Abstract
Transcriptional transactivators (Tat) from human immunodeficiency and equine infectious anemia viruses (HIV and EIAV) interact with their transactivation response elements (TAR) to increase the rates of viral transcription. Whereas the human cyclin T1 is required for the binding of Tat to TAR from HIV, it is unknown how Tat from EIAV interacts with its TAR. Furthermore, Tat from EIAV functions in equine and canine cells but not in human cells. In this study, we present sequences of cyclins T1 from horse and dog and demonstrate that their N-terminal 300 residues rescue the transactivation of Tat from EIAV in human cells. Although human and equine cyclins T1 bind to this Tat, only the equine cyclin T1 supports the binding of Tat to TAR from EIAV. Finally, a reciprocal exchange of the valine for the leucine at position 29 in human and equine cyclins T1, respectively, renders the human cyclin T1 active and the equine cyclin T1 inactive for Tat transactivation from EIAV. Thus, the collaboration between a specific cyclin T1 and Tat for their high-affinity interaction with TAR is a common theme of lentiviral transactivation.
Equine infectious anemia virus (EIAV) and Human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2) are members of the Lentivirus genus of the Retroviridae family (23). All share similarities in genetic organization and biological properties and cause persistent diseases in their host. Like HIV-1, EIAV codes for structural, enzymatic, and regulatory proteins. In the latter category, EIAV codes only for the transactivator Tat and the regulator of viral gene expression (Rev) (15, 23, 27, 28).
Tat proteins from HIV-1 (hTat) and EIAV (eTat) are essential for high levels of viral gene expression and replication. Their functional motifs have been identified (4, 12, 13, 23). The activation domain in hTat contains essential and contiguous N-terminal, cysteine-rich, and core sequences. The RNA-binding domain contains an arginine-rich motif (ARM) followed by C-terminal sequences, which are not essential for its function. Intriguing differences in this arrangement are found in eTat. First, the N-terminal domain of eTat is dispensable for function. Second, its cysteine-rich motif contains only two cysteines rather than the seven found in hTat. Between the core domain and the ARM are five additional residues, which include flanking leucines. The ARM is followed by essential C-terminal sequences, which end in critical dileucines (10, 11, 23). These differences suggested that other sequences in eTat, in addition to cysteine-rich and core domains, participate in the interaction with its cellular coactivator and that nucleic acid-binding strategies might be different between these two viruses (4, 7, 13).
The basic domain in hTat (ARM) binds to the 5′ bulge in TAR from HIV-1 (hTAR), which forms an RNA stem-loop of 59 nucleotides (23). Human cyclin T1 (hT) binds to the activation domain of hTat and to the central loop in hTAR. This tripartite complex has exquisite specificity and affinity for hTAR (31). In sharp contrast, no binding between eTat and its TAR (eTAR) has been demonstrated to date. Nevertheless, eTat should bind to eTAR. This conclusion is based on several observations. First, the secondary structure of eTAR, which contains 25 nucleotides and lacks a 5′ bulge, resembles the longer hTAR (6, 7, 14, 16). Second, mutageneses of eTAR revealed that the stem-loop in eTAR is important for its function (7). Third, eTat could be brought to the EIAV and HIV-1 LTRs by heterologous RNA-tethering systems (12). Fourth, eTat and hTat cross-squelch each other's transcriptional activities (5, 26).
Finally, as has been reported for hTat, eTat transactivation is also restricted to cells from organisms that can be infected by EIAV. For example, hTat functions optimally in human cells and poorly in rodent cells (23, 26). Likewise, eTat transactivates eTAR in equine and canine cells but not in human cells (4, 26). For HIV-1, this species specificity was mapped to a cysteine at position 261 in hT, which is a tyrosine in the murine cyclin T1 (mT). Reciprocal exchanges of this one residue between hT and mT rendered the mutant mTY261C protein active and the mutant hTC261Y protein inactive (3, 18, 19, 21, 25). Cyclins T form the positive transcription elongation factor b complexes (22, 30, 31), which also contain the cyclin-dependent kinase 9 (CDK9) that hyperphosphorylates the C-terminal domain RNA polymerase II, thus facilitating the transition from initiation to elongation of transcription (22).
These findings suggested that horse and dog cyclins T1 (equine and canine cyclins T1 [eT and cT, respectively]) might perform a similar role in eTat transactivation. Indeed, a recent study identified eT (2). Like hT, eT mediated the binding between eTat and eTAR and rescued eTat transactivation in human cells (2). Although N-terminal residues of eT were identified as essential for the interaction between eTat and eTAR and for eTat transactivation in human cells, specific residues in eT that would provide a molecular explanation for the inability of hT to support eTat transactivation in human cells were not identified (2). In this study, we report the isolation of eT and cT. Both cyclins T1 rescued eTat transactivation in human cells. Although hT and eT bound to eTat, only eT supported the interaction between eTat and eTAR. Most importantly, a reciprocal exchange of the valine for the leucine at position 29 in hT and eT, respectively, rendered hT active and eT inactive. Thus, the interaction between a specific cyclin T1, Tat, and TAR represents a common theme of lentiviral transactivation.
