Abstract
PURPOSE
In the United States, a comprehensive national breast cancer registry (CR) does not exist. Thus, care and coverage decisions are based on data from population subsets, other countries, or models. We report a prototype real-world research data mart to assess mortality, morbidity, and costs for breast cancer diagnosis and treatment.
METHODS
With institutional review board approval and Health Insurance Portability and Accountability Act (HIPPA) compliance, a multidisciplinary clinical and research data warehouse (RDW) expert group curated demographic, risk, imaging, pathology, treatment, and outcome data from the electronic health records (EHR), radiology (RIS), and CR for patients having breast imaging and/or a diagnosis of breast cancer in our institution from January 1, 2004, to December 31, 2020. Domains were defined by prebuilt views to extract data denormalized according to requirements from the existing RDW using an export, transform, load pattern. Data dictionaries were included. Structured query language was used for data cleaning.
RESULTS
Five-hundred eighty-nine elements (EHR 311, RIS 211, and CR 67) were mapped to 27 domains; all, except one containing CR elements, had cancer and noncancer cohort views, resulting in a total of 53 views (average 12 elements/view; range, 4-67). EHR and RIS queries returned 497,218 patients with 2,967,364 imaging examinations and associated visit details. Cancer biology, treatment, and outcome details for 15,619 breast cancer cases were imported from the CR of our primary breast care facility for this prototype mart.
CONCLUSION
Institutional real-world data marts enable comprehensive understanding of care outcomes within an organization. As clinical data sources become increasingly structured, such marts may be an important source for future interinstitution analysis and potentially an opportunity to create robust real-world results that could be used to support evidence-based national policy and care decisions for breast cancer.
This institutional data mart is a template for collecting real-world breast cancer outcomes in the United States.
INTRODUCTION
Breast cancer is the most common cancer diagnosed in women and the leading cause of cancer death in women worldwide,1 with estimated global macroeconomic cost of $2 trillion international dollars for 2020-2050, using 2017 prices.2 In the United States, breast cancer treatment accounts for 14% of all cancer costs with 2020 annual expenditure of $29.8 billion. The highest care cost occurs in the final year of life, estimated to be $76,100 per patient.3 Despite the magnitude of expenditures on diagnosis and treatment of this disease, an estimated 43,000 women will die in 2023 in the United States from it.4 Thus, identifying real-world optimal diagnosis and care strategies is critical to reduce individual and societal impacts. Such determinations are difficult because of lack of real-world data comparing different approaches for detection, diagnosis, and treatment. For a comprehensive analysis of breast cancer care in the United States, large data sets are needed, and, as outcomes are dependent on regional variances that include delivery of care and population differences, data sets from multiple institutions and regions are required to understand the status nationally.
CONTEXT
Key Objective
In the United States, what resource contains comprehensive real-world data on breast cancer? Currently no single resource contains this information. Institutions, however, do have all the data needed for their patients. We report a novel breast care research data mart that may be a template for other organizations to evaluate any question related to breast care.
Knowledge Generated
A research breast data mart containing over 500 discrete data elements and free-text reports having relevant information was created from multiple clinical source structures for approximately 500,000 patients. Cancer biology, treatment, and outcome details for approximately 15,000 breast cancer cases were included.
Relevance (J.L. Warner)
-
Disease-specific data marts have the potential to lower the barrier for translational research efforts by researchers and clinicians.*
*Relevance section written by JCO Clinical Cancer Informatics Editor-in-Chief Jeremy L. Warner, MD, MS, FAMIA, FASCO.
In several countries, population-based national registries collect detailed screening, treatment, and outcome data. This enables direct understanding of benefits and costs of screening and treatments in those populations. However, in the United States, screening is voluntary, and no such comprehensive registry exists. Federal law requires states maintain a cancer registry (CR). The Centers for Disease Control (CDC) oversees the National Program of Cancer Registries (NPCR), which encompasses approximately 97% of the population. The National Cancer Institute SEER program includes approximately 48% of the population. Both registries collect detailed information regarding stage at diagnosis, treatments, and outcomes, as does the American Surgical Society's Commission on Cancer (CoC) registry. None collect imaging information. The American College of Radiology's registry, the National Mammography Database, collects detailed imaging history but only has cursory data on stage at diagnosis and no outcome data.
The Breast Cancer Surveillance Consortium (BCSC) includes six active SEER registries collecting screening information and SEER data so is an important comprehensive database. The US Preventative Services Task Force guidelines, updated in 20095 and 2016,6 have been based in part on BCSC data with estimations for population-level outcomes using NCI's Cancer Information and Modeling Network (CISNET). Critics have raised concern that because BCSC registries are in regions with relatively poor care delivery, the data do not reliably predict outcomes of screening, but instead reflects a fractured health care delivery system. Merging of SEER, NPCR, NMD, and BCSC is possible but challenging because of significant privacy concerns regarding data sharing, costs in creating and maintaining a new database, and other factors.
Institutions have invested significantly in electronic storage of medical information in the past several decades. Most have multiple products intended to collect portions of information such as radiology (RIS), pathology (PD), and medical oncology (MD) information systems, electronic health records (EHRs), and CRs. Although each serves a particular purpose for patient care, historically these have been siloed. Recently, institutions have begun to realize the powerful clinical care, quality improvement, and clinical research benefits that internal data warehouses can provide. Large-scale initiatives such as from the Office of the National Coordinator for Health Information Technology (ONC) also are moving health care vendors and organizations toward ever increasing standardization to facilitate information exchange for optimal patient care. This initiative and others will ultimately facilitate easier sharing across institutional data marts for other purposes such as research.
Herein, we present the process involved in creation of a research institutional breast data mart. The mart's purpose is to study innumerable questions related to breast health and care delivery and to be a template for other organizations.
METHODS
Mart Construction
Institutional review board approval was obtained to create and study a Health Insurance Portability and Accountability Act (HIPPA)-compliant research breast data mart within our preexisting research data warehouse (RDW), Neptune. The RDW was the resource for cohort identification and data extraction. It contains atomic layer data stored close to source structures, containing only transformations for deidentification requirements, and extracted from our EHR systems, health insurance claims, and research data. Data are extracted monthly from the source structures for transformation and loading into the RDW, the construction of which has been previously published.7 This design allows the same high level of granularity as the source systems, thereby creating a gold standard that would be lost if the data were manipulated or rolled up in any way. In cases where domain data are pulled from multiple source systems, the domain data are stored in a table specific to that domain and source system, thereby permitting researchers to select the data from the preferred source system. As new elements are adopted by source structures and added to RDW, the mart will be updated as well. Thus, the mart represents an evolving clinical database that will stay current with clinical systems.
A working group of radiology, pathology, surgical, medical, and radiation oncology physicians, the chief informatics research officer, computer scientists, and the cancer registrar itemized a comprehensive list of data elements and their source information system(s) related to patient demographics, imaging, diagnosis, treatments, costs, and outcomes.
