Abstract
Chimeras were previously generated between the ecotropic (Moloney-MuLV) and amphotropic (4070A) SU and TM proteins of murine leukemia virus (MuLV). After passage in D17 cells, three chimeras with junctions in the C terminus of SU (AE5, AE6, and AE7), showed improved kinetics of viral spreading, suggesting that they had adapted. Sequencing of the viruses derived from the D17 cell lines revealed second-site changes within the env gene. Changes were detected in the receptor binding domain, the proline-rich region, the C terminus of SU, and the ectodomain of TM. Second-site changes were subcloned into the parental DNA, singly and in combination, and tested for viability. All viruses had maintained their original cloned mutations and junctions. Reconstruction and passage of AE7 or AE6 virus with single point mutations recovered the additional second-site changes identified in the parental population. The AE5 isolate required changes in the VRA, the VRC, the VRB-hinge region, and the C terminus of SU for efficient infection. Passage of virus, including the parental 4070A, in D17 cells resulted in a predominant G100R mutation within the receptor binding domain. Viruses were subjected to titer determination in three cell types, NIH 3T3, canine D17, and 293T. AE6 viruses with changes in the proline-rich region initially adapted for growth on D17 cells could infect all cell types tested. AE6-based chimeras with additional mutations in the C terminus of SU could infect D17 and 293T cells. Infection of NIH 3T3 cells was dependent on the proline-rich mutation. AE7-based chimeras encoding L538Q and G100R were impaired in infecting NIH 3T3 and 293T cells.
Entry is an integral part of the life cycle of a virus. Pursuant to entry are the initial requirements for interaction with a host cell receptor followed by the events of fusion of the viral and host cell membranes and internalization of the retroviral particle into the host cell. Murine leukemia viruses (MuLVs) are divided into five classes, based on receptor usage, by viral interference assays (52, 53, 57). These classes are ecotropic, amphotropic, polytropic, xenotropic and 10A1. The receptors for the ecotropic, amphotropic, and 10A1 MuLVs have been cloned and denoted MCAT, Pit2, and Pit1 respectively (1, 31, 32, 40, 41, 66). Each has been shown to be sufficient for infection when expressed in nonpermissive cell lines. Both the MCAT and Pit proteins are multiple membrane-spanning transporters; MCAT is a cationic amino acid transporter, whereas Pit1 and Pit2 are inorganic phosphate symporters.
The retroviral env gene encodes the surface (SU) and transmembrane (TM) proteins, required for the binding and entry of virus into the host cell. Sequence alignments of SU proteins in each class reveal a large diversity of amino acid sequences in the N terminus, while the C terminus is generally conserved. The study of functional domains of the envelope proteins has been approached by mutagenesis (2, 7, 12, 22, 23, 29, 37, 46, 56, 60, 61, 63, 64, 68, 70), as well as by construction of chimeric enveloped MuLVs (5, 26, 42, 43, 47). Receptor usage is specified by determinants located in the N-terminal half of SU. The first N-terminal 230 amino acids of the ecotropic MuLV SU protein are sufficient for binding to the ecotropic receptor, MCAT, as demonstrated by studies with chimeras (4, 5, 47, 52), binding studies with soluble truncated SU (4, 14, 27), and interference assays (4, 5, 27). Determinants sufficient for binding to the Pit2 receptor protein are located within the first N-terminal 208 amino acids of the amphotropic 4070A SU protein (4–6, 47).
Hypervariable regions within the receptor binding domain, consisting of complex disulfide-bonded loops, have been identified (5, 34, 35). Recently, the structure of the ecotropic Friend receptor binding domain (RBD) (17) has been solved, and the individual cysteine loops (34, 35) have been defined as VRA, VRB, and VRC. The entire 236-amino-acid sequence folds as a structural domain and helps to explain the lack of viability of envelope chimeras with junctions within this region (5, 26, 42, 47). Particular amino acids within the VRA are critical for binding to the receptors (3, 15, 37) and are located on a protruding nonstructured loop. For 4070A SU, binding to Pit2 requires amino acids 78 to 104 (3, 25, 62). While amino acids Y90 and V91 appear to be critical in the context of chimeric envelope proteins and chimeric receptors (62), these residues alone are not determinants for receptor recognition in the context of the wild-type 4070A envelope protein (25). Additional experiments (62) indicate a role for the VRB in receptor recognition. Amino acids comprising the VRC domain are also located within a helical loop.
The RBD is connected to the conserved C-terminus of SU through the proline-rich region (PRR). The N-terminus of the PRR is conserved among the ecotropic, amphotropic, xenotropic, and polytropic classes of MuLVs, with a consensus sequence of GPR(I/V)PIGPNP (23), and is essential for viral infection (68). The PRR C terminus is variable in length and conservation. The amphotropic PRR is 14 amino acids longer than the ecotropic region. In the cases of polytropic, xenotropic, and 10A1 viruses, the PRR in the SU protein influences interactions with receptors as a determinant for entry (4, 45).
The second envelope protein, TM, is involved in the fusion event of entry and is characterized by a hydrophobic stretch of amino acids at its extracellularly oriented N terminus, a transmembrane domain, and an intravirally oriented cytoplasmic domain. The structure of a limited 55-amino-acid region lacking the fusion peptide of the ectodomain has revealed a trimeric coiled-coil motif consisting of 33 amino acids within Moloney MuLV (M-MuLV) (L516 to L547) (18). The chain reverses direction, forming a small alpha helix consisting of residues L554 to L558, which lies perpendicular to the coiled-coil. Cysteines 555 and 562 are linked by disulfide bonding, while cysteine 563 appears to be a free thiol, presumably available to form a disulfide bond with SU (48, 51).
Since MuLVs are currently one of several candidates being explored for delivery of therapeutic molecules in gene therapy applications, it is important to understand the basic virology of entry. Many details of the entry of MuLVs, including SU-TM protein-protein interactions, are not fully understood. Although the structures of a portion of the ectodomain of TM and the receptor binding domain of ecotropic MuLV have been solved, these offer only a limited picture of the three-dimensional organization of portions of the envelope proteins under very particular conditions. A series of 22 chimeric ecotropic and amphotropic envelope proteins were previously constructed (AE series and EA series) and analyzed for viral spread, host range, viral interference, and syncytium formation (47). This study identifies second-site mutations within the amphotropic/ecotropic (AE series) chimeric enveloped MuLVs which increase viral infectivity. Like mutational approaches with hemagglutin (13, 20, 59), the identification of second-site changes in the envelope proteins of amphotropic/ecotropic chimeras provide insights into regions within SU and TM which interact, expanding our view of the entry process.
MATERIALS AND METHODS
Cell lines and maintenance.
Canine D17 cells and D17/viral producer cell lines were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM) (pH 7.4) supplemented with 2 mM glutamine, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 7.5% fetal bovine serum. A plasmid clone of the ecotropic receptor under the control of a cytomegalovirus promoter (clone 1475) in the pCDNA 3.1 vector backbone (Invitrogen) was generously provided by L. Albritton (University of Tennessee); the pJET cell line was subsequently generated by introducing this plasmid DNA into D17 cells by calcium phosphate transfection as specified by the directions supplied with the Stratagene mammalian transfection kit (no. 200385) and selected and maintained in 400 μg of G418 per ml. NIH 3T3 cells were maintained in DMEM (pH 7.4) supplemented with 10% calf serum, 2 mM glutamine, and 0.1 mg of gentamicin sulfate per ml. 293T cells were maintained in DMEM (pH 7.4) supplemented with 10% calf serum, 2 mM glutamine, 0.1 mg of gentamicin sulfate per ml, and 400 μg of G418 per ml.
DNA and plasmids.
Chimeric proviral DNA clones (47), pNCA-C (19), and pNCA-Am (47) were as previously described. pNCA-Am contains the 4070A envelope within the M-MuLV backbone and is referred to as 4070A in the text and figures. pHIT456, previously described (58), contains the 4070A envelope sequence under the control of a cytomegalovirus promoter and was used with the following modifications. The XhoI site at position 9093 was destroyed by linearizing the plasmid with XhoI, filling in with T4 DNA polymerase, and self-ligation. An NheI site was created at the 3′ end of 4070A env sequence by the insertion of an NheI adapter (oligonucleotide 6452; 5′AATTGGCTAGCC 3′) at the 3′ XbaI site. All DNAs used for transfections were purified by cesium chloride banding. Nucleotide positions are based on the RNA sequence of M-MuLV as defined by to Shinnick et al. (55). The amphotropic envelope sequence is numbered by the system of Ott and Rein (44).
Transfections.
D17 cells were seeded at a density of 1 × 105/60-mm plate or 2 × 105/100-mm plate 24 h prior to transfections. The cells were incubated with 0.5 μg of DNA/60-mm plate or 1 μg of DNA/100-mm plate in the presence of 0.5 mg of DEAE-dextran per ml, 0.9 mM CaCl2, and 0.5 mM MgCl2 at 37°C. After 30 min, the DNA was removed and replaced with fresh medium (39). Transfections testing reconstructed second-site changes were performed at least twice.
RT assays.
The presence of virus was monitored by the release of reverse transcriptase (RT) activity into the culture medium. The medium was collected up to day 40. RT activity was assayed as previously described (21). Virus was collected either from cells transiently transfected with DEAE-dextran at least 40 days posttransfection or from producer cells 10 days postinfection.
Viral DNA isolation and PCR amplifications.
