Abstract
PURPOSE
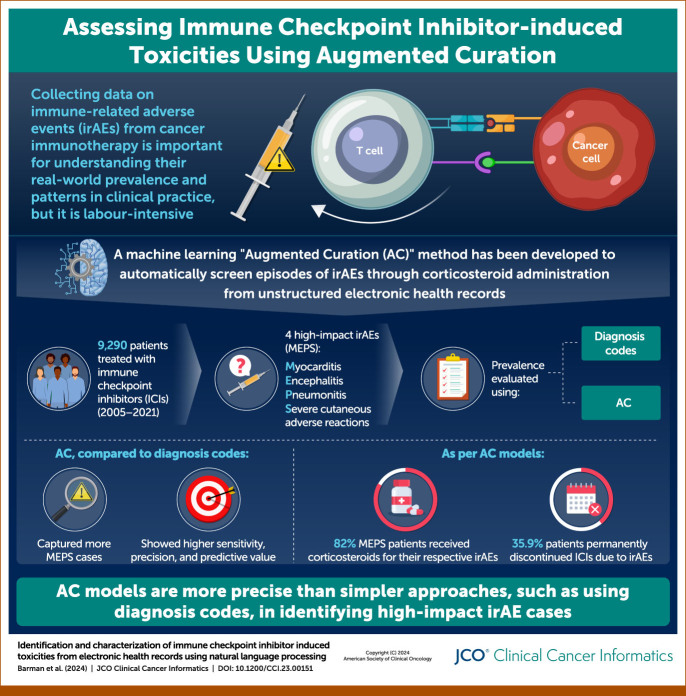
Immune checkpoint inhibitors (ICIs) have revolutionized cancer treatment, yet their use is associated with immune-related adverse events (irAEs). Estimating the prevalence and patient impact of these irAEs in the real-world data setting is critical for characterizing the benefit/risk profile of ICI therapies beyond the clinical trial population. Diagnosis codes, such as International Classification of Diseases codes, do not comprehensively illustrate a patient's care journey and offer no insight into drug-irAE causality. This study aims to capture the relationship between ICIs and irAEs more accurately by using augmented curation (AC), a natural language processing–based innovation, on unstructured data in electronic health records.
METHODS
In a cohort of 9,290 patients treated with ICIs at Mayo Clinic from 2005 to 2021, we compared the prevalence of irAEs using diagnosis codes and AC models, which classify drug-irAE pairs in clinical notes with implied textual causality. Four illustrative irAEs with high patient impact—myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions, abbreviated as MEPS—were analyzed using corticosteroid administration and ICI discontinuation as proxies of severity.
RESULTS
For MEPS, only 70% (n = 118) of patients found by AC were also identified by diagnosis codes. Using AC models, patients with MEPS received corticosteroids for their respective irAE 82% of the time and permanently discontinued the ICI because of the irAE 35.9% (n = 115) of the time.
CONCLUSION
Overall, AC models enabled more accurate identification and assessment of patient impact of ICI-induced irAEs not found using diagnosis codes, demonstrating a novel and more efficient strategy to assess real-world clinical outcomes in patients treated with ICIs.
**Authors request we do not post about the article on social media.
INTRODUCTION
Immune checkpoint inhibitors (ICIs) have revolutionized cancer treatment, yet their use is associated with immune-related adverse events (irAEs), some of which lead to treatment discontinuation and are potentially life-threatening.1 Given the selective nature of pivotal clinical trials, it is important to determine the prevalence and to characterize the patient impact of ICI therapy-associated irAEs in real-world settings. Electronic health records (EHRs) represent a rich, longitudinal source of patient health information that includes structured data (eg, diagnosis codes, laboratory values, medication orders) and unstructured free-text (eg, provider notes, pathology reports, admission/discharge summaries). Studies using diagnosis codes and medication orders have aimed to understand the safety and effectiveness of drugs. However, this approach does not comprehensively illustrate the patient care journey, suboptimally captures concurrent medical conditions, and offers no insight into drug-irAE causality.2-5
CONTEXT
Key Objective
To compare electronic health record data extraction methods for identifying immune checkpoint inhibitor (ICI)–associated immune-related adverse events (irAEs) and to use natural language processing (NLP) approaches to assess patient impact of certain irAEs.
Knowledge Generated
NLP models demonstrated advantages compared with the gold standard (manual curation of patient notes) including higher accuracy, detection of additional irAEs not identified using International Classification of Diseases codes, characterizing ICI-irAE causality, and assessment of patient impact of irAEs.
Relevance (F.P.-Y. Lin)
-
Innovative application of NLP on unstructured data within electronic medical records enhances the curation process of significant irAEs related to the real-world use of ICIs. This can support pharmacovigilance, clinical quality assurance, and research related to treatment toxicities with efficiency, accuracy, and at scale.*
*Relevance section written by JCO Clinical Cancer Informatics Associate Editor Frank P.-Y. Lin, PhD, MBChB, FRACP, FAIDH.
The unstructured notes within the EHR provide significant clinical information that is otherwise not captured by structured data. However, manual review of patient notes presents an arduous task for abstractors at scale. Natural language processing (NLP) techniques, such as augmented curation (AC), are effective tools for rapidly and accurately turning free text into structured data for analysis.6-9
The objectives of this retrospective study were twofold: (1) to compare different EHR data extraction methods for identifying ICI-induced irAEs and the treatments used for their management and (2) to understand clinical outcomes and patient impact of immunotherapy-induced myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions (SCAR), abbreviated as MEPS because of their high patient impact across different organ systems.10 Corticosteroid administration and ICI discontinuation were used as proxies of severity. To assess the accuracy of each data extraction method, all deidentified patient clinical records for each MEPS irAE were manually reviewed. Overall, our results support using AC on unstructured EHRs to identify drug-related adverse events (AEs) and treatment accurately, comprehensively, and at scale in patients receiving cancer therapies.
METHODS
Mayo Deidentified Data and Privacy
nference is the exclusive data analytics engine for Mayo Clinic's Clinical Data Analytics Platform. In partnership with Mayo Clinic (Mayo), nference has access to deidentified data, pursuant to an expert determination in accordance with the Health Insurance Portability and Accountability Act Privacy Rule, representing the entire patient population at Mayo (approximately 9.8 million patient lives).11 These data only exist within Mayo's secure cloud environment. To maintain full confidentiality, the results were only reported as aggregate statistics. Roche had access solely to these results.
Study Population
We performed a retrospective analysis of all patients who received ICIs at Mayo as part of usual clinical practice from July 15, 2005 to October 15, 2021. As of 2019, Mayo instituted a specialty immunotherapy clinic for evaluating patients with confirmed or presumed ICI-associated irAEs, where all providers have clinical trials experience, which includes AE assessment and grading. Patients must have received at least one dose of an ICI (Appendix Table A1) or a combination therapy (two or more ICIs administered within ±7 days of one another in the EHR) at Mayo Clinic. Patients with preexisting irAE diagnosis codes were excluded from analyses in addition to patients with only mentions of receiving ICIs, irAEs, or receiving steroids in their clinical notes. All patients were age 18 years or older and had research authorization on file. Among the resultant cohort of 9,290 patients, individuals on average had 1.28 years of follow-up after ICI administration. This study was approved by the Mayo Clinic Institutional Review Board (22-002906).
