Abstract
Metabolic adaptations in response to changes in energy supply and demand are essential for survival. The mitochondrial calcium uniporter coordinates metabolic homeostasis by regulating TCA cycle activation, mitochondrial fatty acid oxidation and cellular calcium signaling. However, a comprehensive analysis of uniporter-regulated mitochondrial metabolic pathways has remained unexplored. Here, we investigate the metabolic consequences of uniporter loss- and gain-of-function, and identify a key transcriptional regulator that mediates these effects. Using gene expression profiling and proteomic, we find that loss of uniporter function increases the expression of proteins in the branched-chain amino acid (BCAA) catabolism pathway. Activity is further augmented through phosphorylation of the enzyme that catalyzes this pathway’s committed step. Conversely, in the liver cancer fibrolamellar carcinoma (FLC)-which we demonstrate to have high mitochondrial calcium levels- expression of BCAA catabolism enzymes is suppressed. We also observe uniporter-dependent suppression of the transcription factor KLF15, a master regulator of liver metabolic gene expression, including those involved in BCAA catabolism. Notably, loss of uniporter activity upregulates KLF15, along with its transcriptional target ornithine transcarbamylase (OTC), a component of the urea cycle, suggesting that uniporter hyperactivation may contribute to the hyperammonemia observed in FLC patients. Collectively, we establish that FLC has increased mitochondrial calcium levels, and identify an important role for mitochondrial calcium signaling in metabolic adaptation through the transcriptional regulation of metabolism.
INTRODUCTION
Efficient utilization of energy sources based on their availability and cellular needs requires metabolic flexibility. Mitochondrial calcium (Ca2+) signaling plays a central role in metabolic adaptation to acute or prolonged changes in energy demands and metabolite levels1–5. A principal regulator of this signaling pathway is the mitochondrial Ca2+ uniporter (referred to as the uniporter from here on), a Ca2+-selective channel in the inner mitochondrial membrane. The uniporter facilitates bulk entry of Ca2+ into the mitochondrial matrix6,7. This Ca2+ influx contributes to activation of the tricarboxylic acid (TCA) cycle, buffering of cytosolic Ca2+ signaling, or mitochondrial damage and cell death, depending on the amount of Ca2+ that enters the mitochondria8. Thus, the uniporter regulates metabolism, mitochondrial and cytosolic Ca2+ signaling, and cell survival.
Five core proteins constitute the uniporter: three transmembrane proteins (MCU, EMRE and MCUb) and two membrane-associated regulatory subunits (MICU1–2)6,7,9,10. MCU and EMRE are necessary and sufficient to form a functional Ca2+ channel, and MICU1–2 and MCUb regulate uniporter activity10–13. In addition, diverse physiological and pathological stimuli alter uniporter mediated mitochondrial Ca2+ uptake through transcriptional and post-translational mechanisms1,8,14. Both increased and decreased mitochondrial Ca2+ levels are observed in disease-associated states, such as high fat diet-induced obesity15, neurodegeneration16,17, metabolic disorders18 and cancer19. Reduced mitochondrial Ca2+ uptake leads to slower cell proliferation rates20–22, smaller body size23,24, exercise intolerance24, and epigenetic changes25 in mice. Conversely, increased mitochondrial Ca2+ load is most often associated with mitochondrial damage and cell death26. Nevertheless, enhanced uniporter activity is thought to be beneficial under energetic stress1, pointing to a complex regulation of mitochondrial Ca2+ signaling in order to balance cellular metabolic needs.
Despite the importance of the uniporter for metabolic adaptation, a comprehensive analysis of mitochondrial pathways that are regulated by the uniporter is lacking. To better understand the effects of the uniporter on mitochondrial function, we analyzed MCU knockout (KO) mitochondria using proteomics and RNA sequencing. Our results show increased expression of branched-chain amino acid (BCAA) catabolism proteins in HeLa MCU KO cells. Furthermore, a key phosphorylation event that inhibits pathway activity is reduced in these cells, thereby augmenting BCAA catabolism in the absence of uniporter activity.
To determine if these metabolic changes are reversed under conditions of elevated mitochondrial Ca2+, we turned to fibrolamellar carcinoma (FLC), an oncocytic tumor with indications of increased mitochondrial Ca2+ levels in the literature27–29. FLC is a rare liver cancer that primarily affects adolescents and young adults. Recent in-depth analysis of the FLC proteome and transcriptome identified significant differences in tumor mitochondria and metabolism compared to adjacent non-tumor liver samples30, indicating a key role for mitochondria in FLC pathogenesis. Using patient samples and cell models, we show that increased mitochondrial Ca2+ levels is a distinguishing feature of FLC. Furthermore, expression of BCAA pathway proteins is significantly decreased in FLC, a phenotype which is reversed by MCU knockdown. This is consistent with the uniporter playing a central regulatory role in BCAA catabolism. We also report that expression of the transcription factor KLF15, a regulator of multiple BCAA catabolism genes, is suppressed in FLC tumors and is regulated by MCU. This pathologic event has important implications for FLC patients due to KLF15’s role in promoting expression of the urea cycle enzyme ornithine transcarbamylase (OTC)31. Downregulation of OTC is thought to contribute to impaired urea cycle function and subsequently to hyperammonemia and hyperammonemic encephalopathy diagnosed in some late-stage FLC patients32. We observe that, similar to KLF15, OTC expression is suppressed in FLC tumors and is MCU-regulated in cellular models of FLC.
Overall, our results implicate increased uniporter function in FLC and identify BCAA catabolism and the urea cycle as novel uniporter-regulated pathways downstream of KLF15. Based on these findings, we propose that MCU and KLF15 are potential therapeutic targets in the treatment of FLC and its sequelae.
RESULTS
Loss of uniporter slows growth and alters expression of mitochondrial proteins
To better characterize how mitochondria respond to loss of mitochondrial Ca2+ uptake, we generated a HeLa MCU KO cell line as well as a rescue cell line by stable expression of FLAG-tagged MCU in the KO cells. As previously reported, loss of MCU significantly slowed growth, which was remedied by re-expression of MCU (Figs. 1A, 1B)20–22,33. Yet, MCU KO cells did not show a significant decrease in their basal respiration, maximal respiration, ATP production, proton leak, or non-mitochondrial oxygen consumption rates, and they showed only a mild reduction in their spare respiratory capacity (Figs. S1A–G).
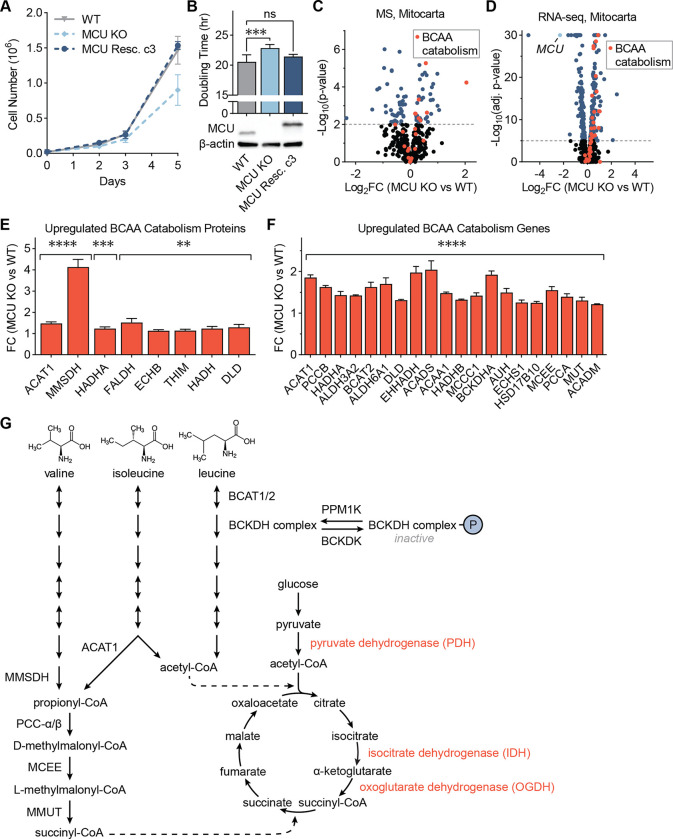
Figure 1: MCU KO cells exhibit growth defects and altered mitochondrial proteome.
(A) Loss of MCU reduces proliferation of HeLa cells. WT, MCU KO, and MCU rescue cells were counted on days 2, 3, and 5 after plating; n=4–6 replicates. (B) HeLa cell doubling times were calculated from cell counts on days 2 to 5 in (A); p-values were determined by one-way ANOVA; expression of MCU and MCU-FLAG were confirmed by Western blot. (C) Volcano plot shows relative abundance of mitochondrial proteins in MCU KO cells compared to WT cells. Red points indicate proteins in the valine, leucine, and isoleucine degradation KEGG pathway; n=5. (D) Volcano plot shows relative abundance of mRNAs coding mitochondrial proteins in MCU KO cells compared to WT cells. Red points indicate genes in the valine, leucine and isoleucine degradation KEGG pathway; MCU is marked in light blue; n=3. (E, F) Fold change of valine, leucine, and isoleucine degradation-associated proteins (E) and genes (F) enriched in MCU KO cells in (C, D); proteins and genes are listed in order of ascending p-value. (G) Schematic of BCAA catabolism and TCA cycle enzymes and metabolites; BCAA catabolism produces acetyl-CoA and succinyl-CoA, which can enter the TCA cycle; Ca2+-regulated enzymes are shown in red. All error bars indicate standard deviation; ** indicates a p-value < 0.01, *** indicates a p-value < 0.001, **** indicates a p-value < 0.0001.
These mild energetic phenotypes have been attributed to the activation of glycolysis and glutamine utilization in MCU KO cells20,33. Whether there are additional metabolic changes that sustain TCA cycle activity and energy production in the absence of uniporter is unclear. To better characterize the effects of MCU loss on the mitochondria, we compared WT and MCU KO mitochondria using proteomics (Fig. 1C) and transcriptomics (Fig. 1D). Gene set enrichment analysis34 of significantly upregulated proteins and genes in MCU KO cells showed a strong enrichment of the branched-chain amino acid (BCAA) catabolism pathway and fatty acid oxidation (FAO) in both datasets (Figs. S1H, S1I). 8 of the 48 proteins in the Kyoto Encyclopedia of Genes and Genomes (KEGG) BCAA catabolism pathway (hsa0028) showed a significant increase in protein levels (Fig. 1E). Similarly, 20 out of 48 KEGG BCAA catabolism genes showed a significant 1.3–2 fold increase in their mRNA levels in MCU KO cells (Fig. 1F).
The three BCAAs – valine, leucine, and isoleucine – cannot be synthesized in human cells and must be acquired from diet. They are either utilized in protein synthesis or catabolized as sources of carbon and nitrogen for metabolic pathways. Their catabolism in the mitochondria generates the TCA cycle intermediates acetyl-CoA and succinyl-CoA (Fig. 1G). Loss of MCU abrogates stimulation of the Ca2+-sensitive TCA cycle enzymes pyruvate dehydrogenase (PDH), isocitrate dehydrogenase (IDH), and oxoglutarate dehydrogenase (OGDH), thereby dampening the production of acetyl-CoA and succinyl-CoA by the TCA cycle. Hence, we conjecture that increased BCAA catabolism in MCU KO cells compensates for lack of Ca2+ stimulation by funneling BCAA catabolic products into the TCA cycle (Fig. 1G). Similarly, increased FAO can generate more acetyl-CoA. Consistent with increased FAO, MCU KO cells were found to contain markedly reduced levels of very long-chain fatty acids (Fig. S1J). This was accompanied by increased levels of acylcarnitines, fatty acid conjugates that are transported to the mitochondria for oxidation (Fig. S1K). These data are consistent with the notion that metabolic changes compensate for loss of MCU. They also identify a previously unappreciated role for BCAA catabolism in MCU KO cells.
BCAA catabolism pathway activity is upregulated by MCU loss
To better understand the relationship between mitochondrial Ca2+ signaling and BCAA metabolism, we monitored expression of select pathway proteins after uniporter perturbation. HeLa cells lacking MCU exhibited a small but consistent increase in levels of BCAA catabolism proteins (Figs. 2A, 2B, S2A). We observed similar changes in HeLa cells that lack EMRE, an essential component of the uniporter (Figs. 2C, 2D, S2B). To establish whether uniporter-mediated BCAA regulation extends to other cell types and species, especially to cells that originate from tissues with high BCAA catabolism rates such as liver, we blotted for pathway proteins in the mouse hepatocyte cell line AML12. RNAi-mediated knockdown of MCU reduced mitochondrial Ca2+ uptake rates (Fig. S2C) and increased the expression of proteins in the BCAA catabolism pathway (Figs. 2E, 2F).
