Abstract
Insulin/IGF signaling (IIS) regulates developmental and metabolic plasticity. Conditional regulation of insulin-like peptide expression and secretion promotes different phenotypes in different environments. However, IIS can also be regulated by other, less-understood mechanisms. For example, stability of the only known insulin/IGF receptor in C. elegans, DAF-2/INSR, is regulated by CHIP-dependent ubiquitination. Disruption of chn-1/CHIP reduces longevity in C. elegans by increasing DAF-2/INSR abundance and IIS activity in adults. Likewise, mutation of a ubiquitination site causes daf-2(gk390525) to display gain-of-function phenotypes in adults. However, we show that this allele displays loss-of-function phenotypes in larvae, and that its effect on IIS activity transitions from negative to positive during development. In contrast, the allele acts like a gain-of-function in larvae cultured at high temperature, inhibiting temperature-dependent dauer formation. Disruption of chn-1/CHIP causes an increase in IIS activity in starved L1 larvae, unlike daf-2(gk390525). CHN-1/CHIP ubiquitinates DAF-2/INSR at multiple sites. These results suggest that the sites that are functionally relevant to negative regulation of IIS vary in larvae and adults, at different temperatures, and in nutrient-dependent fashion, revealing additional layers of IIS regulation.
Keywords: C. elegans, DAF-2, insulin, IGF, ubiquitin, CHIP, starvation, L1 arrest
ARTICLE SUMMARY
Insulin-like signaling plays a critical role in helping animals adapt to different environmental conditions. Differences in abundance of insulin molecules drive differences in insulin signaling, affecting growth, metabolism, and resistance to stressful conditions. Previous work in the roundworm C. elegans showed that targeted degradation of the insulin receptor also regulates insulin signaling. We show here that this process is affected by developmental stage, nutrient availability, and temperature, revealing additional ways that insulin-like signaling is regulated in this valuable animal model.
INTRODUCTION
IIS regulates growth, development, metabolism, stress resistance, and aging in metazoans. In the nematode C. elegans, the sole known insulin/IGF receptor is encoded by daf-2/INSR (Murphy and Hu 2013). DAF-2/INSR signals through a conserved phosphoinositide 3-kinase (PI3K) pathway including AGE-1/PI3K, PDK-1, AKT-1, and AKT-2 to govern nuclear localization and activity of the transcription factor DAF-16/FOXO.
When worms hatch in the absence of food, they remain in a developmentally arrested state in the first larval stage known as L1 arrest (or L1 diapause) (Baugh 2013). Extended starvation during L1 arrest causes developmental abnormalities of the gonad, including germline tumors (Jordan et al. 2019). Worms also arrest development as dauer larvae in the third larval stage in response to adverse environmental conditions including high population density, limited nutrient availability, and high temperature (Hu 2007). IIS regulates L1 arrest and dauer formation, with daf-2/INSR mutants displaying constitutive arrest phenotypes as L1 and dauer larvae (Gems et al. 1998; Baugh and Sternberg 2006) as well as increased starvation resistance, including increased survival of L1 arrest (Muñoz and Riddle 2003; Baugh and Sternberg 2006) and suppression of starvation-induced developmental abnormalities (Jordan et al. 2019). IIS also regulates adult physiology, and disruption of daf-2/INSR substantially increases lifespan and suppresses vitellogenesis (Cynthia Kenyon et al. 1993; Murphy et al. 2003; Depina et al. 2011).
Ubiquitin is a small peptide that is covalently attached to proteins. Ubiquitination often targets proteins for degradation, but it can regulate protein function in other ways as well (Kipreos 2005; Zheng and Shabek 2017). The quality-control E3 ubiquitin ligase CHIP mono-ubiquitinates worm, fly, and human INSR, with DAF-2/INSR being ubiquitinated at multiple sites (Tawo et al. 2017). DAF-2/INSR stability is regulated by the sole worm ortholog of CHIP, encoded by gene chn-1. Disruption of chn-1/CHIP increases DAF-2/INSR abundance in adults, increasing IIS activity and reducing lifespan (Tawo et al. 2017). daf-2(gk390525) is a point mutation resulting in a K1614E amino acid substitution, affecting one of the lysine residues targeted by CHN-1/CHIP-dependent ubiquitination (Tawo et al. 2017). This mutant has reduced lifespan (Tawo et al. 2017; Zhao et al. 2021) and increased vitellogenesis (Kern et al. 2021), consistent with increased IIS activity, causing it to be described as a gain-of-function allele. Although the effects of chn-1/CHIP and DAF-2/INSR ubiquitination have been documented in adults, it is unknown if or how ubiquitination affects IIS during larval development or in conditions where there are large differences in IIS, such as in fed vs. starved animals.
We characterized daf-2(gk390525) and chn-1(by155) phenotypes in L1 arrest and recovery, dauer formation, and in adults. We complemented phenotypic analysis with quantification of DAF-16/FOXO sub-cellular localization as a proxy for IIS activity. We confirm published results showing that this allele causes increased IIS activity in adults. We also show that it increases IIS activity during temperature-dependent dauer formation. However, we demonstrate that it reduces IIS activity during larval development and L1 arrest, behaving like a loss-of-function allele. In contrast to daf-2(gk390525), disruption of chn-1/CHIP increases IIS during L1 arrest, suggesting regulation of DAF-2/INSR through ubiquitination of one or more amino acid residues other than K1614. These results demonstrate that chn-1/CHIP and ubiquitin-dependent regulation of DAF-2/INSR varies during development and in different conditions. They also highlight that daf-2(gk390525) has complex effects on IIS, warranting caution in interpretation of phenotypic analysis.
RESULTS AND DISCUSSION
daf-2(gk390525) behaves as a gain-of-function allele in adults
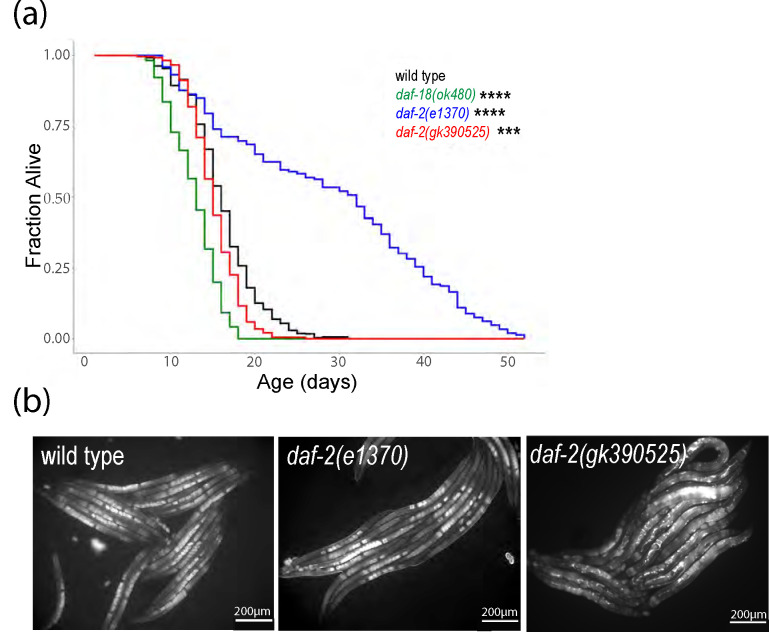
We assayed lifespan for daf-2(gk390525), a reportedly gain-of-function allele (Tawo et al. 2017; Kern et al. 2021; Zhao et al. 2021), daf-2(e1370), a class 2 loss-of-function allele (Gems et al. 1998), and daf-18(ok480), a null allele of a negative regulator (daf-18/PTEN) of AGE-1/PI3K and thus IIS (Ogg and Ruvkun 1998). As expected, daf-2(e1370) had a significant increase in lifespan relative to the wild-type (N2) control, and daf-18(ok480) had a significant decrease (Fig. 1a). daf-2(gk390525) had a relatively small, albeit significant, decrease in lifespan. These results confirm the published gain-of-function behavior of daf-2(gk390525) with respect to lifespan (Tawo et al. 2017; Zhao et al. 2021).
