Abstract
Frailty is a complex trait. Twin studies and recent Genome-Wide Association Studies have demonstrated a strong genetic basis of frailty but there remains a lack of genetic studies exploring genetic prediction of Frailty. Previous work has shown that a single polygenic predictor – represented by a Frailty polygenic score - predicts Frailty, measured via the frailty index, in independent samples within the United Kingdom. We extended this work, using a multi-polygenic score (MPS) approach to increase predictive power. Predictor variables - twenty-six polygenic scores (PGS) were modelled in regularised Elastic net regression models, with repeated cross-validation, to estimate joint prediction of the polygenic scores and order the predictions by their contributing strength to Frailty in two independent cohorts aged 65+ - the English Longitudinal Study of Ageing (ELSA) and Lothian Birth Cohort 1936 (LBC1936). Results showed that the MPS explained 3.6% and 4.7% of variance compared to the best single-score prediction of 2.6% and 2.2% of variance in ELSA and LBC1936 respectively. The strongest polygenic predictors of worsening frailty came from PGS for Chronic pain, Frailty and Waist circumference; whilst PGS for Parental Death, Educational attainment, and Rheumatoid Arthritis were found to be protective to frailty. Results from the predictors remaining in the final model were then validated using the longitudinal LBC1936, with equivalent PGS scores from the same GWAS summary statistics. Thus, this MPS approach provides new evidence for the genetic contributions to frailty in later life and sheds light on the complex structure of the Frailty Index measurement.
Introduction
Frailty is a multifaceted state associated with ageing and it reflects a decline in physiology and resilience to stressors (Kojima, Liljas, & Iliffe, 2019). Though there is yet to be a unified understanding of what frailty is and how to best measure it, one of the most widely used tools is the frailty index [FI] (Donneau et al., 2017; Searle et al., 2008). The FI outlines frailty as a risk index – a cumulative deficit model which totals the number of deficits, such as falls, physical health conditions, grip strength, cognitive functioning, to produce an individualised frailty score - frailty indexes often fluctuate with the number of deficits, the criteria suggest a minimum of 30–40 deficits spanning multiple systems, such as health, cognitive, and psychosocial systems related to ageing (Searle et al., 2008; Blodgett et al., 2015). Higher levels of frailty, reflected by a high frailty score, have been shown to predict adverse outcomes such as hospital stays, disability, and mortality (Blodgett et al., 2015). Other research has shown that frailty is malleable and can be reversed when measured with the FI (Feng et al., 2017; Gordon & Hubbard, 2022). As the number of people aged over 60 is expected to grow from 901 million to 1.4 billion from 2015 to 2030, health-conditions like frailty are expected to rise and more people will be living in poorer health in later life (Lazarus et al., 2018; Beard & Bloom, 2015). There is, therefore, a necessity to understand how omics, such as genetics, can be used to identify those at greater risk and the mechanisms underlying its complex nature.
Whilst research investigating the genetic associations of frailty remains in its infancy, the phenotypic associations of frailty have been widely investigated. A systematic review of factors associated with frailty found that the most common factors are mainly sociodemographic factors, such as age, sex, educational level and socioeconomic status. Other factors included body weight and level of physical activity, psychological factors such as depressive symptoms, diet quality, neighbourhood, health care quality/access to private healthcare insurance (Serra-Prat et al., 2016). Furthermore, cross sectional research investigating phenotypic factors associated with frailty found that loneliness, mobility issues, history of stroke, arthrosis, peripheral vascular disease, cancer, diabetes, hypertension, pain and polypharmacy and drug interactions may predispose an individual to frailty (Sharma, Reddy & Ganguly, 2020).
Methods which measure the heritable risk of developing a disease via combining the risk of different genetic variants to produce a quantitative score – polygenic score [PGS] (Lewis & Vassos, 2020; Lambert et al., 2019) have potential to uncover new information about the molecular underpinnings of this complex multifaceted trait. Through data gathered via Genome Wide Association Studies [GWAS], such as GWAS of complex traits like frailty (Mekli et al., 2018; Atkins et al., 2021; Ye et al., 2023), PGS can be developed for individuals (Lewis & Vassos, 2020). Once developed and modelled alongside social and environmental variables, these scores have the potential to be highly predictive for trait and disease prevention – allowing deeper understand of ageing outcomes and age-related traits and disease, such as Frailty.
Our previous work showed that a PGS for frailty can predict frailty, when measured with the FI, at varying ages in later life in the Lothian Birth cohort 1936 [LBC1936] and the English Longitudinal Study of Ageing [ELSA] (Flint et al., 2023). However, if frailty arises from the cumulative effects of its individual components, as the FI states, then just as the FI includes over thirty components to measure frailty, we can approach the genetic prediction of frailty in a similar manner – a multi-polygenic approach (Krapohl et al., 2018). Due to the polygenic and complex nature of frailty it is informative to aggregate PGS to investigate their joint predictive power, allowing a more rounded understanding of the genetic contributions to frailty and for refined phenotypic prediction. The GWAS of the FI, in the UK Biobank, tested various PGS as predictors of FI in univariate linear regression models and found that PGS for BMI, Inflammatory Bowel Disease [IBD], Waist Hip Ratio [WHR], menarche, grip strength, age at first sexual intercourse, parents’ survival and educational attainment were associated with the FI (Kojima, Liljas, & Iliffe, 2019). However, when modelling genetic variables, which are highly correlated with each other, there is risk of multicollinearity (Altelbany, 2021). Thus, this study utilises multivariate statistical measures with data from the most recent genome wide association studies to create polygenic scores for multiple variables that are expected to relate to the indicators of FI. The training set for model prediction is the larger study, ELSA. To handle multicollinearity, the polygenic scores will be built into a net-elastic regression model to estimate their joint predictive power and order these predictors by their contributing strength to the FI phenotype. Then, to validate the findings from ELSA the PGS that contribute the most to frailty prediction will be tested, longitudinally, in the LBC1936 at ages ~70, ~76 and ~82.