Unintegrated viral DNA was isolated by the method of Hirt (28). Cells were infected in the presence of 8 μg of Polybrene per ml for 2 h and were harvested approximately 48 h postinfection in 100 mM Tris-HCl (pH 7.5) buffer containing 0.6% sodium dodecyl sulfate, 10 mM EDTA, and 1 M sodium chloride. Low-molecular-weight DNA was isolated by subsequent centrifugations and phenol-chloroform extractions. Prior to any analyses, the low-molecular-weight DNA was treated with RNase A and diethylpyrocarbonate. PCRs were performed to amplify the env gene with primers within the M-MuLV pol gene (primer 3807; MuLV bases 4924 to 4938 [5′ gatatacatatgGCCGTTAAACAGGGA3′]) and the 3′ long terminal repeat (primer 6320; MuLV bases 7815 to 7791 [5′ccttaaggCCCCCCTTTTTCTGGAGACTAAATA3′]) by using 4.8 U of Taq polymerase (GIBCO/BRL) and PFU polymerase (recombinant; Stratagene) in a ratio of 20:1, respectively, yielding the full-length 2.9-kb product. One-hundredth to one-fifth of a plate of cell Hirt DNA was used in the PCRs. In some instances, the initial PCR product was reamplified before being sequenced.
Sequencing.
Sequencing of the viral PCR product encoding the env gene amplified from the Hirt DNA was performed with primers spanning the entire env gene and reagents supplied in the Perkin-Elmer Amplicycle kit. Base changes from wild-type M-MuLV ecotropic and 4070A amphotropic envelope genes of MuLV were confirmed by performing sequencing reactions with primers which read the opposite strand. Approximately 99 to 99.8% of the coding sequence was read for each PCR-amplified Hirt DNA.
Reconstruction cloning.
Changes detected by sequencing the viral populations were subcloned into the parental chimeric backbones, replacing the wild-type sequence. Where more than one mutation was detected in a viral population, the mutation was reconstructed singly and in combination within the parental background. All restriction fragments were isolated by gel electrophoresis, purified by a glass-milk method (65), and ligated with T4 DNA ligase. Constructs were screened by restriction digestion to confirm the chimera backbone and by diagnostic sequencing reactions to identify the second-site mutations. Amino acids are denoted by the single-letter code, and the positions are based on Env precursor protein. The following single mutations were reconstructed into parental backbones as described below.
(i) N-terminal change.
The amphotropic 4070A G100R mutation was reconstructed by using a three-fragment ligation of the 1,090-bp SfiI-EcoRI fragment isolated from the PCR amplification of the Hirt DNA (containing G100R), the 1,227-bp EcoRI-BspDI fragment from the parental AE5 or AE7, and the 8,767-bp BspDI-SfiI fragment from the parental AE5 or AE7. The change from the wild-type codon GGG (glycine) to mutant codon aGG (arginine) was sequenced with primer 1559 (5′CCAGTAAGCtTGTCCG3′; bases 463 to 448 of 4070A env). The AE6 population contained a mixture of the wild-type glycine and mutant arginine sequences. An alternative scheme was used to clone the mutation within the AE6 backbone. The 2,771-bp SalI-EcoRI fragment from the AE5/NCAC clone containing the G100R mutation was ligated in the presence of the 6,631-bp HindIII-SalI fragment from AE6 and the 495-bp EcoRI-HindIII fragment from AE6.
(ii) VRB-hinge changes.
The change in the amphotropic G209R detected in the AE5 population was reconstructed into both the AE5 parental DNA and 4070A Env in the pNCA-Am and pHIT456 backbones. The G209R region was introduced into AE5 by exchange of the 2,726-bp SacII-ClaI fragment and into pNCA-Am by exchange of the 1,128-bp SfiI-BspMI fragment. The change from wild-type codon GGG (glycine) to aGG (arginine) was detected with sequencing primer 2846 (5′CCAAGGGGCTACTCGAGGGGG 3′; bases 583 to 603 of 4070A env). The 1,500-bp XhoI-NheI fragment containing the G209R mutation from G209R/pNCA-Am replaced the 1,100-bp XhoI-NheI fragments in the pHIT 456 vector.
(iii) C-terminal changes.
The changes in the amphotropic R302K,G detected in the AE6 construct and the R302S mutation from the AE6/3T3/D17 were reconstructed through ligation of the 495-bp EcoRI-HindIII fragment from the PCR-amplified Hirt DNA (containing R302K/G/S) with the 2,771-bp SalI-EcoRI and the 6,631-bp HindIII-SalI fragments from the parental AE6. The change from wild-type codon AGA (arginine) to AGt (serine) or Gga (glycine) was detected with sequencing primer 2922 (5′CCCCTACAAGTCCAAG3; bases 893 to 908 of 4070A env). The mutant codon AaA (lysine) was not observed in the viral population and was subsequently detected in two of the five isolates sequenced for the presence of the serine mutation. The change in AE6 within the M-MuLV coding region, A419V, was reconstructed by using a three-fragment ligation with the 780-bp HindIII-BspDI fragment from the PCR-amplified Hirt DNA product (containing A419V), the 8,767-bp BspDI-SfiI fragment, and the 1,540-bp SfiI-HindIII fragment from the parental AE6. The change from wild-type codon GCT (alanine) to GtT (valine) was detected with sequencing primer 4784 (5′ggtctagaTCTTTTGTGTCGGTTGGATC3′; bases 7180 to 7158 of M-MuLV) (55).
The change observed in the AE5 construct within the M-MuLV sequence at T392I was reconstructed by ligation of the 420-bp SalI-HpaI fragment from the PCR-amplified Hirt DNA product (containing T392I) and the 8,841-bp HpaI-SacII and 1,826-bp SacII-SalI fragments of the parental AE5. The change from the wild-type codon ACA (threonine) to mutant codon AtA (isoleucine) was detected with sequencing primer 1091 (5′GCTCCAGCCggCTGCTCCGTG3′; bases 6858 to 6878 of M-MuLV) (55).
The alteration in AE7 within M-MuLV at L538Q was reconstructed by ligation of the 700-bp BamHI-BspDI fragment from the PCR-amplified Hirt DNA product (containing L538Q) plus the 8,767-bp BspDI-SfiI and 1,594-bp SfiI-BamHI fragments from the parental AE7. The change from the wild-type codon CTA (leucine) to mutant codon CaA (glutamine) was detected with sequencing primer 4781 (5′ggtctagaTCTGTTAAACAGTCCCTCAAACC3′, bases 7583 to 7561 of M-MuLV) (55).
Multiple mutations, observed in some viral populations, were reconstructed into parental backbones as described below. The amphotropic R302K/G/S (AE6 no. 1 and 3 [see Table 1]) changes and the M-MuLV A419V (AE6 no. 2 [see Table 1]) change observed in independent AE6 populations were combined within one proviral backbone by ligation of the 495-bp EcoRI-HindIII fragment from AE6 R302K, R302G, and R302S mutants with the 6,631-bp SalI-HindIII and the 2,271-bp SalI-EcoRI fragments of the AE6 (no. 2)/A419V construct. The changes from wild-type codon AGA (arginine) to AGt (serine), GgA (glycine), or AaA (lysine) were detected with sequencing primer 2922.
TABLE 1.
Second-site changes in chimeric AE and wild-type virus populations
| Chimera populationa | Amino acidb | Domainc |
|---|---|---|
| AE4 | Ampho G100R* | VRA |
| Ampho E80K* | VRA | |
| AE5d | Ampho G100R | VRA |
| Eco T392I | C terminus | |
| Ampho G209R | VRB-hinge | |
| AE6 no. 1 | Ampho R302K | PRR |
| Ampho R302S | PRR | |
| AE6 no. 2 | Eco A419V | C terminus |
| AE6 no. 3d | Ampho G100R | VRA |
| Ampho R302G | PRR | |
| AE7 | Ampho G100R | VRA |
| Eco L538Q | TM (ectodomain) | |
| AE8 | Ampho G100R* | VRA |
| 4070A | Ampho G100R* | VRA |
| Ampho G541R* | TM (ectodomain) |
Chimeric junctions as defined in the text and in Fig. 1; all viruses were passaged through canine D17 cells.
Amino acid numbering includes the signal sequence; location within the amphotropic or ecotropic portion of the chimera is noted. Ampho, amphotropic; Eco, ecotropic; ∗, not reconstructed.
Domains as defined in text; unless otherwise specified, domains are within the SU protein.
Virus was originally isolated from NIH 3T3 producer cells and subsequently used as inoculum into D17 cells.
The amphotropic G100R and R302G, observed in the AE6 (no. 3) viral population was reconstructed by ligation of the 6,631-bp HindIII-SalI fragment of AE6, the 2,044-bp SalI-EcoRI fragment from the AE5/G100R clone, and the 495-bp EcoRI-HindIII fragment from the AE6 (no. 3) PCR-amplified Hirt DNA product. Changes from the wild-type codons GGG (glycine) and AGA (arginine) to mutant codons aGG (arginine) and gGA (glycine) were detected with sequencing primers 1559 and 2922, respectively.
Reconstruction of the AE7 viral population was performed by ligation of the 1,227-bp EcoRI-BspDI fragment from the AE7/L538Q clone, the 1,093-bp SfiI-EcoRI fragment from the AE7 PCR-amplified DNA product, and the 8,767-bp BspDI-SfiI fragment from parental AE7. Changes from the wild-type codons GGG (glycine) and CTA (leucine) to aGG (arginine) and CaA (glutamine) were detected with sequencing primers 1559 and 4781, respectively.
Two changes observed in the AE5 population (G100R and T392I) were reconstructed within one proviral backbone by ligation of the 1,227-bp EcoRI-BspDI fragment from AE5/T392I clone, the 1,093-bp SfiI-EcoRI fragment from AE5 PCR-amplified Hirt DNA, and the 8,767-bp BspDI-SfiI fragment from the AE5 parent. The presence of the mutant codons aGG (arginine) and AtA (isoleucine) was confirmed by sequencing with primers 1559 and 1091, respectively.
Transient expression, metabolic labeling, and immunoprecipitation.