At outset, the RDW did not contain RIS (ImageCast, General Electric, Waukesha, WI) and CR (METRIQ, Eleckta Inc, Stockholm, Sweden) content. Appropriate leaders were interviewed to understand their databases and data transfer agreements established. For the CR, all persons with an International Classification of Diseases (ICD)-9 or ICD-10 code indicating a breast cancer diagnosis (IDC-9: 174.0-174.9,198.81, 233 and ICD-10: C50.011-50.919, C79.81, D05.90, D05.91, D05.92) who underwent initial therapy at our primary breast care facility (Magee Womens Hospital of University of Pittsburgh Medical Center) were included for initial trial mart construction. North American Association of Central Cancer Registries (NAACCR)–required elements and several site-specific elements were included (Appendix Table A1). All RIS elements in the source structure were incorporated. Data dictionaries were collected for both the CR and RIS.
PD (CoPath Plus, Sunquest, Tucson, AZ) and MD (Aria, Varian Inc, Palo Alto, CA) information were in the RDW before this effort; however, the data exist as free text. For example, synoptic surgical pathology reports that contained detailed information on every cancer had not been discretely identified nor evaluated for potential mining previously. To improve the RDW search for these elements, interviews with the pathology informatics and physician leaders clarified the report constructs. Then, an RDW analysis identified these reports, determined what structured data were available, and what data existed as free text.
Data Collection
In an iterative process, every possible source location for each element was identified and evaluated. When possible, the primary element source was the one most frequently completed, robust, and extractable. As examples, EHR and RIS included family history. We compared element fill rates and specific relationship information. The EHR was selected because of better granularity (eg, EHR had paternal uncle, whereas RIS only contained uncle). In another example, cancer immunohistochemistry elements (ie, estrogen receptor, progesterone receptor and Her-2-neu) were in the PD and CR. The CR was selected as primary as it contained discrete site-specific data elements, whereas the PD contained free text only in the synoptic surgical pathology reports. To mine free text, an algorithm such as natural language processing (NLP) is necessary. All available sources for every element were included in the mart regardless of which was denoted as primary, then elements were sorted and labeled into related groups.
Mart Organization
The RDW was queried for all patients with at least one breast imaging examination (on the basis of RIS examination codes) from January 1, 2004, through December 31, 2020. Each patient was assigned a unique research identifier. Oracle (Oracle, Inc, Austin, TX) was used for the mart construction. Data domains were defined by prebuilt views of grouped elements to extract data denormalized according to RDW requirements. The views were used to add data using an export, transform, load pattern from the RDW to the mart.
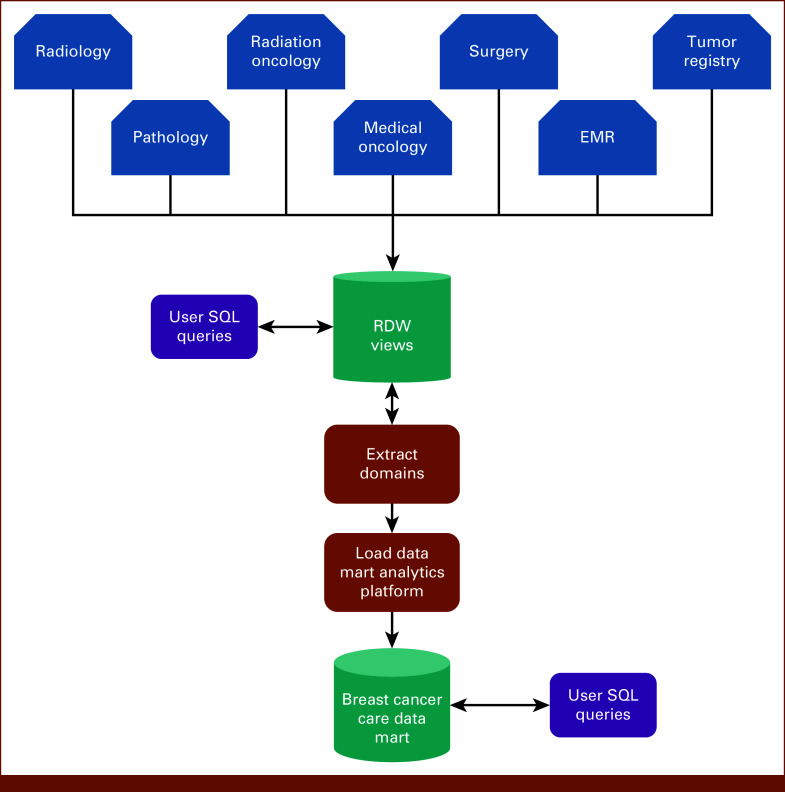
The mart was structured into two cohorts so that domains (except the CR) contained two views. Cohort” view included everyone with an ICD-9 or ICD-10 diagnosis code for breast cancer; control view included everyone with a RIS examination code specific for a breast imaging examination and without these ICD-9/-10 codes. Each Oracle view, imported element, source structure name, and comments from the RDW building team (if needed) were cataloged in the mart for researcher reference. Figure 1 depicts the data flow. Appendix Table A1 lists all mart data elements and views.
FIG 1.
Data flow diagram. Neptune is the name of our research data warehouse. EMR, electronic medical record; RDW, research data warehouse; SQL, structured query language.
Initial Mart Quality Analysis
Structured query language searches sorted the number of examinations by year and compared results against the historic number of examinations performed in the organization to filter and consolidate duplicate records. Valid mammogram dates (ie, January 1, 2004, to January 1, 2021) linked an examination to a CR entry to determine screening interval. Examinations with invalid or unknown dates were removed. Remaining examinations were then sorted by patient research ID. Screening examinations were identified by screening indication examination codes or examinations more than 260 days from the most recent previous examination date. Screen interval of a minimum of 260 days was selected to avoid inclusion of examinations that may have been performed for 6-month short-interval follow-up from the last screen. This generated an imaging history for each patient. Nonduplicates were concatenated, separated with a semicolon then joined to the CR view using patient ID and examination date as links with the CR element date of first contact. Results were pivoted to arrange all examinations for each patient in chronological order, with the associated columns for each examination.
RESULTS
Data Collection, Data Elements, and Domain Mapping
RDW query identified 497,218 unique patients with at least one breast imaging examination and a total of 2,967,364 breast imaging examinations. Electronic medical record query identified 39,860 patients with a diagnosis of breast cancer, based on ICD-9/-10 codes (Table 1). CR query revealed 15,619 patients who received their initial breast cancer treatment at our primary breast care facility.
TABLE 1.