MATERIALS AND METHODS
Cloning of equine and canine cyclins T1.
cDNAs were first synthesized from total RNA, which was extracted from equine dermal or canine D17 cells, by reverse transcription with random hexamer primers. PCRs were performed on these cDNAs with the primers 5′-TACGATCGGATCCATGGAGGGAGAGAGGAAGAAC, 5′-TACGCTGAATTCCATTAAACCTGCAATGGTTGTGTC, and 5′-TACGCTGAATTCCTTAGGAAGGGGTGGAAGTGGTGG. These were designed based on N- and C-terminal sequences of the full-length and the first 300 residues of hT (31). The resulting cDNA fragments were cloned into the pEF-BOS vector in the BamHI and EcoRI sites, resulting in plasmids eT, eT300, and cT300, which were subjected to dideoxynucleotide DNA sequencing. The sequences presented in Fig. 1A were derived from several independent cloning events.
FIG. 1.
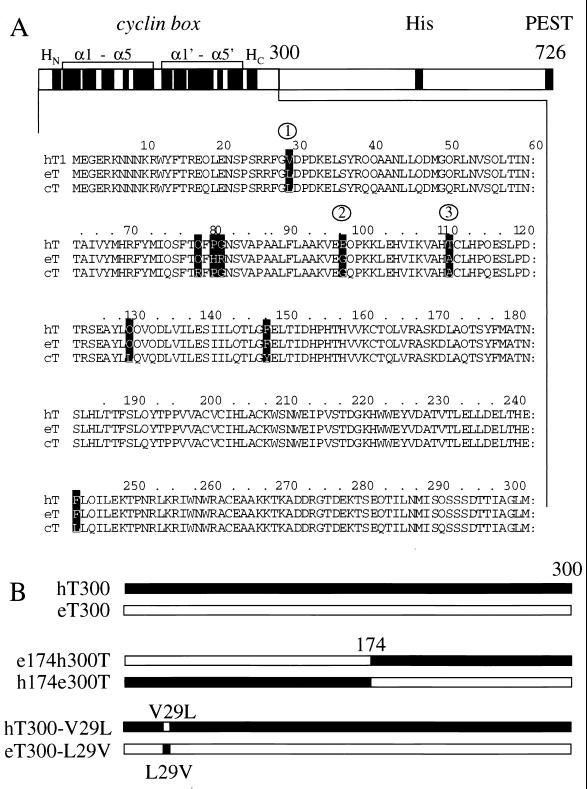
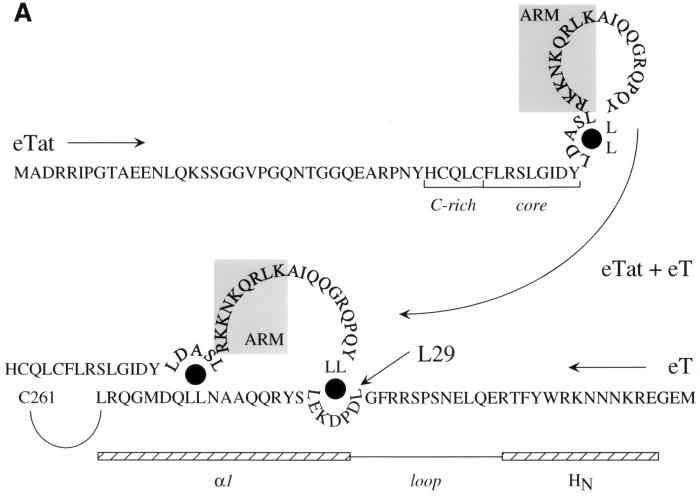
Alignment of the N-terminal 300 residues of hT, eT, and cT and a diagrammatic representation of the cyclins T1 used in this study. (A) Below a schematic diagram of hT726 are presented the N-terminal 300 sequences from hT, eT, and cT (hT300, eT300, and cT300). Twelve solid boxes under the “cyclin box” represent α helices: HN and HC and N- and C-terminal α helices, respectively, and the rest form duplicated cyclin boxes of five α helices each. Histidine-rich (His) and PEST sequences have been described previously (31). Differences between the cyclins T1 are depicted by white letters in black boxes. These cyclins have 98% sequence identity. Of eight changed residues, only three (L29, G96, and A110) are conserved between eT300 and cT300, and they are highlighted by numbers within circles. (B) Schematic representation of hT300 (black) and eT300 (white) chimeras between these cyclins T1 and the mutants hT300-V29L and eT300-L29V proteins used in this study. The hybrid eT174hT300 protein contains the N-terminal 174 residues of eT and the C-terminal 126 residues of hT300. The hybrid hT174eT300 protein contains the reciprocal arrangement of these sequences. The mutant hT300-V29L protein contains a point mutation at position 29 in hT300, where the valine is changed to the leucine found in eT300. The mutant eT300-L29V protein contains the reciprocal exchange, where the leucine at position 29 of eT was mutated to the valine found in hT.