D17 cells were seeded at a density of 3 × 106 in 100-mm plates. A 5-ml volume of serum-free medium was added to the cells prior to the addition of the DNA-Lipofectamine mixture. A 12-μg sample of each DNA per time point was incubated for 15 min with 30 μl of Plus reagent (Gibco Life Technologies) in the presence of 600 μl of serum-free medium supplemented with 0.1 mM MEM non-essential amino acids (Gibco/BRL) and then mixed with an additional 600 μl of serum-free medium plus 54 μl of Lipofectamine for another 15 min before being added to cells. The DNA and medium was removed after 3 h and replaced with 10 ml of complete medium. Cells were washed with phosphate-buffered saline 39 to 40 h after DNA addition and incubated in 5 ml of Hanks balanced salt solution (Gibco/BRL) for 20 min at 37°C. Cells were metabolically labeled with 150 μCi of Tran35S-label (ICN) for 40 min at 37°C. The T = 0 time point is after the 40-min labeling period. Radioactive medium was replaced with 10 ml of complete medium, and the cells were harvested at the 2-, 4-, 6-, and 8-h time points by lysis in 3 ml of ice-cold phosphate lysis buffer (10 mM Na2HPO4-NaH2PO4 [pH 7.4], 100 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% deoxycholate, 1% Triton X-100). Extracts were centrifuged for 3 h at 35,000 rpm in a Beckman Ti50 rotor. Extracts corresponding to half of a 10-cm plate were precleared with normal goat serum (Gibco/BRL) for 2 h at 4°C (20 μl) and then immunoprecipitated with 10 μl of 79S-842 (anti-SU antiserum; National Cancer Institute) and 50 μl of 42-114 (anti-TM antiserum) (50). Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Molecular weight markers were obtained from Bio-Rad.
Time course infection experiments.
Medium was collected from near-confluent viral producer cells after an overnight incubation and assayed for reverse transcriptase activity (21). Based on the reverse transcriptase activity, equal amounts of each virus were applied to host cells in the presence of 8 μg of Polybrene per ml. After 2 h, the viruses were removed and replaced with 10 ml of fresh medium. Beginning on day 0, supernatants were collected and were assayed for virus spreading by measuring the release of RT activity.
Virus titers.
D17/pJET cells were used to generate a lacZ reporter cell line. The ecotropic receptor expressed in these cells allowed infection with virus from the ecotropic CRE cell line packaging the MFG/NB lacZ vector (27). D17/pJET/lacZ cell lines were subsequently infected with the reconstructed chimeric viruses of amphotropic host range (pJET/MFG). Viruses collected overnight from the pJET/MFG cell lines were used to infect 105 NIH 3T3, canine D17, and 293T cells in the presence of 8 μg of Polybrene per ml for 2 h. The cells were stained for lacZ 48 h postinfection (47).
Modeling of the 4070A receptor binding domain.
The ecotropic Friend MuLV receptor binding domain (1aol.pdb) was used as a base molecule for homology modeling of the corresponding region of 4070A by using the program GeneMine Look (version 3). The second-site mutations were substituted into the 4070A structure. The resulting models of the 4070A RBD trimers was based on homology to the Friend MuLV trimer model, in which the VRC of one monomer interacts with the VRB of a second monomer (16, 17). Energy minimizations were performed for both the monomeric and trimeric models of 4070A. The electrostatic surface potentials of the Friend MuLV RBD and model 4070A molecules were analyzed by Grasp (version 1.3). The space-filling renditions of all molecules were generated with the program Sybyl (version 6.5; MIPS3-IRIX6.2).
RESULTS
Generation of chimeric virus producer cell lines: transient expression versus infection.
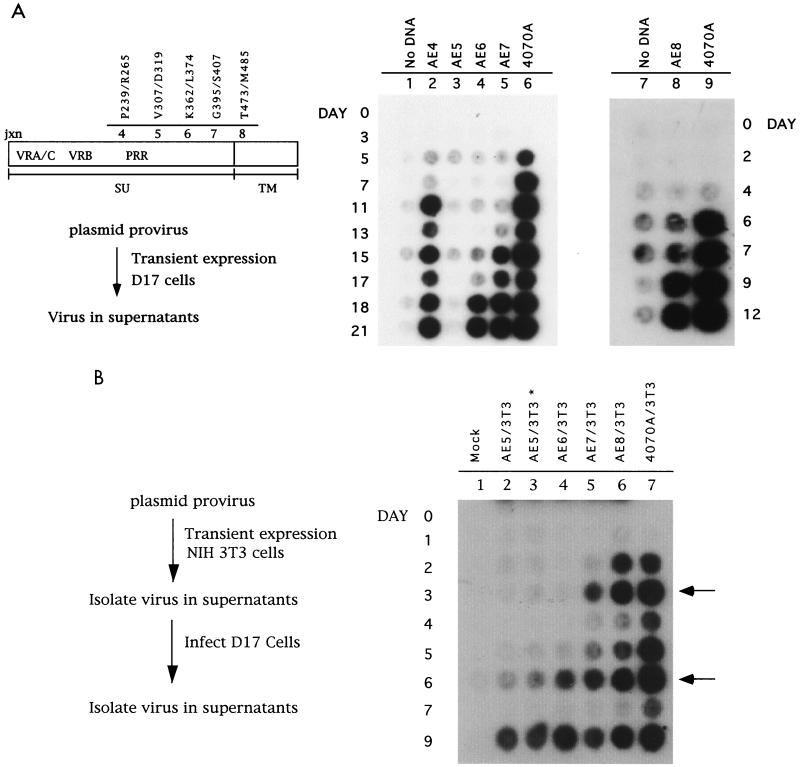
We had previously generated producer cell lines of chimeric amphotropic and ecotropic enveloped MuLVs in both canine D17 cells and NIH 3T3 cells to map host range determinants (47). Viruses which contained the N-terminal half of 4070A SU through the proline-rich region were viable and showed viral interference with the amphotropic virus (47). Virus viability was assessed after transient expression of plasmid DNA introduced into cells with DEAE-dextran. Chimeric viruses with junctions in the C terminus of SU (AE6 and AE7) (Fig. 1A, lanes 4 and 5) were delayed in viral spreading compared to wild-type 4070A in D17 cells (lane 6). Typical delays for these chimeras were 10 to 14 days with respect to wild-type 4070A. In contrast, chimeras with junctions corresponding to the RBD (AE4) (lane 2) and the entire SU protein (AE8) (lane 8) typically showed only a short delay of 1 to 4 days compared to the wild-type 4070A control (lanes 6 and 9). A chimeric virus with a junction just after the PRR, AE5, was not viable in D17 cells if proviral DNA was introduced by using DEAE-dextran (lane 3). However, this chimeric virus was viable in NIH 3T3 cells (47). To examine the block in viral spreading in the D17 cell line, the AE5 virus released from NIH 3T3 cells was tested for the ability to directly infect D17 cells. It was reasoned that using a population of high-titer virus may initiate infection not detected in transient-expression assays. Sequence analysis of the total population of AE5 virus produced in NIH 3T3 cells revealed no second-site changes, and the restriction site used to generate this chimera had been maintained. AE5 virus with equivalent RT activity to wild-type 4070A from NIH 3T3 producer cells was used to infect D17 cells. In addition, a 13-fold excess of AE5 was tested. Both levels of AE5 viruses became RT positive by day 9, representing a delay of approximately 7 days compared to wild-type 4070A (Fig. 1B, lanes 2 and 3).
FIG. 1.
Time course of infection of chimeric AE env series in D17 cells. (A) A 0.5-μg sample of plasmid DNA expressing the chimeric AE envelope proteins and the wild-type control, 4070A, within the MuLV provirus was introduced into 105 cells per 60-mm-diameter plates by the DEAE-dextran method, allowing transient expression of the virus. Supernatant medium was assayed for the presence of RT as described in Materials and Methods. Constructs are indicated at the top of each lane. The number of days after introduction of DNA is indicated on the left. Lanes 7 to 9 show the results of an independent transfection. A linear schematic of SU and TM is shown to the left and indicates regions within SU and TM relative to chimeric junctions, in addition to amino acid joining points in the amphotropic and ecotropic envelope proteins. (B) Chimeric AE and wild-type 4070A-envelope-expressing viruses were collected from NIH 3T3 cell culture supernatants. Equivalent levels of virus, standardized by RT activity, were introduced into D17 cells by infection for 2 h at 37°C. For chimeric virus AE5 (∗), an additional amount of virus (lane 3), representing a 13-fold excess compared to the other samples, was used in the infection. Supernatant medium was assayed for the presence of RT, as described in Materials and Methods. Constructs are indicated at the top of each lane. The number of days after introduction of virus is indicated on the left. Arrows on the right indicate days on which cells were passaged.
AE6, AE7, and AE8 chimeric viruses from NIH 3T3 cells were also introduced into D17 cells by infection (Fig. 1B, lanes 4, 5, and 6, respectively). Each chimeric viral population from NIH 3T3 producer cells was sequenced prior to this infection, and while each had retained the original restriction sites used to generate the chimera, no additional second-site changes were detected. Cells infected with chimera AE6 became positive for RT activity on day 6. This is a delay of 4 days compared with wild-type 4070A virus isolated from NIH 3T3 cells. Thus, AE6 is delayed in D17 cells both by transient transfection of plasmid DNA with DEAE-dextran, and by infection. Chimeric virus AE6 originally derived from NIH 3T3 cells is referred to in future analyses as AE6 no. 3. Chimeric viruses AE7 isolated from infection of NIH 3T3 cells showed a delay in the detection of RT activity in the medium of only 1 day, while AE8 isolated from NIH 3T3 cells showed no delay compared to wild-type 4070A.
Second round of infection with virus from producers generated by transient expression with DEAE-dextran and by infection.
