Abstract
PURPOSE
Few studies have explored the potential for pharmacological interventions to delay disease progression in patients undergoing active surveillance (AS). This preplanned transcriptomic analysis of patient samples from the ENACT trial aims to identify biomarkers in patients on AS who are at increased risk for disease progression or who may derive the greatest benefit from enzalutamide treatment.
PATIENTS AND METHODS
In the phase II ENACT (ClinicalTrials.gov identifier: NCT02799745) trial, patients on AS were randomly assigned 1:1 to 160 mg orally once daily enzalutamide monotherapy or continued AS for 1 year. Transcriptional analyses were conducted on biopsies collected at trial screening, year 1, and year 2. Three gene expression signatures were evaluated in samples collected at screening and in available samples from patients on AS at any time during surveillance (expanded cohort): Decipher genomic classifier, androgen receptor activity (AR-A) score, and Prediction Analysis of Microarray 50 (PAM50) cell subtype signature.
RESULTS
The Decipher genomic classifier score was prognostic; higher scores were associated with disease progression in the expanded cohort and AS arm of the expanded cohort. Patients with higher Decipher scores had greater positive treatment effect from enzalutamide as measured by time to secondary rise in prostate-specific antigen >25% above baseline. In patients treated with enzalutamide, higher AR-A scores and PAM50 luminal subtypes were associated with a greater likelihood of negative biopsy incidence at year 2.
CONCLUSION
This analysis suggests that the Decipher genomic classifier may be prognostic for disease progression in AS patients with low- to intermediate-risk prostate cancer. Higher Decipher and AR-A scores, as well as PAM50 luminal subtypes, may also serve as biomarkers for treatment response.
INTRODUCTION
For patients with low- or favorable intermediate-risk prostate cancer, clinical guidelines now promote active surveillance (AS) as a preferred option in most cases for managing disease progression, with multiple factors influencing the decision for patients on AS to initiate therapeutic intervention.1,2 Patients may choose to undergo AS to avoid the adverse effects associated with treatments from definitive therapy, such as radical prostatectomy, external beam radiation therapy, or brachytherapy.1,2
CONTEXT
Key Objective
To identify potential biomarkers that may be associated with increased risk of disease progression or greater response to enzalutamide therapy in patients with low- to intermediate-risk prostate cancer undergoing active surveillance (AS).
Knowledge Generated
Higher scores for the Decipher genomic classifier were associated with disease progression and greater positive treatment effect from enzalutamide therapy. In those treated with enzalutamide, higher androgen receptor activity (AR-A) score and luminal subtypes in the Prediction Analysis of Microarray 50 (PAM50) cell subtype signature were associated with an increased likelihood of negative biopsy incidence.
Relevance
The results of this study suggest that the Decipher genomic classifier may be prognostic for disease progression in AS patients with low- to intermediate-risk prostate cancer. Higher Decipher scores, greater AR-A scores, and PAM50 luminal subtypes may be indicators of greater treatment response to enzalutamide therapy.
Enzalutamide is an oral androgen receptor inhibitor approved to treat castration-resistant prostate cancer, metastatic hormone-sensitive prostate cancer (also known as metastatic castration-sensitive prostate cancer), and nonmetastatic castration-sensitive prostate cancer (nmCSPC) with biochemical recurrence (BCR) at high risk for metastasis.3,4 ENACT (ClinicalTrials.gov identifier: NCT02799745) was a phase II, open-label, exploratory, randomized clinical trial that assessed the efficacy and safety of enzalutamide monotherapy in the AS population.5 The EMBARK study, which resulted in the US Food and Drug Administration's approval of enzalutamide for treating patients with nmCSPC with high-risk BCR, previously also showed that risk of metastasis or death was significantly lower in the enzalutamide monotherapy group than in the leuprolide-alone group.3,6 In ENACT, patients with clinically localized low- or intermediate-risk prostate cancer on AS treated with oral enzalutamide 160 mg once daily for 1 year had a 46% reduced rate of disease progression versus those who continued on AS (hazard ratio [HR], 0.54 [95% CI, 0.33 to 0.89]; P = .02), although the benefits abated by 1 year after cessation of treatment.5
Few published studies have assessed whether pharmacological interventions may delay disease progression in patients with prostate cancer undergoing AS. Patient biomarker analyses are essential for identifying factors that can help predict which patients with low- or intermediate-risk prostate cancer are most likely to benefit from active treatment. In this analysis, we report the results of a preplanned ENACT study transcriptomic analysis that evaluated the potential prognostic and predictive capabilities of biomarkers to identify patients on AS who may be at greater risk for disease progression and are therefore better suited to active treatment, as well as patients who may have a more profound response to enzalutamide monotherapy.
PATIENTS AND METHODS
Patients and Study Design
In the ENACT trial, patients on AS with clinically localized low- or intermediate-risk prostate cancer were randomly assigned to receive 1 year of enzalutamide therapy or continued AS and then followed for 2 years.5 Transcriptional analyses were conducted on formalin-fixed, paraffin-embedded tissue from enrolled patients who consented to the ancillary biomarker analysis study. Tumor expression profiles were generated from biopsies obtained at screening, year 1, and year 2 (Data Supplement, Fig S1) of ≥0.5 mm of tumor linear length using a clinical grade transcriptome assay (Veracyte, Inc, San Diego, CA).7 Biopsy samples required at least 1 mm of linear tumor length, with the region of interest meant for macrodissection having ≥25% cancer cells and <15% benign cell contamination. Biopsy specimen sectioning was performed centrally and subsequently stored at the University of Michigan until shipped for analysis by Veracyte. Analysis was performed in samples from two cohorts: (1) the analytic cohort comprised samples collected at screening and (2) the expanded cohort further incorporated samples collected at any time during surveillance.
