Abstract
The cascade of herpes simplex virus (HSV) gene expression that results in viral replication begins with the activation of viral immediate-early (IE) genes by the virion-associated protein VP16. VP16 on its own is inefficient at associating with complexes formed on IE gene promoters and depends upon the cellular factor HCF for its activity. In this respect VP16 mimics the host basic leucine zipper (bZIP) protein Luman, which also requires HCF for activating transcription. Our objective is to explore interactions between Luman and HCF and to determine if they play a role in the biology of herpesviruses. In this report we show that in cultured cells ectopically expressed Luman was retained in the cytoplasm, where it colocalized with Calnexin, a protein normally associated with the endoplasmic reticulum (ER). Retention of Luman in the ER depends on a hydrophobic segment of the protein that probably serves as a transmembrane domain. Deletion of this domain changed the intracellular location of Luman so that most of the mutant protein was in the nucleus of cells. While HCF was present in the nucleus of most cells, in cells expressing Luman it was retained in the cytoplasm where the two proteins colocalized. This cytoplasmic association of Luman and HCF could also be demonstrated in neurons in trigeminal ganglia removed from cattle soon after death. Cells in tissue culture that expressed Luman, but not a mutant form of the protein that fails to bind HCF, were resistant to a productive infection with HSV type 1 (HSV-1). We hypothesize that similar Luman-HCF interactions in sensory neurons in trigeminal ganglia result in the suppression of viral replication and the establishment of latency. Interestingly, Luman could activate the promoters of IE110 and LAT, two genes that are critical for reactivation of HSV-1 from latency. This suggests a role for Luman in the reactivation process as well.
Infection for the first time with an alphaherpesvirus leads to active replication of the virus in epithelial cells at the portal of entry. The virus spreads to adjoining cells but also enters the termini of sensory nerves, innervating the site of replication. It is then transported along axons to neuronal cell bodies in sensory ganglia, where it establishes a latent infection. The virus is relatively quiescent during the latent state but is reactivated periodically to replicate and move back down the axons to reinfect epithelial surfaces (reviewed in reference 42).
Herpes simplex virus type 1 (HSV-1) and Bovine herpesvirus type 1 (BHV-1) are alphaherpesviruses that usually cause primary infections in epithelia of the oropharynx and face. The main sites of latency for these viruses are neurons in the trigeminal ganglia (recently reviewed by Jones [18]). During latent infections by HSV-1 and BHV-1, transcripts arising from only a small portion of the viral genomes (LAT or LRG, respectively) can be detected in neurons. The exact role of these transcripts in the establishment of the latent infection and in reactivation from it is unclear. However, evidence suggests that the transcripts themselves or products derived from them are required for the efficient establishment and maintenance of the latent state (7, 9, 12, 45). Paradoxically, these transcripts also appear to play a role in reactivation from latency (3, 4, 10, 17, 38, 39).
In contrast to the latent state, productive infection in epithelial cells by HSV-1 (reviewed in reference 43) and BHV-1 (31) leads to the expression of large number of viral regulatory, enzymatic, and structural proteins. The genes for these proteins are expressed in a regulated cascade, and most genes can be categorized as immediate-early (IE or α), early (E or β), or late (L or γ), depending on the order of their expression during infection. This cascade is initiated when the expression of IE genes is activated by a viral protein brought into the cell as a component of the infecting virion. This protein, VP16 or α gene trans-inducing factor (αTIF), recognizes cis-acting response elements, the octamer-GARAT and TAATGARAT (R is a purine) sequences, in promoters of IE genes (32, 35, 40, 51). Unlike other transcription activators, VP16 does not directly contact its response element but binds to the cellular protein Oct-1 attached to the octamer or TAAT portion of the element (1, 13, 23, 29, 30, 35, 41, 47). Another host protein, HCF, is required for this interaction (13, 19, 56). It is not known why the viruses have evolved to rely on these cellular factors for the initiation of their replicative cycles.