Methods
Target/Independent Sample
The target sample consisted of the observed genotypes (genome-wide SNPs) and phenotypic data from 5955 adults aged 65–99, mean age 72.73 (SD = 7.2, 2740 males), in ELSA. ELSA is a prospective cohort study which is representative of individuals aged over 50 and living in private households in England (Steptoe et al., 2013). Ethics for ELSA have been approved via the South Central – Berkshire Research Ethics Committee (21/SC/0030, 22nd March 2021). PGS for ELSA were acquired through request to the ELSA genetics team (Ajnakina & Steptoe, 2019).
An independent test sample was used to validate the findings longitudinally at ~70, ~76 and ~82 years in LBC1936. LBC1936 is an ongoing longitudinal study of older adults living in the community in Edinburgh and surrounding Lothian areas of Scotland, United Kingdom (Deary et al., 2007). Individuals were initially recruited based on having been part of the Scottish Mental Survey (1947) and have thus far taken part in 6 waves of testing. Data were drawn from Waves 1, 3 and 5 where appropriate frailty data was collected. At Wave 1 there were 1005 older adults mean age 69.58 (SD = 0.83, n = 1091, 548 males), 697 older adults at Wave 3 mean age 76.30 (SD = 0.68, n = 697, 360 male) and 431 older adults at Wave 5 mean 82.06 (SD = 0.53, n = 431, 209 male). Ethical permission was approved from the Multi-Centre Research Ethics Committee for Scotland (Wave 1: MREC/01/0/56), the Lothian Research Ethics Committee (Wave 1: LREC/2003/2/29), and the Scotland A Research Ethics Committee (Waves 2, 3, 4 and 5: 07/MRE00/58). Written consent was obtained from participants at each of the waves. DNA was collected via blood samples from the majority of participants at wave 1 and genotyping was performed using stringent quality control measures (Houlihan et al., 2010).
Predictors
As the PGS for the ELSA study were either publicly available or sought from the genetics team upon our request, we acquired the available PGS that were either variables usually used in the FI or most associated phenotypically with the FI. Twenty-six PGS were selected following these criteria. To validate the ELSA findings the PGS that were significant were then constructed in LBC1936. Quality control processing was done using the R package QCGWAS [20] for GWAS summary statistics and PGS were derived using the same method and software as those in ELSA - PRSice (version 2) polygenic software (Van der Most et al., 2014) - with the raw genotype data from LBC1936. A list of the predictors selected in ELSA and those validated in LBC1936 can be found in supplementary methods.
Outcomes
The FI in ELSA was previously constructed from the ELSA dataset [12]. The index consists of 62 deficits, as shown in supplementary Table S1, and a frailty score was created for participants if data were available for 30 out of the 62 possible deficits. Due to skewness in the data, the FI variable was transformed using a square root transformation.
In LBC1936, the FI was previously constructed in the dataset (Welstead et al., 2022) and contains 30 deficits, including physical, biological, social, psychological, and cognitive deficits, consistent with the FI in ELSA and other research (Searle et al., 2008) – the items in the index can be seen in supplementary Table S2. Deficits were either dichotomised as either 0 (absent) or 1 (present); in some cases, 0.5 was used to represent a partially present deficit or were on a continuous scale (such as walking time) on a scale ranging from 0 to 1. For each individual, the number of deficits present was summed and divided by the total number of deficits. Scores ranged from 0 to 1 – with higher scores indicating higher frailty.
Despite there being more deficits in the FI used in ELSA than in LBC1936, guidelines indicate that as long as a minimum of 30 deficits are used to cover the relevant domains (disability, disease, cognitive functioning) then differences between number of deficits should not be an issue (Searle et al., 2008). Further, the index was calculated in the same way as the LBC1936 index; both LBC1936 and ELSA followed the same guidelines when creating the index. For LBC1936 and ELSA FI scores were standardised to allow comparisons when interpreting the results.
Covariates
Age and sex (the strongest frailty predictors) were controlled in analysis. Four genetic ancestry principal components for LBC1936 and 10 ancestry principal components for ELSA were also controlled for in the analyses to account for population stratification - systematic genetic differences due to ancestry differences. LBC1936 and ELSA only included participants with European ancestry in genotyping.
Single-polygenic models
To evaluate the benefit of a multi-polygenic approach in predicting frailty, single-polygenic linear regression models, with the 26 PGS, were built in ELSA with the FI as the outcome measure for comparison.