Methods of Data Acquisition
The two methods of data acquisition to identify 27 potential irAEs (Appendix Table A2) were (1) diagnosis codes (International Classification of Diseases9/10, health insurance claim, Systematized Nomenclature of Medicine) and (2) AC, which uses neural network-based NLP algorithms to classify the sentiment of toxicities experienced from drugs in the unstructured portions of the Mayo EHR, including but not limited to clinical notes and radiology/pathology reports (Fig 1A).7,8 Diagnosis codes were queried between the first record date of ICI administration and the last record date ≥60 days, to ensure that later-onset irAEs were captured, even after the cessation of the medication.
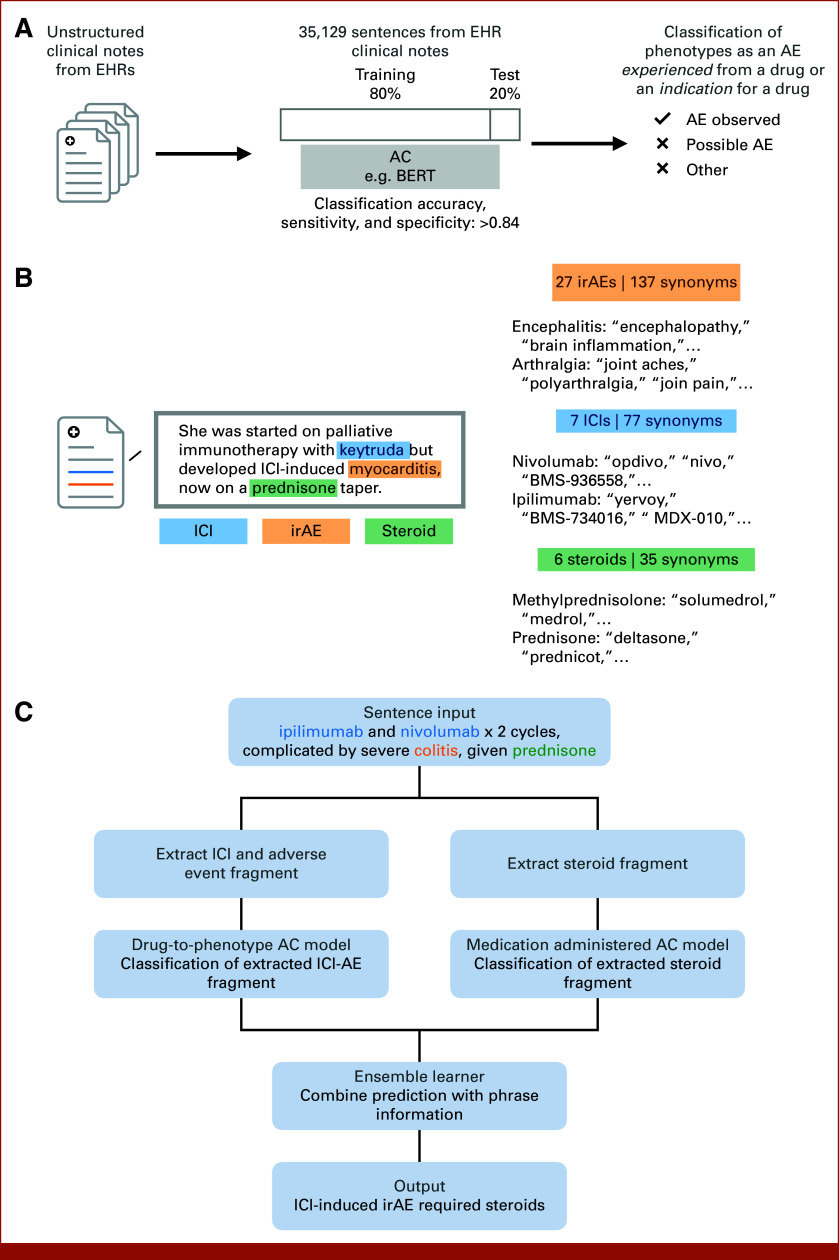
FIG 1.
Overview of study design and methodology. (A) Illustrates the process of AC of the unstructured clinical notes from EHRs. (B) ICI-induced irAE entity recognition, sentiment analysis, and synonym grouping. (C) Description of the ensemble learner approach, parsing sentences into distinct sub-phrases to determine predictions of whether an irAE was caused by an ICI and subsequently treated with a corticosteroid. AC, augmented curation; AE, adverse event; BERT, Bidirectional Encoder Representations from Transformers; EHRs, electronic health records; ICI, immune checkpoint inhibitor; irAE, immune-related adverse event.
Drug-to-Phenotype Model Development
An NLP classification model (drug-to-phenotype model) was developed to determine whether a condition mentioned alongside a drug in the clinical notes was the indication treated by the drug or an adverse event caused by the drug (Fig 1B). Drug indications extracted from the US Food and Drug Administration (FDA) Label database, along with drug-adverse event pairs, obtained from the FDA Adverse Event Reporting System public dashboard, were used to query sentences from clinical notes. These sentences were then labeled and used to fine tune the pretrained SciBERT6 sentence classification model to output one of the following labels: adverse event observed—confirmed toxicity caused by the drug, indication treated—the drug was used to treat the phenotype, other—alternate context (eg, family history of irAE, discussion of research findings, etc). The model was trained on a set of 39,892 sentences from the Mayo EHR using an 80%:20% train:test split. The model achieved an out-of-sample accuracy of 84.8%, with precision and recall values over 85% for both positive and negative sentiment classifications.
Primary Outcome: Classification of irAEs Using the Drug-to-Phenotype Model
For each patient, we applied the drug-to-phenotype model across all eligible notes in our ICI cohort for the seven ICIs and 27 irAEs (Appendix Tables A1 and A2). Patients with at least one note indicating an ICI-induced irAE with an adverse event observed label and a model confidence score of 80% or higher were counted as positive for that irAE.
Manual review of full patient charts was performed on all patients with MEPS detected by either AC and/or diagnosis codes to calculate accuracy. The final MEPS cohort included a set of manually reviewed and confirmed patients from both sources. Performance metric definitions and calculations are detailed in Appendix Table A3. CIs were calculated using 50 bootstrapping runs.
Comparison of ICI-Induced irAEs and Corticosteroid Usage From Different Data Sources and Assessments of Proxies of Severity
After detecting a causal relationship between an ICI and an adverse event, we sought to determine whether the adverse event was treated with a corticosteroid and/or discontinued (Fig 1C). Two methods of data acquisition were used: (1) structured medication orders and (2) an AC approach, specifically a medication administered model. Structured medication orders were pulled from EHR tables containing medications administered while a patient was admitted to the clinic or orders in an outpatient setting. The medication administered model was developed to classify whether a medication mentioned in the clinical notes was received by the patient or simply discussed as a possible therapeutic option. The model was fine-tuned with a set of 13,961 sentences, using an 80%:20% train:test split, and achieved an out-of-sample accuracy of 89%, with precision and recall values over 88% for medication received classification. A full list of corticosteroids and their synonyms are listed in Appendix Table A4. For clinical sentences that only referenced a class of drugs, for example, “patient experienced nivolumab-induced pneumonitis and was treated with corticosteroids,” the general term of corticosteroid was used.
Corticosteroid use for irAEs found using structured medication orders was compared with usage found via the medication administered model, manual review, and visualized using Venn diagrams. Manual chart review for all patients with MEPS was also conducted to determine whether ICI discontinuation was due to MEPS.
