Abstract
The latent membrane protein LMP1 of Epstein-Barr virus (EBV) is often present in EBV-associated malignancies including nasopharyngeal carcinoma and Hodgkin's lymphoma. Previous work demonstrates that the LMP1 gene of EBV is sufficient to transform certain established rodent fibroblast cell lines and to induce the tumorigenicity of some human epithelial cell lines. In addition, LMP1 plays pleiotropic roles in cell growth arrest, differentiation, and apoptosis, depending on the background of the target cells. To examine the roles of LMP1 in cell proliferation and growth regulation in primary culture cells, we constructed a recombinant retrovirus containing an LMP1 gene. With this retrovirus, LMP1 was shown to stimulate the proliferation of primary mouse embryonic fibroblasts (MEF cells). It has a mitogenic activity for MEF cells, as demonstrated by an immediate induction of cell doubling time. In addition, it significantly extends the passage number of MEF cells to more than 30 after retroviral infection, compared with less than 5 for uninfected MEF cells. Furthermore, LMP1 cooperates with a p16-insensitive CDK4R24C oncogene in transforming MEF cells. Our results provide the first evidence of the abilities of the LMP1 gene, acting alone, to effectively induce the proliferation of primary MEF cells and of its cooperativity with another cellular oncogene in transforming primary cells.
Epstein-Barr virus (EBV) is a human gammaherpesvirus commonly carried in the majority of the human population. It is the causative agent for proliferative diseases such as infectious mononucleosis and oral hairy leukoplakia in AIDS patients. It is also implicated in a variety of human malignancies that include Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's lymphoma, nasal T-cell lymphoma, and immunoblastic lymphomas in posttransplant and AIDS patients (39). The virus often adopts latent forms in EBV-associated cancers. In nasopharyngeal carcinoma and Hodgkin's lymphoma, type II latency gene expression was observed. During this type II latency, only three viral proteins are routinely detected: EBV nuclear antigen 1 (EBNA1) and two latent membrane proteins, LMP1 and LMP2. Among the three EBV genes, the LMP1 gene is the only one implicated in cell immortalization and transformation. It is one of the viral genes required for B-cell immortalization, together with the EBNA2, EBNA3a, and EBNA3c genes (8, 15, 22, 23, 48). In addition, LMP1 has been shown to induce the transformation of certain established rodent fibroblast cell lines, including Rat-1 and BALB/c 3T3 (2, 33, 49). Furthermore, LMP1 induces the morphological transformation of the RHEK-1 cell line (12) and the tumorigenicity of epithelial cell lines in severe combined immunodeficient mice (18, 36).
Although EBV is an important oncogenic virus in human, the LMP1 genes appeared to act differently from oncogenic genes of other DNA oncogenic viruses, including simian virus 40 (SV40) large T antigen, adenovirus E1A and E1B, and human papillomavirus (HPV) E6 and E7. There is little evidence suggesting that the LMP1 gene can function similarly to these viral oncogenes or even a cellular oncogene such as ras or myc. Though LMP1 transforms several immortalized rodent fibroblast cell lines and participates in B-cell immortalization, a single LMP1 gene has not been shown to induce continuous proliferation of any primary cell culture. Similarly, although it cooperates with a number of EBV genes in inducing B-cell proliferation, there is no report on the cooperativity of the LMP1 gene with another cellular oncogene in transforming primary cells.
Protein sequence analysis of LMP1 revealed at least three domains: an amino-terminal cytoplasmic domain of 20 amino acid residues, a six-transmembrane domain of 185 amino acid residues, and a carboxy-terminal cytoplasmic domain of 200 amino acid residues (24). The six-transmembrane domain was shown to be required to form cytoplasmic membrane patches. The oligomerization of LMP1 in the cytoplasmic membrane may mimic that of other membrane receptor molecules induced by ligand-receptor interactions, resulting in constitutive activation of the LMP1 signaling pathway (14). The C-terminal cytoplasmic region was shown to interact with two families of proteins, TRAF (tumor necrosis factor [TNF] receptor-associated factor) and TRADD (TNF receptor-associated death domain), through two distinct domains (21, 34). These two domains participate in NF-κB activation (19, 32) and in the transformation of primary B cells with other EBV genes (20, 21). Other recent studies suggest that LMP1 can also induce the activation of c-Jun N-terminal kinase (JNK), resulting in the induction of AP1 activity of the transfected cells (11, 25). However, JNK and AP1 activations were also reported for the TNF signaling pathway responsible for the suppression of apoptosis (1). The roles of JNK and AP1 activation in LMP1-mediated cellular proliferation and transformation remain to be elucidated.
To understand the roles of LMP1 in cell proliferation and growth regulation in primary culture cells, we used a retrovirus to deliver LMP1 in the primary mouse embryonic fibroblast (MEF cell) model system. This approach allowed the integration and stable expression of LMP1 gene in the target cells at a high efficiency, thus permitting continuous analysis of the effect of LMP1 in primary culture cells in the absence of any drug selection. The results of our study indicate that LMP1 alone is sufficient to induce the proliferation of MEF cells. It has a mitogenic activity for MEF cells and is capable of significantly extending their passage numbers in culture. Furthermore, although LMP1 fails to cooperate with the ras oncogene to transform MEF cells, it cooperates with the CDK4R24C oncogene (46) to do so.
MATERIALS AND METHODS
Retroviral plasmids.
A 1.95-kb fragment containing a full-length LMP1 gene (pLMP1 [6]) was removed and inserted into the BamHI site of pcDNA3 to give pcDNA3-LMP1. The LMP1 fragment then was isolated with HindIII and EcoRV digests and cloned into HindIII and ClaI sites of the retroviral vector pLNSX (31) to generate pLNSX-LMP1. Other retroviral plasmids used in the study are pBabe-Ras with an active form of human H-rasV12 cDNA (46) and pWZL-R24C with a p16-insensitive CDK4R24C oncogene (46).
Cell culture and preparation of MEF cells.
Cell culture was carried out in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (GIBCO). PA317 and Bosc23 (American Type Culture Collection) are amphotropic and ecotropic retrovirus packaging cell lines, respectively. To produce MEF cells for the experiment, day 14 embryos were obtained from pregnant BALB/c mice. After removal of the head and red organs, the torso was minced and digested with 0.1% trypsin and 0.1% collagenase for 30 min at 37°C. The cells were seeded onto 10-cm cell culture dishes and split once at 1:4 before being frozen. For retroviral infections, the cells were thawed and split once more prior to infection.