A multiple linear regression model was first fit including sex, age, and ancestry principal components covariates and the outcome (FI) - the null model. Each polygenic score was then added as a predictor to the null model and re-run (the full model). The variance explained by the PGS (PGS R2) is calculated through deducting the R2 of the null model from that of the full model for each of the 26 models.
Multi-polygenic models
The multi-polygenic model was built to test the joint prediction of 26 PGS and rank the prediction of each PGS to the FI outcome. Elastic net regularized regression has been shown to be a useful technique to reduce issues that occur when a magnitude of predictions, such as multiple polygenic scores, lead to overfitting within a traditional multiple linear regression model (Krapohl et al., 2018).
Elastic net regularized regression benefits from two regularization techniques - L1 regularization from LASSO regression and L2 regularization from ridge regression through using variable selection and only retaining variables penalizing coefficients for overfitting. Elastic net regression is particularly valuable for PGS prediction with highly correlated genetic signal, as the elastic net regression method will include all highly correlated variables in a grouping effect. The final coefficients in the model allow for the predictors to be ranked by their contribution of prediction to the outcome. The elastic net was computed using the glmnet and caret R package (Friedman, Tibshirani & Hastie, 2010; Kuhn, 2008).
The model was trained and tested in an 80% train (4764 participants) and 20% (1191) test method in the ELSA dataset with cross validation 4-fold to aid in producing unbiased - repeated cross-validation enhances the model and the final model is then applied to the independent test data set. The final PGS predictors from the ELSA dataset were then created and computed in LBC1936 Waves 1,3 and 5 to compare effects across age and for further validation.
Results
The mean scores for frailty and age in ELSA and LBC1936 Wave 1, 3 and 5 are shown in Table 1. As expected, mean frailty scores increase with age. Table 1 also shows the pairwise correlations of frailty between LBC1936 waves - results confirmed that frailty is relatively stable over time. Correlations were at a minimum moderate, > 0.5 correlation, and most were strong, Pearson’s r > 0.7. Correlations between the predictor variables, PGSs, can be found in supplementary Figures S1 and S2 – the correlation matrices only display the significant correlations and as expected PGS which are expected to share similar mechanisms are moderately-strongly correlated Waist circumference and BMI, Waist circumference and Waist hip-ratio, Educational attainment and General cognitive functioning, Frailty and Chronic pain.
Table 1.
Descriptive statistics for age and FI in ELSA and LBC1936 (raw scores), alongside Pairwise correlations of the FI across Wave 1, 3 and 5 in LBC1936
| N | Mean | SD | Range | LBC1936 Wave 1 | LBC1936 Wave 3 | LBC1936 Wave 5 | |
|---|---|---|---|---|---|---|---|
| ELSA | |||||||
| FI | 5959 | .17 | 0.11 | 0 – 0.68 | |||
| Age | 5959 | 72.73 | 7.2 | 65 – 99 | |||
| LBC1936 Wave 1 | |||||||
| FI | 1005 | .16 | 0.09 | 0 – 0.49 | 1*** | ||
| Age | 1005 | 69.58 | 0.83 | 67.66 – 71.35 | |||
| LBC1936 Wave 3 | |||||||
| FI | 697 | .20 | 0.09 | 0.02 – 0.65 | .71*** | 1*** | |
| Age | 697 | 76.30 | 0.68 | 74.64 – 77.75 | |||
| LBC1936 Wave 5 | |||||||
| FI | 431 | 0.21 | 0.09 | 0.03 – 0.58 | .57*** | .74*** | 1*** |
| Age | 431 | 82.06 | 0.54 | 80.98-83.19 |
Single models - ELSA
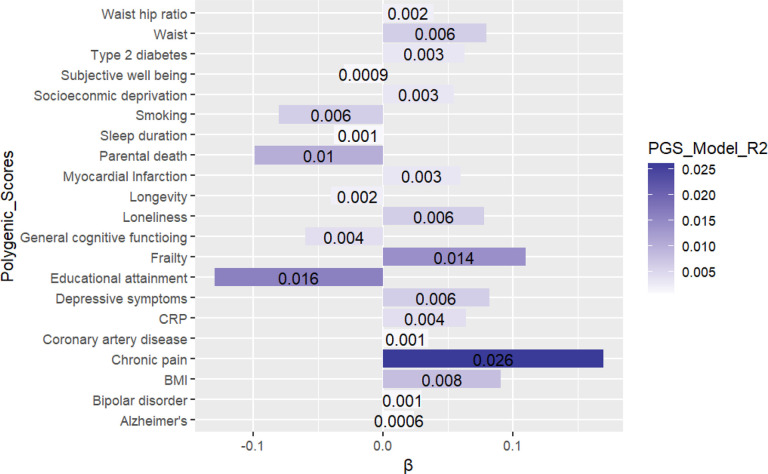
Out of the 26-single score GWAS-PGS models 21 had significant associations with the FI with the best single-score model coming from the 2019 GWAS-PGS of chronic pain - predicting 2.6% of the variance in the FI (β = ‒ 0.16). p < 0.001. The PGS on the right of Figure 1 are those that indicate risk to frailty. For example, those who have genetic predisposition to experience Chronic pain have an increased risk of frailty. Whereas the PGS on the left of Figure 1 are those with protective mechanisms. For example, genetic predisposition to having Parental life length (parents living longer) is associated with a decreased risk of frailty.