Figure 1.
daf-2(gk390525) mutants demonstrate documented IIS gain-of-function phenotypes in adults. A) Adult lifespan was scored daily for three or five biological replicates, and pooled results are presented. See Table S1 for complete results. *** P < 0.001, **** P < 0.0001; log-rank test. B) Representative images of VIT-2::GFP expression were taken in the indicated genetic backgrounds at 200x total magnification. Well-fed animals were imaged during early adulthood, when a single row of embryos was visible in the uterus. To ensure matching stages, wild type and daf-2(e1370) were imaged after 84 hours recovery from L1 arrest, and daf-2(gk390525) was imaged after 72 hours.
DAF-16/FOXO antagonizes vitellogenesis (Murphy et al. 2003; Depina et al. 2011), and IIS promotes vitellogenesis and yolk venting (Kern et al. 2021). We imaged expression of a multi-copy VIT-2 reporter gene in daf-2(e1370) and daf-2(gk390525) gravid adults. VIT-2 and other vitellogenin proteins are synthesized in the intestine and secreted into the body cavity, and oocytes are provisioned through receptor-mediated endocytosis of vitellogenin lipoprotein particles (Perez and Lehner 2019). VIT-2::GFP expression appeared lower in the intestine and body cavity of daf-2(e1370) compared to wild type (Fig. 1b), consistent with reduced IIS and vitellogenesis. However, embryos in utero appeared brighter in daf-2(e1370), consistent with increased vitellogenin provisioning with reduced IIS (Jordan et al. 2019). In contrast, daf-2(gk390525) appeared to have a higher total signal for VIT-2::GFP, with expression conspicuously increased in the body cavity. These results confirm the published gain-of-function behavior of daf-2(gk390525) with respect to vitellogenesis (Kern et al. 2021).
daf-2(gk390525) behaves as a loss-of-function allele in fed and starved larvae
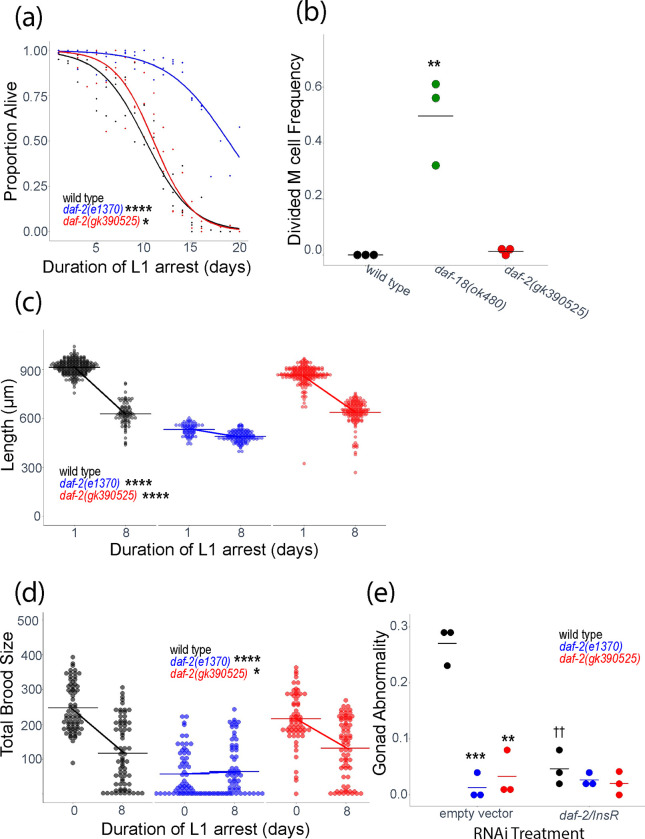
DAF-2/INSR and IIS affect a variety of larval phenotypes, but the effect of daf-2(gk390525) on them is unknown. We assayed multiple phenotypes related to L1 arrest and recovery in daf-2(gk390525) and daf-2(e1370) mutants. IIS regulates survival during L1 arrest. daf-2(e1370) survived L1 arrest significantly longer than wild-type (Fig. 2a), as expected (Muñoz and Riddle 2003; Baugh and Sternberg 2006; Hibshman et al. 2017). However, daf-2(gk390525) also displayed a relatively minor but significant starvation-resistant phenotype, suggesting loss of function during L1 arrest, despite displaying gain-of-function behavior in adults (Fig. 1).
Figure 2.
daf-2(gk390525) mutants display IIS loss-of-function larval phenotypes related to L1 starvation resistance. A) L1 starvation survival was scored daily for three biological replicates. Individual points represent an observation for a population of ~100 animals (median = 97, range = 36–152) in a single biological replicate, and curves were fit with logistic regression. Two-tailed, unpaired variance t-tests were used to compare half-lives between wild type and each mutant. See Table S2 for complete data. B) M cell divisions were scored after 3 days of L1 arrest in the daf-18(ok480) mutant background, and after 8 days for all other genotypes using an hlh-8 reporter gene as an M cell marker in three biological replicates. Horizontal lines represent the mean proportion of animals with at least one M cell division across replicates, and points represent the proportion per replicate (scoring ~100 animals). A one-sided t-test was used to assess significance. See Table S2 for complete survival data. C) Worms were imaged and body length was measured after 48 hours of recovery from 1 (control – arrested L1s plated 24 hr after hypochlorite treatment) or 8 d L1 arrest in three biological replicates. Individual points represent an observation for a single animal. D) Total brood size was scored for ~18 individual worms in each of three biological replicates (median = 18, range = 11–18). Worms recovered from 8 d L1 arrest are compared to unstarved controls (0 days L1 arrest – embryos plated directly with food). Individual points represent individual animals. E) The frequency of adults with gonad abnormalities was scored in previously starved animals (8 days L1 arrest) in three biological replicates. Individual points represent an observation in a population of ~50 individuals in a single biological replicate. Horizontal lines represent the mean abnormality frequency across replicates within the same condition. Homogeneity of variance across conditions and replicates was tested using Bartlett’s test, and if variances were found to not be unequal they were pooled for statistical analysis. Two-tailed, unpaired t-tests with variance pooled were used to compare the frequency of gonad abnormalities between each mutant and wild type (indicated with asterisks) and between each RNAi treatment and empty vector control (indicated with crosses). C, D) A two-factor, linear, mixed-effects model was fit to the data with body length (C) or total brood size (D) as the response variable, interaction between starvation and genotype as the fixed effects, and replicate as the random effect. P-values for interaction terms are reported, assessing starvation-dependent effects of each genotype on phenotype. Horizontal lines represent the mean length per condition across replicates, and diagonal lines connecting the means between conditions within the same genotype represent the effect of starvation. A-E) * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, †† P < 0.01.
