNA transcription assay with different RNA substrates. Results described in this study show that an 18-amino-acid truncation at the C terminus of NS5B did not affect the RdRp activity in vitro. In addition, Mn2+ stimulated the in vitro RdRp activity by a factor of 20-fold compared to that in the presence of Mg2+. Furthermore, several lines of evidence demonstrate that NS5B is able to initiate de novo RNA synthesis in vitro by using either a synthetic nonviral RNA or poly(C) and poly(U) RNA homopolymers as templates.
MATERIALS AND METHODS
Cells and viruses.
Sf21 cells were grown in Sf900 (GIBCO BRL, Gaithersburg, Md.) medium following a standard procedure (44). Recombinant baculoviruses were constructed either by cotransfection with a donor plasmid DNA and BaculoGold DNA (Pharmingen, San Diego, Calif.) or by using the FastBac system (GIBCO BRL) according to manufacturer's specification.
Plasmid construction.
Teresa L. Wright (University of California, San Francisco) kindly provided the serum of an HCV-infected patient. The HCV in the patient's serum was determined to be genotype 1b (T. Wright, personal communication). The viral RNA was directly extracted from the serum with TRIzol reagent (GIBCO BRL). To amplify the NS5B cDNA, a Titan one-step reverse transcription-PCR (RT-PCR) system was used (Boehringer Mannheim, Indianapolis, Ind.). Two oligonucleotides, universal/1b/5B (5′-GCGAATTCGGATCCATGTCAATGTCYTAYACNTGGACRGG-3′) and 1b/5B/SpeI (5′-CGCGCACTAGTTATCATCGGTTGGGGAGCAGGTA-3′), were used as primers. The RT-PCR DNA fragment was then digested with EcoRI and SpeI and inserted into an EcoRI/SpeI-digested pFastBac1 donor plasmid of the BAC-to-BAC baculovirus expression system (GIBCO BRL).
The VED mutant 1b-42 NS5B was created by changing a highly conserved GDD motif to VED in the putative enzyme active site with a specific oligonucleotide and the Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad, Hercules, Calif.). To convert the C-terminus-truncated 1b-42 to a full-length NS5B (designated 1b-42F), a 182-bp DNA fragment between the HindIII (nt 1598 of NS5B) and SpeI (pFastBac 1 vector) sites of the 1b-42 cDNA clone was replaced with the corresponding DNA fragment of the 1b-35 cDNA clone (Table 1). Therefore the stop codon was changed back to the tryptophan codon found in other 1b isolates (7).
TABLE 1.
RdRp activity and sequence comparison of different isolates of 1b NS5B
| Isolate | RdRp activityb | Amino acid differencesa |
|---|---|---|
| 1b-2 | − | V116I, L434P, A471T |
| 1b-6 | − | V116I, K124Q, Q151E, S218A, N231S, C324R, A499T |
| 1b-8 | ++ | Consensus |
| 1b-10 | + | L30S, L111P, V144A, Y276C |
| 1b-21 | + | L31V, R50G, A75G, K79R, K151E, F162L, G549S |
| 1b-29 | − | V116A, L474P, R508H |
| 1b-32 | + | D62N |
| 1b-35 | ++ | Consensus |
| 1b-42 | +++ | K523R, W574amSTOP |
| 1b-47 | ++ | K209N, V338A, A499T |
| BK | + | S46G, Q47L, Q49N, R50K, R98K, G113S, R117N, E131V, A185V, A207T, T213N, T300S, C316N, Q464E, A499V, K510R, R531K, Q544R, W574L |
The amino acid differences were based on the consensus sequences of the 10 clones compared with Sequencer software. For BK, the amino acid differences are to the published BK NS5B sequence (7).
Relative activity levels are indicated by the number of plus signs. −, no activity detected.
To generate D(+) (same polarity as D-RNA) and D(−) (opposite polarity of D-RNA) RNA transcripts, plasmid pRH405 was constructed by insertion of a DNA fragment between the EcoRI and BamHI sites of pT7DCoH (kindly provided by R. DeFrancesco) into a pGEM7Zf(+) vector (Promega, Madison, Wis.).
Preparation of RNA templates.
Poly(A), poly(I), poly(C), and poly(U) homopolymer templates and oligo(dT) primer were purchased from Pharmacia (Piscataway, N.J.). RNA oligomers [poly(A), poly(C), and poly(G)] are 12 nt long and were synthesized by National Biosciences Inc. (Plymouth, Minn.). The D-RNA, D(+) RNA, and D(−) RNA templates were synthesized with a MEGAscript RNA transcription kit (Ambion, Austin, Tex.). The D-RNA and D(+) RNA were transcribed from BamHI-linearized pT7DCoH DNA and pRH405 DNA, respectively, by in vitro transcription with T7 RNA polymerase. D(−) RNA was synthesized from pRH405 DNA digested with EcoRI by in vitro transcription with SP6 RNA polymerase. After extensive treatment with DNase I, all RNA transcripts were purified through phenol-chloroform extraction, ethanol precipitation, and centrifugation in a Chroma Spin 100 column (Clontech, Palo Alto, Calif.). For kinetic studies, RNA templates were further purified by electrophoresis in a 6% polyacrylamide–7 M urea gel. The RNA in the excised gel slice was eluted out by electrophoresis and collected by ethanol precipitation.
To block the 3′ OH group of the RNA, the D(+) RNA template was treated with cordycepin triphosphate (Sigma, St. Louis, Mo.) and poly(A) polymerase (Pharmacia). Briefly, 10 μg of D(+) RNA was incubated with 1 mM cordycepin triphosphate and 4 U of poly(A) polymerase in a 50-μl reaction mixture containing 40 mM Tris-HCl (pH 8), 10 mM MgCl2, 2.5 mM MnCl2, 250 mM NaCl, and 50 μg of bovine serum albumin/ml. After a 3-h incubation at 37°C, the reaction mixture was passed through a Sephadex G-50 spin column (Boehringer Mannheim) to remove the unincorporated cordycepin triphosphate. The RNA was then extracted with phenol-chloroform, ethanol precipitated, and resuspended in TE buffer (10 mM Tris-HCl and 1 mM EDTA).
Expression and purification of NS5B.
Sf21 cells at a density of 106 cells/ml were infected with recombinant baculoviruses at a multiplicity of infection of 5. After 2 to 3 days of incubation at 27°C, cells were harvested, collected by centrifugation, and washed with 1× phosphate-buffered saline (GIBCO BRL). The cell pellet was resuspended in buffer N (20 mM Tris-HCl [pH 7.6], 1 mM EDTA, 10 mM dithiothreitol [DTT], 50% glycerol, 1 mM phenylmethylsulfonyl fluoride). Seven milliliters of buffer N was used for approximately 108 cells. Triton X-100 and NaCl were added to the cell suspension to final concentrations of 2% and 500 mM, respectively. MgCl2 and DNase I were added to final concentrations of 10 mM and 15 μg/ml, respectively. The mixture was stirred at room temperature for 30 min. The cell extract was then transferred to tubes and centrifuged in a Beckman 70 Ti rotor at 40,000 rpm for 30 min at 4°C. The clarified cell lysate was diluted with 0.67 volume of LG buffer (20 mM Tris-HCl [pH 7.6], 1 mM EDTA, 2 mM DTT, 20% glycerol, 0.5% Triton X-100) and incubated with 5 ml of DEAE-Sepharose in LG buffer containing 300 mM NaCl at 4°C for 30 min. DEAE-Sepharose resin was then removed by filtration. The solution was further diluted with 1.5 volumes of LG buffer and then loaded onto a heparin-Sepharose column (10 ml) equilibrated in LG buffer containing 200 mM NaCl. The heparin column was washed with 5 column volumes of LG buffer containing 200 mM NaCl, and the bound proteins were eluted with a 100-ml gradient with 200 mM to 1 M NaCl in LG buffer. Fractions containing NS5B protein were detected by Western blotting with polyclonal antiserum raised against the NS5B protein expressed in E. coli. All fractions containing NS5B protein were combined and diluted with LG buffer to a conductivity equivalent to that of 150 mM NaCl. NS5B protein was further purified through a poly(U)-agarose column (2 ml) equilibrated in LG buffer containing 150 mM NaCl. The column was washed with 5 ml of LG buffer containing 150 mM NaCl and eluted with a 20-ml LG gradient buffer of 150 mM to 1 M NaCl. Fractions containing NS5B were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining. The combined fractions containing NS5B were concentrated by centrifugation in a Centricon Plus-20 device (Amicon/Millipore, Beverly, Mass.) and filtered through a Superdex 75 column (25 ml) or an SP Sepharose HiTrap column (Pharmacia). Finally, fractions containing NS5B protein were combined, aliquoted, and stored at −80°C. The concentration of NS5B protein was determined by fluorimager analysis with Sypro Orange staining (Molecular Probes, Eugene, Oreg.) and quantified with the Storm 860 imager (Molecular Dynamics, Sunnyvale, Calif.) by using known amounts of bovine serum albumin as controls.
In vitro RNA synthesis by NS5B.