Bar charts were created using the software package Seaborn (version 0.12.1), and Venn diagrams were created using the software package Venn (version 0.1.3) in Python.
Statistical Analyses
All study variables, including baseline and outcome measures, were analyzed descriptively. Counts and proportions were provided for categorical variables. Means, standard deviations, and medians were provided for continuous variables.
RESULTS
Table 1 summarizes the demographics and clinical characteristics of the patients in the study cohort who received ICIs at Mayo Clinic (N = 9,290) and the subset who experienced ICI-induced MEPS (n = 314). The median age of patients in both cohorts was 66 years, and most patients (>90%) were White/Caucasian, which is consistent with the overall patient population at Mayo.
TABLE 1.
Patient Demographics
| Demographic | ICI-Treated Cohort at Mayo (N = 9,290) | ICI-Treated MEPS Cohort at Mayo (N = 314) |
|---|---|---|
| Age at first ICI initiation, years | ||
| Mean (SD), median | 64.44 (13.25), 66 | 64.41 (12.80), 66 |
| Sex, No. (%) | ||
| Male | 5,469 (58.9) | 175 (55.6) |
| Female | 3,821 (41.1) | 139 (44.1) |
| Race, No. (%) | ||
| Asian | 173 (1.9) | 2 (0.6) |
| Black or African American | 233 (2.5) | 6 (1.9) |
| Chose not to disclose | 83 (0.9) | 0 (0) |
| Native American/Pacific Islander | 52 (0.6) | 0 (0) |
| Other | 135 (1.5) | 8 (2.5) |
| Unknown | 44 (0.5) | 1 (0.3) |
| White | 8,570 (92.2) | 297 (94.3) |
| Ethnicity, No. (%) | ||
| Hispanic or Latino | 267 (2.9) | 10 (3.2) |
| Not Hispanic or Latino | 8,764 (94.3) | 298 (94.6) |
| Chose not to disclose | 145 (1.6) | 4 (1.3) |
| Unknown | 114 (1.2) | 2 (0.6) |
| Cancer types, No. (%) | ||
| Bladder | 422 (4.5) | 8 (2.5) |
| Breast | 217 (2.3) | 20 (6.4) |
| Colorectal | 187 (2.0) | 4 (1.6) |
| Liver | 254 (2.7) | 6 (1.9) |
| Lung | 1,720 (18.5) | 128 (40.8) |
| Renal | 544 (5.9) | 25 (7.9) |
| Skin | 1,918 (20.6) | 89 (28.3) |
| Other | 1,640 (17.7) | 43(13.7) |
| Unspecified | 2,482 (26.7) | 2 (0.6) |
| Therapy type, No. (%) | ||
| ICI monotherapy | 8,687 (93.51) | 287 (91.4) |
| ICI combination therapy | 786 (8.46) | 39 (12.4) |
Abbreviations: ICI, immune checkpoint inhibitor; MEPS, myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions; SD, standard deviation.
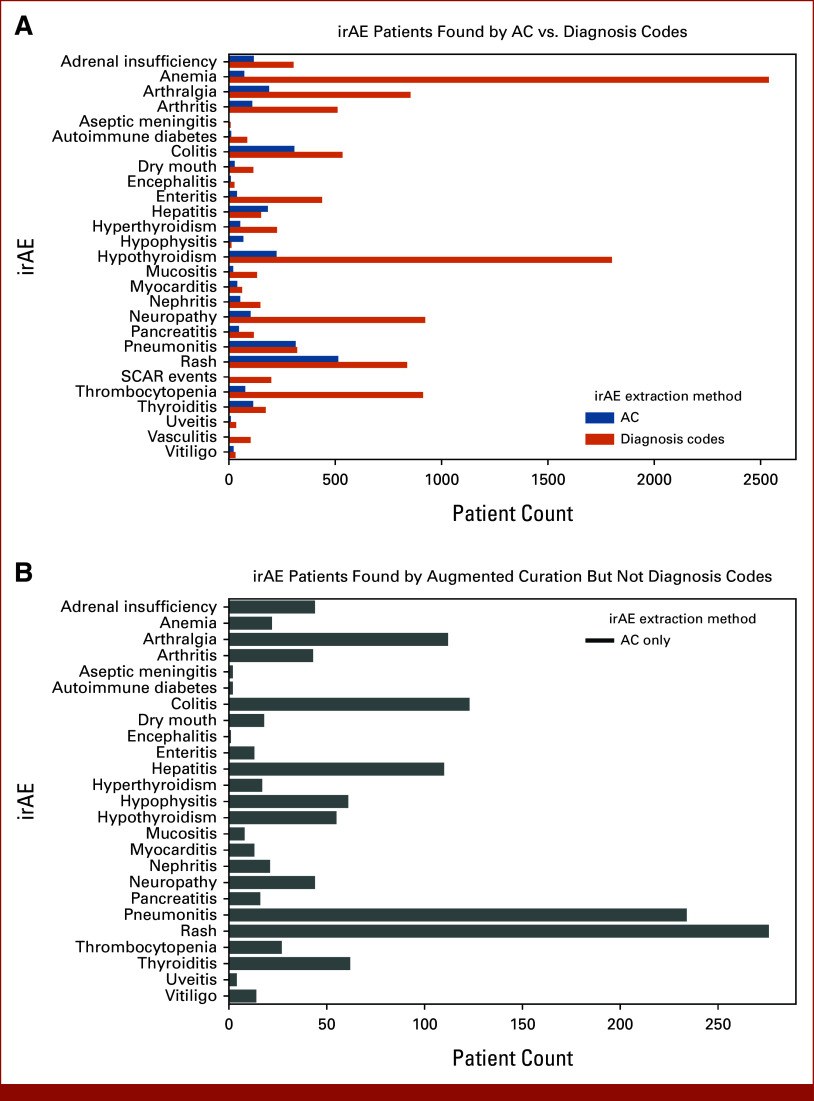
AC identified that 20.8% (n = 1,930) of ICI patients experienced at least one of the 27 irAEs at Mayo, and 5.7% (n = 530) experienced ≥two irAEs (Fig 2A; Appendix Fig A1). Substantially more cases of irAEs, 57.6% (n = 5,348), were identified by diagnosis codes (Fig 2A). However, given the limitations of diagnosis codes, it is difficult to ascertain what proportion of AEs were ICI-induced. For all irAEs, only 20% (n = 1,104) of the patients found using diagnosis codes were also identified by AC (Fig 2B).
FIG 2.
Prevalence counts irAEs from structured diagnosis codes and AC of clinical notes. (A) Comparison of patient counts found by AC model and structured diagnosis codes. (B) Patient counts found by AC models that were not found by structured diagnosis codes (structured diagnosis code FNs). In the absence of information from unstructured data, structured diagnosis codes capture a lot of noise and significantly over-estimate the prevalence of ICI-induced irAEs. AC, augmented curation; FNs, false negatives; ICI, immune checkpoint inhibitor; irAE, immune-related adverse event; SCAR, severe cutaneous adverse reaction.
Among the patients with MEPS identified by diagnosis codes or AC and determined to be true positives following manual chart review (n = 314), the incidence of MEPS of the total ICI population was as follows: myocarditis n = 30 (0.32%), encephalitis n = 13 (0.14%), pneumonitis n = 273 (2.92%), and SCAR events n = 4 (0.04%). Only 24% of patients with MEPS found by diagnosis codes were also identified by AC models (Fig 2B).