Several studies (16, 21, 50) indicate that the ordered cascade of gene expression seen in infected cultured cells does not accompany the reactivation of latent virus. However, the HSV-1 IE protein IE110 (ICP0) may have a specialized role in the reactivation from latent infections. Viruses containing deletions in the gene are impaired in their ability to reactivate from latent infections (25, 48). In addition, in a tissue culture model of latency exogenously supplied IE110 but not another IE protein could induce latent virus to reactivate (44). Since VP16 is not detected in latently infected neurons and is not required for reactivation (46), it is assumed that the expression of IE110 is induced during reactivation by neuronal factors.
The human HCF proteins are derived by the posttranslational processing of a single large precursor (22, 53). The gene for HCF is conserved in a wide variety of metazoan species and, although HCF was initially defined by its role in VP16-Oct1-TAATGARAT complex formation, its ubiquitous nature suggests that it plays a fundamental role in the cellular biology of the metazoa. When cells with a temperature-sensitive mutation in HCF are grown at elevated temperatures, they arrest in the G1/G0 phase of the cell cycle (15). HCF is therefore thought to regulate some aspect of cell replication. However, its role in this process, or other cellular processes, is not known.
Recently, Kristie and coworkers reported that while HCF is located in the nucleus of most cells, it is sequestered in the cytoplasm of neurons of trigeminal ganglia (24). Death of the animal or stimuli that trigger the reactivation of latent herpesviruses leads to the movement of the protein to the neuronal nuclei. These observations suggest that the regulated translocation of HCF to the nucleus may trigger the reactivation of alphaherpesviruses from their latent state.
To determine the role of HCF in normal cellular physiology and in herpesvirus gene expression, we and others have identified a cellular protein which, like VP16, interacts with HCF. This protein, called Luman (27) or L-ZIP (11), is a basic leucine zipper (bZIP) containing transcription activator. In transient-expression assays Luman can activate expression of genes linked to cyclic AMP response elements (CRE). It can also bind these elements in vitro. Luman shares with VP16 the HCF binding motif D/EHXYS/A and activation of CRE-containing promoters by Luman requires binding to HCF through this motif (11, 28).
This report describes interactions between HCF and Luman. We show that in cultured cells ectopically expressed Luman is retained in the cytoplasm, where it colocalizes with Calnexin, a protein normally associated with the endoplasmic reticulum (ER) (37, 52). Retention of Luman in the ER depends on a hydrophobic segment of the protein that probably serves as a transmembrane domain. Deletion of this domain changed the intracellular location of Luman so that most of the mutant protein was in the nuclei of cells. While HCF was present in the nuclei of most cells, in cells expressing Luman it was retained in the cytoplasm. This cytoplasmic association of Luman and HCF could also be demonstrated in neurons in trigeminal ganglia removed from cattle soon after death. Cells in tissue culture that expressed Luman were resistant to a productive infection with HSV-1, presumably because of insufficient HCF in the nucleus to allow for efficient IE gene expression. We hypothesize that similar Luman-HCF interactions in sensory neurons in trigeminal ganglia result in the suppression of viral replication and the establishment of latency. Interestingly, Luman could activate the promoters of the IE110 and LAT genes, suggesting a role for Luman in the reactivation process as well.
MATERIALS AND METHODS
Materials.
All restriction endonucleases, other DNA-modifying enzymes, media for tissue culture, and other reagents (unless stated otherwise) were purchased from Canadian Life Technologies.
Plasmids.