Figure 1.
Bar plot of the single model PGS with the FI as the outcome.
Multi-polygenic models (MPS)
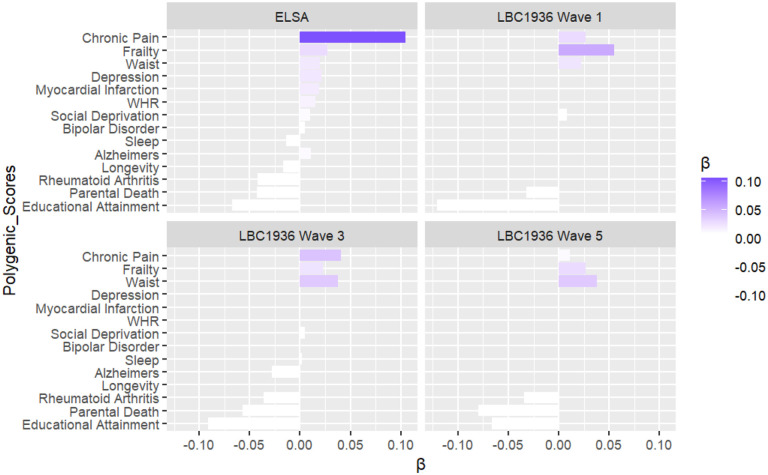
To utilise the benefits of elastic net regression, that is selecting a subset of predictors that are correlated but not redundant to prediction of an outcome, all 26 PGS were modelled into an elastic net regression model with multiple polygenic scores - MPS model. Figure 2 shows the polygenic risk score predictors, with covariates included, ranked in effect with standardized coefficients - with those on the right being PGS which exacerbate/worsen frailty and those on the left PGS which protect against frailty. Much like multiple regression models, standardized coefficients represent the contribution of a standard deviation increase in the predictor, in this case PGS, to change in the outcome, in this case the FI, when all other variables in the model have been adjusted for.
Figure 2.
Bar plot of the MPS, Elastic net regularised regression model, with covariates (age, sex, and ancestry principal components) included.
As Table 2 shows, the MPS models showed that prediction was higher, explaining 3.6% of variance p < 0.001 for the FI in the independent test set (n = 4764) compared to the most predictive single-score model at 2.6% p < 0.001 in ESLA. Out of 26 PGS inputted into the model, 11 came out as the most relevant predictors in ELSA as shown in the first plot in Figure 2 with standardised beta values. Out of 11 PGS all were significant predictors in the single score models apart from PGS for Rheumatoid arthritis in the single-score models. Interestingly, Rheumatoid arthritis emerged as a protective PGS for the FI (β = ‒ 0.043). The other protective PGS were Educational attainment (β = ‒ 0.039) and Parental death PGS (β = ‒ 0.029) PGS for Sleep duration (β = ‒ 0.014) and Longevity (β = 0.017). On the other side, the strongest predictor to risk of frailty in the MPS model was Chronic pain (β = 0.11). Alongside PGS for the FI itself (β = 0.028), Depressive symptoms (β = 0.015), Myocardial infarction (β = 0.015), Waist Hip Ratio (WHR) (β = 0.015) Waist circumference (β = 0.009), Social deprivation (β = 0.005), Alzheimer disease (β = 0.0006), and Bipolar disorder (β = 0.005) were PGS which had risk effects on the FI outcome.
Table 2.
Comparison of variance explained between best single model predictor and MPS in ELSA test sample and LBC1936 Waves 1, 3, and 5.
| Comparison of variance | ||
|---|---|---|
| Single PGS R2 | MPS R2 | |
| Regression output - FI | ||
| ELSA (age ~73) | .026 | .036 |
| Frailty PGS at ~73 | .022 | .047 |
| Frailty PGS at ~76 | .014 | .021 |
| Frailty PGS at ~79 | .019 | .027 |
All analyses controlled for sex and age and population stratification.
LBC1936 Wave 1
The most predictive PGS, found in the elastic net regression model in ELSA, were then modelled in LBC1936 (at Waves 1, 3 and 5). Similar to ELSA, protective PGS included Educational attainment (β = ‒ 0.12) and Parental Death (β = ‒ 0.032). PGS which were associated with risk of frailty included Chronic pain (β = 0.027), Frailty (β = 0.056), Waist circumference (β = ‒ 0.022) and Social Deprivation (β = 0.008).
LBC1936 Wave 3
The third plot in Figure 2 are the results at LBC1936 Wave 3 (Age ~76). Similar to ELSA and LBC1936 Wave 1, protective PGS also included Educational attainment (β = −0.099), Parental death (β = ‒ 0.053) and Rheumatoid arthritis (β = −0.037). However, unlike ELSA, but like LBC1936 Wave 1 PGS for Alzheimer disease (β = ‒ 0.019) came out as a protective PGS for the FI. PGS for Chronic pain and Waist circumference were the most predictive PGS associated with risk of frailty (both β = 0.040), followed by PGSs for Frailty (β = 0.024), Social deprivation (β = 0.005), and PGS for Sleep duration (β = 0.004).