Gene transfer mediated by retrovirus.
To obtain retroviruses, the retroviral plasmids were transfected into the ecotropic retrovirus packaging cell line Bosc23 with Lipofectin (GIBCO). After 15 h of incubation at 37°C, cells were placed in fresh medium and further incubated for 48 h. The virus-containing supernatants were collected at this time and again 12 h later. After filtering through 0.45-μm-pore-size filters (Millipore), the viral stocks were supplemented with Polybrene (8 μg/ml; Sigma). The viruses were then used to infect the amphotrophic viral packaging cell line PA317. After 24 h of infection, the infected PA317 cells were selected with the appropriate drug: puromycin (2 μg/ml) for pBabe, geneticin (500 μg/ml) for pLNSX, or hygromycin (200 μg/ml) for pWZL. Two or three weeks later, the resistant cells were collected and expanded as the virus-producing cell lines. To produce the viral stock, fresh medium was added to the virus-producing cells and incubated for 24 to 48 h. The virus-containing supernatants were collected and filtered as before. After supplementation with Polybrene (4 μg/ml), the supernatants were used fresh as viral stocks or aliquoted and stored at −80°C. Characterization and titration of the viruses were carried out with appropriate drug selections, immunofluorescence, and Western blotting with appropriate antibodies. Most of the viral stocks have titers ranging from 1 × 105/ml to 4 × 105/ml. In a typical experiment, Rat-2 or REF52 cells (5 × 105 per well) were seeded into six-well plates the day prior to infection. A fixed volume of a viral stock was used to infect the target cells for 48 h. LMP1-expressing virus-infected (MEF-LMP1) cells were trypsinized and seeded onto eight-well slides for immunofluorescence analysis by using monoclonal antibody S12, specific for LMP1, to determine the viral titer. Similar results were obtained with a number of mouse fibroblast and human epithelial cell lines. For all subsequent experiments, MEF cells were plated at 2 × 105 cells per well in six-well plates and incubated with an appropriate viral stock overnight. The culture medium was then replaced with another viral stock in the coinfection studies and incubated for another 8 h. The infected MEF cells, regarded as passage 1 (P1), were subcultured in normal medium without drug selection and split 1:4 when they reached confluence.
Cell growth analysis.
For cell growth curves, 2,000 cells were seeded per well into 24-well microtiter plates 24 h after viral infections. At the indicated time points, the cells were removed with trypsin digestion and the number of cells was counted. The experiment was done in triplicate to determine the number of cells at each time point and standard deviation. This experiment was reproduced twice.
Soft agar assay.
For soft agar assay, infected MEF cells at P3 or P6 were seeded into semisolid agarose medium (Dulbecco modified Eagle medium supplemented with 10% fetal calf serum) and low-melting-point agarose (base layer, 0.6%; upper layer, 0.35%) at a density of 4 × 103 cells per well in six-well plates (Nunc). The cells were fed each week with several drops of fresh medium placed directly onto the surface of the upper layer. After 2 to 3 weeks of incubation at 37°C, foci were viewed and counted. Photographs were taken under a microscope for analysis of representative colonies or with a regular camera after staining of the dishes with Giemsa and destaining with 50% ethanol in phosphate-buffered saline.
Immunoblot analysis.
Cells were collected and then washed with ice-cold phosphate-buffered saline and lysed in NP-40 lysis buffer (5). Cell lysates were made and analyzed for protein concentration by the Bradford assay (Bio-Rad). Then 100-μg aliquots of protein lysates were mixed with sample buffer and boiled for 5 min; they were then separated on sodium dodecyl sulfate–10% polyacrylamide gels and transferred to nitrocellulose membranes by electroblotting. Western blotting with a monoclonal antibody against LMP1 (S12) or an antibody against CDK4 (H-303; Santa Cruz) was followed by detection with an ECL (enhanced chemiluminescence) detection kit (Amersham).
Telomerase activity assay.
Telomerase activity was determined by TRAP assay (7). To minimize the effect of telomerase inhibitors that may have been present in the sample extract, each specimen was assayed at two concentrations (1-fold and 10-fold dilutions). For an RNase control, the lysate was preincubated with 0.5 μg of DNase-free RNase at room temperature for 15 min before the TRAP assay.
RESULTS
The LMP1 gene is effectively introduced into MEF cells via retrovirus.
To effectively introduce LMP1 into primary culture fibroblasts and monitor the cells continuously without the selective pressure of drugs, an LMP1-expressing retrovirus (v-LMP1) was generated and evaluated in Rat-2 cells by immunofluorescence and immunoblotting using a specific mouse monoclonal antibody against LMP1, S12 (30) or CS1-4 (42). We estimated that our optimized infection condition routinely resulted in infection of about 50% of the cells. Further study revealed that the v-LMP1 stocks were also capable of effectively infecting mouse fibroblast, human diploid fibroblast (IMR90), and human epithelial cell lines (data not shown).