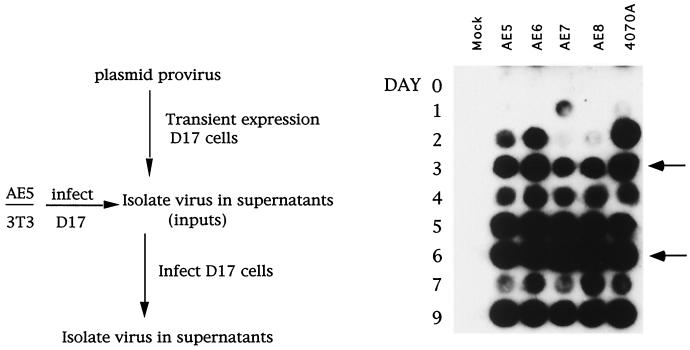
Chimeras which displayed delayed kinetics for viral spreading either in transient-transfection experiments with DEAE-dextran (Fig. 1A) (47) or in infection experiments (Fig. 1B) could have defects in entry and/or assembly. During viral replication, the process of reverse transcription facilitates an adaptive process within retroviruses, resulting in the selection of improved viruses. Viruses which have adapted, in a second round of infection, would display improved kinetics of viral spread compared with the parental construct. RT-positive viruses from D17 producer cells (AE5, AE6, AE7, and AE8) were used in a subsequent infection, each being normalized to wild-type 4070A virus. Similar to the wild-type 4070A control, chimeras AE5 and AE6 became RT positive around day 2 postinfection (Fig. 2). Chimeras AE7 and AE8 were RT positive by day 3 postinfection. AE7 showed no further improvement in the kinetics of viral spread with subsequent infections (data not shown).
FIG. 2.
Time course of infection of chimeric AE and 4070A viruses in D17 cells. Chimeric AE and wild-type viruses were isolated from culture supernatants of D17 cells. RT activity for each supernatant was assayed as described in Materials and Methods. Equivalent amounts of virus, assayed by RT activity, were used to infect D17 cells. Supernatant medium was assayed for the presence of RT. Constructs are indicated at the top, and days after introduction of virus are indicated on the left. Arrows on the right indicate the days on which cells were passaged.
Sequencing of the viral populations and detection of second-site changes.
The results of the time course experiments suggested that the onset of viral spreading in the DEAE-dextran experiment (Fig. 1A) (47) and the infection experiment (Fig. 1B) was the result of an adaptive process. Sequence changes within env were examined, since all of the chimeric proviral constructs contain wild-type M-MuLV gag and pol sequences. Hirt DNAs corresponding to the viral populations, including the wild type, were isolated, PCR amplified, and sequenced. In Table 1, sequencing results are organized by viral population. All viral populations sequenced, except two independent AE6 populations, contained a change at amino acid residue 100 (within the VRA) from a glycine to an arginine. G100R is 5 amino acids from the last cysteine within VRA. All changes in AE4 (G100R and E80K) were within the VRA region. E80K is within the first putative cysteine loop of the SU protein. Replacements of wild-type residues in the AE5 population occurred in the VRB-hinge vicinity (G209R, amphotropic region) and the C terminus of SU (T392I) in addition to G100R (VRA). Two populations of AE6 virus (no. 1 and 3) contained changes at the same amino acid, R302, within the PRR of SU. This amino acid was changed to either a lysine, serine, or glycine. The lysine and serine changes were present within one population of AE6 (no. 1), and the lysine codon was apparent only upon subcloning of the serine mutation. An alternative change seen in a third isolate of AE6 (no. 2) occurs in the C terminus of SU (A419V). Changes in TM occurred in the AE7 population at L538, a residue located within the putative coiled-coil region. The wild-type isolate also contained a change in the ectodomain of TM (G541R).
Reconstruction of second-site changes affects the viability of the parental chimeras.
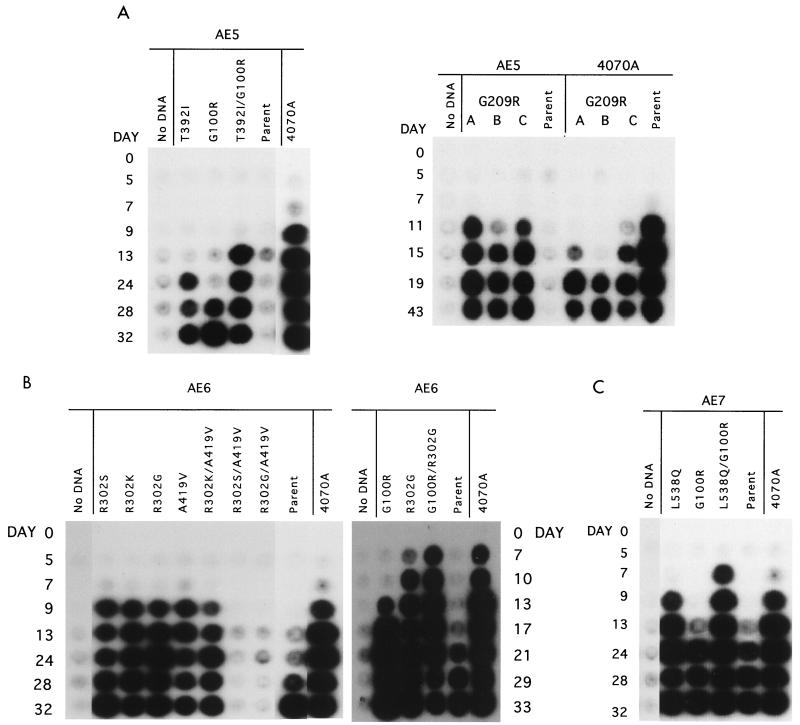
Second-site changes detected in the viral populations were tested for their ability to affect the viability of the parental chimeric viruses. If the sequenced second-site changes improve the viability of the virus, the reconstructed viruses should become RT positive at an earlier time point. Changes detected in chimeric viruses AE4 and AE8 and in the wild-type control (Table 1) could not be tested in this system, because the delay in RT activity is too minimal to see any positive effect. Single and multiple combinations of the second-site changes were reconstructed into the parental chimeric cDNA constructs of AE5, AE6, and AE7. Virus viability was tested by the transient expression of the proviral DNA introduced into cells by DEAE-dextran (Fig. 3). Parental AE5, not viable by DEAE-dextran, could be improved by the replacement of either wild-type residue G100 and T392, becoming RT positive on days 28, and 32 respectively. The presence of both second-site changes in the same protein further improves the viability of AE5, which becomes RT positive by day 13 (Fig. 3A, left). Parental AE5 could also be improved by the replacement of wild-type residue G209, becoming RT positive by days 11 to 15 with kinetics similar to those of wild-type 4070A control for this particular assay (Fig. 3B).
FIG. 3.
Time course of infection of reconstructed AE chimeric env in AE5, AE6, AE7, and 4070A backbones in D17 cells. Mutations detected in isolated AE chimeric viruses (Table 1) were reconstructed into parental AE constructs as described in Materials and Methods. A 1-μg sample of plasmid DNA expressing each AE chimera, including parental AE chimeras, was introduced per 105 cells into 100-mm-diameter plates by using DEAE-dextran. Supernatant medium was assayed for the presence of RT as described in Materials and Methods. Parental constructs are indicated at the top, and mutations are indicated for each lane. The number of days after introduction of DNA is indicated on the left. (A) Reconstruction of mutations in AE5 parental DNA and 4070A (pNCA-Am). (B) Reconstruction of mutations in AE6 parental DNA. (C) Reconstruction of mutations in AE7 parental DNA.
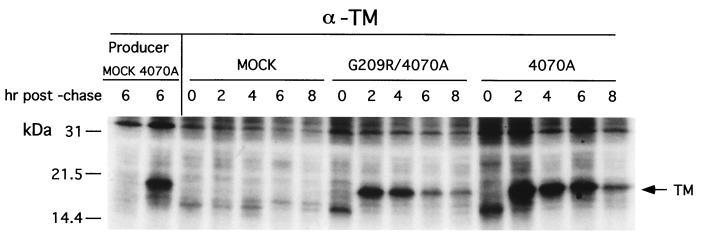
The results indicate that the second-site mutations in AE5 were able to improve the passage of chimeric virus. To test whether the mutations could be beneficial or detrimental to the wild-type 4070A Env, one of the single mutations, G209R, was reconstructed into the NCA-Am (4070A) proviral clone (Fig. 3A, right). Virus viability was tested by transient expression of the proviral DNA introduced into cells by DEAE-dextran (Fig. 3A, right). In contrast to the introduction of G209R into AE5, the introduction of G209R into 4070A impaired viral spread compared to the wild-type control. All replicates of 4070A with the G209R mutation were delayed for the release of RT activity, becoming RT positive between days 15 and 19, compared to a wild-type 4070A control, which was RT positive by day 11. The cause of the delay of 4070A/G209R was further examined. The effect of G209R on the processing and transport of the SU-TM precursor was examined by the release of the processed TM protein. Processing of the precursor SU-TM occurs in the trans-Golgi by cellular proteases. Cells were metabolically labeled, and the TM protein was immunoprecipitated with antibody 42-114 (50). The p15/TM protein was detected after a 2-h chase in both the 4070A and G209R/4070A extracts (Fig. 4). The processing of the precursor SU-TM protein paralleled the release of TM (data not shown). The cause of the delay in 4070A/G209R viral spread therefore occurs after synthesis and processing of the precursor protein.
FIG. 4.
Pulse-chase analysis of envelope proteins after transfection and transient expression. Plasmid DNA encoding G209R/4070A and wild-type 4070A in the pHIT456 backbone was introduced into D17 cells as described in Materials and Methods. At approximately 40 h after introduction of DNA, cells were metabolically labeled with Tran35S-label for 40 min. The zero time point represents cells harvested after the 40-min labeling period. After 40 min, the radioactive medium was replaced with 10 ml of complete medium. Cells were harvested at the times indicated above each lane. Cells were lysed and immunoprecipitated as described in Materials and Methods. A similar pulse-chase analysis was performed on the 4070A/D17 producer line. Molecular mass markers are indicated on the left. Anti-TM (α-TM) antiserum 42-114 (50) was used to immunoprecipitate TM. As a positive control for cleaved TM, a 6-h pulse-chase time point for 4070A/D17 producer cell line is shown to the left of the transiently expressed envelope proteins.