The accuracy of MEPS diagnoses using diagnosis codes was compared with that from AC via manual review of each patient's EHR. The confusion matrix (Table 2) shows the total population as the union of patients with MEPS identified by diagnosis codes or AC.
TABLE 2.
Confusion Matrix Calculations of True-Positive and True-Negative MEPS Found From Structured Diagnosis Codes and AC
| Reviewed MEPS Patient | AC MEPS Positive | AC MEPS Negative | Structured MEPS Positive | Structured MEPS Negative |
|---|---|---|---|---|
| Final MEPS positive (n = 320) | 289 | 31 | 126 | 194 |
| Final MEPS negative (n = 536) | 76 | 460 | 482 | 54 |
Abbreviations: AC, augmented curation; MEPS, myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions.
Performance metrics were computed for ICI-induced MEPS identification by diagnosis codes and AC (Table 3). The following metrics are reported: specificity, sensitivity, positive predictive value (PPV), negative predictive value (NPV), and F1 score (definitions in Appendix Table A3). For these analyses, the manually reviewed MEPS cohort was used as the ground truth. AC has higher sensitivity, precision, F1 score, PPV, and NPV across all MEPS. These results demonstrate how diagnosis codes missed many cases of MEPS that were captured by AC.
TABLE 3.
Performance Metrics of irAEs From Structured Diagnosis Codes and AC Compared With Manually Curated Cohort as Gold Standard for MEPS irAEs
| Data Source | Specificity (95% CI) | Sensitivity (95% CI) | PPV (95% CI) | NPV (95% CI) | F1 Score (95% CI) |
|---|---|---|---|---|---|
| AC | 0.858 (0.828 to 0.882) | 0.903 (0.870 to 0.927) | 0.792 (0.743 to 0.823) | 0.937 (0.912 to 0.953) | 0.844 (0.814 to 0.866) |
| Structured diagnosis codes | 0.101 (0.079 to 0.124) | 0.394 (0.336 to 0.436) | 0.207 (0.175 to 0.248) | 0.218 (0.164 to 0.267) | 0.272 (0.235 to 0.313) |
Abbreviations: AC, augmented curation; irAE, immune-related adverse event; MEPS, myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions; NPV, negative predictive value; PPV, positive predictive value.
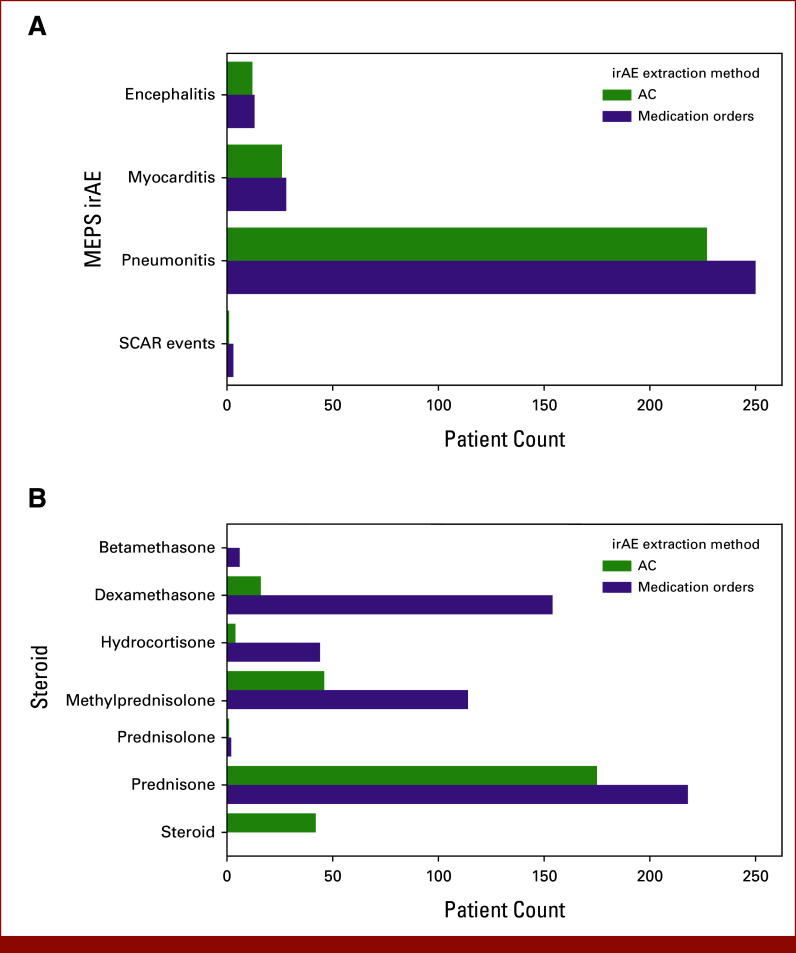
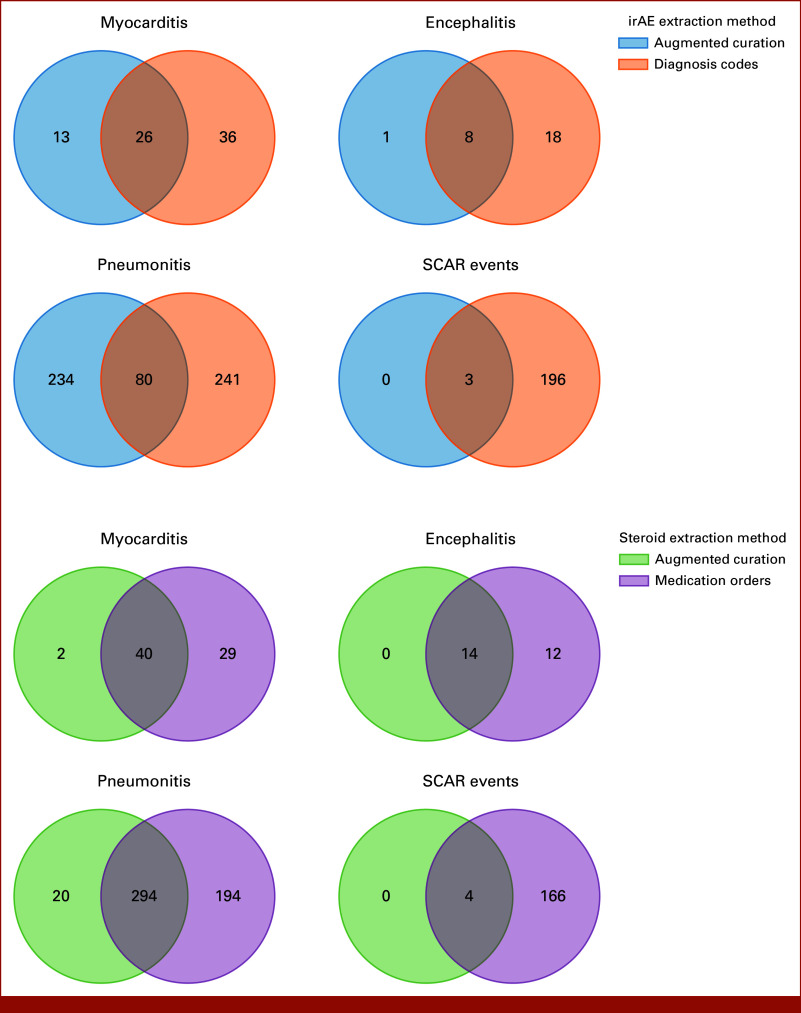
As a proxy of severity, we quantified the number of patients who received corticosteroids or discontinued treatment because of their respective ICI-induced irAE(s). Patients with MEPS were found to receive corticosteroids for their irAEs 82% of the time (Fig 3). Figure 3A shows the number of patients with MEPS who received corticosteroids, identified using either structured medication orders or AC, and did not show any significant differences between the two methods. However, differences were observed when separating corticosteroids by drug type, shown in Figure 3B for patients with pneumonitis as an example. Here, structured medication orders fail to capture causality and therefore overcount medications, for example, dexamethasone, which is also used as an adjunct to chemotherapy and for edema related to brain metastases (Fig 3B). Venn diagrams were generated to compare the varying degrees of overlap between the two data extraction methods across MEPS diagnoses and subsequent corticosteroid administration (Appendix Fig A2). SCAR events had the highest levels of concordance across the two data extraction methods, with 100% of patients found by AC also having relevant diagnosis codes. Pneumonitis events had the lowest overlap, with 25% of the patients found by AC also having relevant diagnosis codes.