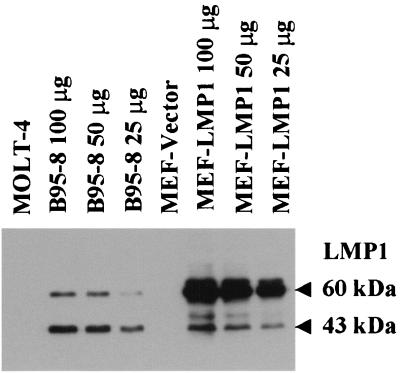
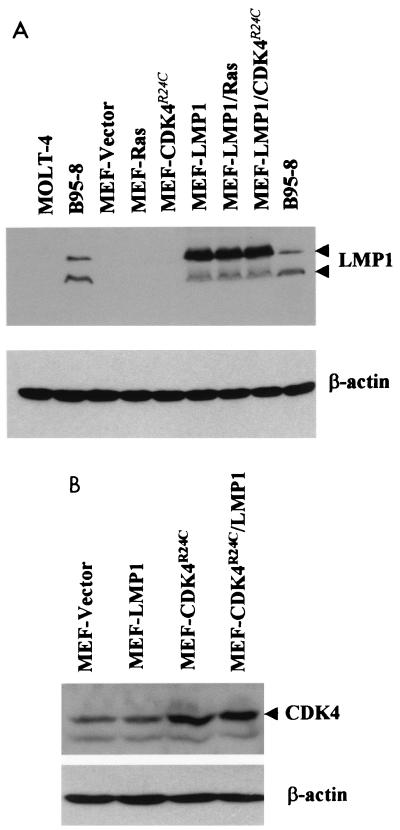
To analyze the effects of LMP1 on primary MEF cells, v-LMP1 was then used to infect the cells. These P1 cells were then cultured in medium without drug selection and split 1:4 upon reaching confluence. After two additional passages, the lysates of cells at P3 were analyzed by Western blotting and immunofluorescence with LMP1 antibody S12. Western blot results revealed two prominent immunoreactive bands with molecular masses of 60 and 43 kDa that were specifically detected from the lysates of v-LMP1-infected MEF cells, identical to those from the lysate of EBV-positive B95-8 cells (42) (Fig. 1). The Western blot result is consistent with previous findings for LMP1 protein. The 60-kDa band is the full-length LMP1, and the smaller 43-kDa band is likely a breakdown product. Both bands are absent in the EBV-negative lymphoblastic leukemia cell line MOLT-4 or in MEF cells infected with a vector retrovirus (v-LNSX). LMP1 proteins in v-LMP1-infected MEF cells are primarily full length. The stability of LMP1 appears to be a feature of the LMP1 gene used in the retrovirus and independent of the types of cells used for the infections (data not shown).
FIG. 1.
Western blot analysis of LMP1 in MEF cells after infection with v-LMP1. MEF cells were infected with either v-LNSX (MEF-Vector) or v-LMP1 (MEF-LMP1) and passaged twice before collected for analysis (P3). B95-8 is an EBV-positive marmoset B-lymphocyte cell line that expresses LMP1, and MOLT-4 is an EBV-negative lymphoblastic leukemia cell line. Cell lysates were made, and 100 μg of protein was loaded in each lane for Western blot analysis unless otherwise indicated. To compare the relative levels of LMP1 in MEF-LMP1 and B95-8 cells, two- and fourfold dilutions of both lysates were made and analyzed. A mouse monoclonal antibody against LMP1, S12, was used for the immunoblot.
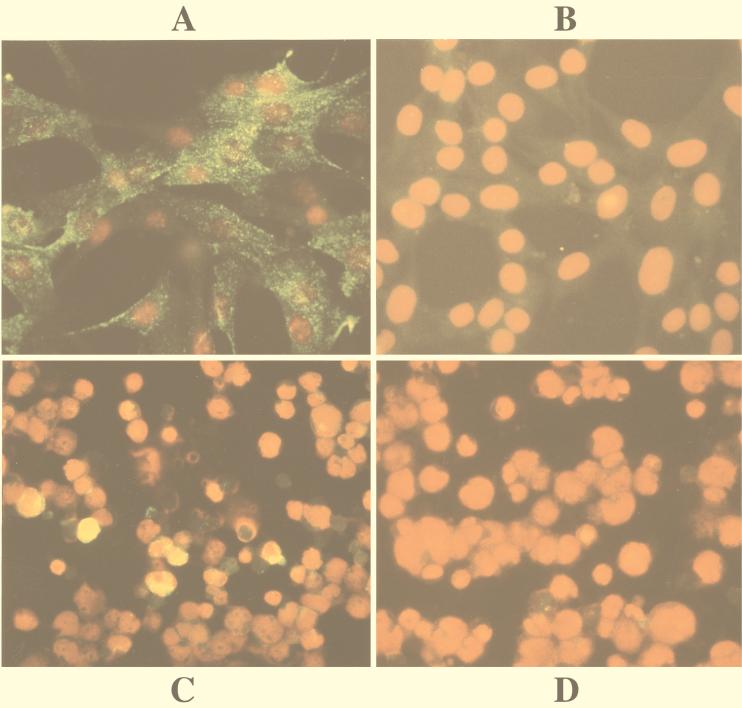
The retrovirus provided a good system for introducing LMP1 into MEF cells. At P3, over 90% of v-LMP1-infected MEF cells expressed LMP1, seen in the cytoplasm or on the cell surface in small aggregations, whereas the v-LNSX-infected MEF cells were negative in the assay (Fig. 2A and B). The total amount of LMP1 in v-LMP1-infected MEF cells was substantially higher than that in B95-8 cells, as indicated by their relative intensities in Western blot analysis of sequentially diluted lysates (Fig. 1). This difference may be partly due to the observation that the majority of v-LMP1-infected MEF cells expressed LMP1 at P3, whereas a significantly smaller percentage of our cultured B95-8 cells were positive for LMP1 in the immunofluorescence assay (Fig. 2C). The negative control MOLT-4 cells were negative for LMP1 (Fig. 2D).
FIG. 2.
Immunofluorescence analysis of v-LMP1-infected MEF cells. Cells were immunostained with LMP1 antibody S12 and counterstained with propidium iodide for nuclei. (A) MEF cells infected with v-LMP1 and subcultured twice (P3) before staining with S12 antibody for LMP1 expression. The green sparkles represent LMP1 proteins localized in cytoplasm or on cytoplasmic membrane. (B) MEF cells infected with control virus v-LNSX (P3). (C) B95-8 cells stained with S12 as the positive control. About 5 to 10% of our cultured cells were positive for LMP1. The positive signal is yellow due to the combined effect of green and red fluorescence. (D) MOLT-4 cells stained with S12 as the negative control.