Demographics of Data Mart Population
| Demographic | Screening Examinations (n = 2,068,144) | Diagnostic Examinations (n = 296,023) | Patients With a Breast Cancer Diagnosis Code (n = 39,860) | Total Population (N = 497,218) |
|---|---|---|---|---|
| Race, No. (%) | ||||
| White | 1,856,144 (90) | 259,973 (88) | 36,412 (91) | 440,001 (88) |
| African American | 154,814 (7) | 25,902 (9) | 2,720 (7) | 39,978 (8) |
| Asian | 18,542 (1) | 3,508 (1) | 281 (1) | 5,732 (1) |
| Other | 3,969 (<1) | 712 (<1) | 67 (<1) | 1,266 (<1) |
| Unknown | 25,675 (1) | 2,928 (1) | 380 (1) | 10,241 (2) |
| Ethnicity, No. (%) | ||||
| Hispanic | 9,189 (<1) | 2,079 (1) | 132 (<1) | 3,381 (1) |
| Non-Hispanic | 1,952,146 (94) | 276,362 (93) | 28,180 (71) | 467,304 (94) |
| Unknown | 106,809 (5) | 17,582 (6) | 1,548 (4) | 26,533 (5) |
The working group defined 331 unique data elements and mapped them to 27 data domains. Because each domain, except the CR, included control and cohort views, the mart contains 589 total elements mapped to 53 Oracle views with an average of 12 elements per view (range, 4-67; Appendix Table A1). Source system contributions included EHR: 17 domains, 34 views, and 311 elements; RIS: nine domains, 18 views, and 211 elements; and CR: one domain, one view, and 67 elements.
Data Curation
Data were curated by consolidating records for a given element that appeared in multiple sources and/or data were missing. For example, the RIS and EHR had menopausal status. The RIS element was the most complete, thus used when available, but when empty, we implemented a classification strategy to establish menopausal status at the time of each imaging examination. The order of analysis was used as a hierarchical decision tool with the first considered most robust and the 6th the least robust. When elements conflicted, related fields were searched to attempt to establish truth. Occasionally, menopausal status was recorded as postmenopausal then later as premenopausal. In this situation, the related RIS element last menstrual period was reviewed and if blank, then EHR drug lists were used to determine if perhaps the patient was on birth control (implying premenopausal status) or prescribed drugs used for cancer treatment in some postmenopausal women (eg, aromatase inhibitors). Thus, all records except 39,123/2,967,364 (1.3%) were assigned premenopausal or postmenopausal with the residual labeled as perimenopausal. Table 2 lists steps and results.
TABLE 2.
Menopausal Status
| Category Hierarchy | Records | Premenopausal | Postmenopausal | Source View | Source Field |
|---|---|---|---|---|---|
| Declared age of menopause | 1,258,195 | 71,327 | 1,186,868 | Hormonal mensa | Age menopause |
| Date of last period | 241,985 | 136,131 | 105,854 | Hormonal mensa | Last menstrual date |
| Listed status in medical record | 302,590 | 161,464 | 141,126 | Hormonal mensa | Menstrual status |
| Medication indicates statusb | 3,448 | 2,009 | 1,439 | Hormonal mensa | Entry name with age of use |
| Age at examination—younger than 46 or older than 54 years | 49,066 | 25,349 | 23,717 | Patient demographics | Date of birth |
| Unknown treated as perimenopausal | 39,123 | NA | NA | NA | NA |
Abbreviation: NA, not applicable.
Hormonal treatments and menstrual data.
Premenopausal if tamoxifen or raloxifene in medication list at the time of examination. Postmenopausal if aromatase inhibitors in medication list at the time of examination.
DISCUSSION
This effort builds on earlier proof-of-concept work in which we matched RIS and CR records of 1,316 patients with breast cancer.8 Although CDC, SEER, BCSC, CoC, and insurance databases such as Optimum each maintain some elements of the patient record, none contain the detail and breadth described herein. By linking patient information from the EHR, CR, RIS, PD, and MD, we created a breast cancer data mart that exists within our organizational RDW. This robust mart has advantages over other existing marts because information on breast imaging procedures and diagnoses are coupled with patient clinical data, treatments, and outcomes.
A strength of real-world data is that they include the source systems with every detail of the medical record. Therefore, evolving treatments, imaging modalities, etc. can be captured at the patient and encounter level to observe outcomes effects and answer important clinical questions. For example, racial disparities in outcomes can be examined at a more granular level and risk assessment can be modeled using a variety of data. In radiology, questions regarding the frequency and modality of screening and the ideal age to start and stop screening can be examined in terms of patient outcomes and economic cost-benefits. In pathology, rare breast cancer subtypes can be extracted for study of the clinical-pathologic features and outcomes to better define these entities. One could also extract granular pathology information including semiquantitative receptor data and automatically compute multivariable models such as Magee Equations to assess clinical outcomes. In breast surgical oncology, the effect of aggressive surgical intervention versus de-escalation could be carefully studied. For example, identification of populations for which surgery or axillary staging could be safely omitted might be possible, including in those receiving neoadjuvant therapies. These can be a cost-effective way of providing outstanding patient care, which can be of interest to many integrated health systems.
Although significant standard structure exists in clinical data sources on the basis of existing standards (eg, HL-7, NAACCR) and federal initiatives (eg, ONC), interpretation of standards, implementation of optional components, robust methodology to establish truth when conflicting source data are present, and a vast amount of information existing as free text all pose future challenges to navigate. For example, because our institution is within an NPCR state, the CR collects all NAACCR-required elements but not all additional SEER-required elements. For example, SEER but not NPCR requires the NAACCR element PR summary. The information is in our pathology data source as free text, thus can be identified using word search or NLP and extracted into the mart.
There are three published reports of breast cancer marts. Nelson and Weerasinghe9 collected data from 2008 to 2011, accumulating 250,968 mammograms for a quality improvement project. Two more recent efforts were created to facilitate research using artificial intelligence. GENERATOR supports breast cancer pathways of care at Gemelli University Hospital in Rome, Italy.10 This mart does not include RIS information, so it is not possible to determine the effects of imaging history or method of cancer detection on patient outcome. The Diagnosis Data Archive at Salah Azaiez University Hospital in Tunisia does contain mode of detection.11 The status of that effort is unclear as no detailed information, such as the number of cases, is reported yet.
Future expansion to include all images, CR from every facility, and financial and genetics information is planned. Given the amount of unstructured clinical data (eg, provider notes, procedural notes, and pathology reports) available across source systems, future work will include incorporating more unstructured data into the mart. This could be accomplished using a tool, such as MetaMap,12 to recognize clinical concepts in unstructured data and map these concepts to the UMLS Metathesaurus.13 Such an approach will allow for the codification of unstructured data, potentially expanding the scope and amount of data in the mart and aiding in the data curation. The size of our mart facilitates use of artificial intelligence, in particular deep learning, to discover new knowledge such as identifying imaging biomarkers to predict tumor response to different treatments.
We experienced several unexpected challenges. Our RIS is based on an older software program, which is harder to mine. We overcame this through a series of meetings between the RDW and RIS leaders to understand data structure details and how to best integrate them. The CR is a network of facility-level registries that migrated to the METRIQ platform at various times over the past several decades. We chose to focus, in this trial analysis, on our primary breast cancer facility as it was comprehensive of the date range and a test for incorporation of the CR more comprehensively into the warehouse. This restriction reduced available cancers for initial analysis to 39% total in the organization.
A limitation of this work is that it comes from a single health care enterprise. Our patient population does not necessarily reflect the distribution of patients across the country in terms of ethnicity, race, and social determinates of health. Nevertheless, our institution is a mixture of academic and community facilities in rural, suburban, and urban settings. It is our hope that our mart will inform other health care enterprises to develop their own mart, and as such, consensus might be achieved on elements for inclusion, thereby facilitating a collection of data marts, which, in turn, may support existing established and burgeoning national endeavors such as NCI's SEER and BOLD, so that real-world data can be studied to inform clinical decisions and national standards. This effort represents only initial internal steps. Much additional work and collaboration is needed within and across institutions and organizations to accomplish such lofty goals.