FIG 3.
Comparison of augmented curation and structured data derived corticosteroid counts for patients with MEPS. (A) Overall steroid counts for patients with MEPS found by AC versus medication orders. (B) Individual steroid counts for patients with pneumonitis. Structured medication orders fail to capture causality and therefore over-count medications such as dexamethasone which is also used as an adjunct to chemotherapy and for edema related to brain metastases. AC, augmented curation; irAE, immune-related adverse event; MEPS, myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions; SCAR, severe cutaneous adverse reaction.
Additionally, 25.6% (n = 70) of patients with pneumonitis and 50% (n = 2) of patients with SCAR discontinued their ICIs after evaluation of toxicity, whereas 100% of patients with encephalitis (n = 13) and myocarditis (n = 30) permanently discontinued their ICI because of their irAE, which is consistent with clinical practice and corresponding guidelines.12
DISCUSSION
The safety profile of ICIs has been well-described in the clinical trial setting.13 However, safety and tolerability of ICIs using RWD is still emerging and presents a need, and an opportunity, to better understand the safety profile in this setting.14,15 The nature of RWD is unique in that it often uses diagnosis codes, intended primarily for billing purposes, and clinical notes in EHRs, which are used for documenting clinical decision making. Claims databases and EHRs have not been traditionally used for safety monitoring, toxicity assessment activities, or to provide causality between drugs and adverse events. This analysis sought to demonstrate how the identification of ICI-induced irAEs could be augmented in an EHR system using a novel approach to identify drug-event pairs with causal inference.
As ICIs are increasingly being used for cancer care, particularly in the adjuvant and neoadjuvant settings, the potential for toxicity must be clearly understood in order for medical oncology providers to properly counsel patients about the balance of risks and benefits of ICI therapy. Moreover, a better understanding of the long-term outcomes of irAE management is critical to optimize practice within an institution. AC represents a rapid, accurate, and scalable means to capture the frequency of irAEs and to determine how they are managed in an RWD setting.
AC allows for extraction of ICI-to-irAE and irAE-to-corticosteroid relationships, which are only documented in clinical notes. AC presents as a more robust method in determining prevalence compared with diagnosis codes. This method showed higher sensitivity, precision, F1 scores, PPV, and NPV across all MEPS compared with diagnosis codes. Evaluating proxies of severity for these irAEs, for example, steroid usage and ICI discontinuation, is important to understand real-world outcomes of ICI-induced toxicities. Establishing a causal link between these drugs and the irAEs is imperative considering these therapeutic regimens could be given for unrelated diagnoses.
Our cohort was robust, with data from 9,290 patients with various solid tumors, across all stages of disease, who were treated with ICIs at Mayo. The prevalence of irAEs identified by AC in our total cohort was 20.8%. This relatively low rate compared with previously published literature may be due to several factors. First, previous studies assessing irAE rates from ICIs have occurred in the clinical trial setting, versus extracted from RWD, and included additional, more prevalent toxicities (eg, fatigue, nausea, etc) that were not included for investigation in the current report.11 In clinical trials, AE data are proactively solicited from patients at regular visits per study protocols, but in the real-world setting, collection of safety data is more dependent on what patients volunteer. Limitations at the model level may also be contributing to this disparity. The current drug-to-phenotype model functions at the sentence level, requiring both entities (ie, the drug and phenotype) to co-occur in the same sentence to be classified. Therefore, if the irAEs are mentioned in the sentence after the mention of the drug, they would not be captured. Additional work is being conducted to refine this approach and capture related entities in neighboring sentences. However, as previously mentioned, patients requiring steroid treatment of irAEs at Mayo Clinic Rochester are referred to the immunotherapy clinic and evaluated by a team of advanced practice providers. Patients seen in this clinic specifically for irAEs undergo rigorous documentation; thus, it would be rare for AC to miss patients treated for irAEs after the clinic's inception.
Additionally, these results are dependent on and limited by the quality of information and the level of detail captured in the clinical notes, which can be subject to incompleteness, vagueness, and bias.12 For example, if drug classes are used instead of specific entities in the notes, for example, “myocarditis was treated with corticosteroids” versus “myocarditis was treated with prednisone,” the level of detail is limited to what is written. Additionally, care received at outside institutions, including laboratory tests and results, were not available for our review. In short, the added utility of NLP accuracy is bounded by the quality and completeness of the documentation in the EHR.
Several qualitative reasons can be identified for diagnosis code over-/under-reporting and the disparity seen between irAE extraction methods. First, the modes of diagnosis and billing considerations play a role in the diagnosis code input by providers. Laboratory-based diagnosis of irAEs (anemia, hypo/hyperthyroidism, and thrombocytopenia) is more likely to trigger diagnosis codes than discussion in the notes because they are automatically coded when the results return. Particularly, anemia and thrombocytopenia are common in oncology patients and are often chronic and unrelated to ICI use (even in patients on ICI therapy). Additionally, if confirmatory imaging is required, a prerequisite diagnosis code is needed. Billing considerations, namely the need to justify tests for treatment, may dictate diagnosis code specification, but may not accurately represent clinical observations. In other circumstances, a symptom is inputted instead of the suspected differential, for example, cough or dyspnea rather than pneumonitis, resulting in undercoding, as shown in Appendix Figure A2, where 25% of the patients with pneumonitis found by AC also have relevant diagnosis codes. Therefore, in the absence of textual evidence for a causal link between the therapy and the AE, over-/under-reporting can occur.
In conclusion, the results from this study suggest that an NLP-based drug-to-phenotype model is a valuable tool to comprehensively capture ICI-induced irAEs. AC identified irAEs not detected by diagnosis codes and helped assess the drug-irAE relationships in unstructured clinical notes. The use of AC to accurately detect key irAEs would allow investigators to leverage the EHR to elucidate the cause and assess severity of specific irAEs in the RWD setting, facilitating a more comprehensive understanding of patient impact.
APPENDIX
FIG A1.