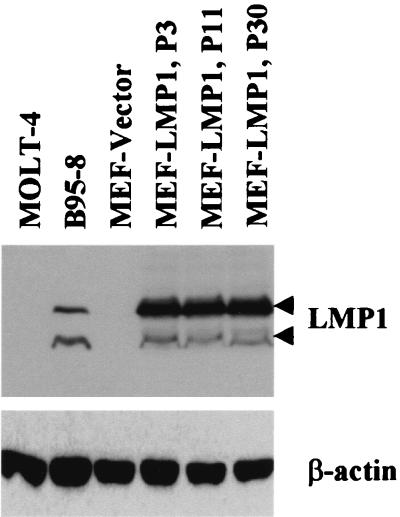
The level of LMP1 expressed in v-LMP1-infected MEF cells was stable for up to 30 passages after infection in the absence of any drug selection, as indicated by Western blot results for LMP1 (Fig. 3). In addition, the immunofluorescence of MEF-LMP1 cells at P24 revealed that virtually all cells were positive for LMP1 (data not shown). Thus, this retroviral system provided an effective way to stably introduce and express LMP1 in primary fibroblasts. The result further suggested the absence of cellular toxicity of LMP in primary MEF cells, as it was expressed in more than 90% of the cells starting from P3 (Fig. 2A), and the level of expression was stable throughout the course of the experiment (Fig. 3). In contrast, there appeared to be a growth advantage for LMP1-expressing MEF cells since only about 50% of the infected cells expressed LMP1 immediately following v-LMP1 infection, at the end of P1. Two passages later (P3), 90% of the MEF were positive for LMP1 in the absence of drug selection. Interestingly, expression of LMP1 was correlated with increasing passage numbers of MEF cells, which normally can be passaged only a few times in culture before reaching senescence.
FIG. 3.
The expression of LMP1 in v-LMP1-infected MEF cells is stable in the absence of drug selection. The cell lysates were obtained from MEF cells infected with v-LNSX (MEF-Vector) or v-LMP1 (MEF-LMP1) at P3, P11, and P30 following infection with v-LMP; 100 μg of protein was loaded in each lane for Western blot analysis with antibody S12. B95-8 is an EVB-positive cell line, and MOLT-4 is an EBV-negative cell line. Western blotting against β-actin was used as the loading control.
LMP1 induces the proliferation of MEF cells associated with extended passage numbers.
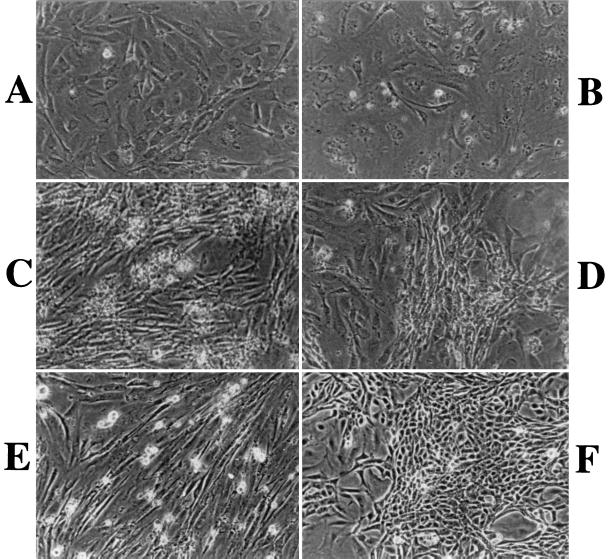
The effects of LMP1 on primary fibroblast cells were studied by infecting MEF cells with v-LMP1 or with v-LNSX as a negative control. The virus-infected cells were subsequently pooled and repeatedly subcultured whenever they reached confluence, which usually took about 1 week at early passages. Immediately after infection with retroviruses, all cell cultures presented the typical morphology of primary fibroblasts (Fig. 4A). The v-LNSX-infected cells had a low plating efficiency and grew slower with each additional passage, similar to the uninfected MEF cells. The cells gradually adopted a flatter and enlarged shape suggestive of an aging process. By P4 (12 population doublings), most v-LNSX-infected cells reached a senescence stage and could not be passaged further (Fig. 4B). The v-LMP1-infected MEF cells, however, behaved very differently from the v-LNSX negative control. Although v-LMP1-infected cells exhibited a morphology and growth rate similar to those of the v-LNSX control during P1, they displayed significant differences in both cellular morphology and proliferating ability at P2 and P3. The cells exhibited a higher plating efficiency and more vigorous growth with an apparently higher growth rate. In addition, the morphology of the cells changed to smaller and a long spindle shape, and the population gradually became relatively homogeneous, as indicated by a photo of MEF at P10 (Fig. 4C). Furthermore, it was clear that MEF-LMP1 cells can be passaged much further than MEF-LNSX cells. While uninfected MEF cells and MEF-LNSX cells were passaged fewer than five times, MEF-LMP1 cells had been subcultured for more than 30 passages and apparently could be readily passaged further. The result that LMP1 induced the proliferation of MEF cells was reproducible in a total of four independent experiments using MEF cells from two sets of mouse embryos over the course of 1 year. While the v-LNSX-infected MEF cells reached senescence before P6 in all experiments, the v-LMP1-infected MEF cells have been consistently passaged further. At present they have undergone 35, 22, 13, and 12 passages in these four experiments, and all maintain vigorous growth. Thus, the LMP1 gene alone is sufficient to induce the extended proliferation of MEF cells by significantly increase their passage numbers in vitro.
FIG. 4.
LMP1 extends the passage number of MEF cells and induces a morphological transformation of the cells in cooperation with CDK4R24C. (A) P1 MEF cells infected with v-LNSX. (B) P4 MEF cells infected with v-LNSX. Cells are flat and enlarged, indicating that they are approaching senescence. This is the last passage that can be routinely obtained with MEF-LNSX cells. (C) P10 MEF cells infected with v-LMP1. Cells are smaller, with a long spindle shape. They have a higher plating efficiency and grow continuously. (D) P10 MEF cells coinfected with v-LMP1 and v-H-rasV12. Their morphology and growth properties are similar to those of v-LMP1-infected cells. (E) P4 MEF infected with v-CDK4R24C. The cells were further passaged once before they stopped dividing. (F) P10 MEF cells coinfected with v-LMP1 and v-CDK4R24C. The cells exhibit a transforming morphology characterized by smaller and shorter cells with a high density. They are the most actively proliferating cells.
LMP1 stimulates the growth rate of MEF cells.