The construction of pcLuman and pGEXLuman (27), pcLuman(1–220) and pcLuman(Y81A) (28), pAB2 (36), and pAB-167 (33), as well as that of pBB8 and pBB15 (2), have been described. pcLuman contains the coding sequence of Luman, as well as an amino-terminal FLAG epitope cloned into the mammalian expression vector pcDNA3 (Invitrogen). pcLuman(1–220) and pcLuman(Y81A) are similar to pcLuman except that pcLuman(1–220) contains only the first 220 amino acids of Luman followed by a termination codon. In pcLuman(Y81A) tyrosine 81, a critical residue in the HCF binding domain of Luman, has been changed to alanine. pGEXLuman contains the coding sequences of Luman linked to the sequences for glutathione S-transferase (GST). Plasmid AB2 contains the promoter sequences of the HSV IE110 gene that lie downstream from the last octamer-GARAT motif linked to the coding sequences of chloramphenicol acetyltransferase (CAT). In pAB2-167 the octamer-GARAT at position −167 in the IE110 promoter has been added to the minimal IE110 promoter of pAB2. The plasmids BB8 and BB15 were obtained from Peter O'Hare. Plasmid BB8 contains 608 bp of DNA upstream from the start of transcription of the HSV-1 latency-associated transcript (LAT), while pBB15 contains 143 bp of this sequence.
In pAB2-CRE1 we changed the putative CRE, AATCGTCA, to AATCGaCt (lowercase letters represent differences from the wild-type sequence) by using the QuickChange site-directed mutagenesis kit (Stratagene). The two complementary oligonucleotides used for the mutagenesis were ATTGGGGGAATCGaCtCTGCCGCCCTT and AAGGGGCGGCAGaGtCGATTCCCCCAAT. Similarly, the complementary oligonucleotides ACCAGCAGCAGCAGCATGTACTCCTCTGAC and GTCAGAGGAGTACATGCTGCTGCTGCTGGT were used to delete the putative transmembrane domain of Luman in pcLuman. The resulting plasmid was called pcLumanΔTm.
Antibodies.
The fusion protein GST-Luman was produced in Escherichia coli BL21(DE3) (Novagen) and purified by using glutathione-Sepharose beads (Pharmacia) as described previously (32). Antibodies were produced at the University of Saskatchewan Animal Resources Centre by immunizing rabbits with about 150 μg of protein in Freund's complete adjuvant. Two weeks after primary immunization the rabbits were given booster injections of 150 μg of protein in Freund's incomplete adjuvant. They were bled for serum 2 weeks later. Control serum samples were obtained from the rabbits before immunization. The animals were treated in accordance with protocols approved by the University of Saskatchewan Animal Care Committee. The anti-Luman serum specifically detects Luman in Western blots of in vitro-synthesized Luman (TnT; Promega) and lysates of transfected mammalian cells.
Rabbit polyclonal antibodies against HCF were obtained from Winship Herr (Cold Spring Harbor Laboratories), and monoclonal antibodies against HSV-1 gC were from Lenore Pereira (University of California, San Francisco). Mouse monoclonal antibodies against the FLAG epitope were purchased from the Sigma Chemical Co., and rabbit antibody against the carboxyl terminus of canine Calnexin was purchased from StressGen Biotechnologies Corp.
Transfection and immunofluorescence.
HeLa or COS7 cells were grown and transfected as described before (6). For immunofluorescence, cells were grown on circular 18-mm-diameter micro-cover glasses (VWR Scientific) in six-well Falcon tissue culture plates (Becton Dickinson). At 40 h after transfection the cover glasses were rinsed once with phosphate-buffered saline (PBS), and the cells were fixed for 20 min in methanol kept at −20°C. The cells were then kept for 20 min in blocking solution (PBS containing 10% newborn calf serum). The cells were incubated for 20 min with primary antibodies appropriately diluted in blocking solution, washed three times in PBS and once in blocking solution, and then incubated with diluted goat anti-rabbit or anti-mouse antibodies tagged with either Alexa488 or Alexa546 (Molecular Probes, Inc.) for 20 min. After being washed as described above, the cover glasses were mounted on glass slides over PBS containing 20% glycerol and sealed with clear nail polish.
The slides were observed by using a Zeiss Axioskop microscope equipped for epifluorescence and the appropriate filters. Images were captured by using Northern Eclipse image analysis software (Empix Imaging, Inc.). Figures for this report were prepared by using Adobe Illustrator 7.0 and Adobe Photoshop 4.0 (Adobe Systems, Inc.).