End Points
The primary end point of this preplanned transcriptomic analysis was time to pathologic or therapeutic disease progression (pathologic disease progression defined as an increase in primary or secondary Gleason pattern of ≥1 or an increase of ≥15% in cancer-positive cores; therapeutic disease progression defined as the earliest occurrence of primary therapy for prostate cancer).5 Secondary endpoints included time to therapeutic disease progression (defined as the earliest occurrence of radical therapy for prostate cancer), incidence of negative biopsy at year 1 and 2, and time to prostate-specific antigen (PSA) progression (defined as the time to first rise in serum PSA >25% above baseline, a rise >25% above nadir, or an absolute increase of >2 ng/mL). Time to secondary rise in serum PSA >25% above baseline was evaluated as a separate end point. The gene expression signatures analyzed included the Decipher genomic classifier (Decipher),8 androgen receptor activity (AR-A) score,9 and the Prediction Analysis of Microarray 50 (PAM50) cell subtype signature.10 The details on the genomic classifiers nominated for analysis are included in the Data Supplement (Table S1). The genomic signatures evaluated in this study (ie, Decipher, AR-A, PAM50) were nominated in a prespecified analysis plan.
Statistical Analyses
Statistical analyses were conducted using a prespecified transcriptomic analysis plan. Cox proportional hazards and logistic regression models were used for time-to-event (time to pathologic disease progression, therapeutic disease progression, and PSA progression) and binary (incidence of negative biopsy at year 1 and 2) end points, respectively. Multivariable (MVA) models were specified to include age, race, time since prostate cancer diagnosis, prostate cancer risk, type of biopsy, and site. Owing to a limited patient sample size, MVA analyses were conducted for all primary and secondary end points, with only age, random assignment arm, and National Comprehensive Cancer Network risk group as covariates.
Prognostic and predictive evaluations of the candidate biomarkers were performed by reporting the main effects and interactions with treatment as estimated from separate MVA models. HRs and odds ratios with 95% CIs are reported for time-to-event and event incidence outcomes, respectively.
The primary analysis was performed using data from patients randomly assigned to either the enzalutamide arm or the AS arm who had provided consent and whose genetic samples passed laboratory quality control screening. Additional sensitivity analyses were performed using data from an additional group of patients randomly assigned to the AS arm who had at least one passing laboratory quality control sample at any time during surveillance. In the analysis of time-to-event end points, these additional samples were included in Cox regression models with left truncation at the time of the nonscreening passing laboratory quality control sample. Two-sided P values below .05 indicate a statistically significant result. Statistical analyses were conducted using R version 4.2.0 (The R Foundation, Vienna, Austria).
RESULTS
In the ENACT study, 227 patients with low- or intermediate-risk localized prostate cancer were randomly assigned.5 The analytic cohort comprised a total of 95 patients who had evaluable screening samples (enzalutamide, n = 49; AS, n = 46; Data Supplement, Fig S1). An additional 26 patients randomly assigned to AS who did not have evaluable screening samples but did have evaluable samples at any time during surveillance were included in the expanded cohort (year 1 biopsy, n = 24; year 2 biopsy, n = 2). Patient and tumor characteristics were generally balanced between the enzalutamide and AS treatment arms (Table 1), except that there were more Black or African American patients in the AS arm (17.4%) than the enzalutamide arm (4.1%; P = .046). About 26% of patients received a magnetic resonance imaging–targeted biopsy, and the median time from diagnosis to random assignment was 3 months. In the expanded cohort, patient and tumor characteristics were similar between treatment arms (Data Supplement, Table S2).
TABLE 1.
Patient and Tumor Characteristics With Genomic Classifier Information by Treatment Arm in the Analytic Cohort
| Parameter | Enzalutamide 160 mg daily (n = 49) | Active Surveillance (n = 46) | P |
|---|---|---|---|
| Age, years, median (range) | 65 (47-84) | 66 (55-86) | .22a |
| Race, No. (%) | |||
| Black or African American | 2 (4.1) | 8 (17.4) | .046b |
| White | 47 (95.9) | 38 (82.6) | |
| Baseline PSA, ng/mL, median (range) | 5.3 (2.1-15.1)c | 6.3 (1.5-16.8)d | .11a |
| Baseline PPC, %, median (range) | 30.8 (7.7-53.9)e | 25.0 (8.3-50)f | .12a |
| NCCN risk group, No. (%) | |||
| Low | 25 (51.0) | 20 (43.5) | .54b |
| Intermediate | 24 (49.0) | 26 (56.5) | |
| Biopsy type, No. (%) | |||
| mpMRI-targeted | 12 (24.5) | 12 (26.1) | .99b |
| Non–mpMRI-targeted | 37 (75.5) | 34 (73.9) | |
| Time from diagnosis to random assignment, months, median (range) | 3.0 (1.0-6.7) | 3.0 (1.3-6.5) | .61a |
| Genomic classifier | |||
| Decipher GC score, median (IQR) | 0.2 (0.1-0.3) | 0.2 (0.1-0.3) | .60a |
| Decipher GC risk group, No. (%) | |||
| Low | 45 (91.8) | 42 (91.3) | .99b |
| Intermediate | 2 (4.1) | 2 (4.3) | |
| High | 2 (4.1) | 2 (4.3) | |
| AR-A score, median (range) | 13.7 (9.8-16.2) | 13.7 (10.7-15.7) | .60a |
| AR-A group, No. (%) | |||
| Average AR-A | 46 (93.9) | 43 (93.5) | .99b |
| Lower AR-A | 3 (6.1) | 3 (6.5) | |
| PAM50 subtype, No. (%) | |||
| Luminal | 27 (55.1) | 24 (52.2) | .84b |
| Luminal A | 11 (22.4) | 9 (19.6) | |
| Luminal B | 16 (32.7) | 15 (32.6) | |
| Basal | 22 (44.9) | 22 (47.8) |
Abbreviations: AR-A, androgen receptor activity; GC, genomic classifier; mpMRI, multiparametric magnetic resonance imaging; NCCN, National Comprehensive Cancer Network; PAM50, Prediction Analysis of Microarray 50; PPC, percent positive core; PSA, prostate-specific antigen.