RdRp reactions were performed in a 10-μl volume containing 20 mM HEPES (pH 8), 1.5 mM MnCl2, 100 mM ammonium acetate, 1 mM DTT, 500 μM GTP, 250 μM ATP and CTP, 10 μM UTP, 0.5 μCi of [α-33P]UTP (NEN, Boston, Mass.), 5 μg of RNA template/ml, 5 to 10 U of RNasin (Promega), and NS5B protein. The RdRp reactions with homopolymer RNA templates were carried out in a 10-μl volume containing 0.5 μCi of α-33P-labeled ribonucleotides and various concentrations of unlabeled ribonucleotides. The reaction mixtures were incubated at 30°C for 2 h. The kinetic analyses of 1b-42 NS5B was done in a 20-μl reaction volume containing 25 ng (10 nM) of D(+) RNA template, 51.2 ng (40 nM) of NS5B protein, 10 u of RNasin, 500 μM (each) ribonucleotides except the limiting ribonucleotide, either 10 μM UTP and 1 to 4 μCi of [α-33P]UTP for ATP, CTP, and GTP or 10 μM CTP and 1 μCi of [α-33P]CTP for UTP. The reaction mixture was incubated at 30°C for 15 min, and the reaction was stopped by addition of 100 mM EDTA. The RNA products were precipitated by addition of trichloroacetic acid (TCA) to 10% and incubated on ice for at least 30 min. The TCA-precipitated RNA was then collected by using a GF/B filter plate with a Filtermate 196 harvester (Packard, Meriden, Conn.) and quantified by a TopCount liquid scintillation counter (Packard). For PAGE analysis of the RNA products, a reaction with a 20-μl reaction mixture was performed except where indicated, followed by treatment with proteinase K, phenol-chloroform extraction, and ethanol precipitation. The RNA products were then analyzed by electrophoresis on a 6% denaturing polyacrylamide–7 M urea gel.
RPA.
The RdRp reaction was performed in a 600-μl volume with 1.68 μg of 1b-42 NS5B protein and 3 μg of D-RNA template. After a 2-h incubation at 30°C, the reaction mixture was treated with proteinase K, extracted with phenol-chloroform, and precipitated with ethanol. The RNA pellet was dissolved in 60 μl of TE buffer and treated with 100 U of RNase T1 (Ambion) in a 100-μl volume containing 50 mM NaCl at 37°C for 1 h. The RNase T1-treated RNA was further treated with proteinase K, extracted with phenol-chloroform, and precipitated with ethanol. An RNase protection assay (RPA) with Ambion's HybSpeed RPA kit was then performed according to the manufacturer's protocol. Briefly, the RNase T1-treated RNA products and 1 μg of either D-RNA, or D(+) RNA, or D(−) RNA were added together, denatured at 95°C for 3 min, and then hybridized at 68°C for 10 min in the HybSpeed hybridization buffer. The RNA hybrids were subsequently treated with RNase A/T1 at 37°C for 30 min. The reaction was stopped by addition of HybSpeed inactivation/precipitation mixture. The protected RNA products were analyzed by electrophoresis on a 6% polyacrylamide–7 M urea denaturing gel.
RNase H digestion.
The condition for RNA synthesis by 1b-42 NS5B was the same as above except that the RdRp reaction was done in a 200-μl volume containing 500 ng of gel-purified D(+) RNA template, 1.5 μg of 1b-42 NS5B enzyme, 500 μM (each) ATP, CTP and GTP, 10 μM unlabeled UTP, and 70 μCi of [α-32P]UTP (NEN) and the reaction mixture was incubated at 30°C for 2 h. After extraction with phenol-chloroform and precipitation with ethanol, half of the sample was used for direct RNase H digestion. The other half of the sample was mixed with fivefold-excess unlabeled D(−) RNA, denatured at 95°C for 5 min, and reannealed in a hybridization buffer (Ambion) at 42°C overnight. The RNAs were then precipitated with ethanol and resuspended in diethyl pyrocarbonate-treated water. For the RNase H assay, a 27-mer oligonucleotide designated oligo/DcOH (5′-CACCTTGAGCACCCACGAATGTGCCGG-3′) was used. The RNase H digestion of the RNA product was done in a 20-μl reaction volume containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT, 10 mM MgCl2, 50 mM KCl, 10 U of RNasin, 0.5 to 2.0 μg of oligo/DCoH, and 4 U of RNase H (GIBCO BRL), and the reaction mixture was incubated at 37°C for 1 h, as reported previously (7). The reaction mixture was then treated with 10 μg of protease K and 0.1% SDS. The RNA was extracted with phenol-chloroform, precipitated with ethanol, and analyzed in a 6% denaturing polyacrylamide–7 M urea gel. The radiolabeled RNA products were autoradiographed and quantified with a Storm PhosphorImager (Molecular Dynamics) (39).
RESULTS
Identification of active NS5B.
HCV NS5B protein is believed to function as a replicase responsible for viral RNA replication, as it has amino acid similarity to other RdRps (41). The RdRp activity of HCV NS5B has recently been demonstrated by an in vitro RNA transcription assay with either non-virus-specific RNA or genomic HCV RNA as templates (7, 36). However, the quasispecies nature of HCV argues for the existence of a mixed virus pool with divergent NS5B sequences that may result in variable RdRp activities. In an effort to isolate an NS5B protein with a high level of RdRp activity, a one-step RT-PCR method was used to directly amplify the NS5B gene from an HCV-infected patient's serum. The HCV from which the NS5B was derived was determined to be genotype 1b (T. Wright, personal communication). The PCR products were then inserted into a pFastBac1 vector, and recombinant baculoviruses containing NS5B cDNAs were generated by following the supplier's instructions (GIBCO BRL). Ten recombinant baculoviruses with independent cDNA clones of NS5B were isolated and used for expression of NS5B proteins. In order to determine RdRp activity, crude extracts of Sf21 cells infected with recombinant baculoviruses were prepared and used in an in vitro RNA transcription assay, as described by Behrens et al. (7). The RNA template used was the same D-RNA as that reported previously (7). Transcription of the D-RNA template by NS5B results in a characteristic hairpin dimer RNA product by a “copy-back” mechanism (7). The RdRp activities of the independent isolates of NS5B were determined by their abilities to produce RNA products, and the RNA products were quantified with a Storm PhosphorImager (39). As a result, one isolate of NS5B, designated 1b-42, was identified as the enzyme with the highest RdRp activity (Table 1). Nearly half of the isolates have no or little RdRp activity, and the remaining isolates have consistently lower activity than 1b-42 (Table 1). Subsequent sequence analysis revealed that the 1b-42 clone contains a nucleotide mutation resulting in the creation of a stop codon and an 18-amino-acid truncation at the C terminus. All the other independent NS5B isolates contain individual different amino acids, but no other deletion mutants were observed (Table 1).
Purification of 1b-42 NS5B.
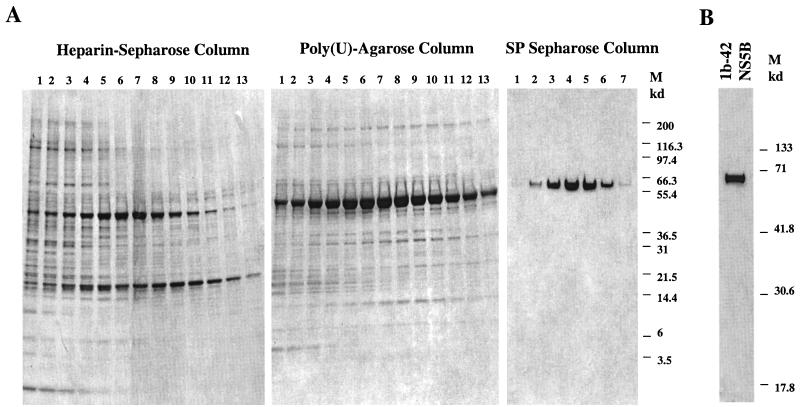
Since the 1b-42 isolate of NS5B had the highest RdRp activity, NS5B 1b-42 was pursued for further purification and characterization of its intrinsic enzymatic properties. In order to purify the 1b-42 NS5B protein, the cell extract derived from Sf21 insect cells infected with recombinant baculovirus was passed through three different types of chromatography columns: heparin-Sepharose, poly(U) Sepharose, and Superdex 75 or SP Sepharose HiTrap columns (Pharmacia). The detailed procedures for the chromatography steps are described in Materials and Methods. The fractions of eluted samples at each chromatography step were analyzed by SDS-PAGE (Novex, San Diego, Calif.). As shown in Fig. 1A, 1b-42 NS5B protein was eluted midway through the gradient of each chromatography step and separated from the majority of contaminants. After three chromatography steps, 1b-42 NS5B was purified to near homogeneity (>90%), as determined by the most sensitive silver staining procedure, SilverXpress (Novex) (Fig. 1B), and the purified enzyme was used in this study. In addition, the purified 1b-42 was examined for contamination of any RNases by incubating it with [α-32P]UTP-labeled D(+) RNA under the RdRp conditions. No RNase contaminants were detected in the purified 1b-42 NS5B (data not shown).
FIG. 1.