To understand real-world benefits, efficiencies and harms of breast cancer screening and treatment in the United States, patient-level linkage of demographic data, imaging history, and results with treatment, cost, and outcomes is needed. Institutions have this information disparately located in many databases. We have demonstrated the creation of a curated data set from disparately located clinical sources into a mart is possible, and we have given a detailed description that should enable any institution to replicate our data mart using their own electronic medical records. Institutional marts may play an important role eventually in understanding real-world outcomes for breast care.
ACKNOWLEDGMENT
The authors thank Jennifer Stalder, Amy Klym, Sharon Winters, Tom Matus, and Sushil Beriwal, MD.
APPENDIX
TABLE A1.
Cancer Registry Data Elements
| Object | Source Table | Data Mart Column | Source Column | Comments |
|---|---|---|---|---|
| Clinical findings CLINICAL_FINDINGS_IE_VW, CLINICAL_FINDINGS_IE_CNTRL_VW |
IMG_Clinical_Finding_IE | PATIENT_STUDY_ID | PERSON_ID | Research identifier of patient |
| Accession_Number | ACCESSION_NUMBER | |||
| Internat_Exam_ID | INTERNAL_EXAM_ID | |||
| Patient_ID | Patient_ID | |||
| Idxrad_Exam_ID | IDXRAD_EXAM_ID | |||
| Clinical_Finding_DE | CLINICAL_FINDING_DE | |||
| Clinical_Finding_CD | CLINICAL_FINDING_CD | |||
| Clinical_Finding_Name | CLINICAL_FINDING_NAME | |||
| Exam_Completed_Date | COMPLETED_DATE | ‘MM/DD/YYYY’ | ||
| Diagnosis DIAGNOSIS_VW, DIAGNOSIS_CNTRL_VW |
Diagnosis | PATIENT_STUDY_ID | PERSON_ID | Research identifier of patient |
| Diagnosis_Code | DX_CODE | |||
| Diagnosis_Type | DX_CODE_REF.CODE_TYPE | Reference lookup for dx code type | ||
| Diagnosis_Name | DX_CODE_REF.CODE_DESCRIPTION | Reference lookup for dx code name | ||
| Diagnosis_From_Date | DX_FROM_DATE | Begin encounter level assignment of DX | ||
| ‘MM/DD/YYYY’ | ||||
| Diagnosis_To_Date | DX_TO_DATE | End encounter level assignment of DX | ||
| ‘MM/DD/YYYY’ | ||||
| Primary_Diagnosis_YN | PRIMARY_DX_IND | ‘Y’ or ‘N’ | ||
| Discharge summary DISCHARGE_SUMMARY_VW, DISCHARGE_SUMMARY_CNTRL_VW |
Enc_Notes, Enc_Notes_Text | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| VISIT.HOSP_ADM_DATE | ||||
| Admission_Date | ‘MM/DD/YYYY HH24:MI:SS’ | |||
| VISIT.START_DATE | ||||
| Discharge_Date | VISIT.HOSP_DISCHG_DATE | ‘MM/DD/YYYY HH24:MI:SS’ | ||
| VISIT.END_DATE | ||||
| Note_Contact_Date | CONTACT_DATE | ‘MM/DD/YYYY’ | ||
| Note_CSN_ID | NOTE_CSN_ID | Unique note identifier | ||
| Line_Num | LINE_NUM | |||
| Note_Text | NOTE_TEXT | |||
| Encounter ENCOUNTER_VW, ENCOUNTER_CNTRL_VW |
Visit | Patient_Study_ID | PERONS_ID | Research identifier of patient |
| Visit_Start_Date | START_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Vist_End_Date | END_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Encounter_Type | CODE_REF.CODE_TITLE | Reference lookup for ‘ENC_TYPE’ | ||
| Facility | CODE_REF.CODE_TITLE | Reference lookup for ‘LOCATION’ | ||
| Appointment_Status | CODE_REF.CODE_TITLE | Reference lookup for ‘APPT_STATUS’ | ||
| Admit_Source | CODE_REF.CODE_TITLE | Reference lookup for ‘ADMIT_TYPE’ | ||
| Hospital_Service | CODE_REF.CODE_TITLE | Reference lookup for ‘HOSPITAL_SERVICE’ | ||
| Patient_Type | CODE_REF.CODE_TITLE | Reference lookup for ‘PATIENT_TYPE’ | ||
| Patient_Class | CODE_REF.CODE_TITLE | Reference lookup for ‘PATIENT_CLASS’ | ||
| Chief_Complaint | CHIEF_COMPLAINT | Free text data capture | ||
| Chief_Complaint_Onset_Date | CHIEF_COMPLAINT_ONSET_DATE | ‘MM/DD/YYYY’ | ||
| Discharge_Disposition | CODE_REF.CODE_TITLE | Reference lookup for ‘DISCHARGE_DISP’ | ||
| Financial_Class | CODE_REF.CODE_TITLE | Reference lookup for ‘FIN_CLASS’ | ||
| Enter edit findings ENTEREDIT_FINDINGS_VW, ENTEREDIT_FINDINGS_CNTRL_VW |
IMG_EnterEdit_Findings | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Accession_Number | ACCESSION_NUMBER | |||
| Org_Code | ORG_CODE | |||
| Internal_Exam_ID | INTERNAL_EXAM_ID | |||
| Patient_ID | PATIENT_ID | Imagecast Patient Identifier | ||
| Composition_Name | COMPOSITION_NAME | |||
| Finding_Location | FINDING_LOCATION | |||
| Finding_Category | FINDING_CATEGORY | |||
| Finding_Rec | FINDING_REC | |||
| Exam_Complete_Date | COMPLETED_DATE | MM/DD/YYYY | ||
| Family history AMILY_HX_VW, FAMILY_HX_CNTRL_VW |
Family_HX | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Line_Num | LINE_NUM | Identify each line in history | ||
| Contact_Date | CONTACT_DATE | ‘MM/DD/YYYY’ | ||
| Medical_HX_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘MEDICAL_HX’ | ||
| Relation_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘RELATION’ | ||
| Hormonal mens HORMONAL_MENS_VW, HORMONAL_MENS_CNTRL_VW |
IMG_Hormonal_Mens | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Accession_Number | ACCESSION_NUMBER | |||
| Internal_Exam_ID | INTERNAL_EXAM_ID | |||
| Patient_ID | PATIENT_ID | |||
| Entry_Name | ENTRY_NAME | |||
| Age_First_Use | AGE_FIRST_USE | |||
| Age_Last_Use | AGE_LAST_USE | |||
| Duration | DURATION | |||