Statistical analysis was conducted using a Mann-Whitney U test.
Statistical analysis was conducted using a Fisher exact test.
Six patients did not have a baseline PSA value available.
Two patients did not have a baseline PSA value available.
Two patients did not have a baseline PPC value available.
One patient did not have a baseline PPC value available.
Tumor molecular characteristics were balanced between study arms (Table 1; Data Supplement, Table S2). More than 90% of patients had low-risk Decipher scores: The median score for the analytic cohort AS arm was 0.18 (IQR, 0.10-0.27) and 0.19 for the enzalutamide arm (IQR, 0.14-0.27). In the analytic cohort, only 6% of patients were classified by AR-A score as having lower AR-A (ie, AR-A <11 as defined in Spratt et al11). Basal tumor subtypes were observed in 48% of patients in the AS arm and 45% of patients in the enzalutamide arm. There was no correlation between Decipher and AR-A and between Decipher and PAM50 (Data Supplement, Fig S2); however, median AR-A score (IQR) was significantly higher (P < .001) in those with luminal PAM50 signatures (14.39 [13.65-14.93]), compared with those with basal PAM50 signatures (13.14 [12.12-13.62]; Data Supplement, Fig S2). Event counts for pathologic and therapeutic disease progression were balanced between treatment arms (Table 2). Compared with the AS arm, the enzalutamide arm had a lower proportion of patients with a PSA rise >25% above baseline and a greater proportion of patients with a negative biopsy at year 2 (Table 2).
TABLE 2.
Event Counts by Analysis Cohort
| End Point | AS Arm (screening samples only), E/N (%) | AS Arm (all available samples), E/N (%) | Enzalutamide Arm, E/N (%) |
|---|---|---|---|
| Pathologic disease progression | 16/46 (34.8) | 20/65 (30.8) | 15/49 (30.6) |
| Therapeutic disease progression | 7/46 (15.2) | 10/66 (15.2) | 7/49 (14.3) |
| PSA rise >25% above baseline | 29/44 (65.9) | 30/51 (58.8) | 19/46 (41.3) |
| Negative biopsy at year 2 | 4/21 (19.0) | 4/37 (10.8) | 9/27 (33.3) |
NOTE. Patients who experienced an event before useable genomic classifiers were available were excluded from the AS (all available samples) column.
Abbreviations: AS, active surveillance; E, number of events; N, number of patients; PSA, prostate-specific antigen.
In the AS arm of the analytic cohort, Decipher score showed a significant positive association with increased rates of pathologic or therapeutic disease progression among the expanded cohort that included additional pathologic samples available at any time during surveillance (MVA HR [95% CI] per 0.1, 1.17 [1.01 to 1.35]; P = .04; Table 3). However, Decipher score did not reach significance for progression when biopsy samples available only at screening were analyzed (MVA HR [95% CI] per 0.1, 1.13 [0.83 to 1.54]; P = .44). There was a nonsignificant correlation between Decipher score and progression among patients in the enzalutamide arm (MVA HR [95% CI] per 0.1, 1.50 [0.91 to 2.48]; P = .11). When considering the analytic cohort, Decipher score at screening displayed a positive association with pathologic or therapeutic disease progression that did not reach statistical significance (MVA HR [95% CI] per 0.1, 1.21 [0.94 to 1.56]; P = .15) and a significant association when including the AS arm of the expanded cohort (MVA HR [95% CI] per 0.1, 1.23 [1.05 to 1.44]; P = .01). Decipher score was also significantly associated with therapeutic disease progression in the analytic cohort (MVA HR per 0.1, 1.51 [1.07 to 2.12]; P = .02; Fig 1; Table 4) and the expanded cohort (MVA HR per 0.1, 1.46 [1.18 to 1.80]; P = .02; Table 4).
TABLE 3.