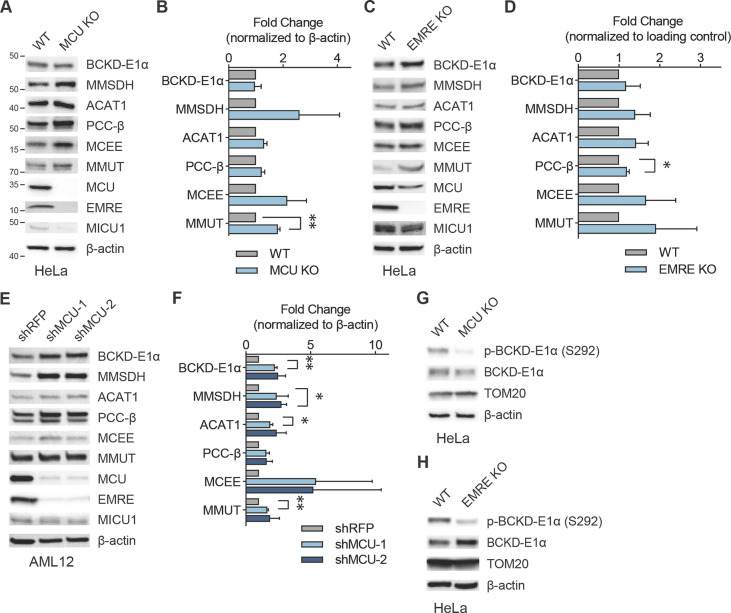
Figure 2: Mitochondrial Ca2+ uniporter regulates BCAA catabolism pathway.
(A, B) Immunoblots of select BCAA catabolism proteins and uniporter components (A) and their quantification (B) in HeLa WT and MCU KO cells show mild but consistent increase in pathway protein expression; n=3. (C, D) Loss of EMRE also causes increased BCAA catabolism protein expression; immunoblots of select BCAA catabolism pathway proteins (C) and their quantification (D) in HeLa WT and EMRE KO cells; n=3. (E, F) Immunoblots of select BCAA pathway proteins (E) and their quantification (F) in AML12 cells after shRNA-mediated MCU knockdown; n=3. (G, H) Immunoblots of phosphorylated and total BCKD-E1α in HeLa WT, MCU KO (G) and EMRE KO cells (H); n=3. Statistical significance determined by one-sample t-test.
The committed step in the BCAA catabolism pathway is the oxidative decarboxylation reaction carried out by the branched-chain α-ketoacid dehydrogenase (BCKDH) complex. The activity of this complex is regulated by phosphorylation and dephosphorylation of the E1α subunit by the branched-chain α-ketoacid dehydrogenase kinase (BCKDK) and mitochondrial protein phosphatase 1K (PPM1K), respectively. Phosphorylation by BCKDK is inhibitory, whereas dephosphorylation by PPM1K activates the complex. We considered the possibility that BCKD-E1α phosphorylation may be Ca2+-regulated, similar to the Ca2+-sensitive phosphorylation of the pyruvate dehydrogenase (PDH) complex in some tissues35,36. Interestingly, in HeLa cells, BCKD-E1α phosphorylation was uniporter-dependent, with a significant reduction in BCKD-E1α phosphorylation in MCU or EMRE KO cells (Figs. 2G, 2H, S2D, S2E). However, in AML12 cells that originate from liver, where PDH phosphorylation is known to be insensitive to mitochondrial Ca2+ levels35, we did not observe a change in phospho-BCKD-E1α levels (Fig. S2F). These data suggest that MCU regulates the activity of the BCAA catabolism pathway both through phosphorylation and transcriptional regulation of pathway enzyme expression in a cell- and tissue-specific manner.
Increased mitochondrial Ca2+ levels are a hallmark of fibrolamellar carcinoma (FLC)
FLC is a rare liver cancer that affects children and young adults. This cancer is characterized by a somatic ~400 kb genomic deletion that fuses the first exon of the DnaJ homolog subfamily B member 1 gene (DNAJB1) to exon 2–10 of the cAMP-dependent protein kinase catalytic subunit alpha gene (PRKACA)37 (Fig. 3A). This deletion event generates a fusion kinase, DNAJ-PKAc, which we refer to as DP from hereon. FLC tumors are oncocytic neoplasms that are characterized by an aberrant number of mitochondria27,28,38. In addition, earlier reports noted the presence of electron dense particles in electron microscopy (EM) images of FLC tumors28. These electron dense particles are consistent with Ca2+ phosphate precipitates that form in the mitochondrial matrix39,40. Increased abundance of these precipitates in FLC mitochondria indicates high levels of mitochondrial Ca2+. We sought to further characterize mitochondrial Ca2+ changes and investigate uniporter function and BCAA metabolism in FLC.
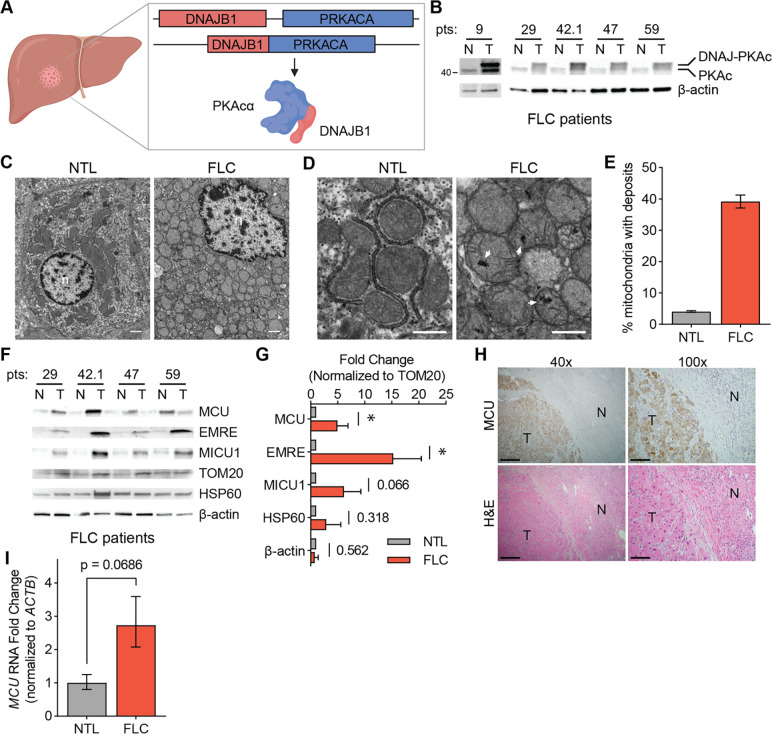
Figure 3: FLC is characterized by increased mitochondrial Ca2+ levels and uniporter expression.
(A) Schematic of FLC liver tumor with heterozygous deletion in chromosome 19 producing the DNAJ-PKAc fusion protein. (B) Immunoblot of lysates from non-tumor (N) and tumor (T) liver from FLC patients shows DP fusion protein expression in the tumor. (C) Electron micrographs at 10,000x magnification of non-tumor (NTL) and tumor (FLC) sections from patient 9; nuclei are labeled n; scale bars = 1 μm. (D) Micrographs of samples shown in (C) at 25,000x magnification; white arrowheads mark representative Ca2+ deposits in the tumor; scale bars = 600 nm. (E) Percentage of mitochondria from FLC patient 9 with Ca2+ deposits; the mean is reported from manual counting of >500 mitochondria per sample by two independent, blinded analysts. (F) Immunoblots of uniporter components and control mitochondrial proteins from paired non-tumor (N) and FLC tumor (T) samples. (G) Pooled quantification of immunoblots in (F) normalized to TOM20 levels; statistical significance determined by one-sample t-test; numbers next to error bars indicate p-values, * indicates a p-value < 0.05. (H) H&E and MCU IHC staining of non-tumor (N) and tumor (T) regions of liver from FLC patient 9; 40x image scale bars are 500 μm and 100x image scale bars are 200 μm. (I) qPCR analysis of MCU RNA expression in paired non-tumor liver and tumors from FLC patients 29, 42.1, 47, and 59. All error bars indicate standard deviation; * indicates a p-value < 0.05
We obtained paired FLC and non-tumor liver (NTL) tissue samples from five patients and confirmed DP expression in the tumors using western blotting with an antibody that detects PKAc (Fig. 3B, Supp Table 1). To investigate mitochondrial Ca2+ levels in FLC, we performed EM analysis on tumor and non-tumor liver from one patient (Figs. 3C, 3D). 39% of FLC mitochondria contained one or more electron dense Ca2+ deposits compared to 4% of mitochondria from non-tumor liver (Fig. 3E). We also analyzed mitochondria from a second patient, for which a control liver sample did not exist, by comparing mitochondria in oncocytic and peri-oncocytic cells from the tumor periphery (Figs. S3A–S3C). This analysis also showed an increased percentage of mitochondria with electron dense particles in oncocytic cells (24%) compared to non-oncocytic cells (10%) (Fig. S3D). EM analysis of tumor and control liver from a hepatocellular carcinoma (HCC) patient did not show more abundant Ca2+ deposits in HCC mitochondria (Figs. S3E–S3G). This suggests that high levels of mitochondrial Ca2+ deposits may be an FLC-specific phenotype.
We next quantified levels of the uniporter proteins MCU, EMRE and MICU1 in FLC and paired NTL control lysates. MCU and EMRE levels were significantly increased in FLC, and MICU1 trended towards higher expression in tumors (Figs. 3F, 3G). The control mitochondrial protein HSP60 did not show a significant difference between tumors and control lysates, suggesting that the increase in uniporter protein expression is specific and is not a result of increased mitochondrial proteome abundance in FLC. Immunohistochemistry (IHC) analysis of tumor and normal liver sections also showed stronger MCU staining in the tumor (Fig. 3H). Quantitative PCR analysis showed that FLC tumors had elevated MCU mRNA levels (Fig. 3I). These data are consistent with higher uniporter expression driving increased mitochondrial Ca2+ levels in FLC.
Cellular FLC models show increased mitochondrial Ca2+ levels
To better understand the role of mitochondrial Ca2+ signaling in FLC metabolism, we turned to clonal AML12 cell lines, c14 and c4, which were previously generated using genome editing41 and used as cellular models of FLC. These clones carry the heterozygous FLC deletion, express a single allele of the fusion kinase DP, and grow faster than WT AML12 cells (Figs. 4A, 4B). The clonal cells also recapitulate FLC-associated signaling events and have been successfully used to investigate FLC biology and therapeutics41,42.
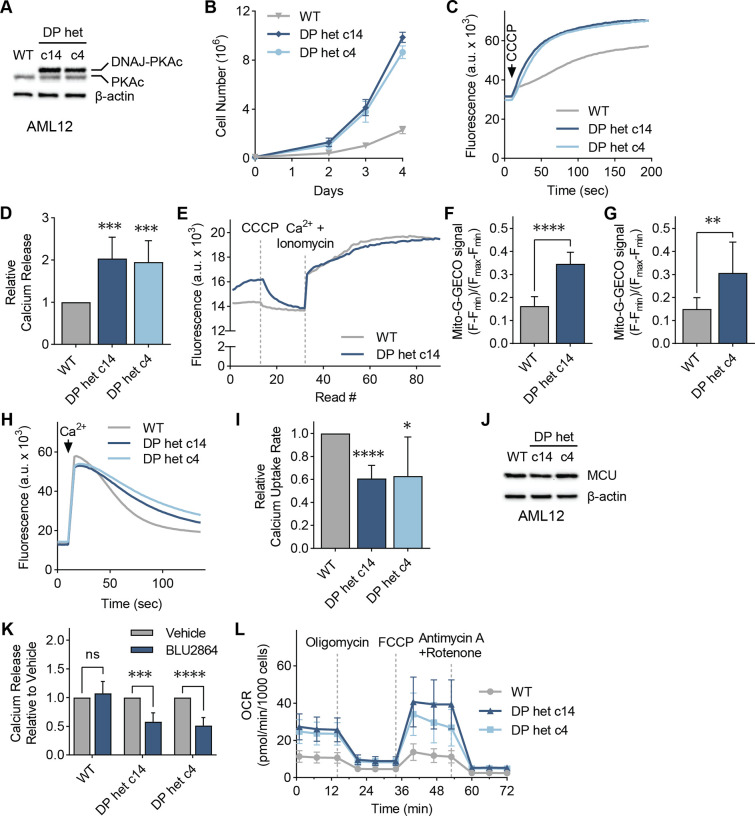
Figure 4: Cellular models of FLC show DP-dependent increase in mitochondrial Ca2+ levels.