Parental AE6 viability could be improved through independent pathways. First, replacement of the wild-type arginine 302 with either lysine, serine, or glycine improved viability of AE6, with kinetics equivalent to those of 4070A. Additionally, replacement of alanine 419 with valine (second-site change detected in an independent population of AE6 no. 2) improved AE6 to be equivalent to 4070A. The possibility that the regions where these two independent changes occurred could interact and/or cooperate was tested. The A419V was reconstructed within AE6 proviral constructs containing R302G, R302S, or R302K. It is important to note that these combinations were not isolated in vivo. Interestingly, juxtaposition of A419V with either R302G or R302S resulted in nonviable virus. Only the conservative R302K remained infectious, with the same kinetics as 4070A in the presence of A419V. G100R tested within the AE6 backbone improves parental viability by almost 2 weeks. In combination with R302G (AE6 no. 3), the virus improved to be equivalent to the wild type.
Parental AE7 kinetics, delayed by almost 2 weeks compared to wild-type 4070A, is improved to wild-type kinetics by the sole replacement of the wild-type residue L538. Replacement of G100 with arginine appears have a similar effect to the parental AE7 control. Similar to AE5, the presence of both second-site changes, G100R and L538Q, improves the viability of the parental chimera. In the assay shown, the constructs containing the L538Q mutation are equal to or better than the control 4070A, while in a second experiment, the L538Q mutant was delayed by 1 day with respect to the wild type (data not shown).
Sequence of the reconstructed viruses after viral spread.
Since reverse transcription is such a dynamic process, it is essential to determine whether the reconstructed virus maintained the cloned second-site changes after viral spread. Sequencing results are shown in Table 2. In general, all viruses had maintained the cloned second-site changes and restriction sites used to generate the parental constructs. Additional second-site changes were acquired in most of the reconstructed viruses bearing second-site changes (underlined in Table 2). In several instances, the cloning of just one second-site change from the original viral population resulted in a virus which had acquired a second-site change which was also present in the original viral population, thus revealing stable sets of changes. All viral populations acquired the G100R change.
TABLE 2.
Comparison of envelope protein sequences of reconstructed chimeric viruses
| Series | Inputa | Post-viral spreadb |
|---|---|---|
| AE5 | T392I (no. 1) | T392I, G100R, G122E, G320R |
| T392I (no. 2) | T392I, G100R, G209R, V341I | |
| G100R | G100R, G51E | |
| G209R (no. 1) | G209R, G100R | |
| G209R (no. 2) | G209R, G100R, G320R | |
| G209R (no. 3) | G209R, G100R, Y347H | |
| T392I, G100R | T392I, G100R, G209R | |
| AE6 | R302K | R302K, G100R (mixed pop.) |
| R302S | R302S, G298R, G100R, L235L (wobble) | |
| R302G | R302G, G100R | |
| A419V | A419V, G100R, Q252R, A264A (wobble) | |
| G100R | G100R, Q252L, G309E | |
| G100R, R302G | G100R, R302G | |
| A419V, R302K | A419V, R302K, G100R | |
| AE7 | G100R | G100R, S291I, L538Q |
| L538Q | L538Q, G100R | |
| G100R, L538Q | G100R, L538Q |
Second-site changes which were reconstructed into the parental proviral backbone.
Second-site changes which were detected by sequencing viruses after the reconstruction experiment (see Materials and Methods); additional changes acquired during viral spread are underlined.
The results for the RT-positive AE5 viral populations reconstructed with the second-site changes, as identified in Table 1, are shown in Table 2, and these include two independent isolates of T392I viruses, three independent isolates of G209R, and one isolate each of G100R and the double combination G100R and T392I. The G100R isolate acquired an additional change in the N-terminal leader domain of SU (G51E). The input of the parent AE5 construct with single or double mutations results in the appearance of a preferential set of limited changes, which include a combination of T392I, G100R, G320R, and G209R. These changes may constitute a stable combination for AE5. It should be noted that T392I, G100R, and G209R were identified in the initial population of AE5 (Table 1). Additional changes have been identified in the reconstructed viruses. These include a change at G122E (VRC domain), G51E (N-terminal leader), and G341I and G347H (C-terminal domain).
The results for the RT-positive AE6 viral populations, reconstructed with the second-site changes identified in Table 1, are shown in Table 2. The combination of G100R and R302G (as isolated in the AE6 no. 3 population, Table 1) is stable in that no additional second-site changes were acquired during viral spread and the reconstruction of R302G alone results in the recovery of G100R. Two other combinations of second-site changes which are stable are R302K/G100R and A419V/R302K/G100R, in that only the G100R is uniformly acquired in these isolates. The R302S isolate acquired an additional change proximal in primary sequence to it (G298R) and a wobble change at L235 (within the receptor binding domain). The G100R isolate acquired additional amino acid changes both within (Q252L) and near (G309E) the PRR. Q252, downstream of the conserved GPRVPIGP PRR, was mutated to arginine in a second independent AE6 reconstructed isolate (AE6/A419V/Q252R).
The results for the RT-positive AE7 viral populations, reconstructed with the second-site changes identified in Table 1, are shown in Table 2. No additional second-site changes were detected in the G100R/L538Q isolate. Reconstruction of the single TM change (L538Q) led to the recovery of the G100R change and the recapitulation of the original AE7 population (Table 1), suggesting that this combination is stable. The G100R isolate acquired the TM ectodomain change seen in the original population and an additional change in the proline-rich region (S291I).
Testing the universality of the second-site changes: effect on viral entry.
The second-site changes were selected by passage of the virus on D17 cells. The universality of the changes in each chimeric virus was tested by determining the titer of each on three different cell lines: NIH 3T3 cells, canine D17 cells (from which they were derived), and human 293T cells. The relative titers of each virus among the three cell types tested can be compared within these experiments but not between different viral stocks. A universal change in the env protein would be supported by titers comparable to those of the wild-type 4070A control in each cell type tested. The results of this analysis are shown in Table 3. The results are organized based on the region of second-site change within the Env protein. AE6 viruses with changes in the PRR could infect all three cell types tested and are biased in their ability to infect cells as follows: D17 < NIH 3T3 cells < 293T cells. AE6 viruses with changes in the C terminus of SU in addition to PRR could also infect all three cell types. The A419V/R302K/G100R combination was created in vitro and was not isolated as a result of viral adaptation. This combination only poorly infects NIH 3T3 cells. Interestingly, the A419V/Q252R/G100R combination infects NIH 3T3 cells better than D17 cells. The AE5/T392I isolate with additional second-site changes in the VRC, PRR, and hinge region was as adept in infecting all three cell types as were the AE6 isolates with the PRR changes. The AE7 isolates containing the TM ectodomain change were able to preferentially infect canine D17 cells; their ability to infect NIH 3T3 cells or 293T cells was markedly reduced (by 5- and 25-fold, respectively).
TABLE 3.
Viral titers
| Chimeraa | Region of changea | Second-site changesb | Titer (% of titer with 4070a)c on:
|
||
|---|---|---|---|---|---|
| Canine D17 | NIH 3T3 | 293T | |||
| AE6 | PRR | R302K; G100R | 96 | 76 | 38 |
| R302S;G298R, G100R | 104 | 72 | 48 | ||
| R302G; G100R | 84 | 58 | 51 | ||
| G100R; Q252L, G309E | 96 | 73 | 53 | ||
| G100R, R302G | 142 | 83 | 67 | ||
| C terminus | A419V; Q252R, G100R | 29 | 44 | 18 | |
| A419V, R302K; G100R | 21 | 6 | 18 | ||
| AE7 | TM | L538Q; G100R | 52 | 10 | 2 |
| L538Q, G100R | 55 | 11 | 4 | ||
| AE5 | C terminus, VRC, VRA | T392I; G320R, G122E, G100R | 119 | 90 | 58 |
Chimeric junctions are as described in the text and in Fig. 1; the common region of second-site change is denoted, occurring in SU unless otherwise noted.
Second-site changes originally subcloned are on the left; changes acquired during viral spread are separated from subcloned changes by a semicolon; underscored residues are those contained within the domain of SU or TM designated in the first column.
Titers are reported as a percentage of 4070A/G100R variant virus titer, scored by counting blue cells (lacZ+); 4070A/G100R viral titers are as follows: 3.5 × 104 (D17 cells), 5.6 × 104 (NIH 3T3 cells), and 2.1 × 104 (293T cells).
DISCUSSION
Viral entry by enveloped viruses is a complex event requiring multiple protein conformations. Recent advances obtained by using X-ray crystallography have provided snapshot pictures of two isolated domains of the envelope proteins of MuLV, namely, the RBD of Friend MuLV (17) and the ectodomain of TM (18). Short of viewing the structure of the complete SU-TM complex before and after receptor binding, interactions within the entire SU-TM monomer and oligomer can only be postulated. Genetic and biochemical data, shown to complement structural data in the identification of functional subdomains of HA (8–11, 67), can provide insights into the functional interactions of the envelope proteins.
Different results for the domains of SU-TM required to yield a functional chimera have been obtained depending on whether viral vectors or replication-competent virus were used. When viral vectors, which can only undergo one round of infection, were used, chimeras which contain junctions at the beginning of the PRR (around junction 4 in this study) yielded virus with normal titers (42). In contrast, vectors with junctions within the C terminus of SU yielded low or no titers (26; L. O'Reilly and M. J. Roth, unpublished data). Chimeric enveloped MuLVs between the ecotropic M-MuLV isolate (E) and the amphotropic 4070A (A) isolate (47) were constructed in a replication-competent virus (AE series and EA series). Although the junctions were initially selected within relatively conserved regions, the results of this study indicate that the chimeric viruses were suboptimal and upon passage have adapted through second-site changes. These changes cannot arise when a viral vector is used, and they can explain the difference between these two systems.