Plasmid constructions.
Plasmid target pUX-E478, which contains the EIAV long terminal repeat (LTR) linked to the chloramphenicol acetyltransferase reporter gene, and the plasmid effector pRS-E-Tat-M, which contains eTat from positions 8 to 75, have been described previously (15). pHIVSCAT and pcDNA3-Tat were also described (17). The hybrid glutathione S-transferase–eTat protein was expressed from pGEX-2TK in Escherichia coli. It was constructed from pRS-E-Tat-M by using appropriate primers with BamHI and EcoRI linkers (Pharmacia, Piscataway, N.J.). For the in vitro transcription and translation, eT300 and hT300 were subcloned into pEF-T7. eT174hT300 and hT174eT300 chimeric proteins were constructed by PCR. Mutagenesis of the leucine and valine residues in eT and hT was performed with the transformer site-directed mutagenesis kit (Clontech). For in vitro synthesis of eTAR RNA, the appropriate oligonucleotide was cloned into pGEM3Z(+). All plasmid constructions were confirmed by DNA sequencing.
Protein purification.
The hybrid GST-eTat protein, the hybrid GST-eT300 and GST-hT300 proteins, and the hybrid mutant GST-hT300-V29L protein were expressed in E. coli DH5α and purified with glutathione-Sepharose beads (Pharmacia) (17, 20). Eluted proteins were examined by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and their concentration was determined with a protein assay kit (Bio-Rad, Hercules, Calif.).
In vitro binding assays.
Protein binding assays for the binding between hybrid GST-eTat protein and eT300 or hT300 were performed as follows. A 5-μg portion of the hybrid GST-Tat protein was incubated at 4°C for 3 h with 35S-labeled eT300 or hT300 transcribed and translated with the rabbit reticulocyte lysate (Promega, Madison, Wis.) in 300 μl of buffer C, consisting of 20 mM HEPES (pH 7.9), 1% Triton X-100, 0.5% Nonidet P-40, 0.3% bovine serum albumin, 2 mM dithiothreitol, 200 mM KCl, 0.5% SDS at 4°C for 3 hr. After binding, GST beads were added to the reaction mixture, which was further incubated for additional 30 min. The beads were washed extensively three times with buffer C containing 1 M KCl (17). Bound proteins were eluted with equal volume of 2× SDS loading buffer, resolved by SDS-PAGE (10% polyacrylamide), and analyzed by autoradiography.
Transient-expression assays.
HeLa cells were cotransfected with pUX-E478 (0.2 μg) and pRS-E-Tat-M (0.2 μg) in the presence of 1 μg of eT300 or cT300 by using Lipofectamine (GIBCO-BRL, Gaithersburg, Md.). For the transfection into CHO cells, pHIVSCAT (0.1 μg) and pcDNA3-Tat (0.1 μg) were cotransfected in the presence of the indicated amounts of eT300 and cT300 (1 μg) (17). Western blot analysis was performed with anti-HA antibodies (8, 9).
Electrophoretic mobility shift assays.
32P-labeled wild-type eTAR was transcribed by T7 from linearized plasmid templates (Ambion, Austin, Tex.). The eTAR probe was incubated with 300 ng of the indicated cyclins T1, which were purified from bacteria as GST fusion proteins and were eluted from the GST beads with 30 mM glutathione followed by dialysis. Binding reactions were performed for 10 min at 30°C in the absence or presence of 80 ng of purified hybrid GST-Tat proteins in 20 μl of buffer E, consisting of 30 mM Tris-HCl (pH 8), 70 mM KCl, 0.01% Nonidet P-40, 5 mM MgCl2, 1 mM dithiothreitol, 13% glycerol, 53 μg of poly(dI-dC) per ml, and 31 μg poly(I-C) per ml (17). A 100-fold excess of cold competitor RNA was used as indicated. RNA-protein complexes were separated on an 8% nondenaturing polyacrylamide gel and subjected to autoradiography.
Nucleotide sequence accession number.
The equine cyclin T1 nucleotide sequence has been submitted to GenBank (accession no. 190905).
RESULTS
Isolation of equine and canine cyclins T1.