| Current_Use_IND | CURRENT_USE_IND | |||
| Never_Use_IND | NEVER_USE_IND | |||
| Age_Menarche | AGE_MENARCHE | |||
| Age_First_Live_Birth | AGE_FIRST_LIVE_BIRTH | |||
| Age_Menopause | AGE_MENOPAUSE | |||
| Age_Hysterectomy | AGE_HYSTERECTOMY | |||
| Age_Right_Ovary_Removal | AGE_RIGHT_OVARY_REMOVAL | |||
| Age_Left_Ovary_Removal | AGE_LEFT_OVARY_REMOVAL | |||
| Parity_Count | PARITY_COUNT | |||
| Pregnancy_Count | PREGNANCY_COUNT | |||
| Last_Menstrual_Date | LAST_MENSTRUAL_DATE | ‘MM/DD/YYYY’ | ||
| Cycle_Phase | CYCLE_PHASE | |||
| Pregnant_IND | PREGNANT_IND | |||
| Menstrual_Status_CD | MENSTRUAL_STATUS_CD | |||
| Exam_Completed_Date | COMPLETED_DATE | ‘MM/DD/YYYY’ | ||
| IMG procedures IMG_PROCEDURES_VW, IMG_PROCEDURES_CNTRL_VW |
IMG_Procedures | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Accession_Number | ACCESSION_NUMBER | |||
| Internal_Exam_ID | INTERNAL_EXAM_ID | |||
| Patient_ID | PATIENT_ID | |||
| Procedure_Side | PROCEDURE_SIDE | |||
| Procedure_Date | PROCEDURE_DATE | ‘MM/DD/YYYY’ | ||
| Procedure_Outcome | PROCEDURE_OUTCOME | |||
| Implant_Side | IMPLANT_SIDE | |||
| Implant_Type | IMPLANT_TYPE | |||
| Exam_Completed_Date | COMPLETED_DATE | ‘MM/DD/YYYY’ | ||
| Laboratory results LAB_RESULT_VW, LAB_RESULT_CNTRL_VW |
Lab_Result | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Result_Date | RESULT_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Component_Name | COMPONENT_NAME | |||
| Result_Value | ORD_VALUE | |||
| Result_Unit | REFERENCE_UNIT | |||
| Reference_Low | REFERENCE_LOW | |||
| Reference_High | REFERENCE_HIGH | |||
| Lab_Result_Status | CODE_REF.CODE_TITLE | Reference lookup for ‘RESULT_STATUS’ | ||
| Specimen_Collected_Date | SPECIMEN_COLLECTED_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Specimen_Received_Date | SPECIMEN_RECEIVED_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Specimen_Type | CODE_REF.CODE_TITLE | Reference lookup for ‘SPECIMEN_TYPE’ | ||
| Specimen_Source | CODE_REF.CODE_TITLE | Reference lookup for ‘SPECIMEN_SOURCE’ | ||
| Laboratory sensitivity LAB_SENSITIVITY_VW |
Lab_Sensitivity | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Contact_Date | CONTACT_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Result_Date | RESULT_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Organism | ORANISM_NAME | |||
| Antibiotic | CODE_REF.CODE_TITLE | Reference lookup for ‘ANTIBIOTIC’ | ||
| Suscept | CODE_REF.CODE_TITLE | Reference lookup for ‘SUSCEPT’ | ||
| Sensitivity_Value | SENSITIVITY_VALUE | |||
| Sensitivity_Units | SENSITIVITY_UNITS | |||
| Sensitivity_Status | CODE_REF.CODE_TITLE | Reference lookup for ‘RESULT_STATUS’ | ||
| Medication fill MED_FILL_VW, MED_FILL_CNTRL_VW |
Med_Fill | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Filled_Date | FILLED_DATE | ‘MM/DD/YYYY’ | ||
| Drug_Name | DRUG_NAME | |||
| Simple_Generic_Name | SIMPLE_GENERIC_NAME | |||
| Drug_Code_Sys | DRUG_CODE_SYS | |||
| NDC | NDC | |||
| Amount | AMOUNT | |||
| Med_Units | MED_UNIT_TXT | |||
| Med_Unit_Strength | MED_UNIT_STRENGTH | |||
| Quantity | QUANTITY | |||
| Days_Supply | DAYS_SUPPLY | |||
| Medication order MED_ORDER_VW, MED_ORDER_CNTRL_VW |
Med_Order | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Order_Date | ORDER_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Medication_ID | MEDICATION_ID | |||
| Medication | MEDICATION_NAME | |||
| Simple_Generic_Name | CODE_REF.CODE_TITLE | Reference lookup for ‘SIMPLE_GENERIC’ | ||
| Pharm_Class | CODE_REF.CODE_TITLE | Reference lookup for ‘PHARM_CLASS’ | ||
| Dose | DOSE | |||
| Med_Units | MED_UNIT_TITLE | |||
| Quantity | QUANTITY | |||
| Refills | REFILLS | |||
| Start_Date | START_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| End_Date | END_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Admin_Route_Title | ADMIN_ROUTE_TITLE | |||
| Frequency | FREQUENCY | |||
| Instructions | INSTRUCTIONS | |||
| Order result ORDER_RESULT_VW, ORDER_RESULT_CNTRL_VW |
Order_Result | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Order_Procedure_ID | ORDER_PROC_ID | |||
| Result_Date | RESULT_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Component_Name | COMPONENT_NAME | COMPONENT_REF | ||
| Result_Value | ORD_VALUE | |||
| Result_Unit | REFERENCE_UNIT | |||
| Reference_Low | REFERENCE_LOW | |||
| Reference_High | REFERENCE_HIGH | |||
| Result_Flag_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘RESULT_FLAG’ | ||
| Result_Status | CODE_REF.CODE_TITLE | Reference lookup for ‘RESULT_STATUS’ | ||
| Specimen_Collected_Date | SPECIMEN_COLLECTED_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Specimen_Received_Date | SPECIMEN_RECEIVED_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Patient data IE PATIENT_DATA_IE_VW, PATIENT_DATA_IE_CNTRL_VW |
IMG_Patient_Data_IE | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Order_Procedure_ID | ACCESSION_NUMBER | |||
| Result_Date | ORG_CODE | |||
| Component_Name | ORG_NAME | |||
| Result_Value | EXAM_CODE | |||
| Result_Unit | EXAM_NAME | |||
| Reference_Low | EXAM_MODIFIER | |||
| Reference_High | EXAM_STATUS | |||