Multivariable Cox Regression Results for Disease Progression in the Analytic Cohort and by Treatment Arm (with and without AS samples)
| Genomic Classifier | Analytic Cohort (screening samples; n = 95) | Expanded Cohort (all samples; n = 114) | AS (screening samples; n = 46) | AS (all samples; n = 65) | Enzalutamide (n = 49) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| Decipher (per 0.1) | 1.21 (0.94 to 1.56) | .15 | 1.23 (1.05 to 1.44) | .01* | 1.13 (0.83 to 1.54) | .44 | 1.17 (1.01 to 1.35) | .04* | 1.50 (0.91 to 2.48) | .11 |
| Enzalutamide v AS | 0.57 (0.27 to 1.17) | .12 | 0.69 (0.34 to 1.38) | .29 | — | — | — | — | — | — |
| Age, years | 0.92 (0.87 to 0.98) | .006* | 0.93 (0.89 to 0.97) | <.001* | 0.93 (0.85 to 1.02) | .10 | 0.94 (0.89 to 1.00) | .06 | 0.90 (0.83 to 0.97) | .007* |
| Intermediate v low PCa risk | 0.74 (0.36 to 1.54) | .42 | 1.00 (0.51 to 1.97) | .99 | 0.49 (0.18 to 1.34) | .16 | 0.85 (0.35 to 2.04) | .71 | 1.31 (0.47 to 3.70) | .61 |
| AR-A (per 1) | 1.01 (0.78 to 1.30) | .93 | 1.06 (0.84 to 1.33) | .62 | 0.91 (0.62 to 1.32) | .62 | 1.05 (0.74 to 1.49) | .80 | 1.02 (0.72 to 1.43) | .92 |
| Enzalutamide v AS | 0.56 (0.27 to 1.16) | .12 | 0.66 (0.33 to 1.31) | .23 | — | — | — | — | — | — |
| Age, years | 0.93 (0.88 to 0.98) | .01* | 0.94 (0.90 to 0.98) | .004* | 0.93 (0.85 to 1.02) | .13 | 0.95 (0.90 to 1.01) | .09 | 0.92 (0.85 to 0.99) | .03* |
| Intermediate v low PCa risk | 0.78 (0.38 to 1.61) | .51 | 1.01 (0.50 to 2.01) | .98 | 0.50 (0.18 to 1.37) | .18 | 0.86 (0.34 to 2.16) | .75 | 1.26 (0.45 to 3.52) | .66 |
| PAM50 basal v luminal | 0.84 (0.41 to 1.72) | .63 | 0.84 (0.43 to 1.63) | .60 | 1.25 (0.41 to 3.81) | .70 | 1.16 (0.46 to 2.89) | .76 | 0.58 (0.17 to 1.92) | .37 |
| Enzalutamide v AS | 0.55 (0.26 to 1.14) | .11 | 0.66 (0.33 to 1.32) | .24 | — | — | — | — | — | — |
| Age, years | 0.93 (0.88 to 0.98) | .01* | 0.94 (0.90 to 0.98) | .002* | 0.94 (0.85 to 1.04) | .21 | 0.96 (0.90 to 1.01) | .14 | 0.93 (0.86 to 1.00) | .06 |
| Intermediate v low PCa risk | 0.77 (0.38 to 1.60) | .49 | 0.99 (0.50 to 1.97) | .99 | 0.52 (0.19 to 1.44) | .21 | 0.86 (0.35 to 2.12) | .75 | 1.21 (0.44 to 3.38) | .71 |
Abbreviations: AR-A, androgen receptor activity; AS, active surveillance; HR, hazard ratio; PAM50, Prediction Analysis of Microarray 50; PCa, prostate cancer.
*Significant P value.
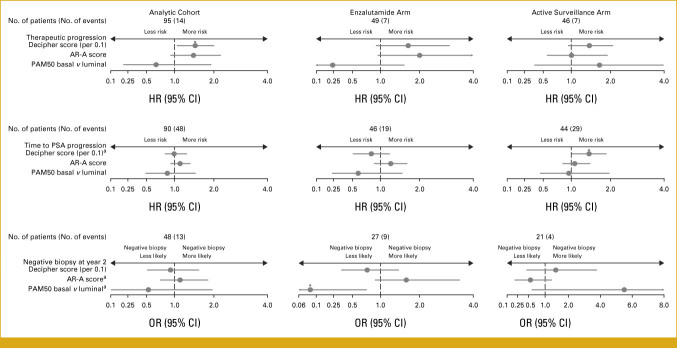
FIG 1.