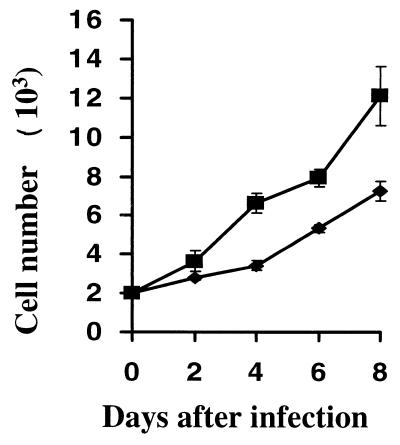
It was apparent that the v-LMP1-infected MEF cells grew faster than the v-LNSX-infected cells starting from P2, one passage away from the initial infections. To analyze the mitogenic effects of LMP1 on MEF cells, we examined the growth rates of MEF cells infected with v-LMP1 or v-LNSX by plating out the cells at the end of P1 into 24-well plates at a density of 2,000 cells per well. The cells were further incubated for growth analysis in a triplicate experiment in the absence of drug selection. At fixed time intervals, the cells were harvested and cells per well were counted for 8 days since the MEF-LMP1 cells nearly reached confluence by day 8. The results indicate that v-LMP1-infected cells grew substantially faster than v-LNSX-infected MEF cells (Fig. 5). The LMP1-expressing cells have a doubling time of about 3 days, whereas the vector control cells double every 4.5 days at this passage. However, it should be noted that the growth of MEF cells in vitro is a dynamic process, and cell growth became slower with each additional passage as though the cells were going through a rapid aging process. The results presented suggest that LMP1 has an immediate mitogenic effect on MEF cells in addition to its ability to significantly extend their passage numbers.
FIG. 5.
LMP1 increase the growth rate of MEF cells. MEF cells were freshly infected with v-LNSX (diamonds) or v-LMP1 (squares). After infection, the cells were removed from the plates with trypsin digestion at the end of P1; approximately 2,000 cells were plated in each well of 24-well plates in triplicate at day 0 and incubated further at 37°C. At the indicated time points, the cells were removed from the plates with trypsin and counted. The average numbers of cells at each time point was determined, and their standard derivations were obtained and plotted.
LMP1 cooperates with CDK4R24C in transforming MEF cells.
Although LMP1 induces the extended proliferation and increased growth rate of MEF cells, the MEF-LMP1 cells were not transformed even after 30 passages, as they preserved contact inhibition for growth on plates and failed to grow in soft agar (data not shown). In fact, most cellular and viral oncogenes are not capable of transforming primary MEF cells alone, and the cooperative transformation of primary murine fibroblast cells between oncogenes is an important feature of them. However, little information is available about the ability of LMP1 to cooperatively transform primary cells with any other cellular oncogene. Such cooperative transformation may be important as an indication of the roles of LMP1 in a multistep process of carcinogenesis, and it may provide a genetic model system with which to delineate the mechanism or pathway involved in LMP1-mediated transformation. To address the role of LMP1 in cooperatively transforming primary rodent fibroblast cells, retroviruses containing constitutive active H-rasV12 (v-H-rasV12 [46]) and p16-insensitive CDK4R24C (v-CDK4R24C [46]) genes were used in the cotransformation assay. A previous experiment indicated that LMP1 could mediate ERK1/2 activation in Rat-1 fibroblasts via a Ras-dependent pathway (40). Activated Ras and ERK1/2 can result in the up-regulation of cyclin D1 expression (28). The second oncogene, CDK4R24C, was originally identified in human melanomas with an arginine-to-cysteine exchange at residue 24 (51). This mutation prevents the binding of CDK4 to its inhibitor p16INK4a. As a result, CDK4R24C forms a complex with cyclin D1, and the CDK4R24C-cyclin D1 complex is resistant to the inhibition of p16INK4a in vivo (3).
As indicated above, a single LMP1 gene did not induce cell transformation. Similarly, the MEF cells coinfected with v-LMP1 and v-H-rasV12 (MEF-LMP1-Ras cells) also had an extended passage number, although they did not present any degree of morphological transformation at P10 in two experiments (Fig. 4D). v-H-rasV12 is functional since it transformed both NIH 3T3 and Rat-1 fibroblasts (data not shown), and we have found that it induces an immediate premature senescence of MEF cells (reference 46 and data not shown). The presence of v-H-rasV12 in MEF-LMP1-Ras cells was verified by the acquired resistance of a large percentage (30 to 50%) of the coinfected cells to puromycin (from pBabe-Ras) at P10. In contrast, both MEF cells (P3) and MEF cells infected with v-LMP1 alone (P10) were sensitive to puromycin.
Interestingly, several cell clones (4 and 19 in two experiments) with distinct morphology emerged only from cells coinfected with v-LMP1 and v-CDK4R24C, displaying an apparent morphological transformation at P3 on cell culture plates, approximately 2 to 3 weeks after coinfection. The cells became shorter and much smaller, and they grew much faster than all other infected cells. By P5, the entire population was largely taken over by these apparently transformed cells. At P10, the entire cell population exhibited a complete morphological transformation (Fig. 4F) that was not seen for cells infected with v-CDK4R24C (Fig. 4E). Although v-CDK4R24C-infected cells exhibited a slightly extended life span, they never exhibited this transforming morphology. In fact, the v-CDK4R24C-infected cells in two experiments were passaged once more (P5) and could not be passaged further. Thus, LMP1 specifically cooperates with CDK4R24C in inducing cellular morphological transformation of primary MEF cells.