(A) Immunoblot of lysates from AML12 WT and clones c14 and c4 using an antibody against PKAc confirm heterozygous DP expression in the clones. (B) Proliferation of cellular models of FLC compared to WT AML12 cells; cells were counted on days 2, 3, and 4 after plating; n=3. (C, D) Representative traces (C) and quantification (D) of mitochondrial Ca2+ release assays in AML12 cells; cells were treated with uncoupler CCCP, and the relative amount of Ca2+ released was quantified using a Ca2+ indicator dye; statistical significance was determined by one-sample t-test; n=10. (E) Representative trace of mitochondrial free Ca2+ levels of AML12 WT and c14 cells quantified using matrix-targeted Ca2+ reporter G-GECO (mito-G-GECO). (F, G) Baseline mito-G-GECO fluorescence normalized to minimum and maximum signals in AML12 WT and c14 (F) or c4 (G) cells; statistical significance determined by Mann Whitney test; n=20–23 (F) and n=12 (G). (H, I) Representative traces (H) and mitochondrial Ca2+ uptake rates in AML12 cells (I); mitochondrial Ca2+ uptake rates were calculated by monitoring Ca2+ clearance in the presence of a Ca2+ indicator dye; statistical significance determined by one-sample t-test; n=9. (J) Immunoblot of MCU shows comparable MCU expression in FLC clones compared to WT controls. (K) Mitochondrial Ca2+ release after cells were treated with 5 μM PKA inhibitor BLU2864 or DMSO for 4 days was measured as in (C); fold change in released Ca2+ is shown relative to DMSO control for each cell line; statistical significance determined by paired t-test, n=7–9. (L) Seahorse extracellular flux analysis in FLC clones compared to WT AML12 cells at baseline and after indicated treatments; n=10–16. All error bars indicate standard deviation; * indicates a p-value < 0.05, ** indicates a p-value < 0.01, *** indicates a p-value < 0.001, **** indicates a p-value < 0.0001
To determine if c14 and c4 clones recapitulate increased mitochondrial Ca2+ levels, we treated digitonin-permeabilized cells with the mitochondrial uncoupler CCCP, which results in release of Ca2+ ions from the mitochondria. Both clones showed increased mitochondrial Ca2+ release compared to WT (Figs. 4C, 4D). Measurement of free matrix Ca2+ levels using mitochondria-targeted G-GECO Ca2+ reporter43 indicated higher resting Ca2+ levels in the FLC clones (Figs. 4E–4G). We also measured mitochondrial Ca2+ uptake rates in digitonin-permeabilized cells. C14 and c4 mitochondria showed decreased Ca2+ uptake rates (Figs. 4H, 4I), despite expressing MCU at comparable levels to WT cells (Fig. 4J). This decrease in Ca2+ uptake rate was not due to reduced mitochondrial membrane potential in FLC clones (Fig. S4A), suggesting that higher concentrations of Ca2+ in the matrix may negatively affect mitochondrial Ca2+ uptake rates. Since FLC tumor growth is dependent on the continued expression of the DP fusion kinase44,45, we tested whether the elevated Ca2+ levels in c14 and c4 mitochondria is also dependent on PKA activity. Interestingly, the PKA inhibitor BLU286446 did not affect the amount of Ca2+ released from WT mitochondria, but caused a significant reduction in the FLC clones (Fig. 4K), with concomitant decreases in PKA substrate phosphorylation in all cell lines (Fig. S4B).
To investigate the consequences of increased mitochondrial Ca2+ levels for oxidative phosphorylation and energy production in FLC, we measured the oxygen consumption rate (OCR) of AML12 cells with the Seahorse extracellular flux analyzer. FLC clones showed increased basal, maximal and non-mitochondrial OCR, as well as increased spare respiratory capacity, proton leak, and ATP production (Figs. 4L, S4C–S4H). These phenotypes are consistent with increased uniporter function in FLC20,47. Based on our data, we conclude that high mitochondrial Ca2+ is a previously underappreciated feature of FLC that is directly coupled to DP activity and alters FLC metabolism.
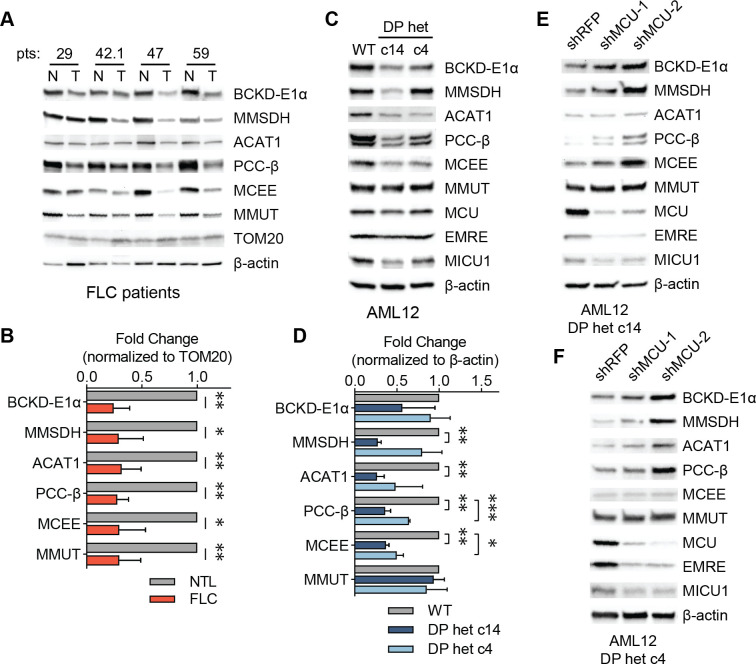
Uniporter regulates expression of proteins involved in BCAA catabolism in FLC
Decreased BCAA catabolism is associated with poor patient outcomes in hepatocellular carcinoma48, however the mechanisms by which cancer cells downregulate this pathway are not known. We asked whether increased uniporter activity in FLC suppresses expression of BCAA catabolism genes, converse to the increase we observed in MCU KO cells. Analysis of tumors from FLC patients showed significantly reduced levels of BCAA catabolism pathway enzymes compared to matching non-tumor liver samples (Fig. 5A), with less than 50% expression in the tumors compared to normal liver when protein levels were normalized to the outer mitochondrial membrane protein TOM20 (Fig. 5B). These trends were also evident in FLC clones c14 and c4 (Figs. 5C, 5D). Knockdown of MCU using two hairpin RNAs in FLC cellular models increased expression of BCAA catabolism pathway enzymes (Figs. 5E, 5F). Interestingly, unlike in MCU KO HeLa cells, we did not observe an MCU-dependent change in phosphorylation of BCKD-E1α in patient lysates or AML12 c14 and c4 cells (Figs. S5A–S5C), suggesting that mitochondrial Ca2+ signaling regulates the pathway’s activity through its effects on gene expression or protein stability in the liver. These results are consistent with previous reports49, and reflect tissue-specific differences in regulation of mitochondrial metabolism by Ca2+ signaling.
Figure 5: Uniporter inhibition alleviates suppressed BCAA catabolism in FLC.
(A) Immunoblots of select BCAA catabolism enzymes from paired non-tumor (N) and FLC tumor (T) lysates. (B) Pooled quantification of protein levels in (A) normalized to TOM20 levels; statistical significance determined by one-sample t-test. (C, D) Immunoblots (C) and quantification (D) of select BCAA catabolism proteins and MCU complex components in AML12 cells. FLC models show decreased expression of BCAA catabolism pathway enzymes; statistical significance determined by one-sample t-test; n=4. (E, F) MCU knockdown increases expression of BCAA catabolism pathway proteins in FLC clones c14 (E) and c4 (F); immunoblots of select pathway proteins and uniporter components are shown. All error bars indicate standard deviation; * indicates a p-value < 0.05, ** indicates a p-value < 0.01, and *** indicates a p-value < 0.001.
Mitochondrial Ca2+ signaling regulates expression of KLF15 and its targets in FLC
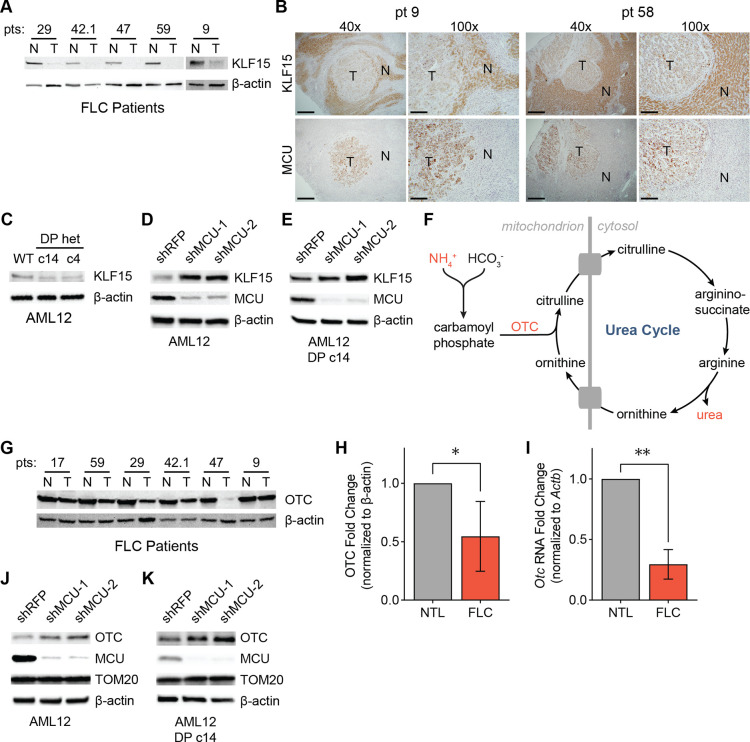
The significant decrease in the levels of BCAA pathway enzymes in FLC suggested that their transcription may be reduced. Krüppel-like factor 15 (KLF15) is an important transcriptional regulator of metabolic gene expression in the liver, including BCAA catabolism and urea cycle genes31,50,51. To investigate whether KLF15 plays a role in FLC metabolism, we first probed for KLF15 in lysates from FLC patients by Western blot. FLC tumors exhibited markedly reduced levels of KLF15 (Fig. 6A). IHC of tissue sections from FLC patients confirmed low KLF15 and high MCU expression in the tumors, with the opposite trend evident in normal liver (Fig. 6B). Likewise, AML12 clones c14 and c4 showed reduced KLF15 expression compared to WT cells (Fig. 6C).
Figure 6: Reduced KLF15 and OTC expression in FLC is regulated by the uniporter.
(A) Immunoblots of KLF15 in paired non-tumor (N) and FLC tumor (T) lysates. (B) IHC of MCU or KLF15 on non-tumor (N) and tumor (T) regions of liver from FLC patients 9 and 58 show high MCU and low KLF15 expression in the tumors; 40x image scale bars are 500 μm and 100x image scale bars are 200 μm. (C) Immunoblot of KLF15 from WT AML12, c14 and c4 lysates. (D, E) Immunoblots of KLF15 from WT AML12 (C) and c14 (D) lysates after MCU knockdown. (F) Schematic of the urea cycle; metabolites and OTC, a mitochondrial protein with reduced levels in FLC, are shown. (G) Immunoblot of OTC from paired non-tumor (N) and FLC tumor (T) lysates. (H) Quantification of OTC levels in (G) relative to β-actin. (I) qPCR analysis of OTC RNA expression in paired non-tumor liver and tumors from FLC patients 29, 42.1, 47, and 59. (J, K) Immunoblots of OTC from WT AML12 (J) and c14 (K) lysates after MCU knockdown. All error bars indicate standard deviation; * indicates a p-value < 0.05 and ** indicates a p-value < 0.01
To better understand the role of uniporter function in KLF15 regulation, we knocked down MCU in AML12 WT and c14 cells. This caused a substantial increase in KLF15 levels (Figs. 6D, 6E), suggesting that mitochondrial Ca2+ signaling negatively regulates KLF15 expression. We reasoned that reduced KLF15 expression may have important metabolic consequences beyond inhibition of BCAA catabolism in FLC, due to the regulation of the urea cycle by KLF15. Defects in the urea cycle can lead to accumulation of ammonia and cause hyperammonemia (Fig. 6F), which can cause neurological damage and death52. Interestingly, KLF15 KO mice suffer from increased blood ammonia levels due to significantly reduced OTC expression and activity in the liver31. Decreased OTC expression has also been identified as a potential cause of hyperammonemia in some FLC patients30. Protein and RNA levels of OTC showed a significant reduction in tumor samples compared to normal liver in our patient cohort (Figs. 6G–6I). Moreover, knockdown of MCU in WT AML12 and c14 cells increased OTC expression (Figs. 6J, 6K), suggesting a previously unknown link between mitochondrial Ca2+ signaling and urea cycle regulation.
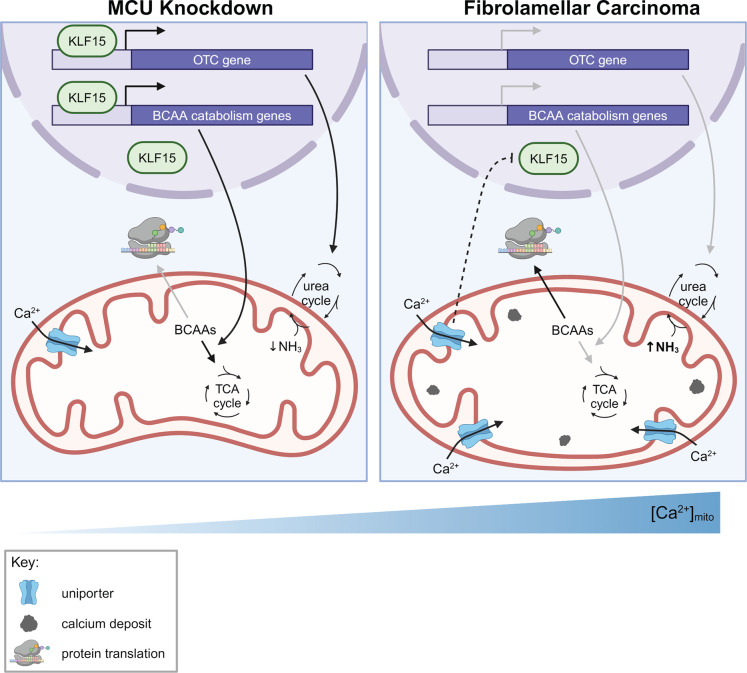
Based on our data, we propose a model in which mitochondrial Ca2+ signaling regulates the BCAA catabolism pathway and the urea cycle through the transcription factor KLF15, thereby coordinating cellular metabolic demands with fuel availability (Fig. 7). In cells with increased mitochondrial Ca2+ levels, such as FLC cells, reduced KLF15 expression leads to reduced expression of BCAA catabolism pathway genes, which conserves essential BCAAs for protein synthesis and promotes cell growth. In cells that do not have bulk mitochondrial Ca2+ uptake (MCU-deficient cells), increased KLF15 and activated BCAA catabolism supplies the TCA cycle with metabolites in the absence of Ca2+ stimulation of TCA enzymes.