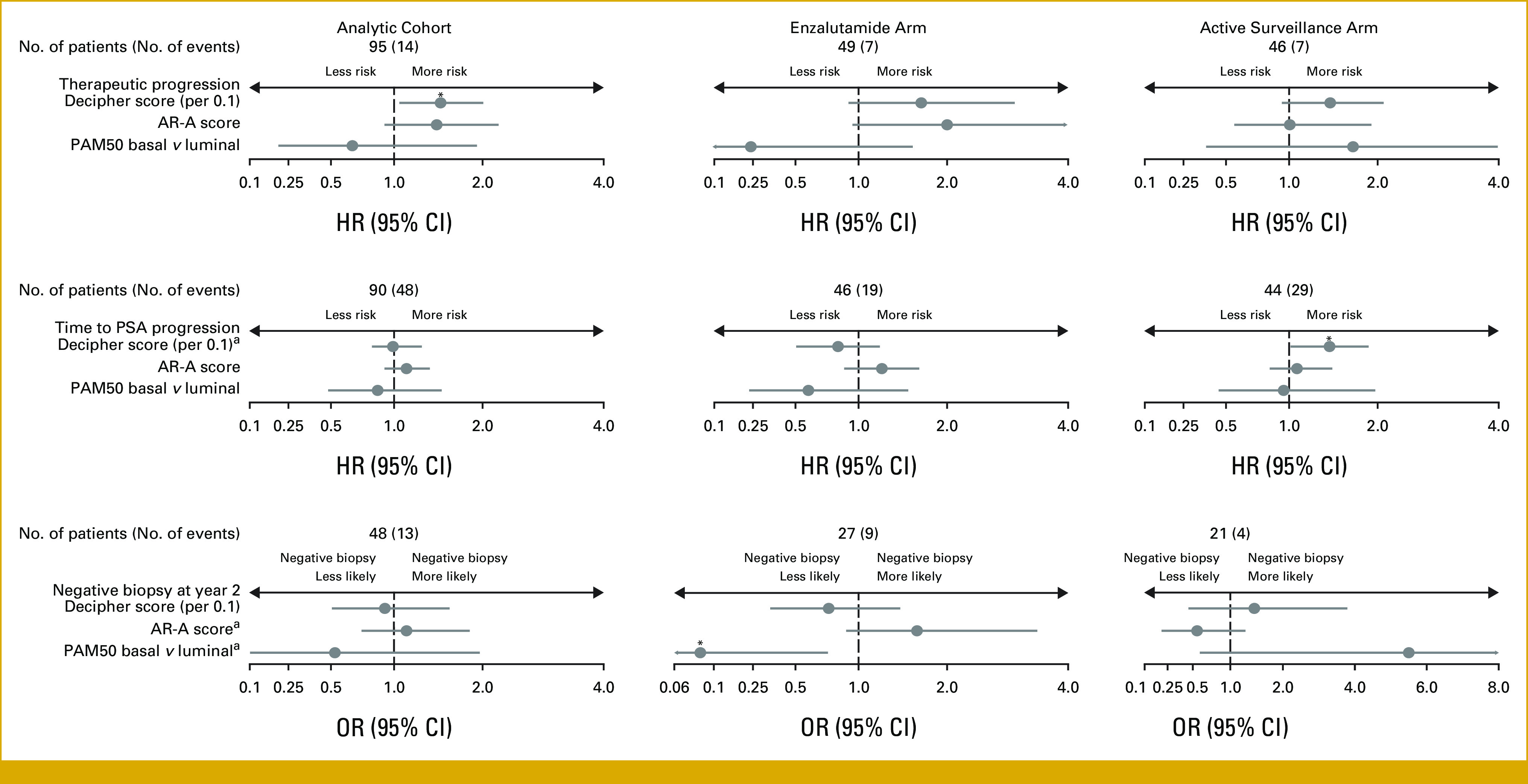
Multivariable analysis association between changes in Decipher scores, AR-A scores, and PAM50 subtype with patient outcomes in the analytic cohort stratified by treatment arm. AR-A, androgen receptor activity; HR, hazard ratio; OR, odds ratio; PAM50, Prediction Analysis of Microarray 50; PSA, prostate-specific antigen. aInteraction P value was significant in the analytic cohort. *Significant P value.
TABLE 4.
Full Multivariable Cox Regression Results for Therapeutic Disease Progression
| Genomic Classifier | Analytic Cohort (screening samples; n = 95) | Expanded Cohort (all samples; n = 115) | AS (screening samples; n = 46) | AS (all samples; n = 66) | Enzalutamide (n = 49) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| Decipher (per 0.1) | 1.51 (1.07 to 2.12) | .02* | 1.46 (1.18 to 1.80) | <.001* | 1.40 (0.92 to 2.12) | .11 | 1.36 (1.11 to 1.66) | .003* | 2.17 (1.00 to 4.71) | .05 |
| Enzalutamide v AS | 0.77 (0.26 to 2.30) | .64 | 0.90 (0.33 to 2.45) | .83 | — | — | — | — | — | — |
| Age, years | 0.95 (0.87 to 1.03) | .19 | 0.95 (0.89 to 1.02) | .18 | 0.97 (0.86 to 1.09) | .59 | 0.98 (0.91 to 1.06) | .69 | 0.91 (0.80 to 1.02) | .10 |
| Intermediate v low PCa risk | 1.18 (0.39 to 3.54) | .77 | 1.67 (0.68 to 4.09) | .27 | 0.78 (0.17 to 3.65) | .75 | 1.51 (0.45 to 5.04) | .50 | 1.93 (0.42 to 8.94) | .40 |
| AR-A (per 1) | 1.43 (0.89 to 2.29) | .14 | 1.38 (0.99 to 1.94) | .06 | 1.03 (0.55 to 1.93) | .93 | 1.11 (0.80 to 1.56) | .53 | 1.90 (0.86 to 4.19) | .11 |
| Enzalutamide v AS | 0.64 (0.22 to 1.90) | .42 | 0.71 (0.28 to 1.81) | .47 | — | — | — | — | — | — |
| Age, years | 0.97 (0.89 to 1.06) | .49 | 0.99 (0.92 to 1.06) | .71 | 0.98 (0.87 to 1.10) | .68 | 1.00 (0.94 to 1.07) | .94 | 0.98 (0.86 to 1.12) | .78 |
| Intermediate v low PCa risk | 1.45 (0.49 to 4.27) | .50 | 1.63 (0.61 to 4.35) | .33 | 0.95 (0.20 to 4.41) | .95 | 1.40 (0.40 to 4.97) | .6 | 1.81 (0.40 to 8.15) | .44 |
| PAM50 basal v luminal | 0.62 (0.21 to 1.87) | .40 | 0.60 (0.21 to 1.70) | .34 | 1.56 (0.29 to 8.47) | .61 | 1.21 (0.36 to 4.07) | .76 | 0.22 (0.02 to 1.90) | .17 |
| Enzalutamide v AS | 0.69 (0.23 to 2.03) | .50 | 0.78 (0.30 to 2.08) | .62 | — | — | — | — | — | — |
| Age, years | 0.96 (0.89 to 1.04) | .34 | 0.98 (0.92 to 1.04) | .46 | 0.99 (0.87 to 1.13) | .90 | 1.01 (0.95 to 1.07) | .81 | 0.97 (0.86 to 1.09) | .64 |
| Intermediate v low PCa risk | 1.35 (0.46 to 3.97) | .59 | 1.56 (0.60 to 4.07) | .37 | 0.93 (0.21 to 4.18) | .92 | 1.38 (0.40 to 4.80) | .61 | 1.73 (0.38 to 7.87) | .48 |
Abbreviations: AR-A, androgen receptor activity; AS, active surveillance; HR, hazard ratio; PAM50, Prediction Analysis of Microarray 50; PCa, prostate cancer.