LBC1936 Wave 5
The fourth plot in Figure 2 are the results at LBC1936 Wave 5 (Age ~82). Similar to ELSA and LBC1936 Wave 1 and Wave 3, protective PGS also included Parental death (β = ‒ 0.080), Educational attainment (β = ‒0.069) and Rheumatoid arthritis (β = ‒0.033). Alzheimer disease was not a significantly protective PGS for the FI at Wave 5. PGS for Waist circumference (β = 0.039) and Frailty (β = 0.033) were the most predictive PGS associated with risk of frailty, followed by the PGS for Chronic pain (β = 0.010).
Discussion
This was the first study to simultaneously use a large number of polygenic scores as genetic instruments to explore the traits that both protect and elevate risks of frailty in independent cohorts (LBC1936 and ELSA) - measured with the FI. PGS that were available from the English Longitudinal Study of Ageing with adequate statistical power were modelled, with Elastic net regularized regression, to investigate each PGS’s capacity to predict frailty in English participants aged 65–99 years of age. The PGS that were found most relevant to Frailty prediction from the ELSA model were then validated, longitudinally, in the Lothian Birth Cohort 1936 at ages ~70, ~76 and ~82. The PGS which replicated across the datasets and associated with increased risk of frailty were PGS for Chronic pain (all datasets/waves), Frailty (all datasets/waves), Waist circumference (all datasets/waves) and Socioeconomic deprivation (ELSA and LBC1936 Wave 1 and 3). Protective PGS that replicated were PGS for Educational attainment (all datasets/waves), Parental death (all datasets/waves) and Rheumatoid arthritis (ELSA, LBC1936 Wave 3 and 5).
In reference to these findings, it may have been expected that the Frailty PGS would be the most powerful predictor across the datasets - as this PGS is based on items within the FI itself (outcome measure); however this was not the case. Chronic pain was the most predictive PGS in the ELSA datasets and at Wave 3 in LBC1936 - with the effect size for PGS chronic pain almost four times that of the PGS for frailty in ELSA. This finding can be supported by research that has found some shared mechanisms between pain and frailty - suggesting that pain may predispose an individual to developing frailty (D’Agnelli et al., 2022). Inflammatory mediators have been found to be important mediating factors in the progression of frailty and during chronic pain. One example is an abnormal increase in oxidative stress which is present in both pain and the ageing process (D’Agnelli et al., 2022). The investigation into chronic pain and frailty may also have clinical applications. For instance, a pain treatment plan has been shown to reverse adverse changes in the immune system and reverse neuro-inflammation and restore pro and anti-inflammatory cytokine balancing in the body (Fehrenbacher et al., 2017). Such a plan might be effective in treating and reversing frailty development. However, when multimorbidity and polypharmacy are present pain treatment (medication treatment) could worsen frailty (D’Agnelli et al., 2022; Franchi et al., 2017). Despite the clear clinical relevance into the pain-frailty dyad, more research is needed to understand the shared aetiology and direction of influence between the two phenotypes, including any interactions with medication.
Like chronic pain, the PGS for Waist circumference was also a strong predictor of frailty and was stronger than the PGS for frailty in LBC1936 Wave 3 and 5. Waist circumference has been found to be a better predictor of frailty risk than BMI measures (Liao et al., 2018) with high waist circumference being an important risk factor for frailty - therefore our results suggest that a genetic predisposition for a high waist circumference puts an individual at risk to higher frailty levels. Previous research has found that higher waist levels were associated with higher frailty scores and this could be, in part, due to genetic predisposition of obesity (Liao et al., 2018) The GWAS for waist circumference included many datasets with varying adult age. Therefore, the genetic variants associated with higher waist circumference which predispose individuals at risk to later frailty may be important in early adulthood. However, it would be important to know whether genetic risk to waist circumference in later life (on a GWAS which only included individuals 65+) still puts individuals at risk to frailty or whether the effect is reversed. It would be important to investigate such age-related nuances as in middle adulthood a lower waist circumference may be associated with lower frailty however in older adult a lower waist circumference may been associated with poor health/frailty.
Alongside discussing the reasons for the PGS which put someone at risk to frailty, it is important to discuss the PGS that have protective associations with the FI across the datasets/waves. In regard to the PGS for Educational attainment, in a similar study to ELSA – the Health and Retirement Study, higher PGS for educational attainment was associated with lower frailty levels, when measured with the FI (Huibregtse et al., 2021). However, the same study found the effects of PGS on educational attainment were not present after 80 years - our study contradicts these findings as the effect for PGS for educational attainment was still present in LBC1936 Wave 5 (age ~82). Although our study did find that the association weakened with advancing age, similar to the Health and Retirement Study findings. It is important to note here that genetics, to a degree, influences education attainment, partly via the personal characteristics that help individuals achieve academically (Mosing et al., 2012). Thus, when we discuss genetic variance of educational attainment, we must reflect on these heritable traits/characteristics, including cognitive traits and personality traits such as conscientiousness that also relate to healthy life styles, money for health care – mainly driven via the association between educational attainment and socioeconomic level (Mosing et al., 2012; Marioni et al., 2016; Hil et al., 2020). The PGS for Educational attainment is different to the other PGS, as many of the others are directly part of the FI. There are applications for social researchers/statisticians here to further understand the relationship between society, educational attainment PGS, and outcomes like frailty.