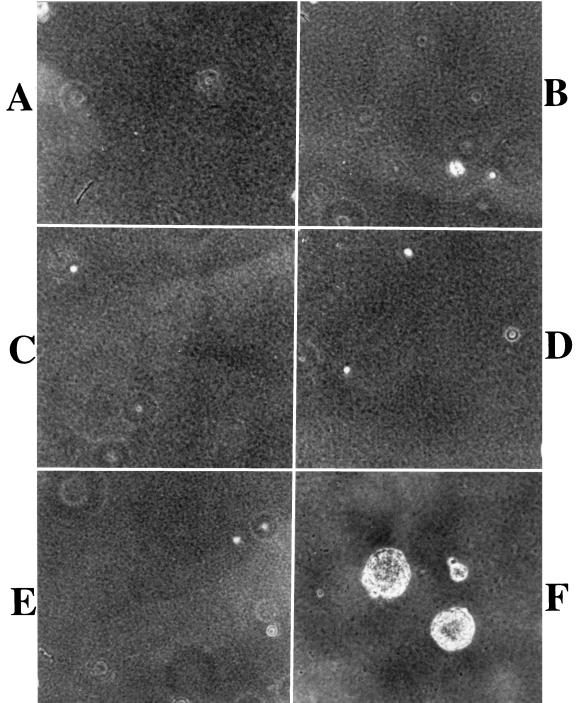
To further examine the transformation properties of cells coinfected with v-LMP1 and v-CDK4R24C, a soft agar assay was performed to demonstrate the anchorage-independent growth of these cells. About 15% (610 and 650 of 4,000 cells plated in two separate experiments) of the cells coinfected with v-LMP1 and v-CDK4R24C at P6 were capable of producing foci in the soft agar assay (Fig. 6F), confirming that they were transformed cells. In contrast, none of the four individual retroviruses, v-LNSX (P3), v-LMP1 (P6), v-H-rasV12 (P3), or v-CDK4R24C (P3), resulted in any focus formation in soft agar, nor did v-LMP1 and v-H-rasV12 coinfection (P6) (Fig. 6A to E). Cells at a different passage (P3) were used in some cases due to the limited passage numbers of the primary fibroblasts since none of the infected MEF cells without v-LMP1 could be passaged beyond P5. Thus, the results of the soft agar assay indicate that LMP1 cooperates with the CDK4R24C gene but not the H-rasV12 gene in transforming MEF cells.
FIG. 6.
Cooperative transformation of MEF cells by LMP1 and CDK4R24C. A soft agar assay was performed with cells infected with v-LNSX (A), v-LMP1 (B), v-H-rasV12 (C), v-LMP1 plus v-H-rasV12 (D), v-CDK4R24C (E), or v-LMP1 plus v-CDK4R24C (F). The cells at P6 were used for cells infected with v-LMP1 (B), v-LMP1 plus v-H-rasV12 (D), and v-LMP1 plus v-CDK4R24C (F). Cells at P3 were used for the other three cultures due to their limited passage numbers (P5 or less). The foci were counted in two independent experiments (see Results).
Like that of v-rasV12, the presence of both v-LMP1 and v-CDK4R24C in MEF cells was confirmed by the acquired resistance of the infected cells to geneticin and hygromycin, respectively. It is interesting that the MEF cells coinfected with v-LMP1 and v-CDK4R24C were the only ones resistant to both geneticin (for v-LMP1) and hygromycin (for v-CDK4R24C [46]) at P10, and they were resistant to both drugs despite the absence of any prior selection. In addition, Western blot results with anti-LMP1 antibody S12 revealed the expression of LMP1 from cell lysates of all v-LMP1-infected MEF cultures (Fig. 7A). Furthermore, an increased level of CDK4 protein could be detected in MEF cells infected with v-CDK4R24C (Fig. 7B). Thus, the results suggested that cointroduction of the EBV LMP1 and cellular CDK4R24C oncogenes resulted in the transformation of primary mouse fibroblasts.
FIG. 7.
Detection of LMP1 and CDK4 in infected MEF cells. (A) Cell lysates were made from MEF cells infected with v-LNSX, v-LMP1, v-H-rasV12, or v-CDK4R24C individually or in combinations as indicated and used for Western blot analysis of LMP1 protein. Controls for LMP1 protein were B95-8 (positive) and MOLT-4 (negative) cells. (B) Cell lysates from MEF cells infected with v-LNSX, v-LMP1, or v-CDK4R24C individually or in combination were analyzed for the expression of CDK4 with a CDK4-specific antibody (H-303; Santa Cruz). Although the endogenous CDK4 is present in the vector-infected cells (MEF-Vector), an increased level of CDK4 can be observed with v-CDK4R24C-infected MEF cells. Western blotting against β-actin was used as the loading control.
MEF cells infected with v-LMP1 were negative for telomerase.
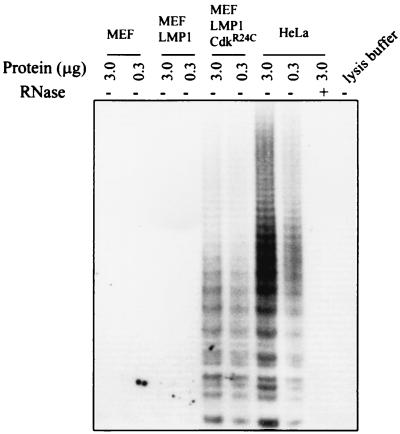
As indicated above, none of the MEF cells infected with v-LNSX, v-H-rasV12, or v-CDK4R24C could be passaged beyond P5 in the experiment presented. Because LMP1 is associated with extended passage numbers of MEF cells, we examined whether the proliferative MEF-LMP1 cells were positive for telomerase. Cell lysates from parent MEF cells and from MEF infected with v-LMP1 at P10 were examined for telomerase activity (Fig. 8). HeLa cell lysate was used as a positive control to indicate the sensitivity of the assay and its specificity after treatment of the lysate with RNase. The result indicated that v-LMP1 alone, although it extended the passage number of MEF cells, was not sufficient to induce telomerase activity. In contrast, the MEF cells coinfected with v-LMP1 and v-CDK4R24C were telomerase positive at P10, consistent with the transformation properties that they acquired. The v-CDK4R24C-infected MEF cells were not examined because they had a limited passage history (P5), one passage longer than that of the v-LNSX cells. The results suggest that although LMP1 efficiently induces the proliferation of MEF cells, additional changes are likely needed for the MEF-LMP1 cells to become immortal.
FIG. 8.
Detection of telomerase activity in uninfected MEF cells and in MEF cells infected with v-LMP1 (P10) or v-LMP1 and v-CDK4R24C (P10). A lysate from HeLa cells was used as the positive control, and an RNase-treated HeLa cell lysate was used as the negative control.
DISCUSSION
In the work presented here, primary BALB/c MEF cells were used to study the proliferation and transformation functions of the EBV transforming gene encoding LMP1. MEF provides a model system free of any genetic mutation that may complicate the results of the study, and it is commonly used as a well-defined system to study the effects of oncogenes in growth stimulation and cell transformation. An LMP1-expressing retrovirus was used to stably introduce LMP1 into MEF cells at a high efficiency and allow us to monitor the entire population of infected MEF cells immediately and continuously throughout the course of the experiment without any drug selection. As a result of this new approach, LMP1 was found to have a mitogenic effect on MEF cells, as indicated by an immediate increase in cell growth rate. In addition, while MEF cells undergo only a few passages, the v-LMP1-infected MEF cells can be readily subcultured for up to 30 passages, indicating that LMP1 stimulates the long-term proliferation of MEF cells. Furthermore, LMP1 cooperates with CDK4R24C in transforming MEF cells, suggesting that LMP1 is capable of cooperatively transforming primary rodent fibroblast cells with a cellular oncogene.