Purification of 1b-42 NS5B. (A) Purification of NS5B through three different types of chromatography columns. Fractions eluted out from each chromatography column were analyzed on 4 to 20% gradient SDS-PAGE gels (Novex), and the proteins were stained by Coomassie brilliant blue R250 (Sigma). The type of chromatography column and fraction numbers are indicated at the top. The protein molecular mass markers (right) are only shown in the gel for fractions from the SP Sepharose HiTrap column. (B) Examination of the purity of 1b-42 NS5B enzyme used in the study. Purified 1b-42 enzyme (2 μg) was analyzed in a 12% SDS–PAGE gel (Bio-Rad) and stained with SilverXpress silver staining kit (Novex). The gel was dried and scanned into a Photoshop program. The protein molecular mass markers are on the right.
Effects of monovalent and divalent ions on the RdRp activity of 1b-42 NS5B.
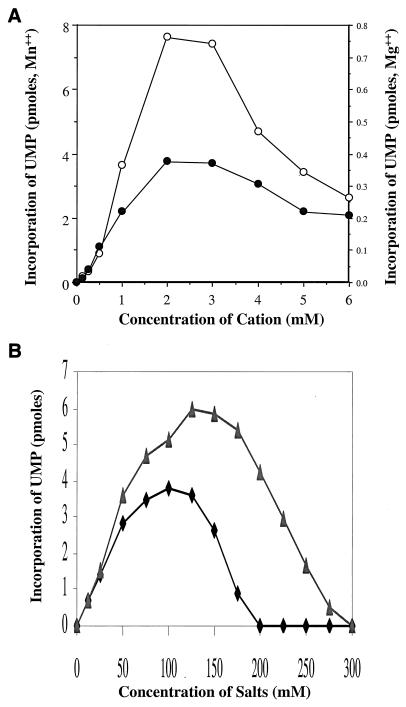
To optimize conditions for in vitro RdRp activity of 1b-42 NS5B, an initial effort was made to examine the effects of different divalent and monovalent ions on RdRp activity. In these experiments, RdRp activity was determined through measurement of the incorporation of [α-33P]UTP with D(+) RNA as a template. As shown in Fig. 2A, the RdRp activity of NS5B is about twenty times higher (7.6 pmol of UMP incorporated) in the presence of manganese (Mn2+) than in the presence of magnesium (Mg2+; 0.38 pmol of UMP incorporated) at 2 mM. In contrast, zinc, cobalt, nickel, and calcium did not support the RdRp activity of 1b-42 in this assay (data not shown). A manganese (Mn2+) concentration of 1.5 mM was used to examine a number of different monovalent salts (Na+, K+, Li+, and NH4+ in their chloridate and acetate forms) for their effects on RdRp activity. All monovalent salts tested here supported the RdRp activity with different ranges of optimal concentrations (data not shown). Results for two of these monovalent ions, KCl and ammonium acetate, are compared in Fig. 2B. Both KCl and ammonium acetate stimulated RdRp activity. However, ammonium acetate produced an approximately 50% greater and more-sustainable RdRp activity over a broader range of concentrations.
FIG. 2.
Effects of divalent and monovalent ions on RdRp activity of NS5B. 1b-42 NS5B (56 ng) was incubated with 25 ng of D(+) RNA template in the presence of 1 μCi of [α-33P]UTP, as described in Materials and Methods. The reaction mixture was incubated at 30°C for 2 h. The incorporation of [α-33P]UTP in picomoles was determined by TCA precipitation and scintillation counting. (A) Effect of Mg2+ and Mn2+ on the RdRp activity of 1b-42 NS5B. RdRp reactions were run at different concentrations of either MnCl2 (○) or MgCl2 (●). There is a 10-fold difference between the y-axis scale for Mn2+ and that for Mg2+. (B) Comparison of 1b-42 RdRp activity in the presence of KCl with that in the presence of ammonium acetate. A concentration of 1.5 mM MnCl2 was used, and concentrations of potassium chloride (⧫) and ammonium acetate (▴) were varied as indicated.
Biochemical characterization of 1b-42.
Specificity and fidelity of nucleotide incorporation is an intrinsic property possessed by all polymerases. To determine the nucleotide selectivity and template specificity of 1b-42, the misincorporation activity for each ribonucleotide was determined with either poly(A) or poly(dA) homopolymer as the template and oligo(dT) as the primer. Manganese is known to affect nucleotide specificity for some polymerases (55). Therefore, both manganese and magnesium were used as the divalent cation in the reaction. The results are summarized in Table 2. The misincorporation activity for the incorrect ribonucleotides is less than 0.1% for ATP and TTP in the presence of magnesium (Mg2+) compared to the correct incorporation of UTP. The misincorporation activities of 1b-42 for CTP and GTP were close to the background level. Like that of the D(+) RNA template (Fig. 2A), the incorporation activity on poly(A)-oligo(dT) by 1b-42 is also greater with Mn2+ than with Mg2+. The increased activity with Mn2+ is also reflected in the greater misincorporation of nontemplated nucleotides by 1b-42. Although Mn2+ may increase the misincorporation activity of 1b-42, it is clear that 1b-42 still retains a high level of nucleotide specificity for the templated nucleotide. When a poly(dA) homopolymer was used as the template and oligo(dT) was used as the primer, however, no significant incorporation of either UTP or TTP was detected (Table 2). These findings clearly indicate that 1b-42 NS5B does not have any activity related to reverse transcriptase, DNA-dependent RNA polymerase, and DNA polymerase, in agreement with the findings reported by Behrens et al. (7). Therefore 1b-42 NS5B is an RdRp. Furthermore, when the highly conserved GDD motif was mutated to VED in the active site of 1b-42 NS5B, no significant RdRp activity was detected (data not shown), demonstrating that incorporation of ribonucleotides was the result of specific RdRp activity associated with 1b-42 NS5B.
TABLE 2.
Misincorporation activity of nucleotides by 1b-42 NS5Ba
| Nucleotide | Substrate | Incorporation (pmol)
|
|
|---|---|---|---|
| Mn2+ | Mg2+ | ||
| UTP | poly(A)-oligo(dT) | 56.8 | 11.9 |
| ATP | poly(A)-oligo(dT) | 0.19 (0.33%) | 0.01 (0.08%) |
| CTP | poly(A)-oligo(dT) | 0.02 (0.035%) | <0.003 (<0.03%) |
| GTP | poly(A)-oligo(dT) | <0.01 (<0.01%) | <0.01 (<0.08%) |
| TTP | poly(A)-oligo(dT) | 0.02 (0.035%) | 0.006 (0.05%) |
| UTP | poly(dA)-oligo(dT) | 0.01 (0.02%) | <0.007 (<0.06%) |
| TTP | poly(dA)-oligo(dT) | <0.006 (<0.02%) | <0.006 (<0.05%) |
RdRp reactions were performed as described in Materials and Methods with 56 ng of 1b-42 NS5B, 5 μM concentrations of the indicated 33P-labeled nucleotides, 100 ng of poly(A) or poly(dA), and 10 ng of oligo(dT). Either 1.5 mM manganese chloride (Mn2+) or 5 mM magnesium chloride (Mg2+) was used in the reaction. Values were calculated from the counts per minute of the TCA-precipitated RNA. Each of the numbers in parentheses represents misincorporation activity compared to the correct incorporation of UTP expressed as a percentage.