Figure 7: Model for regulation of KLF15, BCAAs, and the urea cycle by mitochondrial Ca2+ signaling.
Our data suggest that under conditions of low uniporter function, KLF15 expression increases, leading to increased expression of BCAA catabolism pathway genes and OTC. Increased uniporter activity, as observed in FLC, leads to decreased BCAA catabolism pathway and OTC expression, as a result of the effects of uniporter activity on KLF15. This leads to accumulation of BCAAs that can be used for translation and growth. How the uniporter regulates KLF15 expression is not known.
DISCUSSION
We have uncovered a novel role for the mitochondrial Ca2+ uniporter in maintenance of cellular metabolism through regulation of BCAA catabolism. More specifically, we demonstrate that loss of uniporter function results in increased expression of BCAA catabolism enzymes and activation of the pathway’s committed step through dephosphorylation. Conversely, in models of high mitochondrial Ca2+, BCAA catabolism is transcriptionally suppressed. Based on our findings, we propose that mitochondrial Ca2+ signaling acts as a regulatory switch through which BCAAs are either broken down to support energy production through the TCA cycle or conserved for protein synthesis in accordance with the cell’s needs.
The BCAA catabolism pathway is responsive to nutrients, hormones, and circadian rhythm, and its activity is altered in disease states including type II diabetes, cardiovascular disease, liver disease, and cancer53–55. Here, we identify mitochondrial Ca2+ signaling as a previously unrecognized mechanism by which the catabolism pathway is regulated. Our data also point to tissue-specific differences in Ca2+ regulation of this pathway that are akin to the tissue-specific differences in Ca2+-dependent PDH phosphorylation and TCA cycle regulation56. These variances highlight the importance of studying BCAA catabolism regulation in multiple biological contexts. Further work will be necessary to delineate the contribution of Ca2+ signaling in transcriptional and post-translational regulation of BCAA catabolism across a broader range of cell types and tissues.
The majority of BCAA catabolism occurs in skeletal muscle, adipose tissue, and the liver57. However, the initial conversion of BCAAs to α-ketoacids is not performed in the liver, due to the lack of BCAT1 and BCAT2 expression. Instead, skeletal muscle tissue processes BCAAs from the diet and supplies the liver with α-ketoacids through the bloodstream53. In the current study, most experiments were performed in hepatocyte-derived cell lines without BCAT expression or in frozen liver tissue. As such, we were unable to measure changes in BCAA consumption in these samples. This limited our ability to assess rates of BCAA catabolism directly. Nonetheless, regulation of the pathway components remains sensitive to alterations in cell lines, enabling us to characterize upstream effectors regardless of pathway activity in vitro.
We further demonstrated that the relationship between mitochondrial Ca2+ signaling and BCAA catabolism is conserved in FLC. To date, there are no effective treatments for FLC other than surgical resection, indicating a need to elucidate the underlying mechanisms driving this cancer and identify potential drug targets. The genetic driver of fibrolamellar oncogenesis was identified by Honeyman et al. in 2014 with the discovery of the DNAJ-PKAc fusion kinase37. In the subsequent decade, progress has been made in characterizing downstream signaling changes, however, it remained unclear how DP gives rise to the mitochondrial and metabolic alterations observed in FLC tumors30,41,58. Our results confirm previous reports that FLC mitochondria contain an increased number of Ca2+ deposits28 and demonstrate for the first time that DP-expressing cells have elevated levels of free Ca2+ in their mitochondria. We show that FLC tumors have suppressed expression of many of the proteins responsible for BCAA catabolism and that this phenotype is reversible by inhibiting mitochondrial Ca2+ influx.
By reducing BCAA catabolism, FLC tumors conserve their supply of leucine, valine, and isoleucine. We postulate that these essential amino acids are used to sustain increased rates of protein synthesis, supported by a previous study showing a nearly 2-fold increase in protein translation in DP cells58. Additionally, elevated levels of free BCAAs may have important signaling functions. For example, reduced catabolism of BCAAs in the liver can stimulate mTORC148, and accumulation of α-ketoacids inhibits gluconeogenesis in hepatocytes59. Whether reduced BCAA catabolism alters signaling and directly contributes to metabolic changes in FLC remains to be seen.
BCAA catabolism enzymes are known to be regulated by the transcription factor KLF1551,60. We observe a significant, uniporter-dependent reduction in KLF15 protein levels in DP-expressing cells. Reduction in KLF15 levels has been correlated with tumor growth and poor prognosis in several types of cancer61–66, but to our knowledge, this is the first time KLF15 expression changes have been implicated in liver cancer. KLF15 also regulates genes associated with gluconeogenesis, lipogenesis, metabolism of non-BCAA amino acids, and the urea cycle31,67,68. In line with this, FLC patients can develop an impairment in the urea cycle, resulting in hyperammonemia and, in some cases, hyperammonemic encephalopathy30,69–71. This phenotype has been attributed to reduced expression of the KLF15 target OTC30, which we show can be reversed through knockdown of MCU. Downregulation of OTC may favor tumor growth and survival, as build-up of ammonia in the tumor microenvironment has been shown to inhibit immunosurveillance72–76. Additionally, accumulation of the OTC substrates ammonia and ornithine can support the production of collagen75,77,78. Collagen is a large component of the fibrous bands which are a hallmark of FLC tumors. Fibrosis has also been attributed to downregulation of KLF15 in heart and kidney disease79–81. Furthermore, elevated levels of MCU are associated with increased collagen production and liver fibrosis82. Future investigations should consider a potential link between the uniporter, KLF15 and these pathologic features of FLC.
Our model of FLC signaling asserts that mitochondrial Ca2+ concentrations are increased due to aberrant DP activity and that mitochondrial Ca2+ then suppresses KLF15, thereby regulating expression of BCAA catabolism and urea cycle enzymes. Additional studies are warranted to determine the mechanistic link between DP and accumulation of mitochondrial Ca2+, in particular the upregulation of uniporter proteins. PKA is a multifunctional kinase whose activity and subcellular localization are tightly regulated through A-kinase anchoring proteins (AKAPs)83. dAKAP-1 associates with the outer surface of the mitochondria and orchestrates PKA-dependent local protein synthesis in the perimitochondrial space, directly affecting mitochondrial function84. In contrast to wildtype PKA, the DP kinase shows reduced binding to d-AKAP1 and other AKAPs and consequently exhibits more diffuse cytosolic localization58. Although the mechanism of DP-induced mitochondrial Ca2+ changes remains to be elucidated, a requirement for PKA kinase activity suggests that either reduced PKA localization to the mitochondria or novel PKA substrates targeted by DP lead to mitochondrial Ca2+ changes in FLC. It also remains to be determined how mitochondrial conditions affect either the expression or stability of KLF15 in FLC. However, from these findings, it is clear that Ca2+ signaling is an important component of the bidirectional communication between mitochondria and the rest of the cell.
The MCU/KLF15/BCAA catabolism signaling axis that we identified in FLC may also play a role in other forms of cancer. Based on analysis of the Cancer Genome Atlas (TGCA), the BCAA catabolism pathway was shown to be downregulated in approximately 70% of cancers48. Additionally, both suppressed BCAA catabolism and increased uniporter activity have been reported to correlate with poor patient prognosis in hepatic cancers48,85–87. This raises the intriguing possibility that uniporter-mediated metabolic changes may be a more general mechanism of metabolic regulation in cancer, whereby increased mitochondrial Ca2+ levels stimulate TCA cycle activity, downregulate BCAA catabolism, and conserve essential amino acids for protein synthesis and cell proliferation.
MATERIALS and METHODS
Cell lines and tissue culture
All cell lines were grown in a standard tissue culture incubator (VWR, 75875–212) at 37°C with 5% CO2. HeLa cells were cultured in DMEM (Thermo Fisher Scientific, 11–965-118) supplemented with 10% FBS (VWR, 89510–186), 2 mM GlutaMAX (Thermo Fisher Scientific, 35–050-061), and 100 IU/mL penicillin and 100 μg/mL streptomycin (VWR, 45000–652). HeLa cells were obtained from the Whitehead Institute for Biomedical Research. The HeLa cell line has the following short tandem repeat profile: D5S818 (11, 12); D13S317 (12, 13.3); D7S820 (8, 12); D16S539 (9, 10); vWA (16, 18); TH01 (7); AMEL (X); TPOX (8, 12); CSF1PO (9, 10). This profile is a 94% match to HeLa Cervical Adenocarcinoma (Human) (CCL-2; ATCC) according to the analysis performed by the ATCC database. AML12 cells were cultured in DMEM/F-12 medium (Gibco, 11330057) supplemented with 10% FBS (VWR, 89510–186), 1x insulin-transferrin-selenium (Gibco, 41400045), 40 ng/mL dexamethasone (MP Biomedicals, 0219456180), and 100 IU/mL penicillin and 100 μg/mL streptomycin (VWR, 45000–652). AML12 cells were obtained from ATCC (CRL-2254, lot #70039497) and CRISPR-edited AML12 DNAJ-PKAc clones c14 and c4 were generated and validated by the Scott Lab as described41. All cell lines were tested for mycoplasma using the Genlantis MycoScope PCR Detection Kit (VWR, 10497–508) and were confirmed to be free of mycoplasma contamination.
Generation of knockout and rescue cell lines
Broad Institute gRNA sequences for MCU (TGAACTGACAGCGTTCACGC) and EMRE (GTCTCAGCCAGGTACCGTCG) were each cloned into the pU6T7 vector (Addgene, 71462). 5 million HeLa cells were resuspended in 400 μL DMEM with 2 μg SpCas9 plasmid (Addgene, 71814), 500 ng gRNA plasmid, and 17.5 μg pUC19 plasmid (Addgene, 50005). The cells and DNA were transferred to a cuvette, incubated on ice for 5 min, and electroporated at 200 V with a capacitance of 950 μF and pulse length of 23 msec. Electroporated cells were allowed to grow to confluence on a 10 cm plate before being diluted to 1 cell/200 μL and plated in 96-well plates to obtain single cell clones. HeLa MCU KO and EMRE KO clones were verified by Western blotting, functional assays, and sequencing. To rescue MCU expression, MCU KO clone 18 was infected with lentivirus containing an MCU-FLAG construct with the sequence below. Following resistance marker selection, single cell clones were isolated and screened for MCU expression by Western blot.
MCU-FLAG DNA sequence
ATGGCGGCCGCCGCAGGTAGATCGCTCCTGCTGCTCCTCTCCTCTCGGGGCGGCGGCGGCGGGGGCGCCGGCGGCTGCGGGGCGCTGACTGCCGGCTGCTTCCCTGGGCTGGGCGTCAGCCGCCACCGGCAGCAGCAGCACCACCGGACGGTACACCAGAGGATCGCTTCCTGGCAGAATTTGGGAGCTGTTTATTGCAGCACTGTTGTGCCCTCTGATGATGTTACAGTGGTTTATCAAAATGGGTTACCTGTGATATCTGTGAGGCTACCATCCCGGCGTGAACGCTGTCAGTTCACACTCAAGCCTATCTCTGACTCTGTTGGTGTATTTTTACGACAACTGCAAGAAGAGGATCGGGGAATTGACAGAGTTGCTATCTATTCACCAGATGGTGTTCGCGTTGCTGCTTCAACAGGAATAGACCTCCTCCTCCTTGATGACTTTAAGCTGGTCATTAATGACTTAACATACCACGTACGACCACCAAAAAGAGACCTCTTAAGTCATGAAAATGCAGCAACGCTGAATGATGTAAAGACATTGGTCCAGCAACTATACACCACACTGTGCATTGAGCAGCACCAGTTAAACAAGGAAAGGGAGCTTATTGAAAGACTAGAGGATCTCAAAGAGCAGCTGGCTCCCCTGGAAAAGGTACGAATTGAGATTAGCAGAAAAGCTGAGAAGAGGACCACTTTGGTGCTATGGGGTGGCCTTGCCTACATGGCCACACAGTTTGGCATTTTGGCCCGGCTTACCTGGTGGGAATATTCCTGGGACATCATGGAGCCAGTAACATACTTCATCACTTATGGAAGTGCCATGGCAATGTATGCATATTTTGTAATGACACGCCAGGAATATGTTTATCCAGAAGCCAGAGACAGACAATACTTACTATTTTTCCATAAAGGAGCCAAAAAGTCACGTTTTGACCTAGAGAAATACAATCAACTCAAGGATGCAATTGCTCAGGCAGAAATGGACCTTAAGAGACTGAGAGACCCATTACAAGTACATCTGCCTCTCCGACAAATTGGTGAAAAAGATTCTAGAGGTGGATCTGGTGGATCTGGTGGATCTATGGATTACAAGGATGACGATGACAAG
Proliferation assays
20,000 HeLa cells were plated in 6-well plates on day 0. On days 2, 3, and 5, the media was aspirated, and the cells were washed with PBS (Thermo Fisher Scientific, 20012050), detached with 200 μL trypsin solution (Gibco, 12605–010), and then resuspended in 1.8 mL media for a total volume of 2 mL. The resuspended cells were counted using a Beckman Coulter Z2 Cell and Particle Counter (Beckman Coulter, 383550). 100,000 AML12 cells were plated in 10 cm plates on day 0. On days 2, 3, and 4, the media was aspirated, and the cells were washed with PBS (Thermo Fisher Scientific, 20012050), detached with 2 mL trypsin solution (Gibco, 12605–010), and then resuspended in 4 mL media, for a total volume of 6 mL. The resuspended cells were counted using a Coulter Z2 Cell and Particle Counter (Beckman Coulter, 383550).