*Significant P value.
Other genomic signatures considered in our analysis (AR-A, PAM50) were not prognostic of disease outcomes in the untreated trial population (Table 4; Data Supplement, Tables S3 and S4). Similarly, no significant relationships were observed between the signatures and PSA progression or the incidence of negative biopsy at year 1 among patients in the AS arm (data not shown).
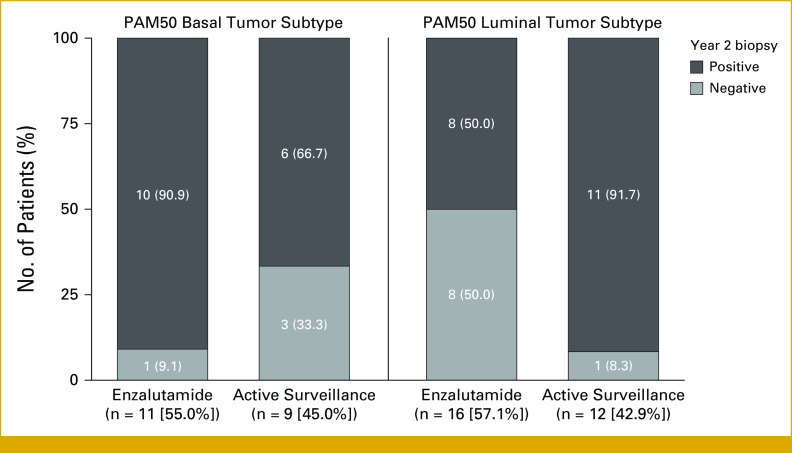
We next examined whether the genomic signatures could identify any factors associated with differential treatment benefits. Patients with higher Decipher scores had greater absolute benefit from enzalutamide as measured by time to secondary rise in serum PSA >25% above baseline (interaction P = .03; Data Supplement, Fig S3 and Table S5). Similarly, higher AR-A scores were associated with greater response to enzalutamide as measured by incidence of negative biopsy at year 2 (interaction P = .04; Data Supplement, Fig S4 and Table S6). Patients with luminal tumor subtypes (as determined by PAM50) who received enzalutamide were also significantly more likely to have a negative biopsy at year 2 (interaction P = .01). In this regard, 50% of patients with luminal tumors treated with enzalutamide had a negative biopsy at year 2 versus only 9% of those with basal tumors treated with enzalutamide (Fig 2).
FIG 2.
Incidence of negative biopsy at year 2 by treatment arm within PAM50 subtype. PAM50, Prediction Analysis of Microarray 50.
DISCUSSION
This preplanned analysis of the ENACT study cohort was conducted to assess the prognostic and predictive capabilities of different transcriptomic biomarkers in determining which patients on AS are at greater risk of disease progression and would be most likely to respond to enzalutamide treatment. Decipher score was significantly associated with pathologic or therapeutic disease progression in the AS arm of the expanded cohort and the full expanded cohort. By contrast, AR-A and PAM50 signatures were not prognostic of disease outcomes in the analytic cohort. There were no significant relationships between the genomic classifiers and PSA progression or incidence of negative biopsy at year 1 among patients in the AS arm of the analytic cohort. Patients with higher Decipher scores had greater response to enzalutamide as measured by secondary rise in PSA >25% above baseline, whereas patients with higher AR-A scores and luminal subtypes per PAM50 showed greater response to enzalutamide as measured by incidence of negative biopsy at year 2. These results suggest that Decipher score, AR-A score, and PAM-50 subtype signatures can be used to identify which patients would derive benefit from enzalutamide treatment.
The results of this analysis demonstrated that Decipher score was similarly prognostic for disease progression but the significance and magnitude of the association was less compared with other studies in which Decipher score was assessed for other oncologic end points.7,12,13 The lower effect size for the Decipher score observed in this analysis was likely due to the limited sample size of the study, as evidenced by the increased significance of association with expansion of the cohort size. Furthermore, the homogeneity of the cohort in terms of both clinicopathologic and genomic risk will by definition reduce the test's performance.