| Result_Flag_Title | SCHEDULED_DATE | ‘MM/DD/YYYY’ | ||
| Result_Status | COMPLETED_DATE | ‘MM/DD/YYYY’ | ||
| Specimen_Collected_Date | FINALIZED_DATE | ‘MM/DD/YYYY’ | ||
| Specimen_Received_Date | BEGIN_DATE | ‘MM/DD/YYYY’ | ||
| Order_Date | ORDER_DATE | ‘MM/DD/YYYY’ | ||
| Exam_Type_ID | EXAM_TYPE_ID | |||
| Exam_Side | EXAM_SIDE | |||
| Exam_Requestor | EXAM_REQUESTOR | |||
| Exam_Type | EXAM_TYPE | |||
| Self_Request_Ind | SELF_REQUEST_IND | |||
| Equipment_Clean_Ind | EQUIPMENT_CLEAN_IND | |||
| First_Mammo_Ind | FIRST_MAMMO_IND | |||
| Last_Screen_Month_CD | LAST_SCREEN_MONTH_CD | |||
| Last_CBE_CD | LAST_CBE_CD | |||
| Last_CBE_Date | LAST_CBE_DATE | ‘MM/DD/YYYY’ | ||
| Last_Activity_Date | LAST_ACTIVITY_DATE | ‘MM/DD/YYYY’ | ||
| Internal_Exam_ID | INTERNAL_EXAM_ID | |||
| Patient_ID | PATIENT_ID | |||
| Patient demographics PATIENT_DEMO_VW, PATIENT_DEMO_CNTRL_VW |
Patient_Demographic | Person_ID | PATIENT_STUDY_ID | Research identifier of patient ‘MM/DD/YYYY’ default to July 1st of birth year |
| Birth_Date | BIRTH_DATE | ‘MM/DD/YYYY’ | ||
| Death_Date | DEATH_DATE | Reference lookup for ‘GENDER’ | ||
| Gender_Title Race_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘RACE’ | ||
| Ethnic_Title | CODE_REF.CODE_TITLE CODE_REF.CODE_TITLE | Reference lookup for ‘ETHNIC_GROUP' | ||
| Pathology PATHOLOGY_VW, PATHOLOGY_CNTRL_VW |
IMG_Pathology | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Bx_ID | BX_ID | |||
| Pathology_Date | PATHOLOGY_DATE | MM/DD/YYYY | ||
| Patient_ID | PATIENT_ID | |||
| Site_Number | SITE_NUMBER | |||
| Side | SIDE | |||
| Lesion_Class | LESION_CLASS | |||
| Lesion_Location | LESION_LOCATION | |||
| Technique | TECHNIQUE | |||
| Comments | COMMENTS | |||
| Pathology findings PATHOLOGY_FINDINGS_VW, PATHOLOGY_FINDINGS_CNTRL_VW |
IMG_Pathology_Findings | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Bx_ID | BX_ID | |||
| Pathology_Date | PATHOLOGY_DATE | MM/DD/YYYY | ||
| Patient_ID | PATIENT_ID | |||
| Finding_ID | FINDING_ID | |||
| Pathology_CD | PATHOLOGY_CD | |||
| Lesion_Class | LESION_CLASS | |||
| Finding_Size_1 | FINDING_SIZE_1 | |||
| Finding_Size_2 | FINDING_SIZE_2 | |||
| Measurement_Type | MEASUREMENT_TYPE | |||
| Histrology_Grade | HISTROLOGY_GRADE | |||
| Nodes_Removed | NODES_REMOVED | |||
| Nodes_Positive | NODES_POSITIVE | |||
| Margin_Status | MARGIN_STATUS | |||
| Estrogen_Receptor | ESTROGEN_RECEPTOR | |||
| Progesterone_Receptor | PROGESTERONE_RECEPTOR | |||
| HER2NEU | HER2NEU | |||
| Stage_T | STAGE_T | |||
| Stage_N | STAGE_N | |||
| Stage_M | STAGE_M | |||
| Stage_Num | STAGE_NUM | |||
| Nipples_Involved | NIPPLES_INVOLVED | |||
| Problem list PROBLEM_LIST_VW, PROBLEM_LIST_CNTRL_VW |
Problem_List | Patient_Study_ID | PERSON_ID | Research Identifier of Patient |
| Diagnosis_Code | DX_CODE | |||
| Diagnosis_Type | DX_CODE_TYPE | |||
| Diagnosis_Name | DX_NAME | |||
| Reported_Date | REPORTED_DATE | ‘MM/DD/YYYY’ | ||
| Onset_Date | ONSET_DATE | ‘MM/DD/YYYY’ | ||
| Resolved_Date | RESOLVED_DATE | ‘MM/DD/YYYY’ | ||
| Resovled_Reason | RESOLVED_REASON | |||
| Status_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘PROBLEM_STATUS’ | ||
| Procedures PROCEDURES_VW, PROCEDURES_CNTRL_VW |
Procedures | PATIENT_STUDY_ID | PERSON_ID | Research identifier of patient Order listed sequence finding first not null value: |
| Procedure_Date | RESULT_DATE, PROC_DATE, ORDER_DATE | ‘MM/DD/YYYY’ | ||
| Procedure_Code | PROC_CODE | |||
| Procedure_Type | PROC_CODE_REF.CODE_TYPE | |||
| Procedure_Name | PROC_CODE_REF.CODE_DESCRIPTION | |||
| Procedure_Location | PROCEDURE_LOCATION | |||
| Order_Date | ORDER_DATE | ‘MM/DD/YYYY’ | ||
| Procedure notes PROCEDURES_NOTES_VW, PROCEDURES_NOTES_CNTRL_VW |
Proc_Notes, Proc_Notes_Text | PATIENT_STUDY_ID | PERSON_ID | Research identifier of patient |
| Order_Date | ORDER_DATE | ‘MM/DD/YYYY’ | ||
| Result_Time | RESULT_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Procedure_Code | PROC_CODE | |||
| Procedure_Name | CODE_DESCRIPTION | PROC_CODE_REF | ||
| Note_CSN_ID | NOTE_CSN_ID | |||
| Line_Num | LINE_NUM | |||
| Note_Text | NOTE_TEXT | |||
| Recommendations RECOMMENDATIONS_VW, RECOMMENDATIONS_CNTRL_VW |
IMG_Recommendation_IE | PATIENT_STUDY_ID | PERSON_ID | Research identifier of patient |
| Accession_Number | ACCESSION_NUMBER | |||
| Internal_Exam_id | INTERNAL_EXAM_ID | |||
| Patient_ID | PATIENT_ID | |||
| Recommend_ID | RECOMMEND_ID | |||
| Recommend_CD | RECOMMEND_CD | |||
| Recommend_Status | RECOMMEND_STATUS | |||
| Recommend_Side | RECOMMEND_SIDE | |||
| Due_Date | DUE_DATE | ‘MM/DD/YYYY’ | ||
| Exam_Completed_Date | COMPLETED_DATE | ‘MM/DD/YYYY’ | ||
| Risk factors RISK_FACTORS_VW, RISK_FACTORS_CNTRL_VW |
IMG_Risk_Factors | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Accession_Number | ACCESSION_NUMBER | |||
| Internal_Exam_id | INTERNAL_EXAM_ID | |||
| Patient_ID | PATIENT_ID | |||
| Risk_Factor_CD | RISK_FACTOR_CD | |||
| Risk_Factor_Name | RISK_FACTOR_NAME | |||
| Risk_Sequence | RISK_SEQUENCE | |||
| Relationship_CD | RELATIONSHIP_CD | |||
| Problem_CD | PROBLEM_CD | |||