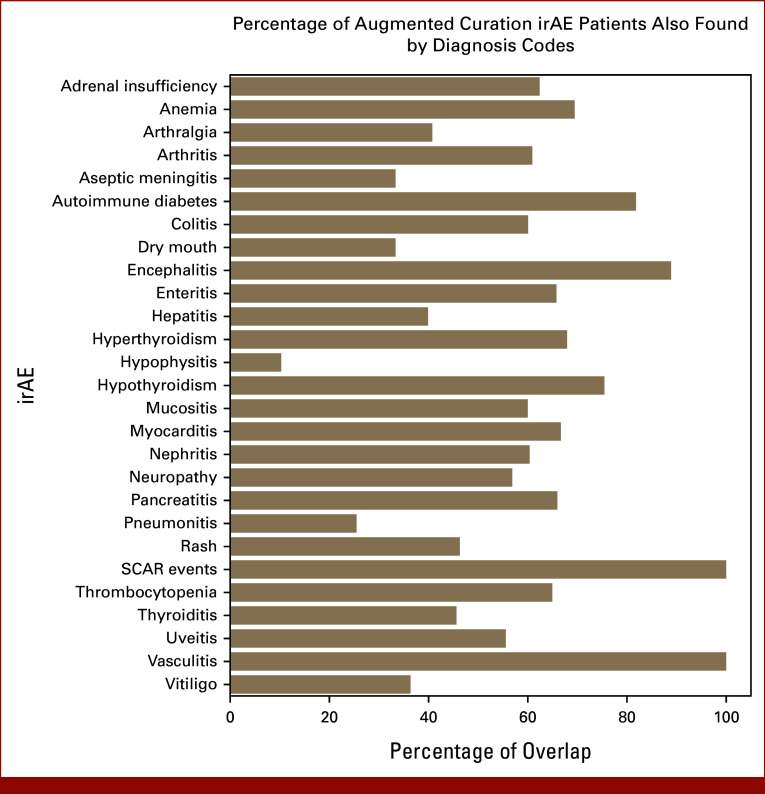
Percentage of patients with irAEs identified from augmented curation who have corresponding diagnosis codes. irAE, immune-related adverse event; SCAR, severe cutaneous adverse reaction.
FIG A2.
Venn diagrams showing the intersections between the two data sources for MEPS diagnoses and corticosteroid administration. The number of patients with the irAE or corticosteroid, according to the data source(s), is given in each section. irAE, immune-related adverse event; MEPS, myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions; SCAR, severe cutaneous adverse reaction.
TABLE A1.
Synonyms Used for Identifying ICI Administration in Structured EHR Tables and Clinical Notes
| Drug Class | ICI | ICI Synonyms |
|---|---|---|
| Anti–PD-1 antibodies | Nivolumab | nivolumab, bms_986298, opdivo_injection, nivo, bms_936658, optivo, nsc_748726, l01xc17, bms_963558, ono_4538, opdivo, nivolumab_opdivo, bms_936558, mdx_1106, nivolumab_bms, bms_93655801 |
| Pembrolizumab | pembrolizumab, anti_pd_1_monoclonal_antibody_mk_3475, keytruda_merck, sch_9000475, pembro, mk3476, sch_900475, mk_3475, lambrolizumab, keytruda, merck_3475, mk3475879 | |
| Cemiplimab | cemiplimab, libtayo, regn2810, cemiplimab_rwlc, anti_pd_1_monoclonal_antibody_regn2810 | |
| Anti–PDL-1 antibodies | Atezolizumab | atezolizumab, ro5541267, anti_pd_l1_monoclonal_antibody_mpdl3280a, tecentric, l01xc32, tecentriq, mpdl328oa, atezolizumab_(mpdl3280a), rg7446, mpdl3280a, mpdl3280, anti_pd_l1, mdpl3280a |
| Avelumab | avelumab, bavencio, msb0010682, anti_pd_l1_monoclonal_antibody_msb0010718c, msb00107, immunoglobulin_g1_lambda1, msb0010718c | |
| Durvalumab | durvalumab, imfinzi, d10808, medi4736, durvalumab_(medi_4736) | |
| Anti-CTLA4 | Ipilimumab | ipilimumab, ipi, bms_734016, mdx_ctla_4, moab_ctla_4, nsc_732442, ipilimumab_yervoy_bms, yervoy, anti_cytotoxic_t_lymphocyte_associated_antigen_4_monoclonal_antibody, mdx_010, mdx_101, monoclonal_antibody_ctla_4, anti_ctla_4_mab_ipilimumab, monoclonal_antibody_mdx_010, cs1002, ibi310 |
Abbreviations: CTLA4, cytotoxic T-cell lymphocyte-4; EHR, electronic health record; ICI, immune checkpoint inhibitor.
TABLE A2.
Structured Diagnosis Codes and Synonyms Used for Identifying irAEs/Potential irAEs
| Sign or Symptom | Diagnosis Codes | Synonyms |
|---|---|---|
| Adrenal insufficiency | 255.5,E27.40,E27.49,E89.6,E27.3 | adrenal_insufficiency, hypoadrenalism, adrenal_gland_hypofunction |
| Anemia | 285.9,D64.9,280.9,D50.9,281.9,285.22,406636013,D64.89,D63.0,285.3,D64.81,285.8,D50.8,283.9,284.89,283.0,285.0,D53.1,D59.9,D59.1,D61.9,283.19,D46.4,2694725011,D61.1,D46.20,D59.0,D64.1,D61.09,D59.2,2694501018,D64.3,D64.2 | anemia, disorders_anemia, anemia_nos, anaemia |
| Arthralgia | M25.511,M25.512,M79.671,719.40,M79.672,M25.50,M25.571,M25.572,M79.662,M79.661,M25.519,M79.673,M25.541,M79.643,M25.542,M25.579,484753013,M79.669,1230691015,M26.629,M26.622,719.41,719.42,719.43,719.44,719.45,719.46,719.47,719.49 | polyarthralgia, arthralgias, polyarthralgias, arthralgia, joint_aches, joint_pain, aches_joint |
| Arthritis | M19.90,274.9,714.0,716.90,714.9,M06.9,M12.9,M06.4,7278014,M16.9,116082011,M10.00,M15.0,M00.9,M05.9,M13.80,M06.80,M13.861,113692012,M13.862,M11.9,M06.00,M11.861,M12.80,M13.811,M13.89,M02.9,M11.831,M13.841,M13.812,M13.851,M11.832,M13.871,M06.041,M13.842,M02.89,M06.042,M11.871,M06.031,M06.032,M06.09,M06.89,M05.79,M05.60,M05.80,M05.70,M05.69,M05.742,M06.849,M05.741,M06.011,M06.839,M06.012,M05.762,711.01,712.30,715.95,715.96,715.97,M05.141 | arthritis, articular_inflammatory_disease, skeletal_joint_inflammation |
| Aseptic meningitis | G03.0 | aseptic_meningitis, acute_aseptic_meningitis |
| Autoimmune diabetes | 250.01,E10.9,E10.65,197984010,E10.649,E10.21,E10.10,E10.40,E10.42,E10.319,E10.621,E10.622,E10.69,E10.22,E10.43,E10.311,E10.59,E10.51,E10.11,E10.3599,E10.3293,E10.3292,E10.3213,250.51,250.61,250.81 | autoimmune_diabetes |
| Colitis | 558.9,K52.9,556.9,K51.90,K52.89,K52.1,K51.00,009.1,K52.3,K52.832,558.1,K52.831,107644019,K52.839,K51.50,106758018,558.2,K51.20,556.6,107643013,K52.0,K52.29,353418019,K51.80,K51.911,K51.918,K52.82,1222282012,K51.914,K51.511,K51.919,K51.811,K51.819 | colitis, colitis_disease, colonic_inflammation, colon_inflammation, colitides |
| Dry mouth | 527.7,K11.7,R68.2 | xerostomia, dry_mouth |
| Encephalitis | G04.90,G04.81,75325010,G04.00 | encephalopathic, brain_inflammation, encephalitis |