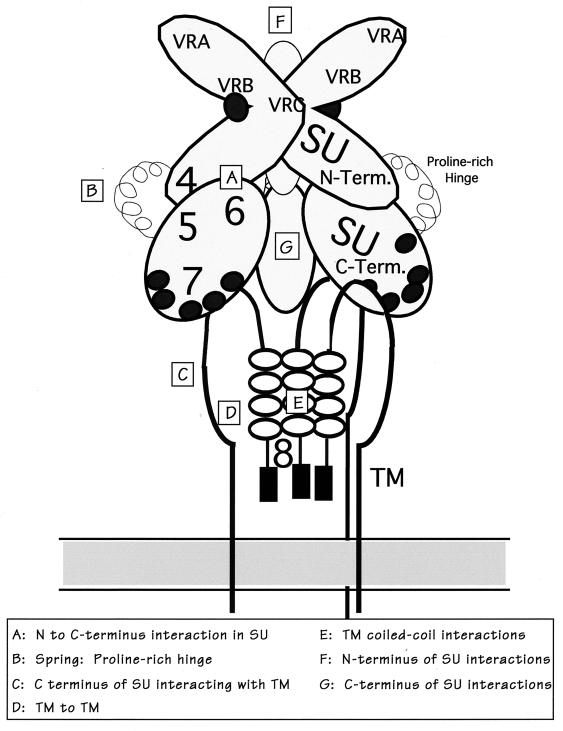
The HA proteins of influenza virus can switch from a native, nonfusogenic structure to fusogenic-active state. The native structure is therefore in a metastable conformation (9). Destabilization of this conformation can be triggered by low pH or other factors such as elevated temperature or addition of urea (9). Maintenance of this metastable conformation would require multiple protein-protein contacts between the two HA proteins. In a similar manner, the nonfusogenic structure of the MuLV envelope proteins would require inter- and intramolecular contacts between the SU and TM proteins. The generation of chimeric proteins could disturb the balance in a spring-loaded mechanism. Figure 5 outlines the potential interactions between the SU and TM proteins, of which at least seven are noted (A to G). The PRR is drawn schematically as a spring which could facilitate the conformational changes required for exposure of the fusion peptide. The positions of the junctions of the chimeras are included in the schematic. Perturbation of any of these interactions could be compensated by strengthening or weakening alternative domains. The results of this study have identified changes which stabilize specific chimeras.
FIG. 5.
Model of interactions of viral envelope proteins SU and TM. A schematic model of potential interactions of SU and TM in their oligomeric form is shown above. The SU protein is depicted as three domains, the N terminus (which includes the VRA, VRB, and VRC domains), the PRR, and the C terminus. The N terminus was drawn based on predictions of trimer packing as proposed by Fass (16), where the VRC of one monomer interacts with the VRB of another monomer. The PRR connects the N and C termini of SU. Glycosylation sites in SU are indicated by solid ovals. TM is depicted with a hidden fusion domain (solid rectangles) and a trimeric coiled-coil motif (open ovals), as discussed in the text (18). Potential interactions which may play a role in the entry and fusion steps of the viral life cycle are denoted by capital letters. Junctions used to construct envelope chimeras are indicated by arabic numbers, with their approximate location in SU and TM.
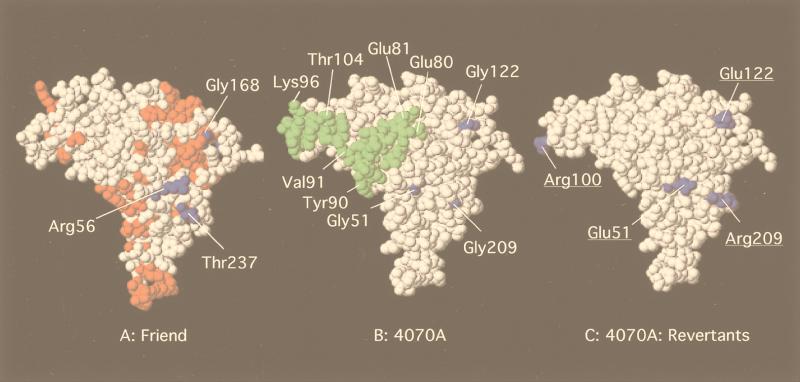
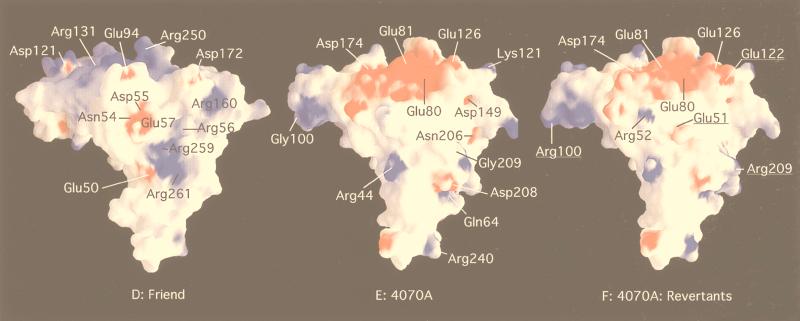
The RBD of the 4070A amphotropic Env was modeled on a computer (Fig. 6B), based on the X-ray crystal structure of the Friend MuLV ecotropic RBD (17) (Fig. 6A). The conserved residues (Fig. 6A) are indicated in red and are not highly surface accessible. The specific changes observed (Fig. 6B and C) and the corresponding residues in Friend MuLV are indicated in blue. Residues 78 to 104 (VRA) in 4070A are indicated in green. Exchange of this region from amphotropic to polytropic sequences results in loss of Pit2 receptor recognition (25). The modeled RBD of 4070A does not differ significantly from the RBD of Friend by a space-filled rendition (Fig. 6A to C). However, by using the program Grasp, the RBDs of Friend MuLV and the modeled 4070A molecules show striking differences in the distribution of surface-accessible charges (Fig. 6D to F). The surface of the Friend MuLV RBD has a large positively charged surface at the top of the domain, corresponding in part to residues Arg 250 (8β/9β), and Arg 131 (VRA). In contrast, this surface of the modeled 4070A is negatively charged, corresponding in part to residues Glu 80 (VRA) Glu 81 (VRA), Glu 126 (VRC), and Asp 174 (VRB). These acidic residues are unique to all amphotropic and polytropic MuLVs. This negatively charged surface of the 4070A RBD is thus composed of residues from many different subdomains. Interestingly, a second-site mutation in AE4 changes E80 to a positively charged lysine. Additional differences in the surface electrostatic potential between the Friend MuLV and 4070A RBD can be noted. In the Friend MuLV RBD, two large positively charged regions are predicted (Arg 160 [VRC] and Arg 259 and 261 [9β]). Mutation of arginine residues in M-MuLV equivalent to Arg 259 and 261 (Arg 256 and 258) resulted in loss of processing of the SU-TM precursor (69) yet maintained viral infectivity.
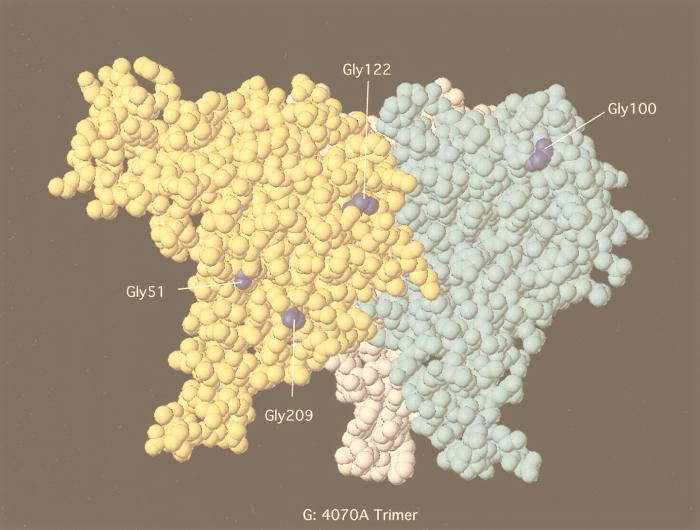
FIG. 6.
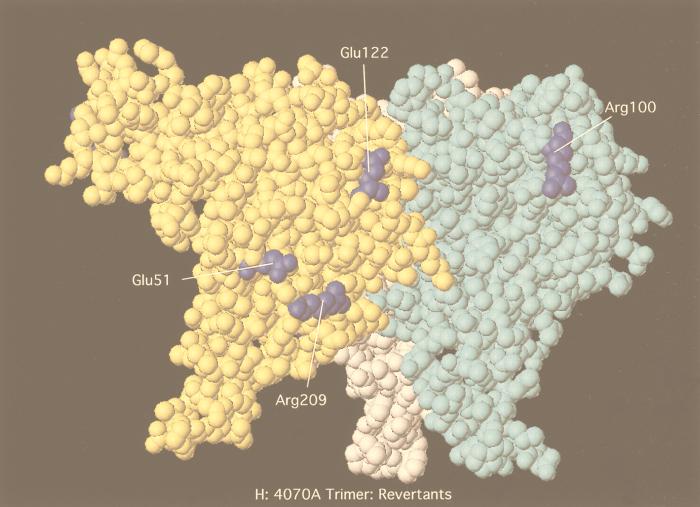
Comparison of the 4070A RBD model and resolved Friend MuLV RBD. Structures corresponding to the RBD of both Friend MuLV (A and D), 4070A (B and E), and 4070A revertants (C and F) are shown. Friend MuLV (1aol.pdb) was solved previously (17); the structure of 4070A was computer modeled based on the Friend MuLV structure, as described in Materials and Methods. Numbering of residues in the Friend MuLV RBD is inclusive of the N-terminal signal peptide (34 amino acids) (17). Structures in A to C are presented as space-filling models. (A) Residues which are conserved between Friend MuLV and 4070A RBD are red; homologous residues in the Friend MuLV RBD to the second-site changes detected in AE chimeras are blue. (B and C) Second-site changes detected in AE chimeras which map to the RBD are blue in the 4070A models; second-site changes are underlined; residues which have been previously studied for a role in receptor binding (3, 25, 62) are green in the 4070A model. (D to F) Modeled structures depict maps of surface charges, analyzed by the Grasp program. Positively charged areas are blue; neutral areas are white; and negatively charged areas are red. (D) Friend MuLV RBD; (E) modeled 4070A RBD; (E) modeled 4070A RBD with second-site revertants (second-site changes are underlined). (G and H) Structures depict the RBD trimer of 4070A as space-filling representations, modeled as described in Materials and Methods. Second-site changes detected in AE chimeras and homologous residues which map to the RBD are depicted in the 4070A models as blue.