Previous studies demonstrated that the transactivation of the HIV-1 LTR by hTat is restricted to primate cells (10, 23). In contrast, eTat cannot transactivate the EIAV LTR in human cells (4, 26). For hTat, this restriction was mapped to hT (31). To determine if eT plays a similar role in eTat transactivation, we isolated eT and eT300 by using total RNA extracted from cells that are permissive for EIAV infection. PCR and appropriate oligonucleotides, based on sequences from the full-length hT and hT300, were used (31). Since eTat functions well in canine D17 cells, cT300 was also isolated from canine cells (23). Figure 1A shows only the N-terminal 300 residues of cyclins T1, since these are sufficient to mediate eTat transactivation (the complete sequence of eT was deposited in GenBank). The N-terminal 300 residues of hT, eT, and cT (hT300, eT300, and cT300) are 98% identical. They contain not only a cyclin box at their N termini but also a conserved cysteine at position 261, which might explain the ability of hTat to function in equine or canine cells (Fig. 1A).
Comparing sequences between hT300, eT300, and cT300 helped us to identify unique residues in eT or cT that might be important for eTat transactivation. Since eTat functions equally well in equine and canine cells, residues that are not shared between eT and cT could be dismissed from consideration. The remaining differences between these cyclins T and hT were good candidates for residues that might be required for eTat transactivation. Indeed, although the first 300 residues contain eight changes between these three cyclins T1 (Fig. 1A, white letters in black boxes), five of them were not conserved between eT and cT. Thus, only three residues, leucine, glutamate, and alanine at positions 29, 96, and 110, respectively (Fig. 1A, numbers in circles) were considered further (see below).
The N-terminal 300 residues in eT support eTat transactivation in human cells.
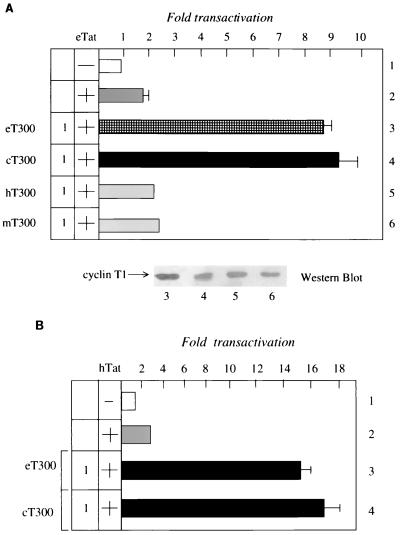
To determine if eT300 and cT300 could rescue eTat transactivation in human cells, we introduced eT300 or cT300 into HeLa cells in the presence of eTat and eTAR. As shown in Fig. 2A, eT300 or cT300, but not hT300 or mT300, increased eTat transactivation fivefold above basal levels in these cells (lanes 3 to 6). Next, we examined the ability of eT300 or cT300 to rescue hTat transactivation in rodent cells. It is known that hTat functions via hTAR in equine or canine cells, suggesting that eT300 and cT300 should support hTat transactivation. As presented in Fig. 2B, both eT300 and cT300 rescued hTat transactivation, up to fourfold above basal levels, in CHO cells, where hTat functions poorly (lanes 3 and 4). This result is expected, since eT300 and cT300 contain the cysteine at position 261, which increases its binding to hTat and hTAR. We conclude that eT300 and cT300 rescue eTat transactivation in human cells and facilitate hTat transactivation in rodent cells.
FIG. 2.
The N-terminal residues in eT and cT support eTat transactivation in human cells and hTat transactivation in rodent cells. (A) The N-terminal 300 residues in eT and cT support eTat transactivation in HeLa cells. The EIAV LTR was expressed alone (pUX-E478; lane 1, white bar) or together with eTat (pRS-E-Tat-M; lane 2, gray bar). To eTat were added effector plasmids coding for eT300 or cT300 (1 μg) (lanes 3 and 4, checked and black bars, respectively), hT300 (lane 5, gray bar) and mT (lane 6, gray bar). The value of the CAT activity of pUX-E478 alone was set to 1. Standard errors of the mean from three independent transfections are shown by errors bars. Western blotting revealed that levels of expression of different cyclins T1 were similar. Numbers under the Western blot correspond to lanes from transient-expression assays. (B) The N-terminal 300 residues in eT and cT support hTat transactivation in CHO cells. The HIV-1 LTR was expressed alone (pHIVSCAT; lane 1, white bar) or together with hTat (pcDNA3Tat; lane 2, gray bar). To hTat were added effector plasmids coding for eT300 or cT300 (1 μg) (lanes 3 and 4, black bars). Values are as in panel A.
eTat binds to the N-terminal 300 residues of eT and hT.