| Person_Age | PERSON_AGE | |||
| Comments | COMMENTS | |||
| Exam_Completed_Date | COMPLETED_DATE | ‘MM/DD/YYYY’ | ||
| Social history alcohol SOCIAL_HX_ALC_VW, SOCIAL_HX_ALC_CNTRL_VW |
Social_HX_Alc | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Contact_Date | CONTACT_DATE | ‘MM/DD/YYYY’ | ||
| Drinks_Per_Week | DRINKS_PER_WEEK | |||
| Drink_Type_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘DRINK_TYPE’ | ||
| Social history tobacco SOCIAL_HX_TOB_VW, SOCIAL_HX_TOB_CNTRL_VW |
Social_HX_Tob | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Contact_Date | CONTACT_DATE | ‘MM/DD/YYYY’ | ||
| Smoking_Use_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘SMOKE_TOB_USE’ | ||
| Tobacco_Use_Per_Day | TOBACCO_PAK_PER_DAY | |||
| Tobacco_Use_Years | TOBACCO_USED_YEARS | |||
| Translate ‘Y’ to ‘Yes’ | ||||
| Cigarettes_Indicator | CIGARETTES_IND | Translate ‘N’ to ‘No’ | ||
| Otherwise NULL | ||||
| Translate ‘Y’ to ‘Yes’ | ||||
| Pipes_Indicator | PIPES_IND | Translate ‘N’ to ‘No’ | ||
| Otherwise NULL | ||||
| Translate ‘Y’ to ‘Yes’ | ||||
| Cigars_Indicator | CIGARS_IND | Translate ‘N’ to ‘No’ | ||
| Otherwise NULL | ||||
| Translate ‘Y’ to ‘Yes’ | ||||
| Snuff_Indicator | SNUFF_IND | Translate ‘N’ to ‘No’ | ||
| Otherwise NULL | ||||
| Translate ‘Y’ to ‘Yes’ | ||||
| Chew_Indicator | CHEW_IND | Translate ‘N’ to ‘No’ | ||
| Otherwise NULL | ||||
| Smokeless_Tob_Use_Title | CODE_REF.CODE_TITLE | Reference lookup for ‘SMOKELESS_TOB_USE’ | ||
| Surgical pathology SURGICAL_PATH_NOTES_VW, Notes SURGICAL_PATH_NOTES_CNTRL_VW |
Proc_Notes, Proc_Notes_Text | PATIENT_STUDY_ID | PERSON_ID | Research identifier of patient |
| Order_Date | ORDER_DATE | ‘MM/DD/YYYY’ | ||
| Result_Time | RESULT_DATE | ‘MM/DD/YYYY HH:MI:SS’ | ||
| Procedure_Code | PROC_CODE | |||
| Procedure_Name | CODE_DESCRIPTION | PROC_CODE_REF | ||
| Note_CSN_ID | NOTE_CSN_ID | |||
| Line_Num | LINE_NUM | |||
| Note_Text | NOTE_TEXT | |||
| Vitals VITALS_VW, VITALS_CNTRL_VW |
Vitals | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| Visit_ID | VISIT_ID | |||
| Weight | WEIGHT | |||
| Weight_Unit | WEIGHT_UNIT | |||
| Height | HEIGHT | |||
| Height_In | HEIGHT_IN | |||
| Height_Unit | HEIGHT_UNIT | |||
| Date_Taken | CONTACT_DATE | |||
| BMI | BMI | |||
| Patient cancer registry PATIENT_CANCER_REGISTRY_VW |
BCC_Cancer_Registry | Patient_Study_ID | PERSON_ID | Research identifier of patient |
| HOSPITAL_ID | HOSPITAL_ID | |||
| SEQUENCE_NUMBER | SEQUENCE_NUMBER | |||
| CLASS_OF_CASE | CLASS_OF_CASE | |||
| CASE_STATUS | CASE_STATUS | |||
| DATE_OF_BIRTH | DATE_OF_BIRTH | Default to July 1st of birth year | ||
| SEX | SEX | |||
| SEX_DESC | SEX_DESC | |||
| RACE_1 | RACE_1 | |||
| RACE_1_DESC | RACE_1_DESC | |||
| SPANISH_HISPANIC_ORIGIN | SPANISH_HISPANIC_ORIGIN | |||
| SPANISH_HISP_ORIG_DESC | SPANISH_HISP_ORIG_DESC | |||
| PRIMARY_PAYOR_DX_DESC | PRIMARY_PAYOR_DX_DESC | |||
| POSTAL_CODE_AT_DX | POSTAL_CODE_AT_DX | |||
| COUNTY_AT_DX | COUNTY_AT_DX | |||
| PRIMARY_SITE | PRIMARY_SITE | |||
| PRIMARY_SITE_DESC | PRIMARY_SITE_DESC | |||
| LATERALITY_DESC | LATERALITY_DESC | |||
| HISTO_BEHAVE_ICDO3 | HISTO_BEHAVE_ICDO3 | |||
| HISTO_BEHAVE_ICDO3_DESC | HISTO_BEHAVE_ICDO3_DESC | |||
| HISTO_BEHAVE_ICDO3_DESC | HISTO_BEHAVE_ICDO3_DESC | |||
| CORRECTED_BEST_STAGE | CORRECTED_BEST_STAGE | |||
| DATE_1ST_CONTACT | DATE_1ST_CONTACT | |||
| COURSE_DATE_1ST_DX | COURSE_DATE_1ST_DX | |||
| COURSE_1ST_DXSTG_PROCFAC_DESC | COURSE_1ST_DXSTG_PROCFAC_DESC | |||
| COURSE_1ST_DXSTG_PROCSUM_DESC | COURSE_1ST_DXSTG_PROCSUM_DESC | |||
| CS_SITE_SPEC_FACTOR_1 | CS_SITE_SPEC_FACTOR_1 | |||
| CS_SITE_SPEC_FACTOR_2 | CS_SITE_SPEC_FACTOR_2 | |||
| CS_SITE_SPEC_FACTOR_15 | CS_SITE_SPEC_FACTOR_15 | |||
| AJCC_CLINICAL_T_DESC | AJCC_CLINICAL_T_DESC | |||
| AJCC_CLINICAL_N_DESC | AJCC_CLINICAL_N_DESC | |||
| AJCC_CLINICAL_M_DESC | AJCC_CLINICAL_M_DESC | |||
| REASON_NO_SURGERY_DESC | REASON_NO_SURGERY_DESC | |||
| MOST_DEFINITIVE_SURG_DATE | MOST_DEFINITIVE_SURG_DATE | |||
| COURSE_1ST_SURG_PRIMSITE_DESC | COURSE_1ST_SURG_PRIMSITE_DESC | |||
| COURSE_1ST_SURG_PRIMSUMM_DESC | COURSE_1ST_SURG_PRIMSUMM_DESC | |||
| COURSE_1ST_SCOPE_LN_SURG_DESC | COURSE_1ST_SCOPE_LN_SURG_DESC | |||
| REGIONAL_NODES_EXAM | REGIONAL_NODES_EXAM | |||
| REGIONAL_NODES_POSITIVE | REGIONAL_NODES_POSITIVE | |||
| COURSE_1ST_SURG_MARG_DESC | COURSE_1ST_SURG_MARG_DESC | |||
| REASON_NO_CHEMO_DESC | REASON_NO_CHEMO_DESC | |||
| COURSE_1ST_CHEMO_DATE | COURSE_1ST_CHEMO_DATE | |||
| COURSE_1ST_CHEMO_FAC_DESC | COURSE_1ST_CHEMO_FAC_DESC | |||
| COURSE_1ST_CHEMO_SUMM_DESC | COURSE_1ST_CHEMO_SUMM_DESC | |||
| REASON_NO_HORMONE_THERAPY | REASON_NO_HORMONE_THERAPY | |||
| COURSE_1ST_HORMONE_RX_DATE | COURSE_1ST_HORMONE_RX_DATE | |||
| COURSE_1ST_HORMONE_FAC_DESC | COURSE_1ST_HORMONE_FAC_DESC | |||
| COURSE_1ST_HORMONE_RX_SUMM | COURSE_1ST_HORMONE_RX_SUMM | |||
| MEDICAL_ONCOLOGY_PHYS_NPI | MEDICAL_ONCOLOGY_PHYS_NPI | |||
| MEDICAL_ONC_PHYS_LASTNAME | MEDICAL_ONC_PHYS_LASTNAME | |||
| MEDICAL_ONC_PHYS_FIRSTNAME | MEDICAL_ONC_PHYS_FIRSTNAME | |||