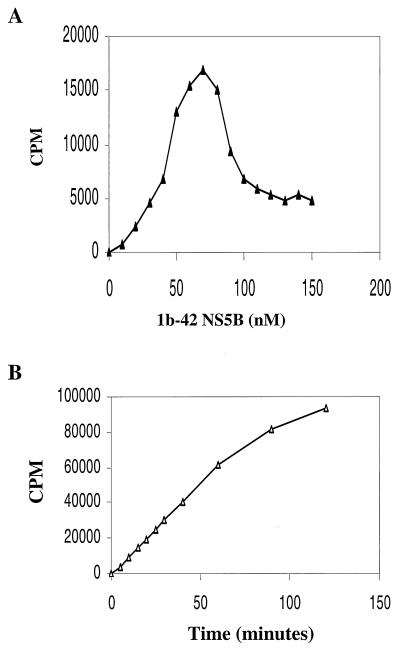
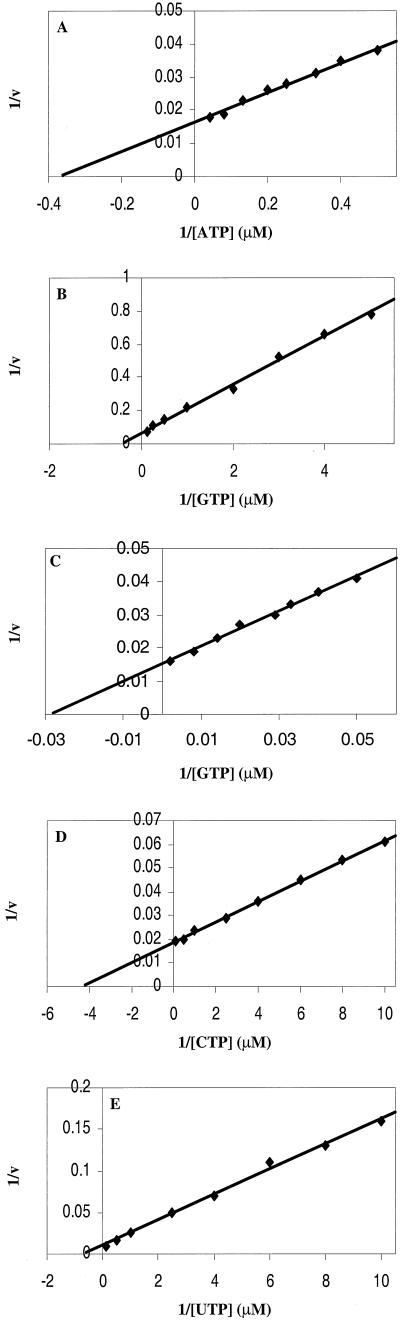
To determine the kinetic constants of the 1b-42 NS5B enzyme, initial efforts were made to optimize the enzyme concentration and to measure the initial velocity of the RdRp reaction of 1b-42 NS5B. Twenty-five nanograms (10 nM) of D(+) RNA template was incubated with increasing concentrations of 1b-42 NS5B enzyme. As shown in Fig. 3A, RNA products synthesized by 1b-42 NS5B increased with increasing amounts of NS5B enzyme up to 70 nM. Further increases of enzyme above 70 nM actually inhibited the RdRp reaction. Inhibition of the RdRp reaction was also found when a constant amount of 1b-42 enzyme and increasing amounts of D(+) RNA template were used (data not shown). The mechanism for substrate inhibition of RdRp transcription is not clear. It is possible that substrate inhibition is due to nonproductive substrate binding since it has recently been reported that NS5B has RNA-binding activity (25, 36). Therefore, the optimal ratio between template and enzyme is about 1 to 7 or less. Based on the enzyme titration curve (Fig. 3A), 40 nM 1b-42 NS5B was used to determine the time course of the RdRp reaction. As shown in Fig. 3B, the RNA products transcribed by 1b-42 NS5B are in linear proportion to the incubation time during the first 60 min. Based on the enzyme titration curve and the time course of the RdRp reaction, 25 ng (10 nM) of D(+) RNA, 51.2 ng (40 nM) of 1b-42 enzyme, and a 15-min incubation time were used for the kinetics analyses of ribonucleotides used by 1b-42 NS5B in an in vitro RNA transcription assay. Results derived from these experiments are presented as double-reciprocal plots (1/v versus 1/[S], where [S] is the nucleotide concentration and v is femtomoles per minute), as shown in Fig. 4. The calculated Km and kcat values are summarized in Table 3. ATP, CTP, and UTP have simple kinetics, whereas GTP has different kinetic constants at low and high concentrations, suggesting complex biphasic kinetics. The biphasic responses observed for GTP resulted in two Km values. The lower value is similar to the Km values for other ribonucleotides, which may reflect ribonucleotide utilization during elongation. The higher Km value for GTP may be the result of de novo initiation of RNA synthesis by 1b-42 NS5B. It has also been shown that Qβ replicase and brome mosaic virus (BMV) replicase display higher Kms for GTP than for other ribonucleotides, as they initiate RNA synthesis by specifically utilizing GTP (10, 29, 53).
FIG. 3.
Determination of substrate concentrations and initial velocity of the RdRp reaction. (A) Titration of 1b-42 NS5B enzyme. Twenty-five nanograms (10 nM) of D(+) RNA was incubated with increasing concentrations of 1b-42 NS5B enzyme in a 20-μl volume containing 500 μM (each) ATP, CTP, and GTP, 10 μM UTP, and 2 μCi of [α-33P]UTP at 30°C for 20 min. The reaction was terminated by addition of 100 mM EDTA. The labeled RNA product was precipitated with 10% TCA on ice for 30 min. The radiolabeled RNA was quantified with a TopCount liquid scintillation counter. The amount of 1b-42 enzyme was plotted against counts per minute. (B) Time course of the RdRp reaction. The assay is the same as that shown in panel A except that 25 ng (10 nM) of D(+) RNA was incubated with 51.2 ng (40 nM) of 1b-42 enzyme. The reaction was stopped by addition of EDTA to 100 mM at each time point (0, 5, 10, 15, 20, 25, 30, 40, 60, 90, and 120 min), and the reaction mixture was kept on ice. The incubation time was plotted against counts per minute.
FIG. 4.
Kinetic analyses of nucleoside triphosphate incorporation on the D(+) RNA template by 1b-42 NS5B. RdRp reactions were performed as described in Materials and Methods with 25 ng (10 nM) of D(+) RNA template, 51.2 ng (40 nM) of 1b-42 enzyme, and increasing concentrations of the limiting ribonucleotide. The incorporation of UMP or CMP was determined by TCA precipitation and liquid scintillation counting as described in Materials and Methods. The limiting ribonucleotide concentrations were as follows: 1.0, 1.25, 1.5, 2.0, 2.5, 3.0, 4.0, 5.0, 7.5, 12.5, 25, and 50 μM ATP (A); 0.14, 0.2, 0.25, 0.33, 0.5, 1.0, 2.0, 4.0, and 8.0 μM GTP (B); 20, 25, 30, 35, 50, 70, 125, and 500 μM GTP (C); 0.1, 0.125, 0.17, 0.25, 0.4, 1.0, 2.0, and 10 μM CTP (D); 0.1, 0.125, 0.167, 0.25, 0.4, 1.0, 2.0, and 10 μM UTP (E).
TABLE 3.
Kinetic constants of 1b-42 NS5B RdRp polymerasea
| Nucleotide | Km (μM) | kcat (min−1) |
|---|---|---|
| ATP | 2.34 ± 0.07 | 0.072 ± 0.001 |
| GTP | 1.85 ± 0.28 | 0.017 ± 0.002 |
| GTP | 38.93 ± 3.86 | 0.086 ± 0.003 |
| CTP | 0.24 ± 0.01 | 0.067 ± 0.001 |
| UTP | 1.51 ± 0.02 | 0.119 ± 0.011 |
Km and kcat values were derived from the slopes and y intercepts shown in Fig. 4. Numbers presented here are mean values (± standard deviation) derived from three independent experiments.
De novo initiation of RNA synthesis on D-RNA and D(+) RNA templates by 1b-42.
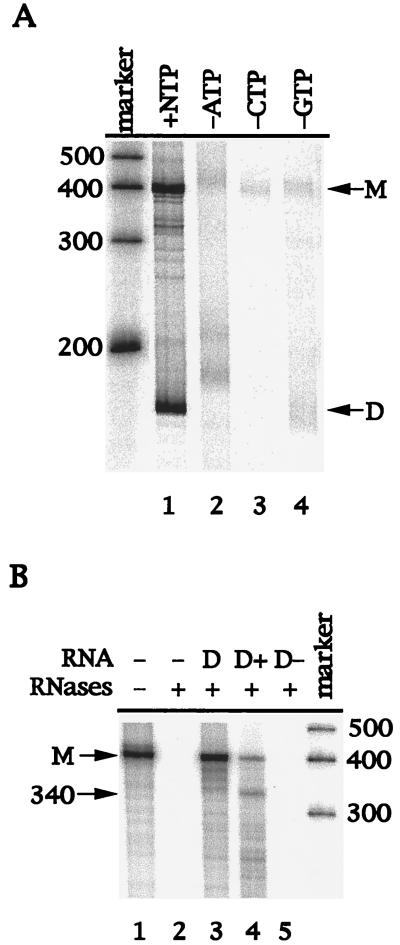
It has been previously reported by Behrens et al. that NS5B produced two different RNA products from the D-RNA template (7). The product identical in size to the input RNA template resulted from a terminal transferase activity associated with NS5B, while the other, faster-migrating product was determined to be a double-stranded hairpin dimer RNA synthesized by NS5B through a copy-back mechanism (7, 15). In this study, D-RNA was also used as a template for in vitro transcription by 1b-42 NS5B. The 1b-42 NS5B protein also synthesized two RNA products on the D-RNA template (Fig. 5A, lane 1). In contrast to the terminal transferase activity seen by Behrens et al. (7), however, synthesis of the RNA monomer by 1b-42 NS5B was dependent on the presence of all four ribonucleotides (Fig. 5A, lanes 2 to 4). Elimination of either ATP (lane 2), CTP (lane 3), or GTP (lane 4) dramatically diminished the synthesis of most monomer RNA and the self-primed hairpin dimer RNA (Fig. 5A). The function of the terminal transferase does not depend on the presence of all other ribonucleotides, and the terminal transferase could add any ribonucleotide to the RNA end, as reported previously (7). Therefore, results from this experiment suggest that synthesis of the RNA monomer is probably the result of de novo initiation by 1b-42 rather than the result of terminal transferase activity.
FIG. 5.
(A) Dependence of RNA synthesis by 1b-42 NS5B on ribonucleotides. The D-RNA template was incubated with 56 ng of 1b-42 NS5B protein in a 20-μl reaction mixture as described in Materials and Methods. The RNA products were analyzed by electrophoresis on a 6% polyacrylamide–7 M urea denaturing gel. The sizes of RNA markers are indicated on the left. M, 384-nt RNA monomer; D, hairpin dimer product. Lanes: marker, Ambion RNA Century marker; 1, presence of all four ribonucleotides; 2, absence of ATP; 3, absence of CTP; 4, absence of GTP. (B) Determination of the RNA polarity by RPA. The RPA was described in Materials and Methods. After the RdRp reaction, RNA products were treated with RNase T1. An aliquot of RNase T1-treated products (lane 1) was hybridized with either no RNA (lane 2), D-RNA (lane 3), D(+) RNA (lane 4), or D(−) RNA (lane 5). After hybridization, the RNA hybrids were further digested by RNases A and T1. The RNA products were precipitated and analyzed by electrophoresis on a 6% polyacrylamide–7 M urea denaturing gel. M, RNA monomer; 340, 340-nt protected fragment. Sizes of the RNA markers are on the right.