Besides the PGS for Educational attainment, PGS for Longevity was also found to be protective against frailty. Previous research has shown that protective mechanisms have been found between parental longevity and physical functioning – a phenotype correlated with frailty (Marioni et al., 2016; Sathyan & Verghese, 2020). Unlike PGS for Educational attainment and Parental longevity, PGS for Rheumatoid arthritis and its relationship with frailty is not well studied or understood. The finding that PGS for Rheumatoid arthritis is a protective PGS for frailty is surprising - as one would expect the inflammation pathways associated with rheumatoid arthritis would put one at risk to frailty. A possible mediation pathway could be the role of anti-inflammatory drugs that people with rheumatoid arthritis may be taking. Some evidence has demonstrated that anti-inflammatory medications may lower levels of inflammatory biomarkers (De Vries et al., 2013) - such medications could also lower the risk of frailty. However, there is mixed data on the association between anti-inflammatory medications and frailty. For example, long-term aspirin use was associated with a 15% lower risk of frailty (Orkaby et al., 2021). Other research has found contradicting findings – such as long-term Nonsteroidal anti-inflammatory agent (NSAID) medication, including aspirin, use being associated with an increased prevalence of frailty, even after consideration of multimorbidity and health behaviours (Beyer et al., 2012). Further research around rheumatoid arthritis, medication use, and frailty is an important avenue of research that is stimulated by the findings of this paper.
The findings from this study could also highlight the issue with the measure of frailty at different ages and biases in sampling. Many of the GWAS come from UK Biobank samples which include younger/middle aged adults who are more likely to be healthy, have higher educational attainment and fewer health conditions - the healthy volunteer effect (Fry et al., 2017). ELSA and LBC1936 may also suffer from the healthy volunteer effect (Mullin et al., 2022). The differences between these cohorts is also important as ELSA is representative of the UK population through its sampling of England (Steptoe et al., 2013). However LBC1936 is unique to an area in Lothian, Scotland, and is less representative of even the Scottish population. In addition, the difference in the PGS Alzheimer results may be due to those who drop out of follow up in LBC1936 not being independent of their dementia status and those people who have genetic risk to Alzheimer disease, for other reasons, may have not developed dementia (yet) and are still in the sample – healthy survivor effect. Lastly, there are issues with measuring frailty with an aggregate/combination of scores – using the FI as an outcome. For example, someone might be more cognitively frail but still be mobile. Such pathways are likely to have different effects, especially genetic effects, which might dilute the findings and miss important subtypes of frailty. The latter has been demonstrated when modelling phenotypic elements of the FI (Johnson et al., 2024) – emphasising the importance of looking at the genetic architecture of the frailty in this way.
Despite the shortcomings, the current study has several strengths. The analysis first being carried out in ELSA and validated longitudinally in LBC1936 increased the validity and generalisability of the findings - helping question and validate any spurious associations. The method also dealt with multicollinearity and overfitting by using Elastic net regularised regression - a method which has been shown to be useful when fitting multiple genetic instruments (Krapohl et al., 2018). Future research might usefully explore the polygenic nature of frailty but with different samples to address the differing strengths of PGS. In particular, replication of our effects in countries other than England and Scotland is needed to understand several mixed findings and direction of PGS effects between ELSA and LBC1936 (e.g., PGS Depression and Alzheimer disease). It may be that interactions with environmental and social factors (e.g., differences in health care service, life adversity) are masking or exacerbating the genetic effect. It may also be valuable to model frailty with different outcomes, such as the Fried Phenotype and/or clinical frailty measures. To understand more about the direction of effects a statistical tool, MiXeR could be utilised (Frei et al., 2019). MiXeR quantifies polygenic overlap irrespective of genetic correlation and allows polygenic scores to be created based on negative and positive effects on an outcome. Given our previous findings and that the literature to date is mixed, a sensitivity analysis modelling anti-inflammatory drugs and pathways as mediators would be useful to further understand the findings.
The findings from this study demonstrate why a multi-polygenic approach is important for highlighting the complex nature of frailty and the somewhat arbitrary way it is divided into phenotypic characteristics. Novel relationships were found between PGS and frailty and this leaves future research to investigate how the genetics of frailty differ with other frailty measures and with advancing age. Lastly, there is an interdisciplinary application from this research – those interested in social determinants of health, such as social statistic researchers, should integrate/control for PGS to assist in removing confounding that often gets misattributed to the environment.
Supplementary Material
Acknowledgments
The LBC1936 is supported by the Biotechnology and Biological Sciences Research Council, and the Economic and Social Research Council [BB/W008793/1], Age UK (The Disconnected Mind Project, which also supported MW), the Milton Damerel Trust, and The University of Edinburgh. SRC is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (221890/Z/20/Z). ELSA is funded by the National Institute on Aging (R01AG017644), and by UK Government Departments coordinated by the National Institute for Health and Care Research (NIHR). No editorial service was provided. Funding Acknowledgements: This research was funded by the Legal & General Group (research grant to establish the independent Advanced Care Research Centre at the University of Edinburgh). The funder had no role in the conduct of the study, interpretation, or the decision to submit for publication. The views expressed are those of the authors and not necessarily those of legal and general.