| Enteritis | 558.9,K52.9,535.60,555.9,K29.80,555.0,K50.90,555.1,K50.00,558.1,K52.0,K50.80,535.61,K29.81 | enterocolitis, duodenitis, ileitis, enteritis, small_intestinal_enteritis, small_intestinal_inflammation, enteritides, small_intestine_inflammation |
| Hepatitis | 070.54,573.3,571.40,571.42,K75.4,K70.10,206586011,K73.9,K75.3,B15.9,B17.10,K71.2,K73.2,K71.3,K75.2,K71.6,B16.9,63183013,K73.8,K73.0,353589015 | hepatitis, liver_inflammation, acute_hepatitis, hepatitides, chronic_hepatitis, chronic_persistent_hepatitis |
| Hyperthyroidism | 242.90,E05.90,E05.00,E05.80,57561015,E05.20,E05.10,2694980014,E05.91,E06.2,E05.81,E05.21,E05.41 | hyperthyroidism, overactive_thyroid, hyperthyroid, hyperthyreosis |
| Hypophysitis | 253.2 | hypophysitis, hypophysitides, pituitary_inflammation, pituitary_gland_inflammation |
| Hypothyroidism | E03.9,68268011,492839019,244.8,E03.8,178809013,91116012,E03.2,137019010,244.9 | hypothyroidism, hypothyreosis, underactive_thyroid, thyroid_deficiency, thyroid_insufficiency |
| Mucositis | 528.00,K12.30,528.01,528.09,K12.32,K12.33,K12.39,K92.81,1782682019,K12.31 | mucositis, mucositides, mucosa_inflammation, mucosal_inflammation |
| Myocarditis | 429.0,I51.4,D86.85,I40.9,I40.1,84844011,I40.8,I41 | myocarditis, myocarditis_nos, inflammatory_cardiomyopathy, myocarditides, myocardial_inflammation, myocardium_inflammation |
| Nephritis | N10,583.81,N12,N05.9,583.89,N11.8,580.89,47940012 | nephritis, nephritis_nos, kidney_inflammation, inflammation_of_kidneys, nephritides |
| Neuropathy | G62.9,355.9,354.2,337.9,353.0,710.9,355.8,351.0,G90.9,356.8,G62.89,355.0,G51.0,053.19,G56.22,G56.21,378.54,G56.13,357.6,1480220018,B02.29,G90.09,G56.11,H46.9,377.39,351.9,G57.01,G56.12,G62.0,G56.23,G57.61,G52.9,G50.9,355.2,G51.9,352.6,G56.31,H47.093,G57.32,H47.012,G57.92,G57.31,G57.91,H47.011,G56.32,H47.013,H47.092,G57.93,H81.22,G99.0,H47.091,G57.00,G52.8,G57.90,G56.91,H47.019,G70.9,G56.92,G56.81,G54.9,357.3,G63,G61.9,G52.7,G62.2,G52.2,G62.81,G57.30,G57.21,G54.3,B02.23,G61.89,G56.93,158470017,G58.0,H47.099,352.4,G61.82,1777429019,G13.0,H49.22,195776019,H49.21,H49.11,35332011,H49.01,H49.12,H49.20,H49.02,G57.23,H49.00,G62.82,345513016,H49.23,H49.13,E09.42,357.9 | polyneuropathy, neuropathy |
| Pancreatitis | 577.0,K86.1,K85.9,K85.90,303630010,K85.10,K85.80,K85.91,K85.8,K85.81,K85.30,K85.31 | acute pancreatitis, pancreatitis, edema_pancreatic_parenchymal, peripancreatic_fat_necrosis, acute_edematous_pancreatitis, pancreatitis_nos, pancreas_inflammation, pancreatitides |
| Pneumonitis | 515,J84.9,J84.89,508.1,508.0,J70.1,516.32,J70.0,314740018,M34.81,J84.114,M05.10,J84.113,J84.17 | pneumonitis, pneumonitides, lung_parenchyma_inflammation |
| Rash | 782.1,R21,709.8,693.0,L27.0,L27.1,2764045019,1226128015 | rash |
| SCAR events | 709.8,709.2,695.89,695.9,L90.5,L53.9,I96,L13.9,L51.9,694.8,L26,L51.1,L51.3 | severe cutaneous adverse reaction, stevens johnson syndrome, sjs, acute generalised exanthematous pustulosis, agep, dress, drug-induced hypersensitivity syndrome, SCAR events, bullous_haemorrhagic_dermatosis, cutaneous_vasculitis, dermatitis_bullous, dermatitis_exfoliative, dermatitis_exfoliative_generalised, bullous_dermatitis, generalized_exfoliative_dermatitis, exfoliative_dermatitis, drug_reaction_with_eosinophilia_and_systemic_symptoms, epidermal_necrosis, erythema_multiforme, erythrodermic_atopic_dermatitis, exfoliative_rash, generalised_bullous_fixed_drug_eruption, oculomucocutaneous_syndrome, sjs_ten_overlap, skin_necrosis, target_skin_lesion, toxic_epidermal_necrolysis, toxic_skin_eruption |
| Thrombocytopenia | 287.5,D69.6,D69.59,287.49,287.31,D69.3,443796011,1227080015,M31.1,294383018,199491013 | thrombocytopenia, thrombocytopenic_disorders, low_platelet_count, thrombopenia, ttp |
| Thyroiditis | 245.2,E06.3,245.9,E06.9,36884017,E06.1,E06.5,E06.0,136211012,E06.4,E06.2,292420015 | thyroiditis, thyroiditis_nos, disease_thyroiditis, thyroid_gland_inflammation |
| Uveitis | 364.3,H20.9,H20.00,H20.13,H20.11,H20.022,H20.021,H30.93,H20.023,360.12,H30.92,H30.91,H30.90,H44.113,206795017,2842394015,H44.112 | uveitis, disease_uveitis, uvea_inflammation, uveitis_nos, uveitides, uveal_inflammation |
| Vasculitis | 447.6,447.8,I77.6,I77.89,417.8,M31.6,I28.8,M31.30,L95.8,L95.9,I67.7,M31.0,D89.1,M31.8,L95.0 | vasculitis, angiitides, angiitis, vasculitis_syndrome |
| Vitiligo | 709.01,L80 | vitiligo, vitiligo_vulgaris |
Abbreviations: irAE, immune-related adverse event; SCAR, severe cutaneous adverse reaction.
TABLE A3.