To confirm that the RNA monomer was synthesized by the RdRp activity of NS5B, an RNase protection assay was used to determine the polarity of the RNA product. If the RNA monomer product resulted from a terminal transferase activity, it would have the same polarity as the input RNA template. Therefore, RNA transcripts with the same [D-RNA and D(+) RNA] or opposite [D(−) RNA] polarity as the input D-RNA template were prepared and used in an RPA. After the RdRp reaction, the excess unhybridized D-RNA template was first removed by treatment with RNase T1. The RNase T1-treated RNA product (monomer) was then hybridized with either D-RNA (input template) or D(+) RNA or D(−) RNA. The RNA hybrids were subsequently treated with RNases A and T1. The unprotected [α-33P]UTP-labeled RNA product would be degraded by RNase digestion. The results are shown in Fig. 5B. As a control, the RNA monomer product without protection and RNase digestion was run in lane 1. The RNA product was completely degraded without the addition of the complementary RNA (lane 2). When D-RNA was used to hybridize with the RNA product, the labeled RNA product was protected from RNase digestion (lane 3), suggesting that the RNA product is complementary to the input RNA template. If the RNA product was protected by D(+) RNA, a 340-nt RNA fragment would be expected following RNase digestion, as D-RNA has a 44-nt sequence at the 5′ end which does not match the sequences of D(+) RNA. As expected, approximately 340-nt RNA was generated following RNase digestion when D(+) RNA was used in the RPA (lane 4). However, the labeled RNA product was not protected from RNase digestion after hybridization with D(−) RNA (lane 5). These findings demonstrate that the RNA product is of complementary polarity to the input RNA template, as a result of polymerization by the RdRp activity of NS5B.
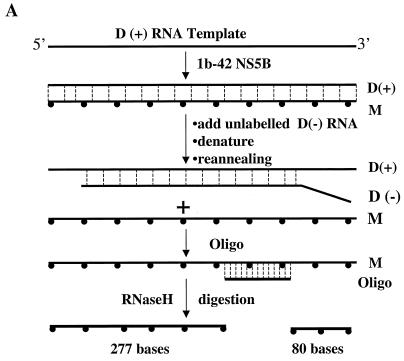
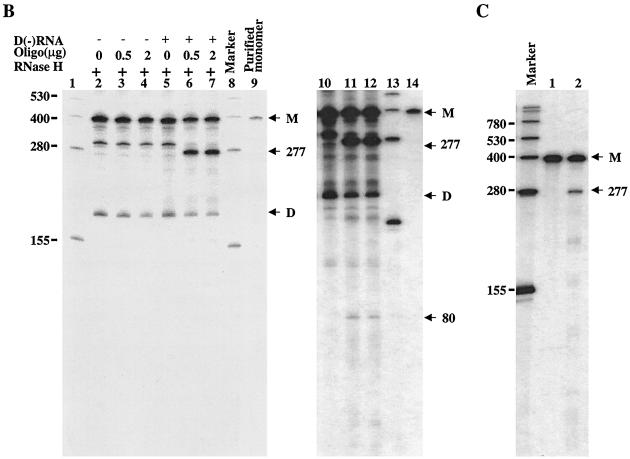
The RNA products used in the above RNase protection assay were first treated with RNase T1 to remove excess input RNA template. The question of whether the complementary RNA product to the input template was converted from the self-primed RNA dimer product as a result of RNase T1 digestion arose. To exclude this possibility and to further verify that the monomer RNA product was indeed synthesized de novo by NS5B enzyme, another independent RNase H digestion assay was developed. As shown in Fig. 6A, the RNA products synthesized by NS5B were incubated with an oligonucleotide complementary to the radiolabeled RNA products and RNase H. The RNase H cleaves the RNA at the region hybridized with the oligonucleotide. As a result, two RNA products with 277 and 80 nt are expected to be generated from the RNA monomer product following the RNase H digestion. As shown in lanes 2 to 4 of Fig. 6B, no products were generated by RNase H digestion. Similar results were obtained when several other denaturing conditions were used (data not shown). This suggests that the RNA duplex formed between the radiolabeled RNA monomer and the excess input RNA template is very tight. We then tried to compete the excess input D(+) RNA template by addition of fivefold-excess unlabeled D(−) RNA (Fig. 6A) prior to RNase H digestion. As described in Materials and Methods, half of the RdRp samples were first mixed with excess unlabeled D(−) RNA, denatured, reannealed, and then subjected to RNase H digestion. As shown in lanes 5 to 7 and 10 to 12 of Fig. 6B, two expected RNA products estimated to consist of 277 (lanes 6 and 7) and 80 nt (lanes 11 and 12) were generated (Fig. 6B). A shorter RNA product (about 300 nt) (lanes 2 to 5 of Fig. 6B) was cleaved even more efficiently by RNase H (lanes 6 and 7 of Fig. 6B). The shorter RNA is less stable and is more easily disassociated from the RNA duplex. Therefore, hybridization of a short RNA product to the oligonucleotide is more efficient. The RNA product shorter than a monomer was probably synthesized de novo by internal initiation of the NS5B enzyme. When the purified RNA monomer product was examined in this assay, it was found that the 277-nt RNA product was also generated (lane 2 of Fig. 6C). These results clearly demonstrate that the RNA monomer product was synthesized de novo by NS5B enzyme.
FIG. 6.
Analyses of RNA products by RNase H digestion. (A) Schematic presentation of the RNase H assay. After the RdRp reaction, RNA samples were mixed with a fivefold excess [to input D(+) RNA template] of unlabeled D(−) RNA, denatured, and reannealed. Hybridization between the input D(+) RNA template and the added in D(−) RNA frees [α-32P]UTP-labeled RNA product, which then hybridizes to the synthetic oligonucleotide and is cleaved by RNase H. Two cleaved RNA products, estimated at 277 and 80 nt, are expected if the monomer RNA product is digested by RNase H. (B) Analyses of RNase H-digested RNA products. RdRp and RNase H reactions were performed as described in Materials and Methods. Half of the sample was directly used for RNase H digestion in the presence of 0 (lane 2), 0.5 (lane 3), and 2.0 μg (lane 4) of oligo/DCoH. The other half was mixed with D(−) RNA, denatured, reannealed, and then digested with RNase H in the presence of 0 (lane 5), 0.5 (lane 6), and 2.0 μg (lane 7) of oligo/DCoH. Lanes 10 to 14 are longer exposures of lanes 5 to 9 to show the smaller digested band. The RNA size markers (left; GIBCO BRL) are shown in lanes 1, 8, and 13. The purified monomer RNA product was run in lane 9 (and is also shown in lane 14) (M). D, self-primed RNA dimer product. Two RNase H-cleaved RNA products with estimated lengths of 277 and 80 nt are indicated on the right. (C) Analyses of purified monomer RNA product by RNase H digestion. After the RdRp reaction, the [α-32P]UTP-labeled monomer RNA product was purified from a 6% polyacrylamide–7M urea gel, eluted out by electrophoresis, and collected by ethanol precipitation. The monomer RNA was then analyzed by RNase H digestion by the same strategy as that for panel A. The RNase H-digested monomer RNA was resolved in a 6% denaturing PAGE–7 M urea gel. The RNA size marker is shown in lane 1. M, monomer RNA product; 277, RNase H-cleaved RNA with 277 nt.
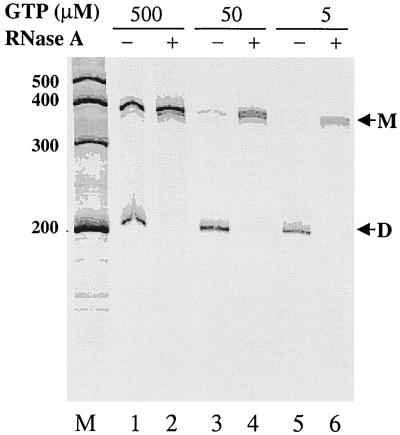
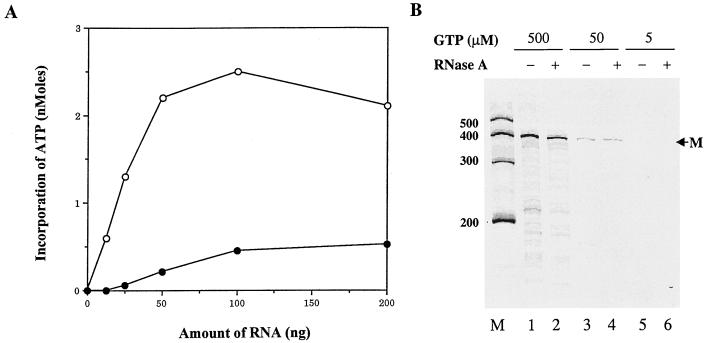
The dimer RNA product derived from the RdRp reaction was confirmed by RNase A digestion. As reported previously, the double-stranded RNA is resistant to RNase treatment and hairpin dimer RNA can be converted to a monomer by the action of RNase A under high-salt conditions (7). The results are shown in Fig. 7. Two RNA products were synthesized by the RdRp activity of NS5B in the presence of either 500 (lane 1), 50 (lane 3), or 5 μM (lane 5) GTP. The faster-migrating RNA was converted to monomer RNA following RNase A digestion (lanes 2, 4, and 6), confirming that the faster-migrating RNA is a hairpin dimer (7). However, synthesis of the RNA monomer product appeared to be proportional to increasing concentrations of GTP, whereas synthesis of the hairpin dimer RNA was not affected by concentrations of GTP. These results are consistent with an earlier suggestion that the higher Km value for GTP may represent the one for initiation and that the lower Km value reflects its Km for elongation (Fig. 4 and Table 3).