Availability of data and material:
Data was obtained from the Lothian Birth Cohort 1936, more information can be found at https://lothian-birth-cohorts.ed.ac.uk/ . The English Study of Ageing data: ukdataservice.ac.uk/datacatalogue//series/series?id=200011. Any further data not found in such sources are available on request.
References
- Ajnakina O., & Steptoe A. (Document Report)
- Altelbany S. (2021). Evaluation of ridge, elastic net and lasso regression methods in precedence of multicollinearity problem: A simulation study. Journal of Applied Economics and Business Studies, 5(1), 131–142. - DOI: 10.34218/JAEBS.5.1.2021.014 [DOI] [Google Scholar]
- Atkins J. L., Jylhävä J., Pedersen N. L., Magnusson P. K., Lu Y., Wang Y., … & Pilling L. C. (2021). A genome‐wide association study of the frailty index highlights brain pathways in ageing. Aging Cell, 20(9), e13459. - DOI: 10.1111/acel.13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard J. R., & Bloom D. E. (2015). Towards a comprehensive public health response to population ageing. The Lancet, 385(9968), 658–661. - DOI: 10.1016/S0140-6736(14)61461-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett J., Theou O., Kirkland S., Andreou P., & Rockwood K. (2015). Frailty in NHANES: Comparing the frailty index and phenotype. Archives of Gerontology and Geriatrics, 60(3), 464–470. - DOI: 10.1016/j.archger.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Beyer I., Njemini R., Bautmans I., Demanet C., & Mets T. (2012). Immunomodulatory effect of NSAID in geriatric patients with acute infection: effects of piroxicam on chemokine/cytokine secretion patterns and levels of heat shock proteins. A double-blind randomized controlled trial (ISRCTN58517443). Cell Stress Chaperones, 17(2), 255–265. - DOI: 10.1007/s12192-011-0309-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries N. M., Staal J. B., Olde Rikkert M. G., & Nijhuis-van der Sanden M. W. (2013). Evaluative frailty index for physical activity (EFIP): a reliable and valid instrument to measure changes in level of frailty. Physical Therapy, 93(4), 551–561. - DOI: 10.2522/ptj.20120147 [DOI] [PubMed] [Google Scholar]
- Feng Z., Lugtenberg M., Franse C., Fang X., Hu S., Jin C., & Raat H. (2017). Risk factors and protective factors associated with incident or increase of frailty among community-dwelling older adults: A systematic review of longitudinal studies. PloS one, 12(6), e0178383. - DOI: 10.1371/journal.pone.0178383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher J. C., Guo C., Kelley M. R., & Vasko M. R. (2017). DNA damage mediates changes in neuronal sensitivity induced by the inflammatory mediators, MCP-1 and LPS, and can be reversed by enhancing the DNA repair function of APE1. Neuroscience, 366, 23–35. - DOI: 10.1016/j.neuroscience.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J. P., Welstead M., Cox S. R., Russ T. C., Marshall A., & Luciano M. (2023). Validation of a polygenic risk score for Frailty in the Lothian Birth Cohort and English Longitudinal Study of Ageing. medRxiv, 2023–04. - DOI: 10.1101/2023.04.03.23288064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei O., Holland D., Smeland O. B., Shadrin A. A., Fan C. C., Maeland S., … & Andreassen O. A. (2019). Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nature Communications, 10(1), 2417. - DOI: 10.1038/s41467-019-10225-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Tibshirani R, Hastie T. (2010). Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software, 33(1), 1–22. - DOI: 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A., Littlejohns T. J., Sudlow C., Doherty N., Adamska L., Sprosen T., … & Allen N. E. (2017). Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. American Journal of Epidemiology, 186(9), 1026–1034. - DOI: 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E. H., & Hubbard R. E. (2022). Frailty: understanding the difference between age and ageing. Age and Ageing, 51(8), afac185. - DOI: 10.1093/ageing/afac185 [DOI] [PubMed] [Google Scholar]
- Hengeveld L. M., Wijnhoven H. A., Olthof M. R., Brouwer I. A., Simonsick E. M., Kritchevsky S. B., … & Visser M. (2019). Prospective associations of diet quality with incident frailty in older adults: the health, aging, and body composition study. Journal of the American Geriatrics Society, 67(9), 1835–1842. - DOI: 10.1111/jgs.16028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. D., Weiss A., Liewald D. C., Davies G., Porteous D. J., Hayward C., … & Deary I. J. (2020). Genetic contributions to two special factors of neuroticism are associated with affluence, higher intelligence, better health, and longer life. Molecular psychiatry, 25(11), 3034–3052. - DOI: 10.1038/s41380-019-0385-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlihan L. M., Davies G., Tenesa A., Harris S. E., Luciano M., Gow A. J., … & Deary I. J. (2010). Common variants of large effect in F12, KNG1, and HRG are associated with activated partial thromboplastin time. The American Journal of Human Genetics, 86(4), 626–631. - DOI: 10.1016/j.ajhg.2010.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L., Guthrie B., Kelly P. A., Anand A., Marshall A., & Seth S. (2024). Frailty or Frailties: Exploring Frailty Index Subdimensions in the English Longitudinal Study of Ageing. arXiv preprint arXiv:2403.00472, 1. - arXiv:2403.00472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima G., Liljas A. E., & Iliffe S. (2019). Frailty syndrome: implications and challenges for health care policy. Risk management and healthcare policy, 12, 23–30. - DOI: 10.2147/RMHP.S168750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S. A., Abraham G., & Inouye M. (2019). Towards clinical utility of polygenic risk scores. Human molecular genetics, 28(R2), R133–142. - DOI: 10.1093/hmg/ddz187 [DOI] [PubMed] [Google Scholar]