Mass spectrometry sample preparation
Five replicates of 500,000 WT and MCU KO HeLa cells were plated in 10 cm plates. After 48 hr, the media was aspirated, and cells were washed with cold PBS (Thermo Fisher Scientific, 20012050) and lysed with 350 μL fresh lysis buffer consisting of 8 M urea (Sigma-Aldrich, U5128), 1 M Tris, 1 M tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Sigma-Aldrich, C4706), and 0.6 M 2-chloroacetamide (CAM, Sigma-Aldrich, C0267). Lysates were sonicated on ice for 10 min with 30 sec on/off pulses at an amplitude setting of 100. Samples were vortexed at 1400 rpm on a thermomixer (Eppendorf) for 30 min at 37 °C. Samples were then centrifuged at 17,000 g for 10 min at 4 °C, and 175 μL of the supernatant was transferred to a new tube. 700 μL cold acetone was added to each sample. 43.75 μL Trichloroacetic acid (TCA, Sigma-Aldrich, T4885) was then added for a final concentration of 5%. The samples were vortexed briefly at 1400 rpm and stored overnight at −20 °C. Samples were centrifuged at 6,000 g for 10 min at 4 °C, and the supernatant was aspirated. 500 μL cold acetone was added to the pellet, and the pellet was sonicated in an ice bath until the pellet was dissolved (5–10 min). The samples were incubated on ice for another 15 min then centrifuged at 6,000 g for 10 min at 4 °C. The supernatant was aspirated, and the pellets were air dried for 2–3 min. Samples were resuspended in 200 μL 8 M urea, 100 mM Tris solution. Samples were vortexed for 1 hr at 37 °C to dissolve the pellet in solution. Protein concentrations were measured with the Pierce 660 nm Protein Assay (Thermo Fisher Scientific, 22660) and 500 μg of protein per sample was used for subsequent mass spectrometry experiments. Samples were diluted 1:1 with 100 mM triethylammonium bicarbonate (TEAB, Sigma, T7408), reducing the urea concentration to 4 M. A stock of 2 μg/μl Lys-C endoproteinase (Fujifilm Wako Chemicals, 121–05063) was added to each sample for an enzyme:protein ratio of 1:100. Samples were vortexed at 1400 rpm for 2 hr at 37 °C. Samples were again diluted 1:1 with 100 mM TEAB, bringing the urea concentration to 2 M. 0.5 μg/μl stock of trypsin (Promega, V5111) dissolved in 40 mM acetic acid was added to each sample for an enzyme:protein ratio of 1:100. Samples were vortexed at 1400 rpm overnight at 37 °C.
The following day, HPLC-grade formic acid (Sigma, 5438040100) was added for a final concentration of 2% by volume. Samples were centrifuged at 11,000 g for 5 min. 5% of the supernatant was used for stage tipping. C18 StageTips were made as described in Rappsilber et al.88. StageTips were washed with sequential washes of 50 μL HPLC-grade methanol (Sigma, 494291), Buffer B, and Buffer A, centrifuging the tips at 1,800 g for 2 min between each wash. Buffer B consisted of 0.1% HPLC-grade trifluoroacetic acid (TFA, Alfa Aesar, AA446305Y) and 80% HPLC-grade acetonitrile (Sigma-Aldrich, 271004). Buffer A consisted of 0.1% TFA and 5% acetonitrile. Samples were loaded on the StageTips, centrifuged at 1,800 g for 2 min, and washed again with Buffer A.
LC-MS/MS-based proteomics and data analysis
Peptides were separated on a Thermo-Dionex RSLCNano UHPLC instrument (Sunnyvale, CA) with 10 cm fused silica capillary columns made in-house with a laser puller (Sutter, Novato CA) and packed with 3 μm reversed phase C18 beads. The LC gradient was 90 min long with 3–35% of solvent B at 200 nL/min. LC solvent A was 0.1% acetic acid and LC solvent B was 0.1% acetic acid, 99.9% acetonitrile. MS data was collected with a Thermo Orbitrap Elite (Thermo Fisher Scientific). Data-dependent analysis was applied using Top15 selection with CID fragmentation. Raw files were analyzed by MaxQuant (version 1.5.2.8) using protein and peptide FDRs of 0.01 and a score minimum of 40 for modified peptides, 0 for unmodified peptides; delta score minimum of 17 for modified peptides, 0 for unmodified peptides. MS/MS spectra were searched against the UniProt human database. MaxQuant search parameters: Variable modifications included Oxidation (M), phosphorylation (STY). Carbamidomethyl (C) was a fixed modification. Max. labeled amino acids was 3, max. missed cleavages was 2, enzyme was Trypsin/P, max. charge was 7, and multiplicity was 3. The initial search tolerance was 20 ppm for FTMS scans and 0.5 Da for ITMS MS/MS scans. Data was further processed using the Perseus software package (version 1.5.2.6), the R environment, Origin Pro 8.0 and Microsoft Excel. GO term enrichment analysis was performed using the ShinyGO 0.76 web application34.
RNA sequencing
500,000 HeLa cells were plated in 6-well plates. After two days, total RNA was isolated using an RNeasy kit (Qiagen, 74106). Samples were sent to Genewiz for sequencing and data analysis, as follows: Samples underwent mRNA enrichment, mRNA fragmentation, and random priming. Genewiz performed first and second strand cDNA synthesis followed by end repair, 5’ phosphorylation, and dA-Tailing. cDNA was sequenced, following adaptor ligation and enrichment by PCR. During analysis, the reads were trimmed and mapped to the Homo sapiens GRCh38 reference genome available89 (https://www.ensembl.org) using the STAR aligner (v.2.5.2b). Hit counts for genes and exons were calculated with featureCounts (Subread package, v.1.5.2). DESeq2 was used for analysis of differential gene expression. p-values and log2 fold changes were determined with the Wald test, and genes with an adjusted p-value < 0.05 and absolute log2 fold change > 1 were considered to be differentially expressed.
Seahorse metabolic flux analysis
HeLa and AML12 cells were seeded at 10,000 or 15,000 cells/well, respectively, in a XFe96 Microplate (Agilent Technologies, 103792–100) and cultured overnight at 37°C in 5% CO2. The Extracellular Flux Assay Kit oxygen probes (Agilent Technologies, 103792–100) were activated in XF calibrant buffer (Agilent, 100840–000) at 37°C in a non-CO2 incubator overnight. Seahorse XF medium was prepared fresh by supplementing XF DMEM Medium, pH 7.4 (Agilent, 103575–100) with 100 mM Pyruvate Solution (Agilent, 103578–100), 1 M Glucose Solution (Agilent, 103577–100), and 200 mM L-Glutamine (Fisher Scientific, BP379–100). The cell culture medium was removed from the plate and the cells were washed once with pre-warmed Seahorse XF medium and replaced with fresh medium a second time. The cells were then incubated in a CO2-free incubator at 37°C for 1 h. Oxygen consumption rate (OCR) measurements were performed with a Seahorse XFe96 Extracellular Flux Analyzer (Agilent, S7800B) using the Cell Mito Stress Test program. The sensor cartridge was loaded with the drugs and inserted into the machine 30 min prior to the flux assay to calibrate the oxygen probes. Following measurements of baseline cellular respiration, respiration was measured after addition of oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), antimycin A, and rotenone with final concentrations of 1 μM. Oxygen consumption rate was analyzed with the Seahorse XF Cell Mito Stress Test Report Generator (Wave Software). OCR measurements were normalized to cell number as determined by the CyQUANT Cell Proliferation Assay Kit (Thermo Scientific, C7026) or DAPI staining for HeLa and AML12 cells, respectively. For the CyQUANT assay, the media was removed from each well and the plates were frozen at −80 °C for >1 hr to lyse the cells. CyQUANT was utilized per manufacturer’s instructions to measure fluorescence intensity (Ex 480 nm, Em 520 nm). For DAPI staining, 40 μL media was left in each well and cells were frozen at −80 °C for >1 hr to lyse the cells. After thawing, 100 μL 1 μg/μL DAPI (Thermo Scientific, 62248) was added to each well for 5 min before measuring DAPI fluorescence (Ex 359 nm, Em 457 nm).
LC-MS-based lipid analysis
Cell pellets containing 10 million HeLa cells per condition were frozen at −80 °C until lipid extraction. Lipid extraction and analysis were carried out as described90. Briefly, cell pellets were resuspended in methanol and sonicated 3×30 sec while on ice. The homogenized solution was centrifuged at 16,900 g for 15 min at 4 °C. 800 μL of supernatant was removed into a fresh 4-dram vial. 1 mL cold methanol was added to the remaining pellet, sonicated, and centrifuged again. This supernatant was combined with the previous extract and dried down using a nitrogen evaporator. LC-MS data was acquired using an Agilent 1260 HPLC with an Agilent 6530 Quadrupole Time-of-Flight mass spectrometer. A Luna C5 reverse-phase column (5m, 4.6 × 50mm, Phenomenex) was used for positive mode and a Gemini C18 reversed-phase column (5m, 4.6 × 50mm, Phenomenex) was used in negative mode. Mobile phase A was 95:5 water:methanol and mobile phase B was 60:35:5 isopropanol:methanol:water for both negative and positive modes. The mobile phases contained additives to aid electrospray ionization and improve ion detection (0.1% formic acid in positive mode; 5 mM ammonium formate, and 0.1% ammonium hydroxide in negative mode). Data acquisition in positive and negative mode used different gradients: ramping from 0% mobile phase B to 100% mobile phase B occurred over 45 min and 65 min for positive and negative mode, respectively. For both methods, the flow rate was set to 0.1 mL/min for the first 5 min at 100% mobile phase A then increased to 0.5 mL/min for the remainder of the run. A dual electrospray ionization source was used; capillary and fragmentor voltages were set to 3500 V and 175 V, respectively. All data were collected in the extended dynamic range mode (m/z = 50 – 1,700). Representative lipid species for each class were targeted by extracting the corresponding m/z for each ion in MassHunter Qualitative Analysis software (version B.06.00, Agilent Technologies). Peak areas were manually integrated and represented as abundance. Relative abundance values were calculated by dividing the abundance of a lipid by the average abundance of that lipid in the wildtype set.
Cell lysate preparation and immunoblots
Cells were grown to 40–80% confluency, then plates were washed with cold PBS (Thermo Fisher Scientific, 20012050) on ice and lysed with either RIPA or triton lysis buffer. RIPA buffer consisted of 50 mM HEPES-KOH, pH 7.4 (Sigma-Aldrich, H3375; Sigma Millipore, 1050121000), 150 mM NaCl (Sigma-Aldrich, 746398), 2 mM EDTA (Sigma-Aldrich, 607–429-00–8), 10 mM sodium pyrophosphate (Sigma-Aldrich, 71501), 10 mM β-glycerophosphate (Sigma-Aldrich, G9422), 1% sodium deoxycholate (Sigma-Aldrich, D6750), 0.1% SDS (Sigma-Aldrich, L4509), and 1% Triton X-100 (Sigma-Aldrich, X100). Triton buffer consisted of 50 mM HEPES-KOH, pH 7.4, 40 mM NaCl, 2 mM EDTA, 10 mM β-Glycerophosphate, 1.5 mM sodium orthovanadate (Sigma-Aldrich, S6508), 50 mM sodium fluoride (Sigma-Aldrich, S7920), 10 mM sodium pyrophosphate, and 1% Triton X-100. Lysis buffers were supplemented with protease inhibitors (Sigma-Aldrich, 11836170001) immediately prior to lysis. Lysates were centrifuged at 17,000 g for 10 min at 4°C. The supernatants were normalized by protein concentration as measured by either a Bradford protein assay (Bio-Rad, 5000205) or Pierce BCA assay (Thermo Fisher Scientific, 23209) and a BioTek Synergy H1 plate reader (Fisher Scientific, 11–120-536). SDS-PAGE samples were prepared from the lysates with 5X reducing sample buffer (pH 6.8) consisting of 10% SDS, 25% 2-mercaptoethanol (Sigma-Aldrich, M3148), 25% glycerol (Sigma-Aldrich, G5516), 50 mM Tris-HCl (Sigma-Aldrich, RDD008), and 0.1% bromophenol blue (VWR, 97061–690).