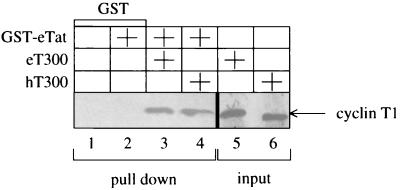
Having established an important role for eT in eTat transactivation, we set out to determine if eTat binds directly to eT300 or hT300. An in vitro protein-binding assay was performed with a hybrid GST-eTat protein, which was incubated with 35S-labeled eT300 or hT300 (Fig. 3). Complexes containing cyclin T1 and hybrid GST-eTat protein were isolated with glutathione-Sepharose beads, washed extensively, separated by SDS-PAGE, and subjected to autoradiography. As shown in Fig. 3, both hT300 and eT300 bind specifically and with similar affinities to the hybrid GST-eTat protein (lanes 3 and 4). These results indicate that the first 300 residues in eT bind to eTat. We conclude that the restriction of eTat transactivation in human cells is not due to decreased binding of eTat to hT.
FIG. 3.
The N-terminal 300 residues in eT and hT bind to eTat. 35S-labeled eT300 or hT300 proteins were incubated with the hybrid GST-eTat protein and selected on glutathione-Sepharose beads. Bound cyclins T1 were separated by SDS-PAGE and subjected to autoradiography (lanes 3 and 4). GST alone was also incubated in the presence or absence of the hybrid GST-eTat protein (lane 1 and 2). The input of each cyclin T1 was equal in all reactions and represented 10% of the amount used for the binding assay (lanes 5 and 6).
Reciprocal exchanges of the valine and leucine at position 29 in hT and eT render hT active and eT inactive for eTat transactivation in human cells.
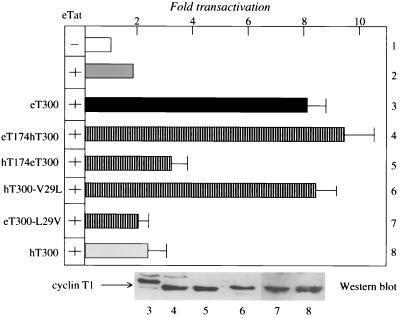
To identify residues in eT300 that are responsible for the inability of hT to support eTat transactivation, we exchanged N- and C-terminal sequences between eT300 and hT300 (Fig. 1B) and examined effects of these chimeras on eTat transactivation in HeLa cells. As shown in Fig. 4, N-terminal sequences from eT300 support eTat transactivation (lane 3). The hybrid eT174hT300 protein contains the N-terminal 174 residues of eT and 126 residues of hT300 and all three conserved changes between cT300, eT300, and hT300 (Fig. 1A). It increased eTat transactivation fivefold above basal levels in HeLa cells (Fig. 4, lane 4). The reciprocal chimera, containing the N-terminal 174 residues of hT and 126 residues of eT300 (hT174eT300), had no effect on eTat transactivation (lane 5). As expected, hT300 protein also had no effect on eTat transactivation (lane 8). Most importantly, a single point mutation in hT300, where the valine at position 29 was changed to leucine, found in eT300 (hT300-V29L), enabled this mutated hT protein to rescue eTat transactivation in human cells (lane 6). In contrast, the reciprocal mutation, where the leucine at position 29 in eT was changed to the valine found in hT (eT300-L29V), did not support eTat function (lane 7). Thus, it is the leucine at position 29 that facilitates the high-affinity interaction between eTat, eT, and eTAR. Similar to the situation with hT, where the cysteine at position 261 is essential for interactions between hTat, hT, and hTAR, the leucine at position 29 in eT is required for eTat transactivation. This leucine at position 29 is a valine in hT, providing a molecular explanation for the inability of hT to support eTat transactivation.
FIG. 4.
A valine-to-leucine change in hT300 rescues eTat transactivation in human cells. Reciprocal substitutions of the valine and leucine at position 29 in hT and eT, respectively, activated hT and inactivated eT for eTat transactivation in HeLa cells. The EIAV LTR was expressed alone (pUX-E478; lane 1, white bar) or together with eTat (pRS-E-Tat-M; lane 2, gray bar) in the presence or absence of the indicated plasmid effectors coding for eT300 (lane 3, black bar), hT300 (lane 8, gray bar), hT300 eT 300 chimeras (lanes 4 and 5, striped bars [see Fig. 1B]), the mutant hT300-V29L protein, where the valine at position 29 was replaced by the leucine residue found in eT (lane 6), and the mutant eT300-L29V protein, where this leucine was replaced by the valine (lane 7). Values are as in Fig. 2A. Western blotting revealed that the levels of expression of different cyclins T1 were similar. Numbers under the blot correspond to lanes from transient-expression assays.
The N-terminal 300 residues of eT support the binding of eTat to eTAR.