Although LMP1 is essential for EBV-mediated B-cell immortalization, this is the first demonstration that the LMP1 gene acting alone is sufficient to induce the proliferation of primary culture cells. The ability of LMP1 to induce cell proliferation may be important in our understanding of EBV-associated malignancies. Previous experiments showed that in addition to a tight association between nasopharyngeal carcinoma and EBV, most of the cancer cells came from clonal expansions of single EBV-infected progenitor cells (38), suggesting that the presence of EBV may provide the initial and fundamental drive in the process leading to the development of nasopharyngeal carcinoma. At present, the ability of a single LMP1 gene to induce the proliferation of primary culture cells is restricted to the model system of murine fibroblasts. It is obviously of interest to examine whether this LMP1 retrovirus is sufficient to induce the proliferation of primary human epithelial cells. It is well known, however, that LMP1 is not sufficient to immortalize primary B cells. Nonetheless, the system of LMP1-induced MEF proliferation should provide a unique basis for addressing roles and mechanisms of LMP1 as the sole EBV gene in cell proliferation and cell cycle regulation.
A variety of different DNA oncogenic viruses, including SV40, adenovirus, and papillomavirus, are capable of immortalizing primary rodent fibroblast cells. Two functions are frequently needed for these viruses to induce the immortalization of primary cells, the inactivation of Rb, and the suppression of p53 activities through direct interactions. SV40 large T antigen effectively induces the immortalization of primary rodent fibroblasts. In addition to its binding to pRb protein, the interaction between T antigen and tumor suppressor p53 is also essential for the immortalization (47, 53). In comparison, both adenovirus E1A and HPV E7, although capable of transforming immortalized rodent fibroblast cell lines, do not effectively immortalize primary cells without the additional partners, adenovirus E1B and HPV E6, respectively (16, 50, 52). The balance between proliferation and abrogation of cell cycle checkpoint is an important feature of these DNA oncogenic viruses. In all of these cases, the viral proteins interfere with the functions of the p53 tumor suppressor to prevent cell cycle checkpoint arrest and apoptosis induced by Rb inactivation. Our data here show that LMP1 is sufficient to effectively induce the proliferation of primary MEF cells, suggesting that LMP1 could have multiple functions in growth stimulation and in prevention of cell cycle arrest or apoptosis induced by abnormal proliferation.
Many studies indicated that the expression of viral or cellular oncogenes often leads to cell cycle arrest, apoptosis, and cell senescence that are in part dependent on p53 and its related cell cycle checkpoints (26, 35). For example, the expression of adenovirus E1A in primary fibroblast cells results in the stabilization of p53 and the induction of apoptosis (10, 29). Therefore, the ability to neutralize p53 is an essential feature of many DNA oncogenic viruses. The p53-binding domain of SV40 T antigen is essential for T antigen to immortalize and transform cells (47, 53). Similarly, both the E1B 55-kilodalton protein of adenovirus and E6 of HPV can inhibit the activity of p53 through direct interactions and, in the case of E6, the degradation of p53 protein (44, 52). Interestingly, the E1B 19-kDa protein is a homologue of Bcl-2 (4), encoded by a gene that was shown to be induced by LMP1 (17). Our results raised the possibility that LMP1 can also affect cell cycle checkpoints in the process of achieving primary cell proliferation. Previous experiments show that LMP1 is capable of inducing the expression of Bcl-2 (17, 43), activating NF-κB activity (19, 32), and up-regulating the expression of an antiapoptotic gene, A20 (27). All of these activities have been linked to suppression of cell apoptosis. Indeed, two studies directly demonstrate the roles of LMP1 in suppressing apoptosis induced by p53 (13, 37). We speculate, based on the ability of LMP1 to induce cell proliferation without cell apoptosis or growth arrest, that the roles of LMP1 in suppressing cell death or checkpoint arrest may also be an important part of LMP1 functions to induce a coordinated cellular proliferation of primary cells. We are currently investigating the roles of LMP1 in altering the regulation of genes involved in cell cycle control and checkpoint regulation.
Although LMP1 induces the proliferation of MEF cells, it is not sufficient to induce telomerase activity. In fact, no telomerase activity was observed in LMP1-induced proliferative MEF cells at P10. This result is consistent with a previous finding that in human B cells transformed with EBV, shortening of telomeres occurred in all EBV-positive populations until chromosomes had little telomeric DNA remaining. At this stage, many clones entered into a proliferative crisis and died, leaving only those with activated telomerase capable of proliferating indefinitely (9).
The exact mechanisms by which LMP1 induces cellular transformation are still under investigation. Two recent studies indicate that LMP1 is involved in induction of JNK (11, 25). The results also indicate that the LMP1-mediated JNK pathway appears to differ from the LMP1-induced NF-κB pathway. However, it is not clear that either pathway can explain the roles of LMP1 during its transformation process. Another recent study reveals that LMP1 can activate the Ras pathway (40). The activated Ras was shown to up-regulate cyclin D1, resulting in the inactivation of Rb through phosphorylation (41). Interestingly, a previous study shows that oncogenic Ras but not Myc can transform primary mouse fibroblast cells deficient for the p16 gene (45), suggesting such a cooperation may be mediated through the p16/CDK4/D1-to-pRb pathway. Analysis of the cooperation between the LMP1 gene and other oncogenes in inducing the transformation of primary fibroblast cells may provide a genetic model system for evaluating the roles of LMP1 and its activated pathways in the process of LMP1-mediated cellular transformation. Our results indicate that the LMP1 gene cooperates with CDK4R24C but not H-rasV12 in transforming MEF cells. The results are consistent with the notion that LMP1 may exert its transformation function through a Ras-related pathway. They are also in agreement with previous studies indicating that Ras readily induces the malignant transformation of MEF cells established from p16INK4a knockout mouse and cooperates with a CDK4R24C mutant in transforming MEF cells (45, 46).