RBD.
Most chimeras passaged through canine D17 cells as well as the wild-type control acquired the G100R change (Tables 1 and 2). G100 is within VRA of the RBD, on the face of the protein which is implicated in direct receptor binding. In our molecular modeling (Fig. 6B), the VRA helix D is absent and G100R is surface accessible. The addition of G100R extends a positively charged surface predicted in the modeling (Fig. 6E and F). Mutation of residues in 4070A near G100 to endogenous 10A1 residues expands the host range to use 10A1 receptor (24). Substitution of the linear sequence between Tyr 94 and Thr 104 (Fig. 6B) with polytropic sequences decreased the titer of amphotropic virus to 4.3% (25). Collectively, our data and that of others (3, 24, 25) support the notion that G100R may be involved in receptor recognition or stabilization of the receptor interaction. This second-site change may be specific for the D17 cell line. Receptor-coreceptor variability between species does exist, and mutations which alter the binding affinity for specific host cell proteins are of great interest. For all chimeras except AE7, the acquisition of the G100R change is sufficient for some low level of virus to initially spread, allowing the selection of additional second-site changes which are advantageous to the virus. Future analysis of G100R will directly monitor binding through fluorescence-activated cell sorter FACS analysis (30).
N-terminal leader.
The leader domain, up to the first cysteine, has not been well characterized. However, alternative ligands for gene therapy applications are frequently inserted within this region (54) and have not yielded productive viral entry, suggesting that interactions mediated by this domain are critical for entry. The histidine residue within the leader domain of ecotropic SU (H41), absent in the resolved structure of the Friend MuLV RBD (17), has been implicated in fusion events (2). An AE5 variant was isolated with a change prior to the first cysteine of the VRA (G51E). This second-site change generates a negative charge in the modeled 4070A RBD where the Friend MuLV RBD contains acidic residues Asp 55 and Glu 57 (Fig. 6D to F).
Base of the RBD.
Quite interestingly, a second charged residue, G209R, which appears in close proximity to the G51E, was independently isolated in AE5 chimeras. G209R is in the vicinity of the I 310 helix by modeling (Fig. 6B). This change switches the hydrophobic residue conserved in amphotropic and polytropic envelope proteins to the polar or positively charged residue found in M-MuLV and the related feline leukemia virus. This charged face of the RBD remains surface accessible, even within the modeled trimer (Fig. 6G and H) (16). In the monomer, the G-to-R change results in a change of both surface topology and charge (Fig. 6E and F). Experiments with G209R substitution in 4070A show that transport and processing are not affected by this substitution (Fig. 4) but that the half-life of the cell-associated TM protein was shorter than that of wild-type 4070A. This phenotype was similar to that found in linker insertions in the C terminus of SU, which resulted in shedding of SU and slow passage of the virus (23). Indirectly, this suggests that SU-TM interactions in the presence of G209R are not stable and is consistent with the slowing of 4070A/G209R relative to the wild-type control in the transient-spreading assay (Fig. 3B). Analysis of the potential of G209R to interact with other envelope protein surfaces would be facilitated by the structure of the full-length SU or SU-TM complex.
RBD multimerization.
One second-site mutation, G122E, has occurred in the VRC domain and has the potential to influence the N-to-N multimerization of the RBD (interaction F in Fig. 5). This is based on a proposal that the VRC region of one monomer of SU interacts with the VRB region of another monomer of SU in the trimeric form of SU-TM (16, 17). This mutation was recovered in one isolate of AE5 upon subcloning the SU C-terminal change T392I. In the computer-generated model of 4070A (Fig. 6B), this residue maps in the vicinity of residues 163 to 168 of the Friend MuLV RBD. By sequence alignment, G168 of Friend MuLV corresponds to G122E. G168 is implicated in structural stabilization of the VRC loop through hydrogen bonding to R160. Additional residues within VRC form the direct contact with VRB of the adjacent monomers. PVC211, a Friend MuLV variant with extended host range resulting in neurodegenerative disease, contains a point mutation E163K within this region of VRC (38). This change in PVC211 is implicated in either binding to receptor or promotion of early conformational changes required for fusion. Analysis of the surface electrostatic potential indicates that the G122E mutation extends the negative charged surface (Fig. 6F) into the VRC region. If this surface charge functions in viral entry, a wider surface may compensate for misaligned chimeric SU trimers.
Hinge and PRR.
Results of both linker insertion mutagenesis (23), deletion (33, 68), and point mutations (33) studies of the PRR indicate this region is involved in stabilizing SU-TM interactions. Two independent mutations of residue Q252, located at the N terminus of the PRR, were isolated in AE6 chimeras (AE6/G100R/G309E/Q252L and AE6/G100R/A419V/Q252R). In related studies, introduction of Q252V/P250I point mutations into 4070A Env resulted in a 5-log-unit decrease in titer and increased shedding of SU (33). In contrast, in this study the AE6 A419V/G100R/Q252R construct contains the highest titer on NIH 3T3 cells. Further investigation of whether the Q252 changes increase shedding within specific AE6 chimeras and their relation to virus viability are under way.
Second-site changes in the PRR could repair the interaction between the N and C termini (interaction A in Fig. 5). Additionally, if the PRR is functioning as a spring or interface between the N and C domains of SU (interaction B in Fig. 5), mutations which generally improve these interactions would improve the virus for entry into all cell types. For AE6, one independent pathway of repair involves changes in the C terminus of PRR through crossover 5. Each of the single changes observed at amino acid position 302 (S, K, and G) could restore the viability of the AE6 chimeric virus to the equivalent of the wild-type control (Fig. 2). AE6 chimeras with second-site changes in or near the PRR (Table 3) infected the D17, NIH 3T3 and 293T cell lines efficiently.
C terminus of SU.
The majority of the chimeras studied in this report contain crossover junctions throughout the C-terminus of SU. The C terminus of SU associates with TM by covalent and noncovalent interactions. Linker insertion mutants in the vicinity of crossover 6 were characterized by early shedding of SU from TM, resulting in decreased syncytium formation with rat XC cells (23). Crossover 6 is immediately C-terminal of the CWLC-conserved motif (56), which is involved in the disulfide linkage with TM (48, 51). Mutagenesis of glycosylation sites within this region (19, 49) indicated that it is critical to the folding of the C terminus of SU, resulting in the block of transport of the precursor SU-TM protein. It would be expected, therefore, that chimeras with junctions at position 6 could require the repair of SU-TM interactions (interaction C in Fig. 5) or within the C-terminal domain, if it functions as a single folding domain, similar to the RBD. It is interesting that in the AE6 series, five (R302K, R302S, R302G, G298R, and G309E) of eight second-site changes (Tables 1 and 2) were in the vicinity of crossover 5 (V307). This region marks the boundary between the PRR and the beginning of the C-terminal half of SU. A second independent pathway of repair of AE6 involving the C terminus was discovered (Tables 1 and 2). The mutation of alanine 419 to valine, a branched residue, suggests that repair of this portion of SU requires the addition of a more rigid component, perhaps stabilizing an internal interaction required for interdomain folding and/or function. The interaction of the C-terminal change with each change at position 302 (Fig. 3) revealed that the only combination which leads to a viable virus is with the conservative lysine. This suggests that regions in the C terminus and near crossover 5 interact.
TM.
Although the majority of chimera junctions and second-site changes occur within SU, second-site changes within TM have the potential to improve SU-TM interactions (interaction C in Fig. 5). Notably, in an AE7 chimera, a crossover within SU yielded changes within the ectodomain of TM at L538Q. Similarly, changes in the ectodomain of TM have also been observed in the EA7 chimeras passaged in D17/pJET cells (O'Reilly and Roth, unpublished). The resolved structure of the ectodomain of ecotropic TM (18) revealed a trimer of TM through a coiled-coil motif (18). Amino acid 538 is at a b position within the potential coiled coil, which is more frequently represented by a glutamine than by a leucine (36). L538 extends outward to the surface of the trimer and thus may be available for interactions with SU. L538Q is located 2 amino acids away from an asparagine residue, which was coordinated to a chloride ion in the structure. It is surprising that the AE7/L538Q virus has a strong bias for infection of D17 cells. For two independent isolates, the titer on 293T cells was 11- to 25-fold lower than on D17 cells (Table 3). This suggests that for the AE7 viruses, a more cell-specific adaptation has occurred. The results for the cell specificity of the AE7/L538Q virus indicate that the L538Q cannot simply affect the universal stabilization of the trimers. Further studies to understand the defects in spreading for the AE7 chimera in divergent cell types are in progress.
A single change in the ectodomain of the wild-type 4070A TM protein was observed (G541R), as well as the G100R change in SU (Tables 1 and 2). The presence of second-site changes in a wild-type control suggest that, as with the AE7 chimeras, changes may be specific for the D17 cell line.
Dynamic interactions of multiple domains are required for envelope protein function.