In wild-type larvae hatched in the absence of food, the M cell does not divide, reflecting developmental arrest (Fig. 2a; Baugh & Sternberg, 2006). In contrast, there were substantial M cell divisions in daf-18(ok480) mutants, as expected with increased IIS (Chen et al. 2022). Over-expression of agonistic insulin-like peptides during L1 starvation drives M cell division (Chen and Baugh 2014), but there were very few M cell divisions in daf-2(gk390525), consistent with this allele not appreciably increasing IIS during L1 arrest (Fig. 2b).
Development is delayed following L1 arrest (Jobson et al. 2015), but reduction of IIS mitigates delay (Olmedo et al. 2020). We used image analysis to assay larval length after 48 hr recovery from L1 arrest. daf-2(e1370) larvae were smaller than wild-type in control conditions (1 d L1 arrest, which is valuable for synchronization), as expected, but there was very little additional effect of 8 d of L1 arrest, reflecting substantial starvation resistance (Fig. 2c). Length of daf-2(gk390525) larvae was only modestly affected in control conditions, and there was a substantial effect of 8 d of L1 arrest on size, but the effect of starvation was significantly dampened compared to wild type, again suggesting loss of function.
Brood size is reduced following extended L1 arrest (Jobson et al. 2015), but disruption of daf-2/INSR function mitigates decreased fecundity (Jordan et al. 2019). daf-2(e1370) brood size was substantially reduced in control conditions (no starvation), as expected, but brood size was not affected by 8 d L1 arrest, again reflecting substantial starvation resistance (Fig. 2d). daf-2(gk390525) brood size was only modestly affected in control conditions, but the effect of starvation was significantly dampened compared to wild type, further suggesting loss of function.
Extended L1 arrest leads to development of germline tumors and other developmental abnormalities in the adult gonad, and disruption of daf-2/INSR suppresses formation of these starvation-induced abnormalities (Jordan et al. 2019, 2023; Shaul et al. 2022). daf-2(e1370) and daf-2 RNAi significantly suppressed formation of starvation-induced abnormalities (Fig. 2e), as expected. daf-2(gk390525) also significantly suppressed development of gonad abnormalities. In addition, daf-2 RNAi of daf-2(gk390525) larvae during recovery from L1 arrest had no effect. These observations further support the conclusion that daf-2(gk390525) functions as a loss-of-function allele during L1 arrest and possibly recovery.
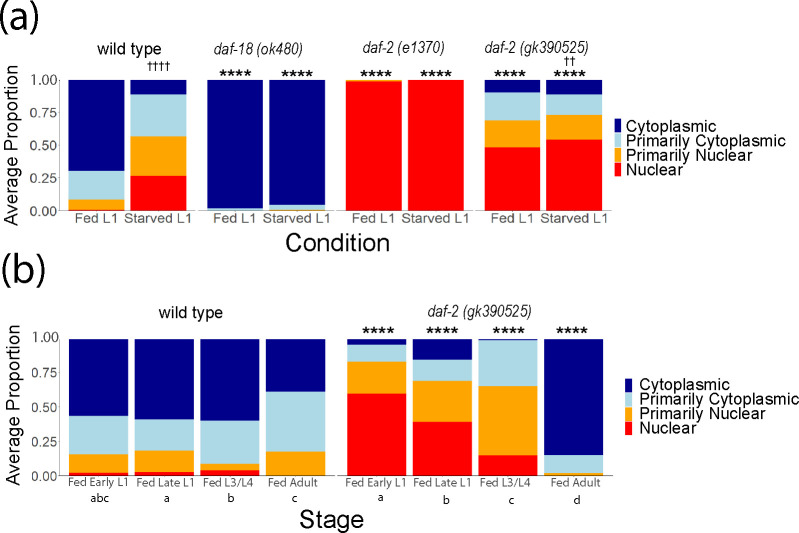
DAF-16/FOXO is the primary transcriptional effector of IIS and is antagonized by DAF-2/INSR and AGE-1/PI3K signaling (Murphy and Hu 2013). When IIS is active (e.g., in fed larvae), DAF-16/FOXO is phosphorylated and cytoplasmic, but when IIS is reduced (e.g., in starved larvae), DAF-16/FOXO translocates to the nucleus and regulates transcription (Henderson and Johnson 2001). We assayed DAF-16::GFP sub-cellular localization as a proxy for IIS activity to complement phenotypic analysis. DAF-16::GFP nuclear localization was significantly increased in starved L1 larvae compared to fed L1 larvae (Fig. 3a), as expected. In addition, DAF-16::GFP was almost entirely cytoplasmic and almost entirely nuclear in both conditions in daf-18(ok480) and daf-2(e1370) larvae, respectively, as expected for constitutively increased and decreased IIS. DAF-16::GFP displayed a modest but significant increase in nuclear localization in daf-2(gk390525) starved L1 larvae compared to fed (Fig. 3a), suggesting reduced environmental responsiveness compared to wild type. Moreover, DAF-16::GFP was significantly more nuclear in daf-2(gk390525) fed and starved L1 larvae compared to wild type in each condition, supporting the conclusion that it reduces DAF-2/INSR function in L1 larvae. Notably, these results extend the phenotypic analysis of L1 arrest and recovery (Fig. 2) to show that daf-2(gk390525) behaves as a loss-of-function allele in starved and fed L1 larvae.
Figure 3.
DAF-16 localization in daf-2(gk390525) reveals a developmental transition from loss-of-function behavior in larvae to gain-of-function behavior in adults. A) DAF-16::GFP sub-cellular localization was scored in fed (18 hr after hypochlorite treatment, or about 6 hr L1 development) and starved (24 hr after hypochlorite treatment, or about 12 hr L1 arrest) L1 larvae. Significant differences within genotype between conditions are indicated with crosses. B) DAF-16::GFP sub-cellular localization was scored throughout postembryonic development (18, 24, 48, and 72 hr after hypochlorite treatment, or about 6, 12, 36, and 60 hr postembryonic development, respectively). Statistically significant differences between stages within the same genotype are indicated with letters (a, b, and c), with members of groups sharing letters not being significantly different from each other (p = 0.05). A, B) Localization was scored as nuclear, primarily nuclear, primarily cytoplasmic, or cytoplasmic in intestinal cells in three biological replicates. Average frequency across replicates for each category is displayed. Cochran-Mantel-Haenszel chi-squared tests were used to compare the distribution of cellular localization between genotypes and conditions. Comparisons between each mutant and wild type within the same conditions are indicated with asterisks. ** P < 0.01, **** P < 0.0001. †† P < 0.01, †††† P < 0.0001
Disruption of DAF-2/INSR ubiquitination at K1614 has different effects on IIS in larvae and adults
daf-2(gk390525) behaves as a gain-of-function allele in adults but a loss-of-function in L1 larvae, leading us to hypothesize that its functional impact on IIS transitions during development. To test this hypothesis, we scored DAF-16::GFP localization over the course of larval development. Fed wild-type animals displayed relatively minor fluctuations in localization throughout development, with DAF-16::GFP being mostly cytoplasmic (Fig. 3b), as expected. In contrast, DAF-16::GFP was primarily nuclear in daf-2(gk390525) fed early L1 larvae, as previously observed (Fig. 3a). In fed late L1 larvae, DAF-16::GFP shifted towards being more cytoplasmic compared to early L1 larvae (Fig. 3b). Likewise, a further shift towards cytoplasmic localization was observed in L3/L4 larvae, and again in adults. Critically, DAF-16::GFP was significantly more nuclear in mutant early L1, late L1, and L3/L4 larvae compared to wild-type, but it was significantly more cytoplasmic in adults. These results demonstrate that the effect of daf-2(gk390525) gradually shifts during development from reducing to increasing IIS, with the transition from behaving as a loss-of-function to gain-of-function allele occurring near the onset of adulthood.