- Kuhn Max (2008). Building Predictive Models in R Using the caret Package. Journal of Statistical Software, 28(5), 1–26 - DOI: 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- Lewis C. M., & Vassos E. (2020). Polygenic risk scores: from research tools to clinical instruments. Genome medicine, 12(1), 1–1. - DOI: 10.1186/s13073-019-0703-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q., Zheng Z., Xiu S., & Chan P. (2018). Waist circumference is a better predictor of risk for frailty than BMI in the community-dwelling elderly in Beijing. Aging clinical and experimental research, 30, 1319–1325. - DOI: 10.1007/s40520-018-0988-y [DOI] [PubMed] [Google Scholar]
- Marioni R. E., Ritchie S. J., Joshi P. K., Hagenaars S. P., Okbay A., Fischer K., … & Nagy R. (2016). Genetic variants linked to education predict longevity. Proceedings of the National Academy of Sciences, 113(47), 13366–13371. - DOI: 10.1073/pnas.1605334113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekli K., Stevens A., Marshall A. D., Arpawong T. E., Phillips D. F., Tampubolon G., … & Pendleton N. (2018). Frailty Index associates with GRIN2B in two representative samples from the United States and the United Kingdom. PLoS one, 13(11), e0207824. - DOI: 10.1371/journal.pone.0207824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosing M. A., Medland S. E., McRae A., Landers J. G., Wright M. J., & Martin N. G. (2012). Genetic influences on life span and its relationship to personality: a 16-year follow-up study of a sample of aging twins. Psychosomatic medicine, 74(1), 16–22. - DOI: 10.1097/PSY.0b013e318235d79b [DOI] [PubMed] [Google Scholar]
- Mullin D. S., Mead G. E., Stallard N., & Metcalf A. K. (2022). Prevalence and predictors of Motoric Cognitive Risk syndrome in a community‐dwelling older Scottish population: A longitudinal observational study. Int J Geriat Psychiatry, 37, gps.5824. - DOI: 10.1002/gps.5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkaby A. R., Yang L., Dufour A. B., & Gaziano J. M. (2021). Association between long-term aspirin use and frailty in men: the Physicians’ Health Study. Journal of Gerontology A Biol Sci Med Sci, 76(6), 1077–1083. - DOI: 10.1093/gerona/glaa269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R., & Deary I. J. (2015). Genetics and intelligence differences: five special findings. Molecular psychiatry, 20(1), 98–108. - DOI: 10.1038/mp.2014.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein E. D., Kunicka J., Kramer N., & Goldstein G. (1994). Modification of cytokine production by piroxicam. J Rheumatol, 21(5), 901–904. - No DOI available. [PubMed] [Google Scholar]
- Sathyan S., & Verghese J. (2020). Genetics of frailty: A longevity perspective. Translational Research, 221, 83–96. - DOI: 10.1016/j.trsl.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P. K., Reddy B. M., & Ganguly E. (2020). Frailty Syndrome among oldest old Individuals, aged≥ 80 years: Prevalence & Correlates. Journal of frailty, sarcopenia and falls, 5(4), 92. - No DOI available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steptoe A., Breeze E., Banks J., & Nazroo J. (2013). Cohort Profile: The English Longitudinal Study of Ageing. International Journal of Epidemiology, 42, 1640–1648. - DOI: 10.1093/ije/dys168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Most P. J., Wezenberg E., Hofman A., Van Greevenbroek M. M., Bergen P. F., Heier M., … & Biobank S. (2014). QCGWAS: A flexible R package for automated quality control of genome-wide association results. Bioinformatics, 30(8), 1185–1186. - DOI: 10.1093/bioinformatics/btt747 [DOI] [PubMed] [Google Scholar]
- Welstead M., Luciano M., Russ T. C., & Muniz-Terrera G. (2022). Heterogeneity of Frailty Trajectories and Associated Factors in the Lothian Birth Cohort 1936. Gerontology, 68, 861–868. - DOI: 10.1159/000522579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Noche R. B., Szejko N., Both C. P., Acosta J. N., Leasure A. C., … & Falcone G. J. (2023). A genome-wide association study of frailty identifies significant genetic correlation with neuropsychiatric, cardiovascular, and inflammation pathways. GeroScience, 1–3. - DOI: 10.1007/s11357-023-00771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data was obtained from the Lothian Birth Cohort 1936, more information can be found at https://lothian-birth-cohorts.ed.ac.uk/ . The English Study of Ageing data: ukdataservice.ac.uk/datacatalogue//series/series?id=200011. Any further data not found in such sources are available on request.