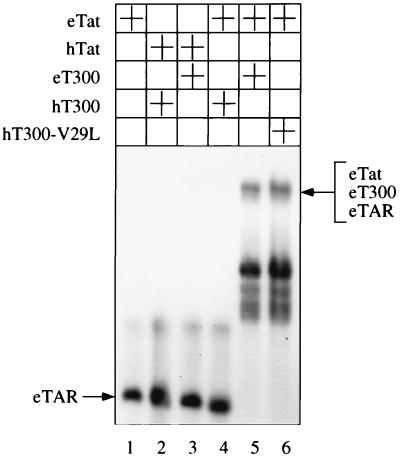
Having established that low levels of eTat transactivation in human cells are not due to a weak interaction between eTat and hT, we examined the ability of eT300, hT300, and the mutant hT300-V29L proteins to support the binding of eTat or hTat to eTAR. Electrophoretic mobility shift assays were performed with GST fusion proteins of cyclins T1 and Tat as well as 32P-labeled wild-type eTAR RNA. As shown in Fig. 5, the hybrid GST-eTat protein alone did not bind to eTAR (lane 1). Moreover, the hybrid GST-hT300 or GST-eT300 proteins did not support the binding between the hybrid GST-hTat protein and eTAR (lanes 2 and 3, respectively). The hybrid GST-hT300 protein also did not mediate the binding between the hybrid GST-eTat protein and eTAR (lane 4). In sharp contrast, the hybrid GST-eT300 or hybrid mutant GST-hT300 proteins, where the valine at position 29 was replaced by the leucine found in eT (hT300-V29L), each supported the binding of the hybrid GST-eTat protein to eTAR (lanes 5 and 6). Thus, it is the inability of hT to support the binding of eTat to eTAR that is responsible for the species-specific restriction of eTat transactivation. We conclude that the first 300 residues in eT are sufficient to mediating the binding of eTat to eTAR.
FIG. 5.
eT300 and hT300-V29L but not hT300 support the binding of eTat to eTAR. In vitro binding reactions were performed between 32P-labeled wild-type eTAR probe and the indicated hybrid GST Tat proteins and the hybrid GST-cyclin T1 proteins. The resulting complexes were resolved on a 8% nondenaturing polyacrylamide gel and subjected to autoradiography. The arrow to the left of the autoradiogram points to eTAR.
DISCUSSION
In this study, we isolated equine cyclin T1 and demonstrated that the N-terminal 300 residues in eT or cT rescue eTat transactivation in human cells. Most importantly, we identified residues in eT or cT that are critical for eTat transactivation. A leucine at position 29, which is a valine in hT, is essential for the ability of eT to mediate the binding between eTat and eTAR. Although the N-terminal 300 residues in hT or eT bind to eTat, only eT300, but not hT300, supports the binding of eTat to eTAR. Indeed, eTat does bind to eTAR! Like hT, where the cysteine at position 261 is essential for the formation of the tripartite complex, the leucine at position 29 in eT is equally responsible for the formation of the corresponding complex in EIAV and for eTat transactivation.
This work is in agreement with a recent report that also identified eT, demonstrating the binding of eTat to eTAR and the rescue of eTat transactivation in human cells (2). Our study identified the additional canine cyclin T1 and demonstrated that its N-terminal 300 residues rescue eTat transactivation in human cells. Moreover, we mapped the leucine at position 29 in eT as an essential residue for eTat transactivation. This single point mutation between eT and hT provides the molecular explanation for the defect of hT in supporting the binding of eTat to eTAR and its function in human cells. The full-length eT protein is of similar size to hT, and its C-terminal extensions do not contribute to eTat transactivation (data not presented). Only eight changes were found between hT and eT or cT, and three of these were conserved between eT and cT (Fig. 1A).
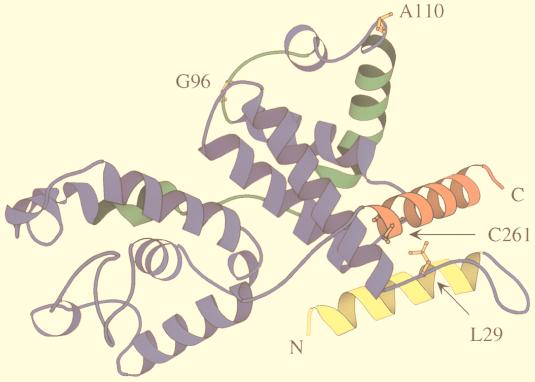
A theoretical model of eT, based on the crystal structure of the related C-type cyclin, cyclin H, also helped us to identify which of these three changes are essential for the complex formation between eTat, eT, and eTAR (Fig. 6) (29). The structure of cyclin H consists of two repeats, containing five α helices, each forming the canonical cyclin box (1). The specificity of the cyclin is provided mainly by two long α helices, which extend the cyclin box at its N and C termini. In cyclin H, these helices pack together against the first repeat on the opposite side of the kinase-binding surface. This model of eT demonstrates that the leucine at position 29 and cysteine at position 261 are in close proximity (Fig. 6). The cysteine is required for the interaction between hTat and hTAR, for the formation of the tripartite complex, and for hTat transactivation (Fig. 6) (3, 18, 19, 21, 25). Here we demonstrate that the leucine in eT at position 29 plays a similar role. In this model, the cysteine at position 261 is located in the C-terminal helix, flanking the α5′ helix. The leucine at position 29 is located in the loop between the N-terminal and α1 helices. The other two conserved changed residues (E96G and T110A) between eT and cT (Fig. 1A) point away from neighboring N- and C-terminal α-helices and are located at the interface that is predicted to bind CDK9 (Fig. 6). eTat is expected to bind to eT at the surface formed by the N- and C-terminal α helices of eT. Thus, the leucine and cysteine at positions 29 and 261, respectively, must be close to each other and form part of this surface (Fig. 6). Although the structure of cyclin T1 has not been solved, this model of eT has a high predictive value for future structure-function studies.