In the AE5 chimera, the designed junction after the PRR could adversely affect multiple SU interactions (interactions A, B, C, F, and G in Fig. 5). The interactions of the PRR are most probably critical for efficient envelope function, since the mere exchange of the larger (14-amino-acid) PRR of 4070A within an ecotropic isolate (68) decreased the viral titer by one-third. Chimeras with changes in the C terminus (AE6/A419V) functioned in all three cell types tested, although each isolate differentially infected NIH 3T3 cells depending on the PRR mutation present. The artificial combination of AE6/A419V/R302K could not infect NIH 3T3 cells well, whereas AE6/A419V plus Q252R, at the N-terminus of the PRR, preferentially infected NIH 3T3 cells. These results highlight the complex interrelationships between the subdomains of Env. Chimera AE5 was not viable in D17 cells when cloned proviral DNA was transiently expressed (47) (Fig. 1A) but can spread, albeit with delayed kinetics, when introduced by high-titer infection (Fig. 1B), suggesting that one defect of this chimera may be related to processing and folding. Attempts to generate a stable cell line producing this chimera were unsuccessful, implying this construct bears multiple defects. The range of second-site changes detected, spanning multiple domains of SU such as VRA, VRC, VRB-hinge, PRR, and C terminus, substantiates these observations (Tables 1 and 2). Indeed, repair of these multiple domains yielded a virus which was highly competent in infecting multiple cell types (Table 3), suggesting that interactions of multiple domains of SU and TM are required for optimal infectivity.
Stable combinations.
Although the retroviral genome is constantly in flux, the independent isolation of specific amino acid changes provides evidence for the positive selection of these changes. For AE5 chimeras, the isolation of the G100R (VRA), G209R (VRB hinge), G320R, and T392I (C terminus) combination occurs through independent events. Similarly, for AE7 chimeras, the isolation of G100R with L538Q occurs after a single round of selection (Tables 1 and 2). The occurrence of this stable set in D17 cells in an AE7 chimera is consistent with the observed lack of further improvement in spreading kinetics in subsequent rounds of infection. Subcloning of one mutation within a population occasionally leads to a new second-site change in the vicinity of a change detected in the original population (AE6 G100R [Table 2]). This data provide insights into the limited number of changes required for the repair of viruses hampered in their ability to spread in a transient-infection assay.
ACKNOWLEDGMENTS
This research is supported by NIH grant R01 CA149932 to M.J.R. L.O. was supported in part by an NIH Predoctoral Training Grant in Cellular and Molecular Biology (grant T32GM-8360) and by a predoctoral award from the State of New Jersey Cancer Commission.
We thank Ujwal Shinde and Shabir Najmudin for their assistance with computer modeling programs Sybyl and Grasp.
REFERENCES
- 1.Albritton L M, Tweng L, Scadden D, Cunningham J M. A putative murine retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J-L, Danos O, Heard J M. Definition of a 14-amino acid peptide essential for the interaction between the murine leukemia virus amphotropic envelope glycoprotein and its receptor. J Virol. 1998;72:428–435. doi: 10.1128/jvi.72.1.428-435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini J-L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini J-L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battini J-L, Rodrigues P, Muller R, Danos O, Heard J M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Goff S P. Point mutations in Moloney murine leukemia virus envelope protein: effects on infectivity, virion association, and superinfection resistance. Virology. 1993;196:748–757. doi: 10.1006/viro.1993.1532. [DOI] [PubMed] [Google Scholar]
- 8.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 9.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr C M, Kim P S. Flu virus invasion: halfway there. Science. 1994;266:234–236. doi: 10.1126/science.7939658. [DOI] [PubMed] [Google Scholar]
- 11.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 12.Colicelli J, Goff S P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 13.Daniels R S, Downie J C, Hay A J, Knossow M, Skehel J J, Wang M L, Wiley D C. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 14.Davey R A, Hamson C A, Healey J J, Cunningham J M. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J Virol. 1997;71:8096–8102. doi: 10.1128/jvi.71.11.8096-8102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey R A, Zuo Y, Cunningham J M. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fass D. Ph.D. thesis. Cambridge, Mass: Massachusetts Institute of Technology; 1997. [Google Scholar]
- 17.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 A resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 18.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 A resolution. Nat Struct Biol. 1996;3:465–468. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 19.Felkner R H, Roth M J. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J Virol. 1992;66:4258–4264. doi: 10.1128/jvi.66.7.4258-4264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godley L, Pfeifer J, Stainhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel J J, Wiley D C. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 21.Goff S P, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants; use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granowitz C, Colicelli J, Goff S P. Analysis of mutations in the envelope gene of Moloney murine leukemia virus: separation of infectivity from superinfection resistance. Virology. 1991;183:545–554. doi: 10.1016/0042-6822(91)90983-i. [DOI] [PubMed] [Google Scholar]
- 23.Gray K D, Roth M. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J-Y, Cannon P M, Lai K-M, Zhao Y, Eiden M V, Anderson W F. Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J Virol. 1997;71:8103–8108. doi: 10.1128/jvi.71.11.8103-8108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J-Y, Zhao Y, Anderson W F, Cannon P M. Role of variable regions A and B in receptor binding domain of amphotropic murine leukemia virus envelope protein. J Virol. 1998;72:9101–9108. doi: 10.1128/jvi.72.11.9101-9108.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han L, Hofmann T, Chiang Y, Anderson W F. Chimeric envelope glycoproteins constructed between amphotropic and xenotropic murine leukemia retroviruses. Somatic Cell Mol Genet. 1995;21:205–214. doi: 10.1007/BF02254771. [DOI] [PubMed] [Google Scholar]
- 27.Heard J M, Danos O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 29.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadan M J, Sturm S, Anderson W F, Eglitis M A. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J W, Closs E I, Albritton L M, Cunningham J M. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature. 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 33.Lavillette D, Maurice M, Roche C, Russell S, Sitbon M, Cosset F-L. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol. 1998;72:9955–9965. doi: 10.1128/jvi.72.12.9955-9965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linder M, Linder D, Hahnen J, Schott H-H, Stirm S. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur J Biochem. 1992;203:65–73. doi: 10.1111/j.1432-1033.1992.tb19828.x. [DOI] [PubMed] [Google Scholar]
- 35.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupus A, VanDyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 37.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda M, Masuda M, Hanson C A, Hoffman P M, Ruscetti S K. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J Virol. 1996;70:8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCutchan J H, Pagano J S. Enhancement of infectivity of SV40 deoxyribonucleic acid with DEAE dextran. J Natl Cancer Inst. 1968;41:351–357. [PubMed] [Google Scholar]
- 40.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan R A, Nussbaum O, Muenchau D D, Shu L, Couture L, Anderson W F. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:4712–4721. doi: 10.1128/jvi.67.8.4712-4721.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nussbaum O, Roop A, Anderson W F. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:7402–7405. doi: 10.1128/jvi.67.12.7402-7405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 Murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park B H, Matuschke B, Lavi E, Gaulton G N. A point mutation in env gene of murine leukemia virus induces syncytium formation and neurologic disease. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peredo C, O'Reilly L, Gray K, Roth M J. Characterization of chimeras between the ecotropic Moloney murine leukemia virus and the amphotropic 4070A envelope proteins. J Virol. 1996;70:3142–3152. doi: 10.1128/jvi.70.5.3142-3152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinter A, Fleissner E. The presence of disulfide-linked gp70-p15(E) complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 49.Pinter A, Honnen W J. O-linked glycosylation of retroviral envelope gene products. J Virol. 1988;62:1016–1021. doi: 10.1128/jvi.62.3.1016-1021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinter A, Honnen W J, Tung J-S, O'Donnell P V, Hammerling U. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology. 1982;116:499–516. doi: 10.1016/0042-6822(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 51.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rein A. Interference groupings of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982;120:251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- 53.Rein A, Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 54.Russell S J, Hawkins R E, Winter G. Retroviral vectors displaying functional antibody fragments. Nucleic Acids Res. 1993;21:1081–1085. doi: 10.1093/nar/21.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 56.Sitbon M, D'Auriol L, Ellerbrok H, Andre C, Nishio J, Perryman S, Pozo F, Hayes S F, Wehrly K, Tambourin P, Galibert F, Chesebro B. Substitution of leucine for isoleucine in a sequence highly conserved among retroviral envelope surface glycoproteins attenuates the lytic effect of the Friend murine leukemia virus. Proc Natl Acad Sci USA. 1991;88:5932–5936. doi: 10.1073/pnas.88.13.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 58.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman M S, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:629–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinhauer D A, Martin J, Lin Y P, Wharton S A, Oldstone M B A, Skehel J J, Wiley D C. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc Natl Acad Sci USA. 1996;93:12873–12878. doi: 10.1073/pnas.93.23.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szurek P F, Yuen P H, Ball J K, Wong P K. A Val-25-to-Ile substitution in the envelope precursor polyprotein, gPr80env, is responsible for the temperature sensitivity, inefficient processing of gPr80env, and neurovirulence of ts1, a mutant of Moloney murine leukemia virus TB. J Virol. 1990;64:467–475. doi: 10.1128/jvi.64.2.467-475.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szurek P F, Yuen P H, Jerzy R, Wong P K Y. Identification of point mutations in the envelope gene of Moloney murine leukemia virus TB temperature-sensitive paralytogenic mutant ts1: molecular determinants for neurovirulence. J Virol. 1988;622:357–360. doi: 10.1128/jvi.62.1.357-360.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas A, Roth M J. Analysis of cysteine mutations on the transmembrane protein of Moloney murine leukemia virus. Virology. 1995;211:285–289. doi: 10.1006/viro.1995.1402. [DOI] [PubMed] [Google Scholar]
- 64.Vahlenkamp T W, Verschoor E J, Schuurman N N M P, VanVliet A L W, Horzinek M C, Egberink H F, Ronde A D. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J Virol. 1997;71:7132–7135. doi: 10.1128/jvi.71.9.7132-7135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Kavanaugh M P, Norith R A, Kabat D. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature. 1991;352:729–731. doi: 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 67.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:424–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 68.Wu B W, Cannon P M, Gordon E M, Hall F L, Anderson W F. Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol. 1998;72:5383–5391. doi: 10.1128/jvi.72.7.5383-5391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zavorotinskaya T, Albritton L M. Failure to cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interactions. J Virol. 1999;73:5621–5629. doi: 10.1128/jvi.73.7.5621-5629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Y, Zhu L, Benedict C A, Chen D, Anderson W F, Cannon P M. Functional domains in the retroviral transmembrane protein. J Virol. 1998;72:5392–5398. doi: 10.1128/jvi.72.7.5392-5398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]