FIG. 7.
Analysis of the RNA hairpin dimer products by RNase digestion. The RdRp reactions were performed with D(+) RNA as the template based on the procedures described previously (7). After the RdRp reaction, half of the sample was treated with RNase A (3 μg/ml) in 500 mM NaCl for 2 h at room temperature (lanes 2, 4, and 6). RNA products were analyzed by electrophoresis on an 8% polyacrylamide–7 M urea denaturing gel. The sizes of the Ambion Century markers are on the left. RNA products: M, monomer; D, hairpin dimer.
To provide more evidence for de novo initiation of RNA synthesis by NS5B, the RNA template was modified by the addition of the chain terminator cordycepin (3′-dATP) at its 3′ end. This modification eliminates the self-priming products as well as the addition of ribonucleotides by the terminal transferase activity. To verify that the D(+) RNA was modified with cordycepin at its 3′ end, the cordycepin-modified D(+) RNA was incubated with [33P]ATP and poly(A) polymerase. Incorporation of [α-33P]ATP into unmodified and cordycepin-modified D(+) RNAs was measured. As shown in Fig. 8A, incorporation of [33P]ATP on cordycepin-modified D(+) RNA was reduced by approximately 90% compared to that on the unmodified D(+) RNA, suggesting that cordycepin treatment efficiently blocked the 3′ end of D(+) RNA. The cordycepin-terminated D(+) RNA was then used in the RdRp reaction. As shown in Fig. 8B, modification of the RNA template with cordycepin at its 3′ end dramatically reduced synthesis of the self-priming hairpin dimer RNA. However, synthesis of the RNA monomer product was not affected by cordycepin modification compared to synthesis of the unmodified RNA template (Fig. 7, lane 1). Although the background was increased by cordycepin treatment, it is not significantly different for untreated (lanes 1, 3, and 5) and RNase-treated RNA products (lanes 2, 4, and 6). In addition, synthesis of the RNA monomer product on the cordycepin-modified RNA template correlates with the increase of GTP concentrations, such as that of the unmodified RNA template (Fig. 7). These results strongly support the conclusion that NS5B can initiate de novo RNA synthesis in vitro.
FIG. 8.
RNA synthesis on the cordycepin-modified D(+) RNA template by 1b-42 NS5B. (A) Verification of cordycepin-terminated D(+) RNA by measuring the incorporation of [33P]ATP catalyzed by poly(A) polymerase. Cordycepin-treated or untreated D(+) RNA was incubated with [33P]ATP and poly(A) polymerase. The incorporation of [33P]ATP catalyzed by poly(A) polymerase was determined by TCA precipitation and scintillation counting. The indicated amounts of D(+) RNA (○) and cordycepin-terminated D(+) RNA (●) were plotted against incorporation of [33P]ATP. (B) RNA synthesis by 1b-42 NS5B with cordycepin-terminated D(+) RNA as the template. RdRp reactions were performed by procedures described previously (7). An aliquot of the reaction products was treated with RNase A (3 ng/μl) in 500 mM NaCl for 2 h at room temperature prior to electrophoresis (lanes 2, 4, and 6). The reaction products were analyzed by electrophoresis on an 8% polyacrylamide–7 M urea denaturing gel. M, position of the 384-nt RNA monomer. Sizes of Ambion RNA Century markers are on the left.
De novo initiation of RNA synthesis by 1b-42 NS5B on homopolymer RNA templates.
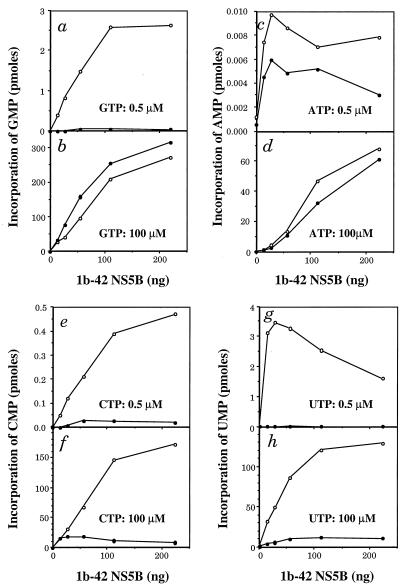
To further examine de novo initiation of RNA synthesis by NS5B, different RNA homopolymers were used as templates for 1b-42 transcription. As reported by others, transcription of homopolymer RNA templates by HCV NS5B is dependent on the presence of oligonucleotide primers (7, 17, 36, 37, 67). Also, higher concentrations of GTP are required for de novo initiation of RNA synthesis, as suggested by results derived from the D-RNA and D(+) RNA templates (Fig. 7 and 8) as well as by studies on Qβ replicase (10). Therefore, the RdRp reactions were performed at low (0.5 μM) and high (100 μM) concentrations of ribonucleotides in the presence or absence of oligonucleotide primers. The results are shown in Fig. 9. At low concentrations of GTP (0.5 μM), synthesis of RNA from poly(C) homopolymer is strongly dependent on the addition of the oligo(G) primer, as almost no RNA was synthesized from poly(C) template in the absence of the oligo(G) primer (Fig. 9a). However, when the concentration of GTP was increased to 100 μM, synthesis of RNA on the poly(C) template takes place in a primer-independent manner, as levels of RNA synthesis in the presence and absence of the oligo(G) primer are virtually identical (Fig. 9b). A similar result was also obtained with ATP incorporation on a poly(U) template (Fig. 9c and d). At a 100 μM concentration of ATP, synthesis of RNA on a poly(U) template without the oligo(A) primer is similar to that with the oligo(A) primer, suggesting that RNA synthesis is primer independent (Fig. 9d). At a 0.5 μM concentration of ATP, however, the RdRp activity of NS5B was extremely low so that no conclusion could be drawn (Fig. 9c). The lower activity of NS5B with the poly(U) template has also been observed by others (36). When both poly(I) (Fig. 9e and f) and poly(A) (Fig. 9g and h) homopolymers were used as templates, synthesis of RNA was found to be strictly dependent on the addition of oligo(C) and oligo(dT) primers, respectively. No RNA was synthesized in the absence of primers no matter whether low (Fig. 9e and g) or high (Fig. 9f and h) concentrations of ribonucleotides were used, suggesting that HCV NS5B may initiate RNA synthesis by using purine but not pyrimidine nucleotides (see Discussion). These results clearly show that NS5B can initiate RNA synthesis without any oligonucleotide primer at high concentrations of either GTP or ATP, confirming the de novo initiation of RNA synthesis by NS5B.
FIG. 9.
NS5B activity on homopolymer RNA templates with or without oligonucleotide primers. RdRp reactions were performed as described in Materials and Methods with 100 ng of homopolymer templates, 10 ng of oligoribonucleotide primer (10 to 1 ratio) (7, 17, 67), and the indicated amounts of 1b-42. [33P]GTP incorporation on poly(C) with (○) or without (●) the oligo(G) primer was determined at either 0.5 (a) or 100 μM GTP (b). [33P]ATP incorporation on poly(U) with (○) or without (●) oligo(A) primer was determined at either 0.5 (c) or 100 μM ATP (d). [33P]CTP incorporation on poly(I) with (○) or without (●) oligo(C) primer was determined at either 0.5 (e) or 100 μM CTP (f). [33P]UTP incorporation on poly(A) with (○) or without (●) oligo(dT) primer was determined at either 0.5 (g) or 100 μM UTP (h). Incorporation of ribonucleotide was determined by scintillation counting after TCA precipitation.
DISCUSSION
Several recent studies have experimentally verified that HCV NS5B is an RdRp (41) that catalyzes ribonucleotide polymerization on heteropolymer as well as homopolymer RNA templates in vitro (7, 17, 35–38, 67). In an attempt to further reveal the intrinsic properties of HCV NS5B, cDNA of NS5B was directly amplified from HCV viral RNA extracted from an HCV-infected patient's serum and used to construct recombinant baculoviruses. Recombinant NS5B proteins were subsequently purified from insect cells infected with recombinant baculoviruses, and RdRp activities were examined in vitro. These efforts have resulted in several active NS5B proteins with various levels of RdRp activity (Table 1). The identified amino acid differences among different isolates likely account for the inactivity or variable activities of NS5B proteins in catalyzing in vitro RNA synthesis (Table 1). However, efforts were not made in this study to determine the effects of the specific amino acid differences on RdRp activity. In addition, nearly half of the NS5B isolates cloned from a patient's serum were found to be inactive or weakly active in the in vitro RdRp assay. The low level of HCV replication in vivo may be associated with a large portion of inactive or weakly active NS5B proteins. These data also show that NS5B proteins with high RdRp activity exist in the virus pool and that they can be identified by examining multiple isolates. The 1b-42 described here possesses the highest RdRp activity among all isolates evaluated. It was therefore purified to >90% homogeneity (Fig. 1), and subjected to extensive characterization in a RdRp assay in vitro. Results derived from characterization of 1b-42 NS5B have led to several interesting findings.