Samples were heated to 95 °C for 5 min and then brought to room temperature. Samples were mixed by pipetting and loaded onto 4–15% gradient Mini-PROTEAN TGX gels (Bio-Rad, 4561086). Following gel electrophoresis in Tris-Glycine-SDS running buffer (Boston BioProducts, BP-150), proteins were transferred to ethanol-activated 0.2 μm PVDF membranes (Bio-Rad, 1620174) using the BioRad Mixed Molecular Weight Protein Transfer setting (7 min, 1.3 A, 25 V) on the Trans-Blot Turbo Transfer System (Bio-Rad, 1704150) with Trans-Blot Turbo Transfer Buffer (Bio-Rad, 10026938). After transfer, membranes were washed in 20% ethanol for ~2 min and checked for equal loading and even transfer by Ponceau stain (Sigma-Aldrich, P7170). Membranes were then incubated with 1% wt/vol BSA (Sigma-Aldrich, A2153) in TBST with 1% Tween-20 (Boston Bio-Products, IBB-180) at room temperature for 1–4 hr. Membranes were incubated overnight with primary antibody and 1% BSA in TBST at 4 °C. All membranes were then washed with TBST three times, 5 min each, and incubated with secondary antibody and 1% BSA in TBST for 1 hr using a BlotCycler W5 (Precision Biosystems). Membranes were washed 4 times, 5 min each, with TBST. Membranes were developed using Clarity Western ECL substrate (Bio-Rad, 170–5060) or Clarity Max Western ECL substrate (Bio-Rad, 1705062). Immunoblots were imaged with a iBrightCL1000 imager (Life Technologies, A23749) and quantified using the Image Studio Lite software, version 5.2 (LI-COR Biosciences). The following antibodies and dilutions were used: 1:3000 rabbit ACAT1 antibody (ABclonal, A13273); 1:3000 rabbit BCKDHA antibody (ABclonal, A21588); 1:5000 mouse β-actin antibody (Cell Signaling Technology, 3700); 1:5000 rabbit EMRE antibody (Bethyl Laboratories, A300-BL19208); 1:3000 rabbit HSP60 antibody (Cell Signaling Technology, 12165); 1:3000 rabbit KLF15 antibody (ABclonal, A7194); 1:3000 rabbit MCEE antibody (ABclonal, A14430); 1:3000 rabbit MCU antibody (Cell Signaling Technology, 14997); 1:3000 rabbit MICU1 antibody (ABclonal, A21948); 1:3000 rabbit MMSDH antibody (ABclonal, A3309); 1:3000 rabbit MMUT antibody (ABclonal, A3969); 1:1000 mouse OTC antibody (Invitrogen, PAS-28197); 1:3000 rabbit PCCB antibody (ABclonal, A5415); 1:3000 rabbit phospho-BCKDH-E1α, human S292/mouse S293 antibody (Cell Signaling Technology, 40368); 1:1000 rabbit phospho-R-X-S*/T* (Cell Signaling Technology, 9621); 1:3000 rabbit PKA-Cα antibody (BD Biosciences, 610980); 1:3000 rabbit TOM20 antibody (Cell Signaling Technology, 42406); 1:10,000 HRP-linked α-rabbit secondary antibody (Cell Signaling Technology, 7074); 1:10,000 HRP-linked α-mouse secondary antibody (Cell Signaling Technology, 7076).
Isolation of patient samples
Fresh non-tumor liver (NTL) and FLC tumor samples were procured from patients undergoing liver resection for FLC. Prior to surgery, written-informed consent was obtained for tissue donation under a research protocol approved by the University of Washington Institutional Review Board (IRB) (#1852) and Seattle Children’s Hospital IRB (#15277). All research samples were de-identified. Clinical details for each patient are outlined in Supplemental Table 1. NTL and FLC samples were processed and transferred to the laboratory on ice within 1 hr of tissue excision. Samples were stored at −80 °C prior to lysate preparation.
Lysis and sample preparation of patient samples
Tissues were lysed with protease inhibitor-supplemented RIPA buffer using a TissueRuptor homogenizer (Qiagen, 9002755). Tissues were homogenized until they were a uniform consistency. Samples were then centrifuged at 17,000 g for 10 min at 4 °C and prepared for SDS-PAGE with 5X reducing sample buffer.
Generation of shRNA constructs
shRNA sequences were designed by the RNAi Consortium (TRC) of the Broad Institute. Forward and reverse oligos encoding the shRNA were annealed to form dsDNA. 100 pmol forward oligo and 100 pmol reverse oligo were resuspended in 25 μL 1x NEBuffer 2 (NEB, B7002S). The solution was incubated in a 95 °C water bath for 4 min then slowly cooled (~0.5 °C/min) to room temperature. The pLKO.1 vector (Addgene, 10878) was digested with AgeI-HF (NEB, R3552) and EcoRI-HF (NEB, R3101) and ligated with the oligo solution according to the NEB Quick Ligase (NEB, M2200) protocol. The ligation mixture was transformed into XL10-Gold competent cells (Agilent, 200315). Resulting plasmids were verified by sequencing.
Mouse shMCU-1, forward oligo:
CCGGGATCCGAGATGACCGTGAATCCTCGAGGATTCACGGTCATCTCGGATCTTTTTG;
reverse oligo:
AATTCAAAAAGATCCGAGATGACCGTGAATCCTCGAGGATTCACGGTCATCTCGGATC.
Mouse shMCU-2, forward oligo:
CCGGTAGGGAATAAAGGGATCTTAACTCGAGTTAAGATCCCTTTATTCCCTATTTTTG;
reverse oligo:
AATTCAAAAATAGGGAATAAAGGGATCTTAACTCGAGTTAAGATCCCTTTATTCCCTA.
shGFP control, forward oligo:
CCGGCCACATGAAGCAGCACGACTTCTCGAGAAGTCGTGCTGCTTCATGTGGTTTTTG;
reverse oligo:
AATTCAAAAACCACATGAAGCAGCACGACTTCTCGAGAAGTCGTGCTGCTTCATGTGG
shRFP control, forward oligo:
CCGGGCTCCGTGAACGGCCACGAGTCTCGAGACTCGTGGCCGTTCACGGAGCTTTTTG;
reverse oligo:
AATTCAAAAAGCTCCGTGAACGGCCACGAGTCTCGAGACTCGTGGCCGTTCACGGAGC
Lentivirus production and transduction
1 million HEK293T cells (from the Whitehead Institute for Biomedical Research) were plated in 6 cm plates. The following day, cells were transfected. 100 ng p-CMV-VSV-G (Addgene, 8454), 900 ng psPax2 (Addgene, 12260), and 1 μg viral plasmid were diluted in 6 μL X-tremeGENE 9 DNA transfection reagent (Sigma-Aldrich, 6365787001) and 150 μL DMEM and incubated at room temperature for 15 min before being added to the cells. After 36–48 hr, the virus-containing medium was filtered through a 0.45 μm sterile filter (VWR, 28145–505) and stored at −80 °C until use. 200,000 HeLa or 100,000 AML12 cells were plated in 6-well plates. The following day, 200 μL of the virus-containing media and 8 μg/mL polybrene (Sigma-Aldrich, H9268) was added to the cells. Infected cells were selected for 48 hr with 2 μg/mL puromycin. For assays measuring transcriptional changes in the BCAA catabolism pathway, shMCU-infected cells were cultured for 2–3 weeks prior to harvesting.
RNA isolation, cDNA synthesis, qPCR
AML12 cells were grown in 6-well plates to ~80% confluency in triplicate. Tumor and non-tumor liver samples were stored in RNALater solution at −80 °C prior to RNA extraction. RNA was isolated with the RNeasy Kit (Qiagen, 74106). For liver samples, ~30 mg of tissue was homogenized using a TissueRuptor homogenizer (Qiagen, 9002755) according to the kit protocol. 100 ng of RNA was used for cDNA synthesis using the Maxima First Strand cDNA synthesis kit (Thermo Fisher Scientific, K1641) according to kit instructions. The resulting cDNA was diluted 1:30 in dH2O, and either 1 or 2 μL of diluted cDNA was used for qPCR using Applied Biosystems TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific, 44–445-57) in a 384-well plate (Thermo Fisher Scientific, 43–098-49) using a QuantStudio 5 RT-PCR System (Thermo Fisher Scientific). The following Taqman probes were used: human ACTB (Thermo Fisher Scientific, 4331182-Hs01060665_g1), human MCU (Thermo Fisher Scientific, 4331182-Hs00293548_m1), and human OTC (Thermo Fisher Scientific, 4331182-Hs00166892_m1).
Immunohistochemistry
Fresh human FLC tissue from liver resections were fixed in formalin and paraffin embedded. 4 μm thick sections were cut and placed on glass slides. Slides were stained with Hematoxylin and Eosin (H&E) using a H&E staining kit (Vector Laboratories, H-3502) according to supplied protocol. For immunohistochemistry staining, slides were deparaffinized, rehydrated, and washed with TBST. After antigen retrieval in 10 mM sodium citrate (pH 6) and quenching of endogenous peroxidase activity with 3% H2O2, samples were blocked with 2.5% normal horse serum (NHS) before incubation with primary antibodies overnight at 4°C. The following primary antibodies were used: 1:300 KLF15 rabbit antibody (ABclonal, A7194) and 1:250 MCU rabbit antibody (Cell Signaling Technologies, 14997). Negative controls were treated with 2.5% NHS without primary antibodies. Signals were processed using the ImmPRESS HRP Horse Anti-Mouse (Vector Laboratories, MP-7402) and Anti-Rabbit (Vector Laboratories, MP-7401) Peroxidase Kits and ImmPACT DAB Substrate Kit Peroxidase (Vector Laboratories, SK-4105) according to manufacturer’s protocol. Slides were counterstained with Hematoxylin QS, dehydrated, and mounted with VectaMount Express Mounting Medium.
Ca2+ uptake in permeabilized cells
Protocol is adapted from Sancak et al.10. Cell medium was changed 1–2 hr prior to harvesting. Cells were detached with trypsin (Gibco, 12605–010) and resuspended in cell medium. 1 million HeLa cells or 0.5 million AML12 cells were centrifuged at 800 g for 3 min. Cell pellets were washed with PBS (Thermo Fisher Scientific, 20012050), centrifuged at 800 g for 3 min, and resuspended in 150 μL KCl buffer. KCl buffer consists of 125 mM KCl (Sigma-Aldrich, 793590), 2 mM K2HPO4 (Sigma-Aldrich, P3786), 1 mM MgCl2, (Sigma-Aldrich, M8266), and 20 mM HEPES, pH 7.2 (Sigma-Aldrich, H3375) supplemented fresh with 5 mM glutamate (Sigma-Aldrich, G1251), 5 mM malate (Sigma-Aldrich, M7397), and 1 μM Oregon Green 488 Bapta-6F (Invitrogen, O23990). 0.01% or 0.005% digitonin (Thermo Fisher Scientific, BN2006) was added for HeLa and AML12 cell assays, respectively. Cells were transferred to a black 96-well plate (Greiner Bio-One, 655076). Fluorescence (Ex 485/20 nm, Em 520/20 nm) was monitored every 2 sec for 136 sec at room temperature using a BioTek Synergy H1 microplate reader (Fisher Scientific, 11–120-536) before and after injection of 50 μM CaCl2 (Sigma-Aldrich, 746495). Ca2+ uptake rates were calculated using the linear fit of uptake data points between 20 and 30 sec after Ca2+ injection. A maximum of eight samples were assayed together, including one wildtype control per assay run.
Ca2+ release from permeabilized cells
Cell medium was changed 1–2 hr prior to harvesting. Cells were detached with trypsin (Gibco, 12605–010) and resuspended in cell medium. 1 million HeLa cells or 0.5 million AML12 cells were centrifuged at 800 g for 3 min. Cell pellets were washed with PBS (Thermo Fisher Scientific, 20012050), centrifuged at 800 g for 3 min, and resuspended in 150 μL KCl buffer. KCl buffer consists of 125 mM KCl (Sigma-Aldrich, 793590), 2 mM K2HPO4 (Sigma-Aldrich, P3786), 1 mM MgCl2, (Sigma-Aldrich, M8266), and 20 mM HEPES, pH 7.2 (Sigma-Aldrich, H3375) supplemented fresh with 1 μM Oregon Green 488 Bapta-6F (Invitrogen, O23990). 0.01% or 0.005% digitonin (Thermo Fisher Scientific, BN2006) was added for HeLa and AML12 cell assays, respectively. Cells were transferred to a black 96-well plate (Greiner Bio-One, 655076). Fluorescence (Ex 485/20 nm, Em 520/20 nm) was monitored every 2 sec for 196 sec at room temperature using a BioTek Synergy H1 microplate reader (Fisher Scientific, 11–120-536) before and after injection of 1 μM CCCP (Cayman Chemical Company, 25458) in KCl buffer. Ca2+ release was calculated using the absolute difference between fluorescence reads prior to CCCP injection (average of six values, 0–10 sec) and 3 min after injection (average of six values, 186–196 sec). A maximum of eight samples were assayed together, including one wildtype control per assay run. For PKA kinase inhibition assays, AML12 cells were cultured for 4 days with 5 μM BLU2864 or DMSO vehicle control. Cells were passaged on day 2, and fresh medium and drug were added. Ca2+ release experiments with BLU2864 were performed with ~0.5 million cells, however BLU2864-treated cells formed clumps and could not be counted precisely. Data were instead normalized by protein concentration. Following Ca2+ release, 1 μL Triton X-100 (Sigma-Aldrich, X100) was added to each well, and the solution was pipetted vigorously. Protein concentration was then determined by Bradford assay (Bio-Rad, 5000205).