Disruption of DAF-2/INSR ubiquitination at K1614 has different effects on IIS at different temperatures
Unlike L1 arrest, dauer arrest occurs in the L3 stage in larvae that had at least some food (enough to develop into dauer larvae) but experienced adverse conditions such as high population density, limited nutrient availability, and/or high temperature (Hu 2007). Multiple signaling pathways regulate dauer formation, including IIS. In conditions of high IIS, larvae proceed through larval development into reproductive adults, but with low IIS they arrest as dauer larvae. Given the ecological significance of dauer development and the fact that dauer formation is one of the most profound consequences of reduced IIS, we wondered how daf-2(gk390525) affects dauer formation. The comfort range for C. elegans development is 15°C to 25°C, and wild-type larvae can develop as dauers at 27°C. We did not observe dauer formation in wild type, daf-18(ok480), daf-2(e1370), or daf-2(gk390525) at 20°C (Fig. 4a), as expected. However, at 25°C daf-2(e1370) displayed significant dauer formation, as expected, though daf-2(gk390525) did not. At 27°C, wild type displayed modest but reproducible dauer formation and daf-2(e1370) formed 100% dauers, as expected. However, daf-18(ok480) did not form dauers at 27°C, consistent with increased IIS, as expected. Likewise, dauer formation was significantly suppressed at 27°C in daf-2(gk390525) compared to wild type, suggesting increased IIS. These results suggest that daf-2(gk390525) behaves like a gain-of-function allele in larvae at 27°C, despite its loss-of-function behavior in larvae at 20°C.
Figure 4.
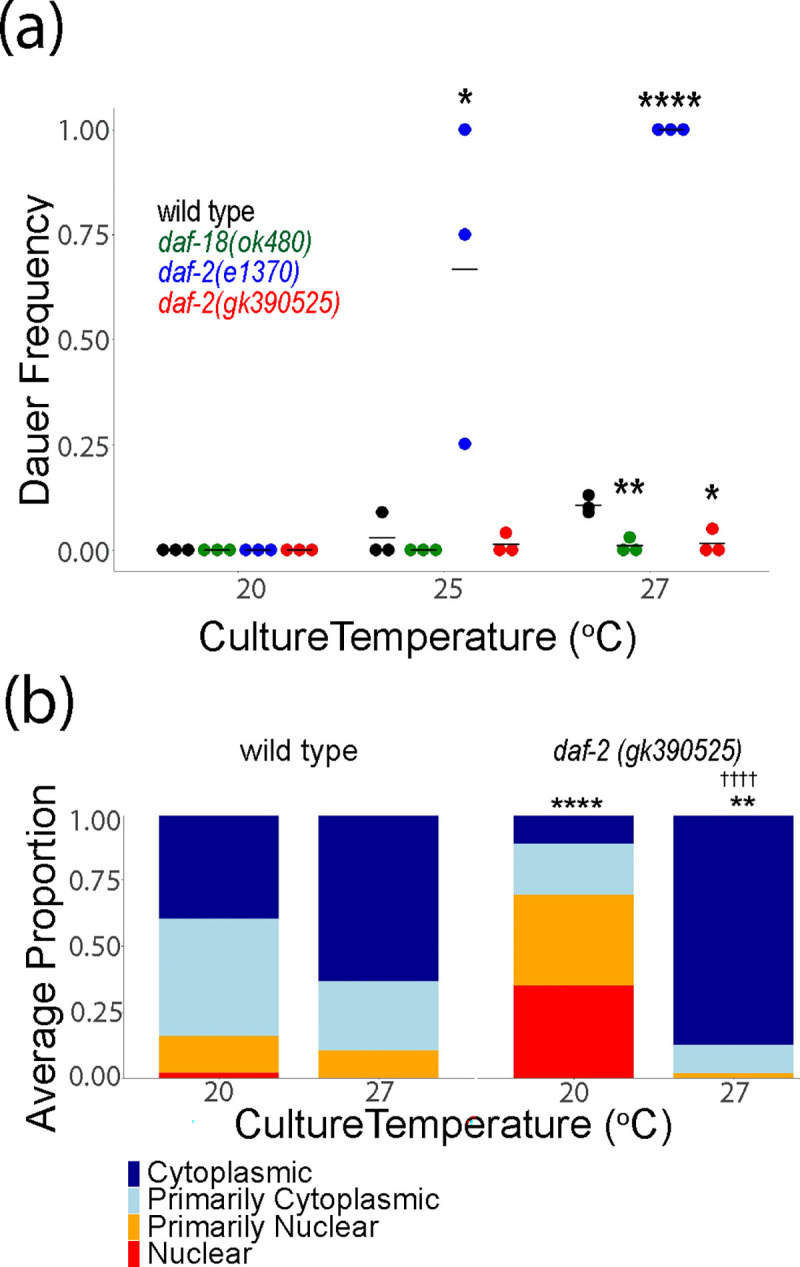
daf-2(gk390525) mutants display IIS gain-of-function phenotypes in larvae at elevated temperature. A) The frequency of dauer larvae was scored at two different temperatures in three biological replicates. Each individual point represents an observation for a population ~100 individuals in a single biological replicate. Horizontal lines represent mean dauer frequency across replicates within the same condition. Two-tailed, unpaired, pooled variance t-tests were used to compare the frequency of dauers between genotypes at each temperature. B) DAF-16 sub-cellular localization was scored in three biological replicates in L1 larvae grown at 20°C or 27°C. Localization was scored as nuclear, primarily nuclear, primarily cytoplasmic, or cytoplasmic in intestinal cells in three biological replicates. Average frequency across replicates for each category is displayed. Cochran-Mantel-Haenszel chi-squared tests were used to compare the distribution of cellular localization between genotypes, with asterisks indicating significance. Significant differences within genotype between conditions are indicated with crosses. †††† P < 0.0001. A, B) * P < 0.05, ** P < 0.01, **** P < 0.0001.
The decision to develop as a dauer larva is made largely based on assessment of environmental conditions in the L1 stage (Schaedel et al. 2012). To determine if the effects of daf-2(gk390525) on temperature-dependent dauer formation reflect temperature-dependent effects on IIS, we scored DAF-16::GFP localization in early L1 larvae cultured at 20°C or 27°C. We did not observe a significant difference in localization in wild-type animals (Fig. 4b), but we did observe a significant difference in daf-2(gk390525), with DAF-16::GFP being predominantly nuclear at 20°C, as seen before (Fig. 3), and predominantly cytoplasmic at 27°C. Critically, the effects of daf-2(gk390525) compared to wild type were significant at each temperature, with increased and decreased nuclear localization at 20°C and 27°C, respectively. These results support the conclusion that whether daf-2(gk390525) behaves as a loss or gain-of-function allele in larvae depends on temperature.