Confusion Matrix Variable Definitions
| Metric | Definition |
|---|---|
| TPs | Sum of positive predictions across all MEPS irAEs on the basis of the classification method and manual review |
| TNs | Sum of negative predictions across all MEPS irAEs on the basis of the classification method and manual review |
| FNs | Sum of negative predictions across all MEPS irAEs on the basis of the classification method but positive on the basis of manual curation |
| FPs | Sum of positive predictions across all MEPS irAEs on the basis of the classification method but negative on the basis of manual curation |
| Specificity (TN rate) | TN/(TN + FP) = (number of true-negative assessment)/(number of all negative assessment) |
| Sensitivity (TP rate, recall) | TP/(TP + FN) = (number of true-positive assessment)/(number of all positive assessment) |
| PPV (precision) | TP/(TP + FP) = (number of true-positive assessments)/(number of all positive calls) |
| NPV | TN/(TN + FN) = (number of true-negative assessments)/(number of all negative calls) |
| F1 score | (2 × TP)/(2TP + FP + FN) |
Abbreviations: FN, false negative; FP, false positive; irAE, immune-related adverse event; MEPS, myocarditis, encephalitis, pneumonitis, and severe cutaneous adverse reactions; NPV, negative predictive value; PPV, positive predictive value; TN, true negative; TP, true positive.
TABLE A4.
Synonyms Used for Identifying Corticosteroid Administration in Structured EHR Tables and Clinical Notes
| Drug | Synonyms |
|---|---|
| Prednisone | prednisone, deltasone, prednicot, prednison, orasone |
| Prednisolone | prednisolon, desowen, desonide |
| Hydrocortisone | cortifoam, cortaid, scalpicin, hytone, westcort, cortef, vytone, cortisol, hydrocortison, hydrocortizone, anusol |
| Dexamethasone | dexameth, intensol, decort, decan, decadron, zema |
| Methylprednisolone | Solu-medrol, solumedrol, medrol |
| Betamethasone | betamethasone |
| Corticosteroid | glucocorticoid, corticocorticosteroid, steroid |
Abbreviation: EHR, electronic health record.
PRIOR PRESENTATION
Presented in part at the Society for Immunotherapy of Cancer annual meeting, Boston, MA, November 8-12, 2022, and ASCO Breakthrough, Yokohama, Japan, August 1-3, 2023.
SUPPORT
Supported by F. Hoffmann-La Roche/Genentech.
AUTHOR CONTRIBUTIONS
Conception and design: Hannah Barman, Sriram Venkateswaran, Antonio Del Santo, Tyler E. Wagner, Rajat Mohindra
Administrative support: Tyler E. Wagner
Collection and assembly of data: Hannah Barman, Unice Yoo, Eli Silvert, Krishna Rao
Data analysis and interpretation: Hannah Barman, Sriram Venkateswaran, Antonio Del Santo, Unice Yoo, Eli Silvert, Krishna Rao, Bharathwaj Raghunathan, Lisa A. Kottschade, Matthew S. Block, G. Scott Chandler, Joshua Zalis, Tyler E. Wagner, Rajat Mohindra
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Hannah Barman
Employment: nference
Stock and Other Ownership Interests: nference
Sriram Venkateswaran
Employment: Roche
Patents, Royalties, Other Intellectual Property: Method for determining process variables in cell cultivation processes
Travel, Accommodations, Expenses: Roche
Antonio Del Santo
Stock and Other Ownership Interests: Roche
Unice Yoo
Employment: nference
Eli Silvert
Employment: nference
Stock and Other Ownership Interests: nference
Research Funding: nference
Travel, Accommodations, Expenses: nference
Krishna Rao
Employment: Bergen Anesthesia Group
Bharathwaj Raghunathan
Employment: nference
Lisa A. Kottschade
Consulting or Advisory Role: Immunocore
Matthew S. Block
Research Funding: Merck (Inst), Immune Design (Inst), Genentech/Roche (Inst), Marker, Inc (Inst), Bristol Myers Squibb (Inst), Pharmacyclics (Inst), Transgene (Inst), Sorrento Therapeutics (Inst), TILT Biotherapeutics, Alkermes, nference, Regeneron, Viewpoint Molecular Targeting
Uncompensated Relationships: Sorrento Therapeutics, Viewpoint Molecular Targeting, TILT Biotherapeutics
G. Scott Chandler
Employment: Roche/Genentech
Stock and Other Ownership Interests: Roche/Genentech
Research Funding: Roche/Genentech
Patents, Royalties, Other Intellectual Property: Patent pending as co-inventor of novel biomarker work performed as Roche employee
Travel, Accommodations, Expenses: Roche/Genentech
Joshua Zalis
Employment: nference
Stock and Other Ownership Interests: nference
Tyler E. Wagner
Employment: nference, Anumana, Inc
Stock and Other Ownership Interests: nference, Anumana, Inc
Patents, Royalties, Other Intellectual Property: Patents and patents pending for technologies developed by nference, Inc
Rajat Mohindra
Employment: Roche, Novartis
Stock and Other Ownership Interests: Roche, Novartis
Research Funding: Roche
Patents, Royalties, Other Intellectual Property: Patent pending as coinventor of novel biomarker work performed as Roche employee
Travel, Accommodations, Expenses: Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Albandar HJ, Fuqua J, Albandar JM, et al. : Immune-related adverse events (irAE) in cancer immune checkpoint inhibitors (ICI) and survival outcomes correlation: To rechallenge or not? Cancers (Basel) 13:989, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng Z, Zhao Y, Sun M, et al. : Rich text formatted EHR narratives: A hidden and ignored trove. Stud Health Technol Inform 264:472-476, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen K, Soguero-Ruiz C, Oyvind Mikalsen K, et al. : Analysis of free text in electronic health records for identification of cancer patient trajectories. Sci Rep 7:46226, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav P, Steinbach M, Kumar V, et al. : Mining electronic health records (EHRs). ACM Comput Surv 50:1-40, 2018 [Google Scholar]
- 5.Velupillai S, Suominen H, Liakata M, et al. : Using clinical natural language processing for health outcomes research: Overview and actionable suggestions for future advances. J Biomed Inform 88:11-19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltagy I, Lo K, Cohan A: SciBERT: A pretrained language model for scientific text, in Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). Hong Kong, China, Association of Computational Linguistics, 2019, pp. 3615-3620
- 7.Pawlowski C, Venkatakrishnan AJ, Ramudu E, et al. : Pre-existing conditions are associated with COVID-19 patients' hospitalization, despite confirmed clearance of SARS-CoV-2 virus. EClinicalMedicine 34:100793, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner T, Shweta F, Murugadoss K, et al. : Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. Elife 9:e58227, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Belouali A, Shah NJ, et al. : Automated identification of patients with immune-related adverse events from clinical notes using word embedding and machine learning. JCO Clin Cancer Inform 10.1200/CCI.20.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang DY, Salem J, Cohen JV, et al. : Fatal toxic effects associated with immune checkpoint inhibitors: A systematic review and meta-analysis. JAMA Oncol 4:1721-1728, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murugadoss K, Rajasekharan A, Malin B, et al. : Building a best-in-class automated de-identification tool for electronic health records through ensemble learning. Patterns (N Y) 2:100255, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnaud-Coffin P, Maillet D, Gan HK, et al. : A systematic review of adverse events in randomized trials assessing immune checkpoint inhibitors. Int J Cancer 145:639-648, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Brahmer JR, Lacchetti C, Thompson JA, et al. : Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American society of clinical oncology clinical practice guideline summary. JCO Oncol Pract 14:247-249, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Reynolds KL, Arora S, Elayavilli RK, et al. : Immune-related adverse events associated with immune checkpoint inhibitors: A call to action for collecting and sharing clinical trial and real-world data. J Immunother Cancer 9:e002896, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing Y, Yang J, Johnson DB, et al. : Harnessing big data to characterize immune-related adverse events. Nat Rev Clin Oncol 19:269-280, 2022 [DOI] [PubMed] [Google Scholar]