TEM of FLC patient liver
Tissue fixation was performed with 4% glutaraldehyde in 0.1 M sodium cacodylate buffer, then stored overnight at 4 °C. The tissue was then washed 5 × 5 min in buffer at room temperature and post fixed in buffered 2% osmium tetroxide on ice for 1 hr. This was followed by 5 washes in ddH2O, then en bloc staining in 1% uranyl acetate (aqueous), overnight at 4 °C. The following day the tissue was washed 5 × 5 minutes in ddH2O then dehydrated in ice cold 30%, 50%, 70%, and 95% ethanol, then allowed to come to room temperature. This was followed by 2 changes of 100% ethanol and two changes of propylene oxide. The tissue was then infiltrated in a 1:1 mixture of propylene oxide:Epon Araldite resin for 2 hr followed by two changes of fresh Epon Araldite, 2 hr each change. It was then placed in flat embedding molds and polymerized at 60 °C overnight. Fixed samples were sliced to a thickness of 80 nm and were imaged on a JEOL1230 Transmission Electron Microscope (TEM) operated at 80 kV.
Quantification of Mitochondria and deposits from EM images
Images of patient liver tissue taken at 10,000x magnification were analyzed by two independent, blinded researchers. First, mitochondria were identified in each image. Mitochondria were characterized as structures with a well-defined, curved border, typically 0.5–1 μm in diameter, with a mottled, gray matrix and at least one visible cristae line. Confirmed mitochondria were examined for the presence or absence of black, electron-dense deposits. Deposits were defined as speckles that were both darker and thicker in diameter than cristae membrane lines. With FLC patient 42.2, for which no non-tumor liver sample was available, cells from the tumor periphery were classified as oncocytic or peri-oncocytic. Cells with a circular nucleus and dispersed mitochondria were classified as peri-oncocytic; cells with misshapen nuclei and abundant cytoplasmic mitochondria were classified as oncocytic.
TMRM measurements
500,000 AML12 cells were centrifuged at 800 g for 3 min, washed with 1 mL PBS (Thermo Fisher Scientific, 20012050), and centrifuged again at 800 g for 3 min. The supernatant was aspirated and the cell pellet was resuspended in 150 μL KCl buffer (for composition, see Ca2+ uptake protocol above) supplemented with 500 nM tetramethylrhodamine methyl ester (TMRM, Thermo Fisher Scientific, I34361) and 0.005% digitonin (Thermo Fisher Scientific, BN2006). Cell suspension was transferred to a black 96-well plate (Greiner Bio-One, 655076). Fluorescence measurements (Ex 540 nm, Em 590 nm) were taken across three periods using a BioTek Synergy H1 microplate reader (Fisher Scientific, 11–120-536). The first measurements were taken every second for 1 min to establish a baseline. Then 500 mM glutamate (Sigma-Aldrich, G1251) and 5 mM malate (Sigma-Aldrich, M7397) stocks were injected for final concentrations of 5 mM, and a second reading was taken every second for 1 min to obtain the minimum fluorescence, corresponding with maximal mitochondrial membrane potential. Finally, mitochondrial membrane potential was dissipated with the addition of 1 μM CCCP (Cayman Chemical Company, 25458), and reads were taken every second for 2 min to obtain the maximum fluorescence. Relative mitochondrial membrane potential was reported as the difference between the maximum (average of data from 294–304 sec) and minimum TMRM fluorescence measurements (average of data from 172–182 sec).
Generation of mito-GECO constructs and matrix Ca2+ measurements
The pLMM1 vector was generated from pLYS191 by replacing the CMV promoter with the EF1α promoter. G-GECO1.143 was cloned into either pLYS1 or pLMM1 with an N-terminal mitochondrial targeting sequence taken from cytochrome C subunit 8A (COX8). AML12 cells were transduced with mito-G-GECO lentiviral constructs generated using pLYS1 (WT and c14) or pLMM1 (c4) and selected with puromycin as described above. 80,000 AML12 wildtype, c14, and c4 cells expressing mito-G-GECO were cultured in 12-well plates for 2 days until reaching 90–95% confluency. Medium was then aspirated and replaced with 500 μL phenol red-free DMEM/F-12 (Thermo Scientific, 21041025) supplemented with 10% FBS (VWR, 89510–186) and 100 IU/mL penicillin and 100 μg/mL streptomycin (VWR, 45000–652) for 1.5 hr. Fluorescence measurements (Ex 480 nm, Em 520 nm) were obtained with a BioTek Synergy H1 microplate reader (Fisher Scientific, 11–120-536) at 37 °C. To establish baseline, fluorescence was measured every 10 sec for 2 min. Then CCCP was added to a final concentration of 50 μM (Cayman Chemical Company, 25458) to each well, and fluorescence was measured to determine minimum fluorescence. Finally, 5 μL of 1mM stock of ionomycin (Sigma-Aldrich, I3909) and 10 μL of 1M stock of CaCl2 were added, and fluorescence was measured to establish maximum fluorescence. To correct for potential differences in mito-G-GECO expression across cell lines, baseline results were reported as a percentage of the difference between maximum and minimum values. To calculate the baseline fraction, minimum fluorescence is subtracted from baseline fluorescence over minimum fluorescence subtracted from maximum fluorescence. For each measurement, baseline, minimum, and maximum values were determined manually.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to the patients who donated samples to be used in research. We are also grateful to Dr. Vishal Gohil and for his thoughtful discussion of this project. We thank Edward Parker for processing patient samples for electron microscopy and his help with image acquisition, Dr. Rong Tien for sharing their KLF15 antibody and plasmids, Dr. Lucas Sullivan for help with metabolomics consultation and feedback on experimental design. Several figures were created with tools from BioRender.com. This work was supported by the following funding sources: R35 GM136234 (Y.S.), Pew Charitable Trusts (Y.S.), 1F31AG072716-01A1 (M.J.S.M), NSF MCB2314338 (G.E.A.G), P30 CA015704 (R.S.Y, H.L.K), DOD W81XWH1910544 (R.S.Y, H.L.K).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Balderas E. et al. Mitochondrial Ca2+ uniporter stabilization preserves energetic homeostasis during Complex I impairment. Nat Commun 13, 2769 (2022). 10.1038/s41467-022-30236-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gherardi G. et al. Loss of mitochondrial Ca2+ uniporter rewires skeletal muscle metabolism and substrate preference. Cell Death Differ 26, 362–381 (2019). 10.1038/s41418-018-0191-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huo J. & Molkentin J. D. MCU genetically altered mice suggest how mitochondrial Ca(2+) regulates metabolism. Trends Endocrinol Metab (2024). 10.1016/j.tem.2024.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong J. Q. et al. The mitochondrial Ca2+ uniporter underlies metabolic fuel preference in skeletal muscle. JCI Insight 3 (2018). 10.1172/jci.insight.121689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomar D. & Elrod J. W. Metabolite regulation of the mitochondrial Ca2+ uniporter channel. Cell Ca2+ 92, 102288 (2020). 10.1016/j.ceca.2020.102288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baughman J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial Ca2+ uniporter. Nature 476, 341–345 (2011). 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Stefani D., Raffaello A., Teardo E., Szabo I. & Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial Ca2+ uniporter. Nature 476, 336–340 (2011). 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacEwen M. J. S. & Sancak Y. Beyond the matrix: structural and physiological advancements in mitochondrial Ca2+ signaling. Biochem Soc Trans 51, 665–673 (2023). 10.1042/BST20220317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plovanich M. et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate Ca2+ handling. PLoS One 8, e55785 (2013). 10.1371/journal.pone.0055785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancak Y. et al. EMRE is an essential component of the mitochondrial Ca2+ uniporter complex. Science 342, 1379–1382 (2013). 10.1126/science.1242993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csordas G. et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metab 17, 976–987 (2013). 10.1016/j.cmet.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacs-Bogdan E. et al. Reconstitution of the mitochondrial Ca2+ uniporter in yeast. Proc Natl Acad Sci U S A 111, 8985–8990 (2014). 10.1073/pnas.1400514111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J. C. et al. MICU1 Serves as a Molecular Gatekeeper to Prevent In Vivo Mitochondrial Ca2+ Overload. Cell Rep 16, 1561–1573 (2016). 10.1016/j.celrep.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patron M., Sprenger H. G. & Langer T. m-AAA proteases, mitochondrial Ca2+ homeostasis and neurodegeneration. Cell Res 28, 296–306 (2018). 10.1038/cr.2018.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arruda A. P. et al. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat Med 20, 1427–1435 (2014). 10.1038/nm.3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konig T. et al. The m-AAA Protease Associated with Neurodegeneration Limits MCU Activity in Mitochondria. Mol Cell 64, 148–162 (2016). 10.1016/j.molcel.2016.08.020 [DOI] [PubMed] [Google Scholar]
- 17.Walters G. C. & Usachev Y. M. Mitochondrial Ca2+ cycling in neuronal function and neurodegeneration. Front Cell Dev Biol 11, 1094356 (2023). 10.3389/fcell.2023.1094356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S. et al. MCU-complex-mediated mitochondrial Ca2+ signaling is impaired in Barth syndrome. Hum Mol Genet 31, 376–385 (2022). 10.1093/hmg/ddab254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchi S., Giorgi C., Galluzzi L. & Pinton P. Ca(2+) Fluxes and Cancer. Mol Cell 78, 1055–1069 (2020). 10.1016/j.molcel.2020.04.017 [DOI] [PubMed] [Google Scholar]
- 20.Fernandez Garcia E. et al. The mitochondrial Ca(2+) channel MCU is critical for tumor growth by supporting cell cycle progression and proliferation. Front Cell Dev Biol 11, 1082213 (2023). 10.3389/fcell.2023.1082213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koval O. M. et al. Loss of MCU prevents mitochondrial fusion in G(1)-S phase and blocks cell cycle progression and proliferation. Sci Signal 12 (2019). 10.1126/scisignal.aav1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H. et al. AMPK-mediated activation of MCU stimulates mitochondrial Ca(2+) entry to promote mitotic progression. Nat Cell Biol 21, 476–486 (2019). 10.1038/s41556-019-0296-3 [DOI] [PubMed] [Google Scholar]
- 23.Liu J. C. et al. EMRE is essential for mitochondrial Ca2+ uniporter activity in a mouse model. JCI Insight 5 (2020). 10.1172/jci.insight.134063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan X. et al. The physiological role of mitochondrial Ca2+ revealed by mice lacking the mitochondrial Ca2+ uniporter. Nat Cell Biol 15, 1464–1472 (2013). 10.1038/ncb2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombardi A. A. et al. Mitochondrial Ca2+ exchange links metabolism with the epigenome to control cellular differentiation. Nat Commun 10, 4509 (2019). 10.1038/s41467-019-12103-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer T. M. & Murphy E. Role of Mitochondrial Ca2+ and the Permeability Transition Pore in Regulating Cell Death. Circ Res 126, 280–293 (2020). 10.1161/CIRCRESAHA.119.316306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig J. R., Peters R. L., Edmondson H. A. & Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer 46, 372–379 (1980). [DOI] [PubMed] [Google Scholar]
- 28.Farhi D. C., Shikes R. H. & Silverberg S. G. Ultrastructure of fibrolamellar oncocytic hepatoma. Cancer 50, 702–709 (1982). [DOI] [PubMed] [Google Scholar]
- 29.Lloreta J., Vadell C., Fabregat X. & Serrano S. Fibrolamellar hepatic tumor with neurosecretory features and systemic deposition of AA amyloid. Ultrastruct Pathol 18, 287–292 (1994). 10.3109/01913129409016302 [DOI] [PubMed] [Google Scholar]
- 30.Levin S. N. et al. Disruption of proteome by an oncogenic fusion kinase alters metabolism in fibrolamellar hepatocellular carcinoma. Sci Adv 9, eadg7038 (2023). 10.1126/sciadv.adg7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeyaraj D. et al. Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab 15, 311–323 (2012). 10.1016/j.cmet.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakral N. & Simonetto D. A. Hyperammonemic encephalopathy: An unusual presentation of fibrolamellar hepatocellular carcinoma. Clin Mol Hepatol 26, 74–77 (2020). 10.3350/cmh.2018.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young M. P. et al. Metabolic adaptation to the chronic loss of Ca(2+) signaling induced by KO of IP(3) receptors or the mitochondrial Ca(2+) uniporter. J Biol Chem 298, 101436 (2022). 10.1016/j.jbc.2021.101436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge S. X., Jung D. & Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020). 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang B. et al. Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J Biol Chem 273, 17680–17688 (1998). 10.1074/jbc.273.28.17680 [DOI] [PubMed] [Google Scholar]