FIG. 6.
Model of eT based on the known crystal structure of cyclin H. Two repeats of five α helices form conserved cyclin boxes (blue and green boxes). N- and C-terminal α helices, which are important for the specificity of the cyclins, are depicted in yellow and red, respectively. They are located opposite the proposed kinase binding site (green), which was derived from the crystal structure of the CDK2-cyclin A pair (1). The cysteine at position 261 (orange) of cyclin T1 is found in the C-terminal α helix. The leucine at position 29 is located in the loop between the N-terminal and α1 helices, which is near the cysteine at position 261. They are part of the surface that binds to Tat. Arrows point to the leucine and cysteine at positions 29 and 261, respectively. The structure was drawn with MOLSCRIPT (24).
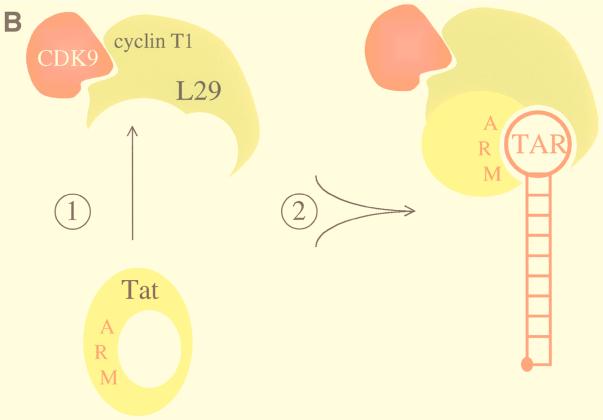
The identity of an important leucine in eT might resolve a conundrum of eTat. Several differences between eTat and hTat had been noted (10, 11, 23). Importantly, eTat contains two separated leucines upstream of the ARM and a pair of dileucines at its C terminus (11). Two groups of leucines arranged similarly are also found in eT (Fig. 7A). They could coordinate intermolecular binding between eTat and eT. In this scenario, free eTat could form an intramolecular leucine bridge, which would block the access of its ARM to eTAR (Fig. 7A, top). Only upon a conformational change, possibly exchanging the intramolecular for the intermolecular interaction between eTat and eT, both of which involve leucines, would the ARM becomes accessible for the binding to eTAR (Fig. 7A, bottom). This proposed model emphasizes the importance of the four leucine residues in eTat and explains, for the first time, the drastic effects of mutations involving these leucines in eTat (11). Since hTat binds equally well to eT (Fig. 3), this leucine at position 29 might not be critical for the binding between eT and eTat. However, since eTat has only two cysteines, rather than the seven in hTat, it might need additional contacts with eT via these “leucine bridges.” These leucines in eT could also dictate the formation of the correct combinatorial surface of eTat and eT for the binding of eTat to eTAR (Fig. 7B). In a simplistic way, the leucine and cysteine at positions 29 and 261, respectively, can then be viewed as anchor residues for the proper combinatorial properties of the two proteins. Nuclear magnetic resonance spectroscopy and/or crystallography, together with further mutagenesis of eT at and eT, should reveal the validity of this model.
FIG. 7.
Model for the interaction between eTat and eT and the formation of the tripartite complex. (A) The interaction between eTat and eT requires the leucine at position 29 in eT. We propose that eTat does not bind to eTAR because its ARM is occluded by intramolecular interactions involving leucines at positions 50, 54, 74, and 75. Mutations in these residues abolish eTat transactivation (11). Upon binding to eT, intermolecular interactions between these leucines in eTat and leucines at positions 29 (arrow), 35, 44, and 45 in eT open the ARM so that eTat can now interact with eTAR. Shown are N-terminal sequences of eTat and eT in the opposite orientation (arrows from eTat and eT). Also marked are the cysteine-rich and core sequences in eTat. The ARM is contained within gray boxes. Also shown are putative N-terminal and α1 helices of eT (striped bars), which are separated by a loop. Black circles mark proposed leucine bridges. (B) Proposed model for the binding of eTat and eT to eTAR. Upon binding of eTat (yellow) to eT (green, step 1), a combinational surface is formed, consisting of the ARM in eTat and basic residues upstream of the cysteine at position 261 in eT. This surface then binds to eTAR (red line) and recruits CDK9 (red) to the EIAV LTR (step 2). The leucine at position 29 in eT is essential for the intermolecular interaction between eTat and eT. The occluded ARM is represented by a closed circle in eTat. The stem and central loop in eTAR contain 10 paired and 4 unpaired nucleotides, respectively.