chn-1/CHIP antagonizes IIS during L1 arrest
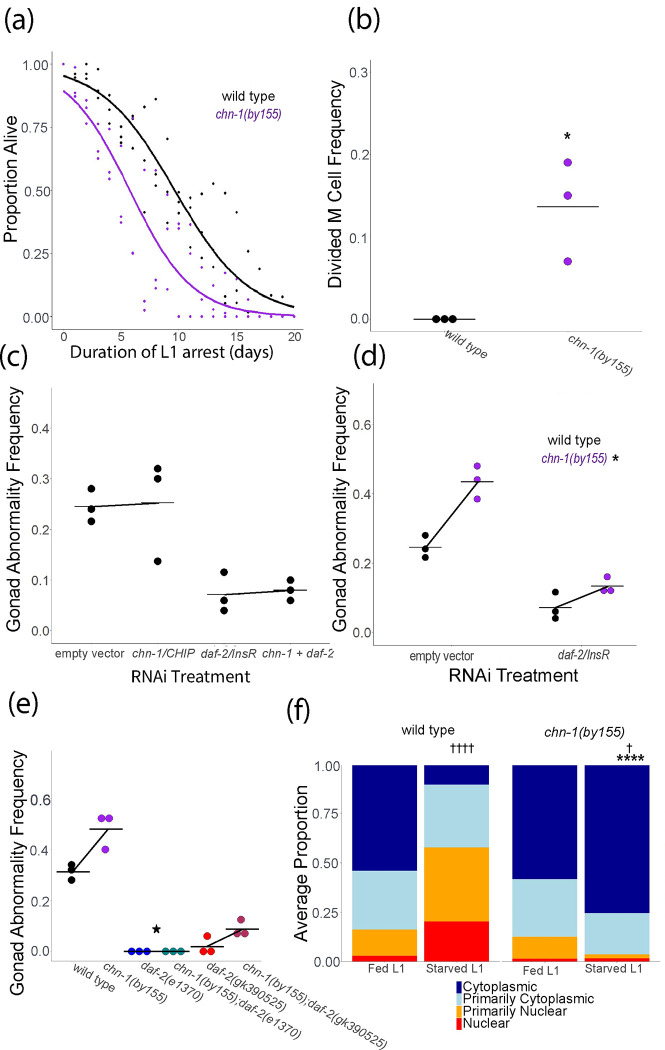
The opposite effects of daf-2(gk390525) on IIS in larvae and adults suggests that DAF-2/INSR K1614 is not ubiquitinated in L1-stage larvae. However, the protein is potentially ubiquitinated at other residues (Tawo et al. 2017) , raising the question of whether CHN-1/CHIP regulates IIS in larvae as it does in adults. We interrogated the function of chn-1/CHIP during L1 arrest to address this question. Although daf-2(gk390525) increased starvation survival during L1 arrest (Fig. 2a), chn-1(by155) decreased survival, though the effect was only marginally significant (p = 0.09, Fig. 5a). In addition, although daf-2(gk390525) had little effect on M cell divisions during L1 arrest (Fig. 2b), chn-1(by155) displayed an arrest-defective phenotype with a significant proportion of animals having M cell divisions (Fig. 5b). These results suggest that chn-1/CHIP negatively regulates DAF-2/INSR activity during L1 arrest.
Figure 5.
chn-1/CHIP antagonizes IIS during L1 arrest. A) L1 starvation survival was scored daily for three biological replicates. Individual points represent an observation for a population of ~100 animals in a single biological replicate (median = 97, range = 36–152), and curves were fit with logistic regression. Two-tailed, unpaired variance t-tests were used to compare half-lives between wild type and each mutant. P = 0.09. See Table S2 for complete data. B) M cell divisions were scored after 8 days of L1 starvation using an hlh-8 reporter gene as an M cell marker in three biological replicates. Horizontal lines represent the mean proportion of animals with at least one M cell division across replicates, and points represent the proportion per replicate (scoring ~100 animals). A one-sided t-test was used to assess significance. See Table S2 for complete survival data. C) Wild type worms were recovered from L1 arrest on empty vector, chn-1, daf-2, or chn-1 + daf-2 RNAi food. P interaction = 0.99. D) Wild type or chn-1 mutants were recovered from L1 arrest on empty vector or daf-2 RNAi food. P interaction = 0.017. E) Worms of the indicated genotype were recovered on empty vector RNAi food (standard food for this assay). P interaction for chn-1(by55) x daf-2(e1370) = 0.034; P interaction for chn-1(by55) x daf-2(gk390525) = 0.38. C-E) The frequency of adults with starvation-induced gonad abnormalities was scored at the onset of egg laying in worms that were starved for 8 d as L1 larvae. Three biological replicates were included. A two-way ANOVA was used to analyze epistasis between each pair of perturbations, and the P-value for a non-linear interaction (epistasis) between the two perturbations is reported. Individual points represent abnormality frequency in a population of ~50 individuals as a single biological replicate. Horizontal lines represent the mean abnormality frequency across replicates within the same condition, and diagonal lines connecting the means represent the effect of perturbations through RNAi or mutation. B, D, E) * P < 0.05. F) DAF-16::GFP sub-cellular localization was scored in fed (18 hr after hypochlorite treatment, or about 6 hr L1 development) and starved (24 hr after hypochlorite treatment, or about 12 hr L1 arrest) L1 larvae. Localization was scored as nuclear, primarily nuclear, primarily cytoplasmic, or cytoplasmic in intestinal cells in three biological replicates. Average frequency across replicates for each category is displayed. Cochran-Mantel-Haenszel chi-squared tests were used to compare the distribution of cellular localization between genotypes and conditions. Significant differences between the mutant and wild type within the same conditions are indicated with asterisks, and significant differences between conditions within genotype are indicated with crosses. † P < 0.05, †††† P < 0.0001.
We extended our phenotypic analysis of chn-1/CHIP to include starvation-induced gonad abnormalities, and we used epistasis analysis to determine if the effects of chn-1/CHIP depend on daf-2/INSR. We assayed development of gonad abnormalities at egg-laying onset in adults that were starved for 8 d as L1 larvae and then recovered on RNAi food targeting chn-1/CHIP, daf-2/INSR, or both (Fig. 5c). daf-2/INSR RNAi suppressed abnormalities, as expected (Fig. 2e), but chn-1/CHIP RNAi had no appreciable effect (Fig. 5c). We did not observe a significant interaction between chn-1/CHIP and daf-2/INSR in a two-factor model, but the lack of an effect of chn-1/CHIP alone makes it impossible to interpret epistasis. It is possible that RNAi did not reduce chn-1/CHIP function enough to elicit a phenotype, or it may be that chn-1/CHIP functions during L1 arrest and this assay interrogates gene function during recovery. In contrast, mutation of chn-1/CHIP significantly increased the frequency of starvation-induced abnormalities (Fig. 5d), suggesting that CHN-1/CHIP negatively regulates IIS in larvae, though it is unclear from this if it functions during L1 arrest, recovery, or both. daf-2/INSR RNAi during recovery partially suppressed abnormalities in the chn-1(by155) background, and there was a significant interaction between chn-1(by155) and daf-2/INSR RNAi (Fig. 5d), suggesting epistasis (non-additivity). daf-2(e1370) completely suppressed abnormalities in the chn-1(by155) background (Fig. 5e), revealing complete epistasis. However, daf-2(gk390525) only partially suppressed abnormalities in the chn-1(by155) background, and there was not a significant interaction between the two mutations (Fig. 5e; see legend), suggesting additive function, or lack of epistasis. In summary, relatively strong loss of daf-2/INSR function, resulting from RNAi or the e1370 allele, resulted in epistasis between daf-2/INSR and chn-1/CHIP, consistent with CHN-1/CHIP targeting DAF-2/INSR in larvae. However, epistasis was not observed with daf-2(gk390525), suggesting the K1614E substitution interferes with ubiquitination in this context.