- 36.Pezzato E., Battaglia V., Brunati A. M., Agostinelli E. & Toninello A. Ca2+ - independent effects of spermine on pyruvate dehydrogenase complex activity in energized rat liver mitochondria incubated in the absence of exogenous Ca2+ and Mg2+. Amino Acids 36, 449–456 (2009). 10.1007/s00726-008-0099-5 [DOI] [PubMed] [Google Scholar]
- 37.Honeyman J. N. et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 343, 1010–1014 (2014). 10.1126/science.1249484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasparre G., Romeo G., Rugolo M. & Porcelli A. M. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta 1807, 633–642 (2011). 10.1016/j.bbabio.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 39.Wolf S. G. et al. 3D visualization of mitochondrial solid-phase Ca2+ stores in whole cells. Elife 6 (2017). 10.7554/eLife.29929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strubbe-Rivera J. O. et al. The mitochondrial permeability transition phenomenon elucidated by cryo-EM reveals the genuine impact of Ca2+ overload on mitochondrial structure and function. Sci Rep 11, 1037 (2021). 10.1038/s41598-020-80398-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnham R. E. et al. An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma. Elife 8 (2019). 10.7554/eLife.44187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinh T. A. et al. Hotspots of Aberrant Enhancer Activity in Fibrolamellar Carcinoma Reveal Candidate Oncogenic Pathways and Therapeutic Vulnerabilities. Cell Rep 31, 107509 (2020). 10.1016/j.celrep.2020.03.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y. et al. An expanded palette of genetically encoded Ca(2)(+) indicators. Science 333, 1888–1891 (2011). 10.1126/science.1208592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumayer C. et al. Oncogenic Addiction of Fibrolamellar Hepatocellular Carcinoma to the Fusion Kinase DNAJB1-PRKACA. Clin Cancer Res 29, 271–278 (2023). 10.1158/1078-0432.CCR-22-1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyota A. et al. Novel protein kinase cAMP-Activated Catalytic Subunit Alpha (PRKACA) inhibitor shows anti-tumor activity in a fibrolamellar hepatocellular carcinoma model. Biochem Biophys Res Commun 621, 157–161 (2022). 10.1016/j.bbrc.2022.07.008 [DOI] [PubMed] [Google Scholar]
- 46.Davis M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29, 1046–1051 (2011). 10.1038/nbt.1990 [DOI] [PubMed] [Google Scholar]
- 47.Garbincius J. F. et al. MCU gain- and loss-of-function models define the duality of mitochondrial Ca2+ uptake in heart failure. bioRxiv (2023). 10.1101/2023.04.17.537222 [DOI] [Google Scholar]
- 48.Ericksen R. E. et al. Loss of BCAA Catabolism during Carcinogenesis Enhances mTORC1 Activity and Promotes Tumor Development and Progression. Cell Metab 29, 1151–1165 e1156 (2019). 10.1016/j.cmet.2018.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowker-Kinley M. M., Davis W. I., Wu P., Harris R. A. & Popov K. M. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J 329 ( Pt 1), 191–196 (1998). 10.1042/bj3290191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., Li L. L. & Du Y. Kruppel-like factor 15 in liver diseases: Insights into metabolic reprogramming. Front Pharmacol 14, 1115226 (2023). 10.3389/fphar.2023.1115226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao D. et al. Glucose promotes cell growth by suppressing branched-chain amino acid degradation. Nat Commun 9, 2935 (2018). 10.1038/s41467-018-05362-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auron A. & Brophy P. D. Hyperammonemia in review: pathophysiology, diagnosis, and treatment. Pediatr Nephrol 27, 207–222 (2012). 10.1007/s00467-011-1838-5 [DOI] [PubMed] [Google Scholar]
- 53.Lo E. K. K. et al. The Emerging Role of Branched-Chain Amino Acids in Liver Diseases. Biomedicines 10 (2022). 10.3390/biomedicines10061444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mann G., Mora S., Madu G. & Adegoke O. A. J. Branched-chain Amino Acids: Catabolism in Skeletal Muscle and Implications for Muscle and Whole-body Metabolism. Front Physiol 12, 702826 (2021). 10.3389/fphys.2021.702826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong Y., Jiang L. & Li T. Aberrant branched-chain amino acid catabolism in cardiovascular diseases. Front Cardiovasc Med 9, 965899 (2022). 10.3389/fcvm.2022.965899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klyuyeva A., Tuganova A., Kedishvili N. & Popov K. M. Tissue-specific kinase expression and activity regulate flux through the pyruvate dehydrogenase complex. J Biol Chem 294, 838–851 (2019). 10.1074/jbc.RA118.006433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGarrah R. W. & White P. J. Branched-chain amino acids in cardiovascular disease. Nat Rev Cardiol 20, 77–89 (2023). 10.1038/s41569-022-00760-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lauer S. M. et al. Recruitment of BAG2 to DNAJ-PKAc scaffolds promotes cell survival and resistance to drug-induced apoptosis in fibrolamellar carcinoma. Cell Rep 43, 113678 (2024). 10.1016/j.celrep.2024.113678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishi K. et al. Branched-chain keto acids inhibit mitochondrial pyruvate carrier and suppress gluconeogenesis in hepatocytes. Cell Rep 42, 112641 (2023). 10.1016/j.celrep.2023.112641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun H. et al. Catabolic Defect of Branched-Chain Amino Acids Promotes Heart Failure. Circulation 133, 2038–2049 (2016). 10.1161/CIRCULATIONAHA.115.020226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanyomse Q. et al. KLF15 suppresses tumor growth and metastasis in Triple-Negative Breast Cancer by downregulating CCL2 and CCL7. Sci Rep 12, 19026 (2022). 10.1038/s41598-022-23750-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang N. et al. Circular RNA MTO1 Inhibits the Proliferation and Invasion of Ovarian Cancer Cells Through the miR-182–5p/KLF15 Axis. Cell Transplant 29, 963689720943613 (2020). 10.1177/0963689720943613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Liu J., Xi S., Pan X. & Fang X. Exploring the Mechanism of KLF15 on the Biological Activity and Autophagy of Gastric Cancer Cells based on PI3K/Akt/Mtor Signaling Pathway. Comb Chem High Throughput Screen (2024). 10.2174/0113862073255591231213053101 [DOI] [PubMed] [Google Scholar]
- 64.Wang Z. H. et al. Circular RNA circFBXO7 attenuates non-small cell lung cancer tumorigenesis by sponging miR-296-3p to facilitate KLF15-mediated transcriptional activation of CDKN1A. Transl Oncol 30, 101635 (2023). 10.1016/j.tranon.2023.101635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu K. Y. et al. The functions and prognostic value of Kruppel-like factors in breast cancer. Cancer Cell Int 22, 23 (2022). 10.1186/s12935-022-02449-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun C. X. et al. MiR-181a promotes cell proliferation and migration through targeting KLF15 in papillary thyroid cancer. Clin Transl Oncol 24, 66–75 (2022). 10.1007/s12094-021-02670-1 [DOI] [PubMed] [Google Scholar]
- 67.Gray S. et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab 5, 305–312 (2007). 10.1016/j.cmet.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haldar S. M. et al. Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proc Natl Acad Sci U S A 109, 6739–6744 (2012). 10.1073/pnas.1121060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chapuy C. I., Sahai I., Sharma R., Zhu A. X. & Kozyreva O. N. Hyperammonemic Encephalopathy Associated With Fibrolamellar Hepatocellular Carcinoma: Case Report, Literature Review, and Proposed Treatment Algorithm. Oncologist 21, 514–520 (2016). 10.1634/theoncologist.2015-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sethi S. et al. Hyperammonemic encephalopathy: a rare presentation of fibrolamellar hepatocellular carcinoma. Am J Med Sci 338, 522–524 (2009). 10.1097/MAJ.0b013e3181bccfb4 [DOI] [PubMed] [Google Scholar]
- 71.Surjan R. C., Dos Santos E. S., Basseres T., Makdissi F. F. & Machado M. A. A Proposed Physiopathological Pathway to Hyperammonemic Encephalopathy in a Non-Cirrhotic Patient with Fibrolamellar Hepatocellular Carcinoma without Ornithine Transcarbamylase (OTC) Mutation. Am J Case Rep 18, 234–241 (2017). 10.12659/ajcr.901682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bell H. N. et al. Microenvironmental ammonia enhances T cell exhaustion in colorectal cancer. Cell Metab 35, 134–149 e136 (2023). 10.1016/j.cmet.2022.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spinelli J. B. et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 358, 941–946 (2017). 10.1126/science.aam9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y. et al. beta-Catenin Activation Reprograms Ammonia Metabolism to Promote Senescence Resistance in Hepatocellular Carcinoma. Cancer Res 84, 1643–1658 (2024). 10.1158/0008-5472.CAN-23-0673 [DOI] [PubMed] [Google Scholar]
- 75.De Chiara F. et al. Ammonia Scavenging Prevents Progression of Fibrosis in Experimental Nonalcoholic Fatty Liver Disease. Hepatology 71, 874–892 (2020). 10.1002/hep.30890 [DOI] [PubMed] [Google Scholar]
- 76.Domagala J. et al. Ammonia inhibits antitumor activity of NK cells by decreasing mature perforin. bioRxiv, 2023.2011.2020.567708 (2023). 10.1101/2023.11.20.567708 [DOI] [Google Scholar]
- 77.Barbul A. Proline precursors to sustain Mammalian collagen synthesis. J Nutr 138, 2021S–2024S (2008). 10.1093/jn/138.10.2021S [DOI] [PubMed] [Google Scholar]
- 78.Harada D. et al. Oral administration of l-ornithine increases the content of both collagen constituting amino acids and polyamines in mouse skin. Biochem Biophys Res Commun 512, 712–715 (2019). 10.1016/j.bbrc.2019.03.147 [DOI] [PubMed] [Google Scholar]
- 79.Mao L. et al. Histone Deacetylase 11 Contributes to Renal Fibrosis by Repressing KLF15 Transcription. Front Cell Dev Biol 8, 235 (2020). 10.3389/fcell.2020.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian Y. et al. KLF15 negatively regulates cardiac fibrosis by which SDF-1beta attenuates cardiac fibrosis in type 2 diabetic mice. Toxicol Appl Pharmacol 427, 115654 (2021). 10.1016/j.taap.2021.115654 [DOI] [PubMed] [Google Scholar]
- 81.Wang B. et al. The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. J Mol Cell Cardiol 45, 193–197 (2008). 10.1016/j.yjmcc.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li S. et al. Nobiletin mitigates hepatocytes death, liver inflammation, and fibrosis in a murine model of NASH through modulating hepatic oxidative stress and mitochondrial dysfunction. J Nutr Biochem 100, 108888 (2022). 10.1016/j.jnutbio.2021.108888 [DOI] [PubMed] [Google Scholar]
- 83.Colledge M. & Scott J. D. AKAPs: from structure to function. Trends Cell Biol 9, 216–221 (1999). 10.1016/s0962-8924(99)01558-5 [DOI] [PubMed] [Google Scholar]
- 84.Gabrovsek L. et al. A-kinase-anchoring protein 1 (dAKAP1)-based signaling complexes coordinate local protein synthesis at the mitochondrial surface. J Biol Chem 295, 10749–10765 (2020). 10.1074/jbc.RA120.013454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li C. J., Lin H. Y., Ko C. J., Lai J. C. & Chu P. Y. A Novel Biomarker Driving Poor-Prognosis Liver Cancer: Overexpression of the Mitochondrial Ca2+ Gatekeepers. Biomedicines 8 (2020). 10.3390/biomedicines8110451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ren T. et al. MCUR1-Mediated Mitochondrial Ca2+ Signaling Facilitates Cell Survival of Hepatocellular Carcinoma via Reactive Oxygen Species-Dependent P53 Degradation. Antioxid Redox Signal 28, 1120–1136 (2018). 10.1089/ars.2017.6990 [DOI] [PubMed] [Google Scholar]
- 87.Ren T. et al. MCU-dependent mitochondrial Ca(2+) inhibits NAD(+)/SIRT3/SOD2 pathway to promote ROS production and metastasis of HCC cells. Oncogene 36, 5897–5909 (2017). 10.1038/onc.2017.167 [DOI] [PubMed] [Google Scholar]
- 88.Rappsilber J., Mann M. & Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906 (2007). 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 89.Yates A. D. et al. Ensembl 2020. Nucleic Acids Res 48, D682–D688 (2020). 10.1093/nar/gkz966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Millner A. et al. Ceramide-1-Phosphate Is Involved in Therapy-Induced Senescence. ACS Chem Biol 17, 822–828 (2022). 10.1021/acschembio.2c00216 [DOI] [PubMed] [Google Scholar]
- 91.MacEwen M. J. et al. Evolutionary divergence reveals the molecular basis of EMRE dependence of the human MCU. Life Sci Alliance 3 (2020). 10.26508/lsa.202000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.