It was found that Mn2+ stimulates the RdRp activity of 1b-42 by a factor of 20-fold compared to Mg2+. Both manganese (Mn2+) and magnesium (Mg2+) have been used previously as divalent metal ions to support the RdRp activity of HCV NS5B (17, 36). An earlier study showed that Mn2+ and Mg2+ stimulated the NS5B RdRp activity to similar degrees when poly(C)-oligo(G) was used as a substrate (36). Results derived from our study, however, revealed that Mn2+ produced 20-times-greater RdRp activity than Mg2+ with D(+) RNA as the substrate. These results confirmed a recent finding that a fourfold-higher RdRp activity was observed in the presence of Mn2+ than in the presence of Mg2+ (17). Mn2+ is known to affect the activity of polymerases in a number of different ways. It lowers the Km for the template and increases the processivity of DNA polymerase β (63). Also, it is required by the poly(A) polymerase of vaccinia virus (43). In addition, it has been reported to enhance the initiation of RNA synthesis by BMV RdRp (53). Furthermore, it was found that Mn2+ increased the misincorporation activity of DNA polymerases (55) and that it can change the substrate specificity of E. coli DNA polymerase I (61). Both the template specificity and the nucleotide selectivity of 1b-42 NS5B are retained under conditions used in our study. However, the mechanism by which Mn2+ stimulates the RdRp activity of HCV NS5B remains to be determined.
It has been recently reported that NS5B with a 21-amino-acid deletion at the C terminus is as active as the full-length protein in catalyzing ribonucleotide polymerization (17, 67). Even C-terminal deletions of up to 63 amino acids do not seem to impair the in vitro RdRp activity (17, 36). The 1b-42 NS5B used in our study has an 18-amino-acid truncation at the C terminus caused by a newly created stop codon (Table 1). Therefore, we also examined if the C-terminal 18 amino acids of NS5B have any effects on in vitro RdRp activity under the conditions used in this study. A full-length NS5B, designated 1b-42F, was restored, expressed, and purified to >80% homogeneity (data not shown). The RdRp activity of 1b-42F was then compared to that of 1b-42 with D(+) RNA as a template. It was found that the C-terminal 18-amino-acid truncation did not affect the in vitro RdRp activity of NS5B (data not shown). Although the C-terminal portion of NS5B appeared to be dispensable for in vitro RdRp activity, its function(s) in vivo remains to be determined. The hydropathy profile of NS5B revealed a highly hydrophobic domain consisting of 21 amino acids at the C terminus (17, 67). A membrane-anchoring domain was also found in this region (24, 67). Deletion analysis of NS5B confirmed the hydrophobic nature of the C-terminal 21 amino acids, as the truncated protein became much more soluble when expressed in E. coli (17, 67). Expression of the truncated NS5B protein in mammalian cells revealed that the C-terminal truncation altered the localization pattern of the NS5B protein (67). The full-length NS5B was found to localize exclusively at the perinuclear membrane (24), whereas the truncated NS5B localized to both the nucleus and cytoplasm (67). Therefore, the C-terminal anchoring domain is probably responsible for association with the perinuclear membrane. Membrane association may be important for formation of the replication complex. Moreover, a recent study suggests that the C-terminal hydrophobic domain also interacts with the NS5A protein (67). These findings suggest that the C-terminal portion of NS5B, while not essential for in vitro RdRp activity, may play an important role in the replication of the viral RNA genome in vivo.
Finally, our results demonstrated that NS5B not only catalyzed the elongation of RNA synthesis by either self-priming to copy back the RNA template or by extending the existing primer, but also initiated RNA synthesis de novo. De novo initiation of RNA synthesis has been reported for other viral RNA polymerases (10, 30, 53), including the recombinant RdRp of bovine viral diarrhea virus (BVDV), also a member of the Flaviviridae family. However, de novo initiation of RNA synthesis has not been previously demonstrated for HCV NS5B. Several studies have shown that HCV NS5B can catalyze the elongation of an RNA or a DNA primer by using RNA homopolymer templates (17, 36, 67). Primer-independent RNA synthesis by NS5B was also reported but was determined to be the result of self-priming or copy-back of the RNA template (7, 36).
Several lines of evidence described in this study demonstrate that HCV NS5B has an intrinsic ability to initiate de novo RNA synthesis in vitro. When previously used D-RNA was tested in the RdRp assay (7, 15), two different RNA products were generated by 1b-42 NS5B (Fig. 5 and 7). Consistent with previous findings, the faster-migrating RNA product was determined to be a hairpin dimer RNA, as it could be converted to a monomer by RNase digestion under high-salt conditions (Fig. 7) (7). The hairpin dimer RNA product was likely synthesized by a self-priming or a copy-back mechanism, as suggested previously (7). Contrary to the previous findings (7), however, the RNA monomer product was transcribed by 1b-42 NS5B from an RNA template through a de novo initiation mechanism rather than by terminal transferase activity (7). This conclusion is strongly supported by substantial evidence derived from several independent assays. First, the appearance of the RNA monomer is strictly dependent on the existence of all four ribonucleotides (Fig. 5A), in contrast to terminal transferase activity, where only one ribonucleotide is needed. Elimination of any ribonucleotide drastically diminished the synthesis of the monomer RNA product as well as the self-primed hairpin dimer. Second, the RNase protection assay demonstrated that the RNA monomer product was complementary to the input RNA template (Fig. 5B). The RNA product would be expected to be of the same polarity as the RNA template if catalyzed by terminal transferase activity. Third, the RNase H digestion experiment demonstrated that the monomer RNA product was synthesized de novo by 1b-42 NS5B, as two expected RNA products were generated from the monomer RNA product following RNase H digestion. The monomer RNA product could not be converted from the self-primed RNA dimer, as no other RNases were used in the RNase H assay and no detectable RNase contaminants were associated with the purified 1b-42 enzyme (data not shown). Finally, blocking the 3′ OH group of the RNA template with cordycepin did not affect the synthesis of the RNA monomer by 1b-42 NS5B, whereas it nearly eliminated the synthesis of the hairpin dimer product (Fig. 8B). These results clearly demonstrated that the RNA monomer product was polymerized by de novo initiation. Moreover, incorporation of GTP or ATP on the poly(C) and poly(U) homopolymer templates, respectively, in the absence of oligo(G) and oligo(A) primers is nearly identical to that in the presence of oligomer primers when high concentrations (100 μM) of GTP and ATP were present (Fig. 9b and d). The primer-independent RNA synthesis on poly(C) and poly(U) templates is suggestive of de novo initiation. However, ribonucleotide polymerization with poly(C) and poly(U) homopolymer templates is primer dependent at low concentrations (0.5 μM) of GTP and ATP, respectively, suggesting that higher concentrations of GTP and ATP are required for de novo initiation of RNA synthesis in vitro (Fig. 9a and c). The finding that higher concentrations of GTP correlate with increased synthesis of the RNA monomer product (Fig. 7 and 8B) further supports this possibility. However, incorporations of CTP and UTP on poly(I) and poly(A) homopolymer templates, respectively, are strongly dependent on the existence of oligo(C) and oligo(dT) primers, regardless of the concentration of ribonucleotide (Fig. 9e to h). These results indicate that HCV NS5B can probably only use GTP and ATP for de novo initiation of RNA synthesis. Furthermore, findings derived from kinetics analyses of 1b-42 NS5B also support the de novo initiation of RNA synthesis by NS5B (Fig. 4 and Table 3). The biphasic kinetics derived from GTP utilization resulted in two different kinetic values (Table 3). The higher Km values might result from the binding sites for GTP, which might resemble initiator sites of E. coli RNA polymerase (48) and BVDV NS5B (30). The inability to detect biphasic kinetics with ATP may be due to maximal de novo initiation already occurring with the saturating concentration of GTP (500 μM) used in the kinetics experiments.
De novo initiation of RNA synthesis appears to be an intrinsic property of HCV NS5B, as NS5B proteins derived from BK strain (7), 1b-47 (Table 1), and genotype 1a (unpublished data) are all capable of synthesizing RNA on a poly(C) RNA homopolymer template in the absence of the oligo(G) primer at a high GTP concentration (100 μM) (unpublished results). This may provide an explanation of why de novo initiation by NS5B was not previously demonstrated (7, 36). It is also possible that the terminal transferase activity present in their NS5B preparations might mask the de novo initiation activity (7, 36). NS5B proteins used in this study (Fig. 5) do not have significant terminal transferase activity. The requirement of high concentrations of GTP is probably critical for de novo initiation of RNA synthesis by HCV NS5B in vitro, and the concentrations of ribonucleotides used by others might not be high enough for de novo initiation (36). A recent study found that high concentrations of GTP stimulated in vitro RNA synthesis up to 100-fold (38). Nevertheless, the de novo initiation of RNA synthesis by NS5B in vitro is likely the mechanism used by HCV for viral replication in vivo. Ribavirin in combination with interferon is currently used as a therapy for hepatitis C, although the mechanism of inhibition is not known (11). However, ribavirin has been reported to deplete the GTP pool inside cells (42). It would be interesting to determine if depletion of GTP by ribavirin is responsible for inhibition of the initiation of RNA synthesis.