We complemented phenotypic analysis of chn-1/CHIP by assaying DAF-16::GFP sub-cellular localization. Mutation of chn-1/CHIP did not affect DAF-16::GFP localization in fed L1 larvae (Fig. 5f). However, mutation of chn-1/CHIP significantly increased cytoplasmic localization in starved L1 larvae, suggesting that CHN-1/CHIP provides conditional, nutrient-dependent regulation of IIS in L1-stage larvae. In conclusion, our results suggest that CHN-1/CHIP antagonizes IIS during L1 arrest, as in adults, but not in fed L1 larvae. However, in contrast to adults, they also suggest that an amino acid other than K1614 is the functional site of DAF-2/INSR ubiquitination during L1 arrest.
Conclusions
This work reveals developmental and conditional regulation of IIS activity via ubiquitination of DAF-2/INSR. We show that daf-2(gk390525), which has one of several CHN-1/CHIP-dependent DAF-2 ubiquitination sites mutated, acts as a gain-of-function allele in adults and larvae cultured at elevated temperature, but that it acts as a loss-of-function allele in fed and starved larvae cultured at 20°C. In contrast, chn-1(by155) increases IIS activity during L1 arrest, as in adults, but not in fed L1 larvae. Furthermore, epistasis analysis suggests CHN-1/CHIP targets DAF-2/INSR during L1 arrest, suggesting that it ubiquitinates an amino acid not disrupted by daf-2(gk390525). Overall, our results suggest that CHN-1/CHIP ubiquitinates DAF-2/INSR, enforcing negative regulation of IIS, in starved L1 larvae, larvae cultured at elevated temperature (27°C), and in adults. However, we found no evidence that CHN-1/CHIP ubiquitinates DAF-2/INSR in fed larvae at 20°C, though it is possible that a different ubiquitin ligase targets DAF-2/INSR in this context. daf-2(gk390525) increases IIS activity in adults because disruption of ubiquitination leads to elevated DAF-2/INSR protein levels (Tawo et al. 2017), but it is unclear why this allele reduces IIS activity in larvae. We speculate that the mutation disrupts DAF-2/INSR function independent of its effect on ubiquitination, such that the net effect is increased function in contexts where K1614 is subject to ubiquitination but decreased function in contexts where it is not. Though biochemical and phenotypic evidence suggests that the DAF-2 K1614 residue acts as a site for mono-ubiquitination leading to degradation (Tawo et al. 2017), we cannot rule out other functional roles for ubiquitination at this site. Researchers should interpret phenotypes resulting from daf-2(gk390525) carefully since it has complex, context-dependent effects on IIS. In summary, CHN-1/CHIP-dependent ubiquitination of DAF-2/INSR is not unitary but instead modifies IIS in different ways depending on conditions.
MATERIALS AND METHODS
Published strains
| Strain | Genotype | Description | Source |
|---|---|---|---|
| N2 | Wild type | ||
| CB1370 | daf-2(e1370) | Substitution: P1465S, CCA to TCA(Kimura et al. 1997) | CGC |
| IC166 | daf-18(ok480) | Deletion: 956bp (Barstead et al. 2012) | Ian Chin-Sang - Queen’s University |
| GA1990 | daf-2(gk390525) | Substitution: K1614E, AAA to GAA (Tawo et al. 2017) | David Gems - University College London |
| BR2823 | chn-1(by155) | Deletion: 989bp (Hoppe et al. 2004) | CGC |
| RT130 | pwIs23 [VIT-2::GFP] | Multi-copy transgene (Balklava et al. 2007) | CGC |
| OH16024 | daf-16(ot971[DAF-16::GFP]) | Endogenous fusion (Aghayeva et al. 2020) | Oliver Hobert – Columbia University |
| LRB455 | daf-16(ot971[DAF-16::GFP]); daf-18(ok480) | Cross: IC166 X OH16024 (Chen et al. 2022) | Ryan Baugh – Duke University |
| PD4667 | ayIs7 [hlh-8p::gfp + dpy-20(+)] | hlh-8 transcriptional reporter for M cell identification | CGC |
| LRB477 | daf-18(ok480); ayIs6[hlh-8p::GFP + dpy-20(+)] | Cross IC166 X PD4667 (Chen et al, 2022) | Ryan Baugh – Duke University |
Generated strains
| Strain | Genotype | Description | Source |
|---|---|---|---|
| LRB537 | pwIs23 [VIT-2::GFP]; daf-2(e1370) | Cross: RT130 X CB1370 | This work |
| LRB607 | pwIs23 [VIT-2::GFP]; daf-2(gk390525) | Cross: RT130 X GA1990 | This work |
| LRB562 | daf-16(ot971[DAF-16::GFP]); daf-2(gk390525) | Cross: OH16024 X GA1990 | This work |
| LRB602 | daf-16(ot971[DAF-16::GFP]); daf-2(e1370) | Cross: OH16024 X CB1370 | This work |
| LRB608 | daf-16(ot971[DAF-16::GFP]); chn-1(by155) | Cross: Oh16024 X BR2823 | This work |
| LRB441 | chn-1(by155); daf- 2(e1370) | Cross: BR2823 X CB1370 | This work |
| LRB442 | chn-1(by155); daf- 2(gk390525) | Cross: BR2823 X GA1990 | This work |
| LRB664 | daf-2(gk390525); ayIs6[hlh-8p::GFP + dpy-20(+)] | Cross: BR2823 X PD4667 | This work |
| LRB665 | chn-1(by155); ayIs6[hlh-8p::GFP + dpy-20(+)] | Cross: GA1990 X PD4667 | This work |
Worm maintenance
All worms were maintained at 20°C on Nematode Growth Medium (NGM) plates seeded with E. coli OP50. Worms were well-fed for at least five generations before experimental use. With the exception of scoring starvation-induced gonad abnormalities, experiments were carried out on NGM media seeded with OP50. All experiments were conducted at 20°C unless otherwise noted.
Lifespan
Seven L4 larvae were picked and allowed to lay eggs overnight (~16 hours) before being removed to obtain a synchronized population. Twenty-five progeny in the L4 stage were picked per 10 cm plate for two total plates per biological replicate for each strain used. Day 1 was defined as the first day of adulthood. Worms were transferred to fresh plates daily throughout egg-laying. Worms were determined to be dead if not moving and/or unresponsive to gentle prodding with a transfer pick every 24 hr. Worms that crawled off the plate, died, burrowed or otherwise were not confirmed as dead were censored. Kaplan-Meier estimations, Log-Rank tests, and other statistics were calculated using the online OASIS 2 application (Han et al. 2016).
Fluorescence imaging of vit-2 reporter
A synchronized population of worms was obtained through a timed egg lay, by picking seven L4 larvae and allowing them to lay eggs overnight (~16 hours) and then removing them. Progeny were grown to early adulthood during which a single row of embryos was observed in the gonad and a few unhatched embryos were seen on the lawn. Approximately 20–40 animals were picked into 10 mM levamisole on 4% noble agar pads. Worms were imaged at a total magnification of 100X using a AxioImager compound microscope (Zeiss) and an AxioCam 506 Mono camera. Fiji was used for basic image processing, including rotating, cropping, file conversion, etc.
L1 starvation culture preparation
Seven L4 larvae were picked onto 10 cm NGM plates seeded with OP50. After 96 hours at 20°C, plates were washed with S-basal medium and hypochlorite treated to obtain embryos (Stiernagle 2006). However, for starvation cultures used for gonad abnormality scoring, bleach plates were prepared by picking eight early adults (just after the onset of egg laying) and were cultured for 72 hours prior to hypochlorite treatment. Embryos were transferred to virgin S-basal (lacking ethanol and cholesterol) at a density of one embryo per microliter of media and maintained at 20°C on a tissue-culture roller drum.