It is worth noting that the cooperative transformation process with v-LMP1 and v-CDK4R24C may be different from v-LMP1-induced proliferation. The first sign of morphological transformation required 2 to 3 weeks with more passages, and it was found for only a very small percentage of infected cells analyzed for focus formation on plates (4 and 19 in two experiments). Alternatively, it is possible, although not very likely based on the viral titers, that only a small percentage of cells have both viruses and expressed both proteins. Nonetheless, the transformed cells induced by LMP1 and CDK4R24C quickly took over the entire population. The LMP1- and CDK4R24C-transformed MEF cells present a distinct morphology that is characteristic of the transformed cells. In addition, they form foci in the soft agar assay, indicative of anchorage-independent growth. Furthermore, they are positive for the telomerase activity that is absent in the proliferative MEF-LMP1 cells. In sum, EBV is a DNA oncogenic virus, and LMP1 appears to be capable of inducing the proliferation of primary MEF cells and cooperatively transforming them with another cellular oncogene.
ACKNOWLEDGMENTS
We are very grateful to F. A. Grasser for the LMP1 plasmid and D. Thorley-Lawson for antibody S12 against LMP1. We thank J. M. Nicholls for critical reading of the manuscript.
This work was supported by RGC and Croucher grants to L. Cao.
REFERENCES
- 1.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Baichwal V R, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- 3.Bartkova J, Lukas J, Guldberg P, Alsner J, Kirkin A F, Zeuthen J, Bartek J. The p16-cyclin D/Cdk4-pRb pathway as a functional unit frequently altered in melanoma pathogenesis. Cancer Res. 1996;56:5475–5483. [PubMed] [Google Scholar]
- 4.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 5.Cao L, Faha B, Dembski M, Tsai L H, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen H F, Kevan-Jah S, Suentzenich K O, Grasser F A, Mueller-Lantzsch N. Expression of the Epstein-Barr virus latent membrane protein (LMP) in insect cells and detection of antibodies in human sera against this protein. Virology. 1992;190:106–115. doi: 10.1016/0042-6822(92)91196-2. [DOI] [PubMed] [Google Scholar]
- 7.Cheng R Y, Yuen P W, Nicholls J M, Zheng Z, Wei W, Sham J S, Yang X H, Cao L, Huang D P, Tsao S W. Telomerase activation in nasopharyngeal carcinomas. Br J Cancer. 1998;77:456–460. doi: 10.1038/bjc.1998.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Counter C M, Botelho F M, Wang P, Harley C B, Bacchetti S. Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein-Barr virus-transformed human B lymphocytes. J Virol. 1994;68:3410–3414. doi: 10.1128/jvi.68.5.3410-3414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 11.Eliopoulos A G, Young L S. Activation of the c-Jun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 12.Fahraeus R, Rymo L, Rhim J S, Klein G. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature. 1990;345:447–449. doi: 10.1038/345447a0. [DOI] [PubMed] [Google Scholar]
- 13.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 16.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 18.Hu L F, Chen F, Zheng X, Ernberg I, Cao S L, Christensson B, Klein G, Winberg G. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene. 1993;8:1575–1583. [PubMed] [Google Scholar]
- 19.Huen D S, Henderson S A, Croom-Carter D, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 20.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi K M, Kieff E K. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempkes B, Spitkovsky D, Jansen-Durr P, Ellwart J W, Kremmer E, Delecluse H J, Rottenberger C, Bornkamm G W, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 25.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 27.Laherty C D, Hu H M, Opipari H M, Wang F, Dixit V M. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 28.Lavoie J N, L'Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 29.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 30.Mann K P, Staunton D, Thorley-Lawson D A. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell T, Sugden B. Stimulation of NF-κ B-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moorthy R K, Thorley-Lawson D A. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of Rat-1 fibroblasts. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 35.Mowat M R. p53 in tumor progression: life, death, and everything. Adv Cancer Res. 1998;74:25–48. doi: 10.1016/s0065-230x(08)60764-2. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson L J, Hopwood P, Johannessen I, Salisbury J R, Codd J, Thorley-Lawson D, Crawford D H. Epstein-Barr virus latent membrane protein does not inhibit differentiation and induces tumorigenicity of human epithelial cells. Oncogene. 1997;15:275–283. doi: 10.1038/sj.onc.1201187. [DOI] [PubMed] [Google Scholar]
- 37.Okan I, Wang Y, Chen F, Hu L F, Imreh S, Klein G, Wiman K G. The EBV-encoded LMP1 protein inhibits p53-triggered apoptosis but not growth arrest. Oncogene. 1995;11:1027–1031. [PubMed] [Google Scholar]
- 38.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 39.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 40.Roberts M L, Cooper N R. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology. 1998;240:93–99. doi: 10.1006/viro.1997.8901. [DOI] [PubMed] [Google Scholar]
- 41.Roussel M F. Key effectors of signal transduction and G1 progression. Adv Cancer Res. 1998;74:1–24. doi: 10.1016/s0065-230x(08)60763-0. [DOI] [PubMed] [Google Scholar]
- 42.Rowe M, Evans H S, Young L S, Hennessy K, Kieff E, Rickinson A B. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J Gen Virol. 1987;68:1575–1586. doi: 10.1099/0022-1317-68-6-1575. [DOI] [PubMed] [Google Scholar]
- 43.Rowe M, Peng-Pilon M, Huen D S, Hardy R, Croom-Carter D, Lundgren E, Rickinson A B. Upregulation of bcl-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-κB activation and to induction of cell surface markers. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 45.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 46.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 47.Srinivasan A, Peden K W, Pipas J M. The large tumor antigen of simian virus 40 encodes at least two distinct transforming functions. J Virol. 1989;63:5459–5463. doi: 10.1128/jvi.63.12.5459-5463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe S, Kanda T, Yoshiike K. Human papillomavirus type 16 transformation of primary human embryonic fibroblasts requires expression of open reading frames E6 and E7. J Virol. 1989;63:965–969. doi: 10.1128/jvi.63.2.965-969.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Buschenfelde K H, Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 52.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 53.Zhu J, Rice P W, Gorsch L, Abate M, Cole C N. Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J Virol. 1992;66:2780–2791. doi: 10.1128/jvi.66.5.2780-2791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]