ACKNOWLEDGMENTS
We thank Michael Armanini for expert secretarial assistance.
M.G. is supported by an EMBO fellowship. This work was supported by the Japanese Foundation for AIDS Prevention and the Howard Hughes Medical Institute.
REFERENCES
- 1.Andersen G, Busso D, Poterszman A, Hwang J R, Wurtz J M, Ripp R, Thierry J C, Egly J M, Moras D. The structure of cyclin H: common mode of kinase activation and specific features. EMBO J. 1997;16:958–967. doi: 10.1093/emboj/16.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Highly divergent lentiviral Tat proteins activate viral gene expression. Mol Cell Biol. 1999;19:4592–4599. doi: 10.1128/mcb.19.7.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll R, Martarano L, Derse D. Identification of lentivirus Tat functional domains through generation of equine infectious anemia virus/human immunodeficiency virus type 1 tat gene chimeras. J Virol. 1991;65:3460–3467. doi: 10.1128/jvi.65.7.3460-3467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll R, Peterlin B M, Derse D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans-activators. J Virol. 1992;66:2000–2007. doi: 10.1128/jvi.66.4.2000-2007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho M, Derse D. Physical and functional characterization of transcriptional control elements in the equine infectious anemia virus promoter. J Virol. 1993;67:2064–2074. doi: 10.1128/jvi.67.4.2064-2074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho M, Derse D. Mutational analysis of the equine infectious anemia virus Tat-responsive element. J Virol. 1991;65:3468–3474. doi: 10.1128/jvi.65.7.3468-3474.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D, Peterlin B M. The HIV transactivator Tat binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:1–12. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derse D, Carroll R, Cavalho M, DaSilva L, Martarano L. Functional topography of lentivirus Tat proteins defined by domain switching. In: Haseltine W A, Wong-Staal F, editors. Genetic structure and regulation of HIV. New York, N.Y: Raven Press; 1991. pp. 251–261. [Google Scholar]
- 11.Derse D, Newbold S H. Mutagenesis of EIAV Tat reveals structural features essential for transcriptional activation and TAR element recognition. Virology. 1993;194:530–536. doi: 10.1006/viro.1993.1291. [DOI] [PubMed] [Google Scholar]
- 12.Derse D, Carvalho M, Carroll R, Peterlin B M. A minimal lentivirus Tat. J Virol. 1991;65:7012–7015. doi: 10.1128/jvi.65.12.7012-7015.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse D, Dorn P, DaSilva L, Martarano L. Structure and expression of the equine infectious anemia virus transcriptional trans-activator (tat) Dev Biol Stand. 1990;72:39–48. [PubMed] [Google Scholar]
- 14.Derse D, Dorn P L, Levy L, Stephens R M, Rice N R, Casey J W. Characterization of equine infectious anemia virus long terminal repeat. J Virol. 1987;61:743–747. doi: 10.1128/jvi.61.3.743-747.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorn P, DaSilva L, Martarano L, Derse D. Equine infectious anemia virus Tat: insights into the structure, function, and evolution of lentivirus trans-activator proteins. J Virol. 1990;64:1616–1624. doi: 10.1128/jvi.64.4.1616-1624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorn P L, Derse D. cis- and trans-acting regulation of gene expression of equine infectious anemia virus. J Virol. 1988;62:3522–3526. doi: 10.1128/jvi.62.9.3522-3526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujinaga K, Cujec T P, Peng J, Garriga J, Price D H, Grana X, Peterlin B M. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujinaga K, Taube R, Wimmer J, Cujec T P, Peterlin B M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann C, Rice A. Specific interactions of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov D, Kwak Y T, Nee E, Guo J, Garcia-Martinez L F, Gaynor R B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA. J Mol Biol. 1999;288:41–56. doi: 10.1006/jmbi.1999.2663. [DOI] [PubMed] [Google Scholar]
- 22.Jones K A. Taking a new TAK on Tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 23.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis P J. MOLSCRIPT, a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 25.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 Tat. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 26.Madore S J, Cullen B R. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maury W. Regulation of equine infectious anemia virus expression. J Biomed Sci. 1998;5:11–23. doi: 10.1007/BF02253351. [DOI] [PubMed] [Google Scholar]
- 28.Noiman S, Gazit A, Tori O, Sherman L, Miki T, Tronick S R, Yaniv A. Identification of sequences encoding the equine infectious anemia virus tat gene. Virology. 1990;176:280–288. doi: 10.1016/0042-6822(90)90254-o. [DOI] [PubMed] [Google Scholar]
- 29.Peitsch M C. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 30.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]