Higher AR-A scores were associated with greater response to enzalutamide, a finding that is consistent with another report that showed that tumors with low AR-A scores were less sensitive to treatments such as androgen-deprivation therapy (ADT) and docetaxel.9 Similarly, the increased response to enzalutamide observed in patients with the PAM50 luminal subtype aligns with the results of a study by Zhao et al,10 which showed that the PAM50 luminal subtype was associated with a greater response to AR inhibition and ADT.
The main limitation of this preplanned analysis was the small sample size, which limits the clinical implications of the results. Specifically, lower numbers of patients with negative biopsy data, intermediate/high Decipher scores, and lower AR-A scores may have skewed their respective analyses. Another limitation to this analysis is that approximately half of the biopsy specimens did not undergo transcriptomic analysis. This occurred because of lack of submission of samples for this exploratory end point and that samples intended for genomics were previously depleted in other testing (ie, immune-histochemistry).
In conclusion, this preplanned ENACT trial biomarker analysis demonstrates the value of the Decipher score, AR-A score, and PAM50 genomic classifiers in identifying patients undergoing AS who are most likely to benefit from enzalutamide treatment. Future studies will aim to validate the utility of these genomic classifiers in a larger patient cohort.
ACKNOWLEDGMENT
We would like to thank Dr Matthew R. Cooperberg for his assistance with interpreting the study data and reviewing the manuscript. Medical writing and editorial assistance were provided by Terrance Ku, MSc, and Nicholas Strange, BA, from Complete HealthVizion, IPG Health Medical Communications, Inc, and funded by the study sponsors.
PRIOR PRESENTATION
Presented at the 2022 Congress of the European Society for Medical Oncology, Paris, France, September 9-13, 2022.
SUPPORT
Supported by Astellas Pharma Inc and Pfizer Inc, the co-developers of enzalutamide.
DATA SHARING STATEMENT
Upon request, and subject to certain criteria, conditions, and exceptions, Astellas will provide access to anonymized patient-level data from completed Astellas-sponsored phase III to IV interventional clinical studies conducted for products and indications which have been approved in any country and for studies conducted for terminated compounds. Approval must have been granted by the agencies of the main regions, the US, the EU, and Japan. If approval is sought in only one or two regions, approval must have been granted by those agencies. Where available, the following anonymized patient-level data and information are provided for each clinical trial: raw data set, analysis ready data set, protocols with any amendments or addenda, annotated case report form, statistical analysis plan, data set specifications, and clinical study report. Additionally, data may be available on request. Researchers may request access to anonymized participant-level data, trial-level data, and protocols from Astellas-sponsored clinical trials at www.clinicalstudydatarequest.com. For the Astellas criteria on data sharing see https://clinicalstudydatarequest.com/Study-Sponsors/Study-Sponsors-Astellas.aspx.
AUTHOR CONTRIBUTIONS
Conception and design: Kenneth K. Iwata, Dina Elsouda, John Hairston, David Russell, Elai Davicioni, Neal D. Shore, Edward M. Schaeffer
Administrative support: Elai Davicioni, Edward M. Schaeffer
Provision of study materials or patients: Elai Davicioni
Collection and assembly of data: David Russell, Elai Davicioni, Edward M. Schaeffer
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Ashley E. Ross
Honoraria: Bayer, Janssen Biotech, Blue Earth Diagnostics, Decipher Biosciences, Tempus, Astellas Pharma, Myovant Sciences, Lantheus Medical Imaging, AstraZeneca
Consulting or Advisory Role: Astellas Pharma, BillionToOne
Speakers' Bureau: Tempus
Open Payments Link: https://openpaymentsdata.cms.gov/physician/40486
Kenneth K. Iwata
Employment: Astellas Pharma
Research Funding: Astellas Pharma
Patents, Royalties, Other Intellectual Property: Patent Number 11833140
Travel, Accommodations, Expenses: Astellas Pharma
Dina Elsouda
Employment: Astellas Pharma
Research Funding: Astellas Pharma
John Hairston
Employment: Astellas Pharma, Courante Oncology
David Russell
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Elai Davicioni
Employment: GenomeDx, Decipher Biosciences, Veracyte
Leadership: GenomeDx, Decipher Biosciences, Veracyte
Stock and Other Ownership Interests: GenomeDx, Decipher Biosciences, Veracyte
Patents, Royalties, Other Intellectual Property: Cancer Diagnostics Using Biomarkers 20140066323
Travel, Accommodations, Expenses: GenomeDx, Veracyte
James A. Proudfoot
Employment: Veracyte
Stock and Other Ownership Interests: Veracyte
Neal D. Shore
Employment: GenesisCare
Leadership: Photocure, Alessa Therapeutics
Consulting or Advisory Role: Bayer, Janssen Scientific Affairs, Dendreon, Tolmar, Ferring, Medivation/Astellas, Amgen, Pfizer, AstraZeneca, Myovant Sciences, Astellas Pharma, AbbVie, Merck, Bristol Myers Squibb/Sanofi, Boston Scientific, Clovis Oncology, Exact Imaging, FerGene, Foundation Medicine, CG Oncology, InVitae, MDxHealth, Myriad Genetics, Nymox, Propella Therapeutics, Genzyme, Sanofi, Sesen Bio, Exact Sciences, Genesis Cancer Care, Pacific Edge Biotechnology, Phosphorus, Urogen Pharma, Speciality Networks, Peerview, Clarity Pharmaceuticals, Lantheus Medical Imaging, Lilly, Photocure, Telix Pharmaceuticals, Tempus, Vaxiion, AIkido Pharma, Arquer Diagnostics, Asieris Pharmaceuticals, Guardant Health, Vessi Medical, ImmunityBio, Incyte, Minomic, NGM Biopharmaceuticals, Nonagen Bioscience, Novartis, PlatformQ Health, Profound Medical, Promaxo, Protara Therapeutics, Fize Medical