L1 starvation survival
Survival during L1 arrest was scored by plating a 100 μL aliquot from a starvation culture on a seeded plate just off the lawn. The number of animals plated was scored by counting the number of L1 larvae in the 100 μL aliquot. After 48 hours the number of animals that survived was scored by counting the number of live worms on the plate. The frequency of animals that survived was calculated by dividing the number of animals that survived by the number of animals plated. Survival was scored daily starting from day 1, 24 hours after preparation of the starvation culture (hypochlorite treatment). Statistical comparisons were made using quasi-binomial logistic regression with the proportion of live worms as the response variable and the duration of L1 arrest as the explanatory variable. Regression was used to estimate half-lives which were subjected to a two-tailed unpaired t-test to compare genotypes.
M cell divisions
M cells were identified with a GFP reporter (strain PD4667 ayIs7 [hlh-8p::gfp]). Arrested L1 larvae were prepared as described above. Three or 8 days after hypochlorite treatment (see figure legend), they were mounted on 4% agarose pads and viewed at 400x total magnification on a Zeiss AxioImager compound microscope, and the number of M cells was scored in each of ~100 larvae.
Growth rate
500 L1 larvae starved in L1 arrest for 8 days or 1 day (actually ~12 hr, given ~12 hr to complete embryogenesis after hypochlorite treatment) as a control were plated and cultured for 48 hours at 20°C. Worms were then washed with S-basal and transferred to clean unseeded NGM plates. 50–100 worms were imaged using a Zeiss SteREO Discovery.V20 stereo microscope and an AxioCam MrM camera at 20X total magnification for worms starved for 1 day, and 30X total magnification for worms starved for 8 days. The Fiji plugin WormSizer was used to measure worm length (Moore et al. 2013). A linear mixed-effects (lme) model was fit using body length as the response variable, the interaction between starvation condition and genotype as the fixed effects, and experimental replicate as the random effect (R: lme(length~strain*day, random = ~1|replicate, data = wormsizer)
Brood size
~50 L1 larvae starved in L1 arrest for 8 days or 1 day (actually ~12 hr, given ~12 hr to complete embryogenesis after hypochlorite treatment) as a control were plated and allowed to grow for 48 hours at 20°C. 18 larvae were then singled onto fresh 6 cm plates. Worms were transferred to fresh plates daily, and the number of progeny was counted after 48 hours until the number of progeny laid in a day reached zero. Worms that arrested after singling, crawled off the plate, died, burrowed or otherwise were not able to lay a complete brood for reasons other than sterility were censored. Total brood size was calculated by adding all days of egg laying. A linear mixed-effect model was fit to the data as described for growth rate however using total brood size as the response variable.
RNA interference
E. coli HT115 was used for RNAi by feeding. The chn-1 RNAi bacterial strain was obtained from the Ahringer RNAi library. The daf-2 RNAi bacterial strain is from the Cynthia Kenyon Lab (Dillin et al. 2002). Empty vector RNAi bacteria carried the L4440 plasmid. Frozen stocks were streaked onto Luria-Bertani (LB) plates with carbenicillin (carb) (100 mg/ml) and tetracycline (tet) (12.5 mg/ml), and single colonies were transferred to 1 mL of LB with carb and tet at the same concentrations. After 16 hours of incubation at 37°C while shaking, 100 μL of culture was transferred to 5 mL of Terrific Broth (TB) with carb (50mg/mL) and incubated for 16 hours at 37°C while shaking. After incubation, cultures were centrifuged at 4000 rpm for 10 minutes and resuspended in S-complete medium with 15% glycerol and aliquoted for freezing. 15 μL from single-use frozen stocks was used to seed lawns on NGM with carb and IPTG plates which were spread to cover ~60% of the plate surface and allowed to grow at room temperature overnight.
Gonad abnormalities
~150 arrested L1 larvae were plated onto NGM + Carb + IPTG plates seeded with E. coli HT115 carrying the indicated RNAi plasmid or the L4440 plasmid as an empty vector control. HT115 carrying L4440 was used as food in all experiments where gonad abnormalities were assayed even when RNAi was not part of the experimental design. Worms were cultured until early adulthood, which varied by genotype. daf-2(e1370) mutants are slow growing and recovered worms were cultured for 96 hours. N2 worms developed at approximately the same rate across RNAi treatments and were scored approximately 72 hours after plating L1s. Both daf-2(gk390525) and chn-1(by155) developed at about the same rate as N2 and were also scored after approximately 72 hours. Worms were then washed from plates with S-basal including 10 mM levamisole and transferred to 4% noble agar pads on a microscope slide. Worms were viewed at 200X total magnification using Nomarski microscopy on a Zeiss AxioImager compound microscope. Worms with proximal germ cell tumors or uterine masses, as described in Jordan et al, 2019, were classified as abnormal. Worms with other, relatively rare abnormalities, were censored, unlike Jordan et al, 2019. Approximately 50 animals were scored per condition, and abnormality frequency was calculated by dividing the number of abnormal worms by the total number of worms scored. Bartlett’s test was used to test homogeneity of variance across replicates and conditions. If variance was not found to be different across these groups (p > 0.05), then two-tailed, unpaired, pooled variance t-tests were used to compare the frequency of abnormalities across pairs of conditions or genotypes. If Bartlett’s test indicated that variance significantly differed across groups, then the same t-tests were performed except variance was not pooled across groups. A two-way ANOVA was used for two-factor comparisons in the case of double mutants, combining mutants and RNAi, and double RNAi treatments, and the p-value for the interaction between factors was used to assess epistasis.
Dauer formation
Seven well-fed L4 larvae were transferred to fresh plates and cultured for 20 hours at the indicated temperature. Adults were removed and plates were returned to the indicated growth temperatures for 48 hours. Dauers were identified visually by morphology (narrow body, elongated) and increased refractive index. The number of dauers on the plate was then counted, and the frequency of dauers was calculated by dividing the number of dauers by the total population size. Statistical comparisons were made similarly to those described for gonad abnormalities. Bartlett’s test was used to test for equal variances and where variances were not unequal, pooled variance unpaired t-tests were used to compare dauer frequency between conditions. If variances were unequal then variance was not pooled.
DAF-16/FOXO sub-cellular localization
Larvae in L1 arrest were obtained as described above, and they were cultured for 18 hr after hypochlorite treatment before being imaged (it takes approximately 12 hr to complete embryogenesis in these conditions; ~6 hr L1 arrest). Synchronized, fed early L1 larvae were obtained by hypochlorite treating gravid worms, plating embryos directly onto food, and culturing them for 18 hours (~6 hr feeding). Late L1 larvae were obtained by plating embryos directly onto food and culturing them for 24 hours (~12 hr feeding). L3/L4 were cultured for 48 hours (~36 hr feeding), and adults were cultured for 72 hours (~60 hr feeding). Timed egg lays were used to obtain synchronized populations of L1 larvae at 20 and 27°C to mimic conditions used in the dauer-formation assay - seven L4s were picked to a fresh, seeded plate and cultured for 24 hours before being removed. The progeny were washed into 1.5 mL Eppendorf tubes with 1 mL of S-basal. Worms were centrifuged at 3000 rpm for 60 seconds then transferred by pipetting 2 μL of volume from the pellet to a 4% noble agar pad. Slides were visualized at 1000x total magnification for early and late L1 larvae, and 400X total magnification for L3/L4s and adults, using a Zeiss AxioImager compound microscope. Sub-cellular localization was scored with four categories ranging from completely nuclear to completely cytoplasmic, as previously described (Chen et al. 2022). To minimize confounding environmental effects on DAF-16 localization, worms were scored for only the first three minutes after being transferred to slides. A Cochran-Mantel-Haenszel Chi-squared test was used to perform pairwise comparisons between genotypes and conditions for the distribution of DAF-16/FOXO subcellular localization categories.