Initiation of nucleotide polymerization is the most complex process of a polymerase cycle. Polymerases use a variety of strategies for initiation, including protein priming, oligonucleotide extension, and de novo initiation. The protein priming mechanism is used by DNA polymerase of adenovirus (49), the reverse transcriptase of hepadnaviruses (3, 62), and the RNA-dependent RNA polymerase of poliovirus (VPg protein) (45). For retroviruses, synthesis of viral DNA is initiated by extending either the tRNA primer for the minus-strand DNA (58) or oligoribonucleotides of the viral RNA genome generated by RNase H activity for the plus-strand DNA syntheses (52). De novo initiation with ATP or GTP has been documented for DNA-dependent RNA polymerases (28), primases (35), Qβ replicase (10), BMV replicase (29, 53), and more recently for BVDV NS5B (30). HCV NS5B appears to be similar to the RdRps of BMV, Qβ virus, and BVDV in its ability to initiate de novo RNA synthesis on single-stranded RNA templates. However, the RdRp activities of BMV and Qβ virus initiate RNA synthesis in a replicase complex associated with other proteins (1, 8, 46), which may account for the high degree of template specificity (9, 21). Although NS5B could initiate RNA synthesis de novo in the absence of other viral proteins, it does not discriminate against nonviral RNA templates in vitro. Therefore, determination of other factors (e.g., viral or cellular proteins and cis signal sequences) conferring the specificity of NS5B for viral RNA replication in vivo will greatly help in understanding HCV replication. Sequence analyses revealed that both the 5′ UTR and 3′ UTR are highly conserved, and it has been speculated that they contain the signal sequences required for viral RNA replication (12, 13, 33, 57, 66). It has also been found that NS5B interacts with other viral proteins (67). Recently, at least two cellular proteins, PTB and p35, were found to specifically interact with the HCV 3′ UTR (26, 39, 60). Whether these proteins or other viral and cellular proteins and cis elements are involved in conferring the specificity of NS5B for HCV replication in vivo remains to be determined. Determination of the replicase complex for HCV replication will inevitably provide a unique anti-HCV target(s) for drug discovery.
ACKNOWLEDGMENTS
G.L. and R.K.H. are co-first authors.
We thank Teresa Wright for kindly providing us with the HCV-infected patient's serum and R. De Francesco (Italy) for plasmid pT7DCoH. We are grateful to Jin-Hua Sun, Min Gao, David Standring, Brian Terry, Moneesh Chatterjee, and Al Torri for stimulating discussions and to Mark Krystal and Al Torri for critical reading of the manuscript.
REFERENCES
- 1.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J, Mast E E. The epidemiology of viral hepatitis in the United States. Gastroenterol Clin N Am. 1994;23:437–455. [PubMed] [Google Scholar]
- 3.Bartenschlager R, Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 1988;7:4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analysis of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartenschlager R. Molecular targets in inhibition of hepatitis C virus replication. Antivir Chem Chemother. 1997;8:281–301. [Google Scholar]
- 7.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenthal T. Qβ RNA replicase and protein synthesis elongation factors EF-Tu and EF-Ts. Methods Enzymol. 1979;60:628–638. doi: 10.1016/s0076-6879(79)60059-9. [DOI] [PubMed] [Google Scholar]
- 9.Blumenthal T, Carmichael G G. RNA replication: function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 10.Blumenthal T. Qβ replicase template specificity: different templates require different GTP concentrations for initiation. Proc Natl Acad Sci USA. 1980;77:2601–2605. doi: 10.1073/pnas.77.5.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brillanti S, Garson J, Foli M, Whitby K, Deaville R, Masci C, Miglioli M, Barbara L. A pilot study of combination therapy with ribavirin plus interferon alfa for interferon alfa-resistant chronic hepatitis C. Gastroenterology. 1994;107:812–817. doi: 10.1016/0016-5085(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 12.Brown E A, Zhang H, Ping L-H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukh J, Purcell R H, Miller R H. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choo Q-L, Kuo G, Weiner A J, Oberby L R, Bradley S W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 15.De Francesco R, Behrens S-E, Tomei L, Altamura S, Jiricny J. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 1996;275:58–67. doi: 10.1016/s0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- 16.Failla C, Tomei L, De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrari E, Wright-Minogue J, Fang J W S, Baroudy B M, Lau J Y N, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francki R I, Fauquet C M, Knudson D L, Brown F. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Arch Virol. 1991;S2:223–233. [Google Scholar]
- 19.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haruna I, Spiegelman S. Specific template requirement of RNA replicases. Proc Natl Acad Sci USA. 1965;54:579–587. doi: 10.1073/pnas.54.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1035–1058. [Google Scholar]
- 24.Hwang S B, Park K-J, Kim Y-S, Sung Y C, Lai M M C. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology. 1997;227:439–446. doi: 10.1006/viro.1996.8357. [DOI] [PubMed] [Google Scholar]
- 25.Ishii K, Tanaka Y, Yap C C, Aizaki H, Matsuura Y, Miyamura T. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology. 1999;29:1227–1235. doi: 10.1002/hep.510290448. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T, Tahara S M, Lai M M C. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen S E, Buch L B, Nierlich D P. Nucleoside triphosphate termini from RNA synthesized in vitro by Escherichia coli. Science. 1969;164:1067–1070. doi: 10.1126/science.164.3883.1067. [DOI] [PubMed] [Google Scholar]
- 29.Kao C C, Sun J-H. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao C C, Del Vecchio A M, Zhong W. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 31.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 33.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 35.Kornberg A, Baker T. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman; 1992. pp. 275–297. [Google Scholar]
- 36.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann V, Roos A, Korner F, Koch J O, Bartenschlager R. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology. 1998;249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann V, Overton H, Bartenschlager R. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J Biol Chem. 1999;274:10807–10815. doi: 10.1074/jbc.274.16.10807. [DOI] [PubMed] [Google Scholar]
- 39.Luo G-X. Cellular proteins bind to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology. 1999;256:105–118. doi: 10.1006/viro.1999.9639. [DOI] [PubMed] [Google Scholar]
- 40.Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yosida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- 41.Miller R H, Purcell R H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller W E G, Maidhof A, Taschner H, Zahn R K. Virazole (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide): a cytostatic agent. Biochem Pharmacol. 1977;26:1071–1075. doi: 10.1016/0006-2952(77)90246-5. [DOI] [PubMed] [Google Scholar]
- 43.Nevins J R, Joklik W K. Isolation and partial characterization of the poly(A) polymerases from Hela cells infected with vaccinia virus. J Biol Chem. 1977;252:6939–6947. [PubMed] [Google Scholar]
- 44.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 45.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 46.Quadt R, Jaspars E M J. Purification and characterization of brome mosaic virus RNA-dependent RNA polymerase. Virology. 1990;178:189–194. doi: 10.1016/0042-6822(90)90393-6. [DOI] [PubMed] [Google Scholar]
- 47.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 48.Reddy P S, Chatterji D. Evidence for a pyrimidine-nucleotide-specific initiation site (the i site) on Escherichia Coli RNA polymerase. Eur J Biochem. 1994;225:737–745. doi: 10.1111/j.1432-1033.1994.00737.x. [DOI] [PubMed] [Google Scholar]
- 49.Rekosh D M K, Russell W C, Bellet A J D, Robinson A J. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11:283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- 50.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 51.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 52.Smith J K, Cywinski A, Taylor J M. Initiation of plus-strand DNA synthesis during reverse transcription of an avian retrovirus genome. J Virol. 1984;49:200–204. doi: 10.1128/jvi.49.1.200-204.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun J-H, Adkins S, Faurote G, Kao C C. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 54.Suzich J A, Tamura J K, Palmer-Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabor S, Richardson C C. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor J M, Hsu T W. Reverse transcription of avian sarcoma virus RNA into DNA might involve copying of the tRNA primer. J Virol. 1980;33:531–534. doi: 10.1128/jvi.33.1.531-534.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van de Sande J H, Loewen P C, Khorana H G. Studies on polynucleotides: A further study of ribonucleotide incorporation into deoxyribonucleic acid chains by deoxyribonucleic acid polymerase I of Escherichia coli. J Biol Chem. 1972;247:6140–6148. [PubMed] [Google Scholar]
- 62.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 63.Wang T S-F, Korn D. Specificity of the catalytic interaction of human DNA polymerase β with nucleic acid substrates. Biochemistry. 1982;21:1597–1608. doi: 10.1021/bi00536a021. [DOI] [PubMed] [Google Scholar]
- 64.Wimmer E, Goldbach R. Viral genetics. Curr Opin Genet Dev. 1992;2:59–60. [Google Scholar]
- 65.World Health Organization. WHO concerns on hepatitis C. Lancet. 1998;351:1415. [Google Scholar]
- 66.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimorura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 67.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 68.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]