Speakers' Bureau: Janssen, Bayer, Dendreon, Astellas Pharma, AstraZeneca, Clovis Oncology, Pfizer, Guardant Health, Merck, Foundation Medicine
Research Funding: AbbVie, Amgen, Astellas Pharma, AstraZeneca, Bayer, Bristol Myers Squibb/Pfizer, Boston Scientific, Clovis Oncology, Dendreon, Exact Imaging, Ferring, Foundation Medicine, InVitae, Janssen, MDxHealth, Merck, Myovant Sciences, Myriad Genetics, Nymox, Pfizer, Sanofi, Sesen Bio, Tolmar, CG Oncology, DisperSol, FORMA Therapeutics, Guardant Health, Jiangsu Yahong Meditech, Novartis, Pacific Edge, POINT Biopharma, Propella Therapeutics, SeaGen, MT Group, Theralase, Veru, Zenflow, Advantagene, Aragon Pharmaceuticals, Endocyte, Exelixis, FKD Therapies, Genentech, Istari Oncology, Medivation, OncoCellMDx, ORIC Pharmaceuticals, Palette Life Sciences, Plexxikon, RhoVac, Steba Biotech, Urogen Pharma, Urotronic, US Biotest, Vaxiion
Expert Testimony: Ferring
Other Relationship: Photocure, Alessa Therapeutics
Edward M. Schaeffer
Consulting or Advisory Role: AbbVie, Pfizer, Lantheus Medical Imaging
No other potential conflicts of interest were reported.
REFERENCES
- 1.National Comprehensive Cancer Network : NCCN Clinical Practice Guidelines in Oncology: Prostate cancer version 3.2022. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf [DOI] [PubMed]
- 2.Parker C, Castro E, Fizazi K, et al. : Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 31:1119-1134, 2020 [DOI] [PubMed] [Google Scholar]
- 3.US Food and Drug Administration : XTANDI highlights of prescribing information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/213674s010,203415s022lbl.pdf
- 4.Astellas Pharma US Inc; Pfizer Inc : XTANDI summary of product characteristics. https://www.ema.europa.eu/en/documents/product-information/xtandi-epar-product-information_en.pdf
- 5.Shore ND, Renzulli J, Fleshner NE, et al. : Active surveillance plus enzalutamide monotherapy vs active surveillance alone in patients with low-risk or intermediate-risk localized prostate cancer: The ENACT randomized clinical trial. JAMA Oncol 8:1128-1136, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freedland SJ, de Almeida Luz M, De Giorgi U, et al. : Improved outcomes with enzalutamide in biochemically recurrent prostate cancer. N Engl J Med 389:1453-1465, 2023 [DOI] [PubMed] [Google Scholar]
- 7.Nguyen PL, Huang HCR, Spratt DE, et al. : Analysis of a biopsy-based genomic classifier in high-risk prostate cancer: Meta-analysis of the NRG Oncology/RTOG 9202, 9413, and 9902 phase 3 randomized trials. Int J Radiat Oncol Biol Phys 116:521-529, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erho N, Crisan A, Vergara IA, et al. : Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 8:e66855, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spratt DE, Alshalalfa M, Fishbane N, et al. : Transcriptomic heterogeneity of androgen receptor activity defines a de novo low AR-active subclass in treatment naive primary prostate cancer. Clin Cancer Res 25:6721-6730, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao SG, Chang SL, Erho N, et al. : Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol 3:1663-1672, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spratt DE, Zumsteg ZS, Pei X, et al. : Predictors of castration-resistant prostate cancer after dose-escalated external beam radiotherapy. Prostate 75:175-182, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vince RA Jr, Jiang R, Qi J, et al. : Impact of Decipher Biopsy testing on clinical outcomes in localized prostate cancer in a prospective statewide collaborative. Prostate Cancer Prostatic Dis 25:677-683, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jairath NK, Dal Pra A, Vince R Jr, et al. : A systematic review of the evidence for the decipher genomic classifier in prostate cancer. Eur Urol 79:374-383, 2021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Upon request, and subject to certain criteria, conditions, and exceptions, Astellas will provide access to anonymized patient-level data from completed Astellas-sponsored phase III to IV interventional clinical studies conducted for products and indications which have been approved in any country and for studies conducted for terminated compounds. Approval must have been granted by the agencies of the main regions, the US, the EU, and Japan. If approval is sought in only one or two regions, approval must have been granted by those agencies. Where available, the following anonymized patient-level data and information are provided for each clinical trial: raw data set, analysis ready data set, protocols with any amendments or addenda, annotated case report form, statistical analysis plan, data set specifications, and clinical study report. Additionally, data may be available on request. Researchers may request access to anonymized participant-level data, trial-level data, and protocols from Astellas-sponsored clinical trials at www.clinicalstudydatarequest.com. For the Astellas criteria on data sharing see https://clinicalstudydatarequest.com/Study-Sponsors/Study-Sponsors-Astellas.aspx.