Acknowledgements
This work was supported by the National Institutes of Health (R01GM117408 and R01GM143159, L.R.B.). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We would also like to thank WormBase and the Alliance of Genome Resources. We would also like to thank Kinsey Fisher and Rebecca Liu for generating strains used in this work.
Data Availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Table S1 and S2 contain replicate-level data for the lifespan and starvation survival analyses in Figures 1a, 2a, and 5a. Strains are available upon request.
Bibliography
- Aghayeva U., Bhattacharya A., and Hobert O., 2020. A panel of fluorophore-tagged daf-16 alleles. microPublication Biol. 2020: 2019–2020. 10.17912/micropub.biology.000210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., and Grant B. D., 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9: 1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Barstead R., Moulder G., Cobb B., Frazee S., Henthorn D., et al. , 2012. Large-scale screening for targeted knockouts in the caenorhabditis elegans genome. G3 Genes, Genomes, Genet. 2: 1415–1425. 10.1534/g3.112.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., and Sternberg P. W., 2006. DAF-16/FOXO Regulates Transcription of cki-1/Cip/Kip and Repression of lin-4 during C. elegans L1 Arrest. Curr. Biol. 16: 780–785. 10.1016/j.cub.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Baugh L. R., 2013. To grow or not to grow: Nutritional control of development during Caenorhabditis elegans L1 Arrest. Genetics 194: 539–555. 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., and Baugh L. R., 2014. Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev. Biol. 394: 314–326. 10.1016/j.ydbio.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Chen J., Tang L. Y., Powell M. E., Jordan J. M., and Baugh L. R., 2022. Genetic analysis of daf-18/PTEN missense mutants for starvation resistance and developmental regulation during Caenorhabditis elegans L1 arrest. G3 Genes, Genomes, Genet. 12. 10.1093/g3journal/jkac092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon Cynthia, Chang Jean, Gensch Erin, Rudner Adam, and Tabtlang Ramon, 1993. A C. elegans mutant that lives twice as long as wild type. Nat. 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Depina A. S., Iser W. B., Park S. S., Maudsley S., Wilson M. A., et al. , 2011. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 11: 11. 10.1186/1472-6793-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Crawford D. K., and Kenyon C., 2002. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science (80-. ). 298: 830–834. 10.1126/science.1074240 [DOI] [PubMed] [Google Scholar]
- Gems D., Sutton A. J., Sundermeyer M. L., Albert P. S., King K. V., et al. , 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150: 129–155. 10.1093/genetics/150.1.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. K., Lee D., Lee H., Kim D., Son H. G., et al. , 2016. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7: 56147–56152. 10.18632/oncotarget.11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., and Johnson T. E., 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11: 1975–1980. 10.1016/S0960-9822(01)00594-2 [DOI] [PubMed] [Google Scholar]
- Hibshman J. D., Doan A. E., Moore B. T., Kaplan R. E., Hung A., et al. , 2017. daf-16/FoxO promotes gluconeogenesis and trehalose synthesis during starvation to support survival. Elife 6: 1–29. 10.7554/eLife.30057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Cassata G., Barral J. M., Springer W., Hutagalung A. H., et al. , 2004. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell 118: 337–349. 10.1016/j.cell.2004.07.014 [DOI] [PubMed] [Google Scholar]
- Hu P. J., 2007. Dauer. WormBook 1–19. 10.1895/wormbook.1.144.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson M. A., Jordan J. M., Sandrof M. A., Hibshman J. D., Lennox A. L., et al. , 2015. Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics 201: 201–212. 10.1534/genetics.115.178699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan J. M., Hibshman J. D., Webster A. K., Kaplan R. E. W., Leinroth A., et al. , 2019. Insulin/IGF Signaling and Vitellogenin Provisioning Mediate Intergenerational Adaptation to Nutrient Stress. Curr. Biol. 29: 2380–2388.e5. 10.1016/j.cub.2019.05.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan J. M., Webster A. K., Chen J., Chitrakar R., and Ryan Baugh L., 2023. Early-life starvation alters lipid metabolism in adults to cause developmental pathology in Caenorhabditis elegans. Genetics 223. 10.1093/genetics/iyac172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern C. C., Townsend S. J., Salzmann A., Rendell N. B., Taylor G. W., et al. , 2021. C. elegans feed yolk to their young in a form of primitive lactation. Nat. Commun. 12. 10.1038/s41467-021-25821-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. D., Tissenbaum H. A., Liu Y., and Ruvkun G., 1997. Daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science (80-. ). 277: 942–946. 10.1126/science.277.5328.942 [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., 2005. Ubiquitin-mediated pathways in C. elegans. WormBook 1–24. 10.1895/wormbook.1.36.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M. J., and Riddle D. L., 2003. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163: 171–180. 10.1093/genetics/163.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Lieb J. D., Bargmann C. I., Kamath R. S., et al. , 2003. Genes that act downstream of DAF-16 to influence C. elegans lifespan. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Murphy C. T., and Hu P. J., 2013. Insulin/insulin-like growth factor signaling in C. elegans. WormBook 1–43. 10.1895/wormbook.1.164.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S., and Ruvkun G., 1998. The C. elegans PTEN Homolog, DAF-18, Acts in the Insulin Receptor-like Metabolic Signaling Pathway is a member of the insu-lin/insulin-like growth factor family of receptors (Kimura et al., 1997). Activation of the DAF-2 signaling cascade is necessary for. Mol. Cell 2: 887–893. [DOI] [PubMed] [Google Scholar]
- Olmedo M., Mata-Cabana A., Jesús Rodríguez-Palero M., García-Sánchez S., Fernández-Yañez A., et al. , 2020. Prolonged quiescence delays somatic stem cell-like divisions in Caenorhabditis elegans and is controlled by insulin signaling. Aging Cell 19: 1–13. 10.1111/acel.13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M. F., and Lehner B., 2019. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21: 143–151. 10.1038/s41556-018-0242-9 [DOI] [PubMed] [Google Scholar]
- Schaedel O. N., Gerisch B., Antebi A., and Sternberg P. W., 2012. Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 10: 1–18. 10.1371/journal.pbio.1001306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul N. C., Jordan J. M., Falsztyn I. B., and Baugh L. R., 2022. Insulin/IGF-dependent Wnt signaling promotes formation of germline tumors and other developmental abnormalities following early-life starvation in Caenorhabditis elegans . Genetics 223: 1–11. 10.1093/genetics/iyac173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans. WormBook 1–11. 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawo R., Pokrzywa W., Kevei É., Akyuz M. E., Balaji V., et al. , 2017. The Ubiquitin Ligase CHIP Integrates Proteostasis and Aging by Regulation of Insulin Receptor Turnover. Cell 169: 470–482.e13. 10.1016/j.cell.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang B., Marcu I., Athar F., Wang H., et al. , 2021. Mutation of daf-2 extends lifespan via tissue-specific effectors that suppress distinct life-limiting pathologies. Aging Cell 20: 1–14. 10.1111/acel.13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., and Shabek N., 2017. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86: 129–157. 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Table S1 and S2 contain replicate-level data for the lifespan and starvation survival analyses in Figures 1a, 2a, and 5a. Strains are available upon request.