| REASON_NO_RADIATION_DESC | REASON_NO_RADIATION_DESC | |||
| DATE_RT_STARTED | DATE_RT_STARTED | |||
| DATE_RT_ENDED | DATE_RT_ENDED | |||
| COURSE_1ST_RADIATION_FAC | COURSE_1ST_RADIATION_FAC | |||
| COURSE_1ST_RT_MODALITY_SUMM | COURSE_1ST_RT_MODALITY_SUMM | |||
| COURSE_1ST_RT_VOLUME_SUMM | COURSE_1ST_RT_VOLUME_SUMM | |||
| COURSE_1ST_RT_REG_DOSE_SUMM | COURSE_1ST_RT_REG_DOSE_SUMM | |||
| COURSE_1ST_RX_SUMMARY | COURSE_1ST_RX_SUMMARY | |||
| RADIATION_ONC_PHYS_NPI | RADIATION_ONC_PHYS_NPI | |||
| RADIATION_ONC_PHYS_LASTNAME | RADIATION_ONC_PHYS_LASTNAME | |||
| RADIATION_ONC_PHYS_FIRSTNAME | RADIATION_ONC_PHYS_FIRSTNAME | |||
| CHEMOTHERAPY_TEXT | CHEMOTHERAPY_TEXT | |||
| DATE_LAST_CONTACT | DATE_LAST_CONTACT | |||
| VITAL_STATUS | VITAL_STATUS | |||
| CANCER_STATUS_DESC | CANCER_STATUS_DESC | |||
| DATE_1ST_RECURRENCE | DATE_1ST_RECURRENCE | |||
| TYPE_1ST_RECURRENCE_DESC | TYPE_1ST_RECURRENCE_DESC |
SUPPORT
Supported by National Institutes of Health (P30CA047904).
AUTHOR CONTRIBUTIONS
Conception and design: Margarita L. Zuley, Jonathan Silverstein, Robert M. Nishikawa
Provision of study materials or patients: Jonathan Silverstein, Adam M. Brufsky
Collection and assembly of data: Margarita L. Zuley, Jonathan Silverstein, Durwin Logue, Richard S. Morgan, Adam M. Brufsky, Robert M. Nishikawa
Data analysis and interpretation: Margarita L. Zuley, Durwin Logue, Rohit Bhargava, Priscilla F. McAuliffe, Adam M. Brufsky, Andriy I. Bandos, Robert M. Nishikawa
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Margarita L. Zuley
Employment: North American Dental Partners
Leadership: North American Dental Partners
Stock and Other Ownership Interests: Pfizer, AstraZeneca/Merck
Consulting or Advisory Role: Bayer Health, Bracco Diagnostics
Speakers' Bureau: Applied Radiology
Research Funding: Hologic (Inst), Hologic (Inst), Bayer Health (Inst)
Rohit Bhargava
Consulting or Advisory Role: GE Healthcare
Speakers' Bureau: AstraZeneca
Adam M. Brufsky
Consulting or Advisory Role: Pfizer, Genentech/Roche, Agendia, Novartis, Lilly, Puma Biotechnology, Merck, Myriad Pharmaceuticals, Eisai, Seagen, Daiichi Sankyo/Lilly, Onc Live, Michael J. Hennessy Associates, Gilead Sciences, General Electric
Research Funding: Roche/Genentech (Inst), AstraZeneca/Daiichi Sankyo (Inst), Merck (Inst), Novartis (Inst), Gilead Sciences (Inst), Lilly (Inst), Puma Biotechnology (Inst)
Expert Testimony: Pfizer
Andriy I. Bandos
Consulting or Advisory Role: Hologic
Robert M. Nishikawa
Research Funding: Hologic, iCAD, Inc
Patents, Royalties, Other Intellectual Property: Royalties from patents that have been licensed to Hologic Inc
Uncompensated Relationships: MaidData, Corp, iCad, Inc
No other potential conflicts of interest were reported.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. : Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209-249, 2021 [DOI] [PubMed] [Google Scholar]
- 2.Chen S, Cao Z, Prettner K, et al. : Estimates and projections of the global economic cost of 29 cancers in 204 countries and territories from 2020 to 2050. JAMA Oncol 9:465-472, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Cancer Institute : Cancer Trends Progress Report. Financial Burden of Cancer Care. https://progressreport.cancer.gov/after/economic_burden [Google Scholar]
- 4.Siegel RL, Miller KD, Wagle NS, et al. : Cancer statistics, 2023. CA Cancer J Clin 73:17-48, 2023 [DOI] [PubMed] [Google Scholar]
- 5.US Preventive Services Task Force : Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 151:716-236, 2009. W-236 [DOI] [PubMed] [Google Scholar]
- 6.Siu AL, US Preventive Services Task Force : Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 164:279-296, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Visweswaran S, McLay B, Cappella N, et al. : An atomic approach to the design and implementation of a research data warehouse. J Am Med Inform Assoc 29:601-608, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuley ML, Nishikawa RM, Lee CS, et al. : Linkage of the ACR National Mammography Database to the Network of State Cancer Registries: Proof of concept evaluation by the ACR National Mammography Database Committee. J Am Coll Radiol 16:8–14, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson HD, Weerasinghe R: Actualizing personalized healthcare for women through connected data systems: Breast cancer screening and diagnosis. Glob Adv Health Med 2:30-36, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marazzi F, Tagliaferri L, Masiello V, et al. : GENERATOR Breast DataMart-The novel breast cancer data discovery system for research and monitoring: Preliminary results and future perspectives. J Pers Med 11:65, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ammar MB, Ayachi FL, Ksantini R, et al. : Data warehouse for machine learning: Application to breast cancer diagnosis. Proced Comput Sci 196:692-698, 2022 [Google Scholar]
- 12.MetaMap—A Tool for Recognizing UMLS Concepts in Text. https://lhncbc.nlm.nih.gov/ii/tools/MetaMap.html [Google Scholar]
- 13.Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/index.html [Google Scholar]