Assays for CAT were performed by using an enzyme-linked immunosorbent assay kit (Boehringer Manneheim) as recommended by the manufacturer.
Trigeminal ganglia.
Trigeminal ganglia were scavenged from cattle euthanized because of terminal illness at the Veterinary Teaching Hospital, Western College of Veterinary Medicine, or at the Veterinary Infectious Diseases Organization. Ganglia were removed, trimmed, embedded in O.C.T. Compound (Tissue-Tek), and frozen immediately in liquid nitrogen. Sections (6 μm) were cut on a Micron cryostat and fixed in methanol. The sections were either stained with hematoxylin and eosin by using routine procedures or were processed for immunofluorescence as outlined above for cells grown on cover glasses.
RESULTS
Location of Luman in cells transiently expressing the protein.
To determine the location of Luman in cells, we transfected HeLa cells with pcLuman, a mammalian expression vector containing the coding sequences for Luman, linked to the FLAG epitope. Transfected cells were then probed with mouse anti-FLAG antibodies, rabbit anti-Luman antibodies, or a combination of the two antibodies. Bound antibodies were visualized with the appropriate fluorescence (Alexa)-labelled anti-mouse or anti-rabbit antibodies. In a transfected culture ca. 5 to 10% of the cells showed bright fluorescence with either antibody. Cells transfected with a control plasmid or Luman expressing cells stained with control serum showed no fluorescence (not shown).
Transfected cultures contained cells that displayed a range of fluorescence intensity presumably reflecting differences in the amount of Luman expressed. In all cells in which Luman was visible, most of the fluorescence was located in the cytoplasm in a “feathery” pattern (Fig. 1A). Occasionally, we saw more-intense staining in small structures located just outside the nucleus. The pattern of staining was the same when either anti-Luman or anti-FLAG antibodies were used. When the antibodies were used simultaneously, with anti-mouse Alexa488 and anti-rabbit Alexa546, the pattern of red and green fluorescence was completely superimposed (not shown). This established that the anti-Luman antibody was specific for Luman. It also suggested that although Luman had originally been obtained from a HeLa cDNA library (27), these cells contain little endogenous Luman protein.
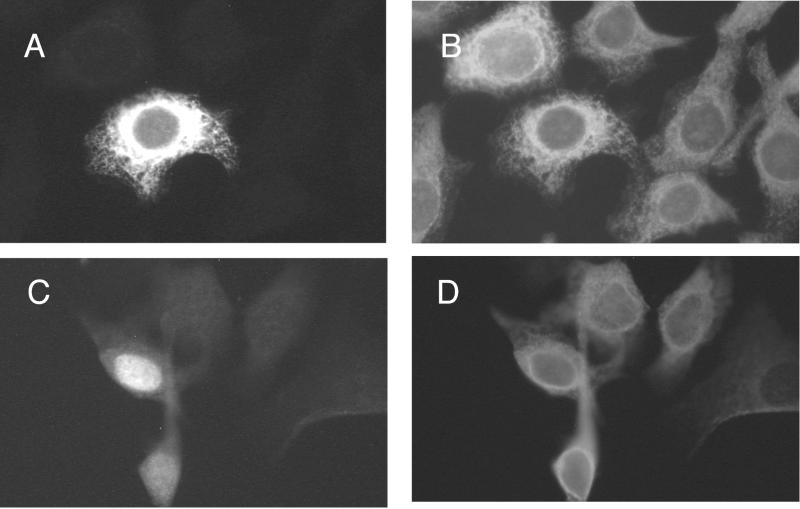
FIG. 1.
Location of Luman and Calnexin in transfected cells. HeLa cells were transfected with either pcLuman (A and B) or pcLumanΔTm (C and D). Cells were stained 48 h later with a mixture of anti-FLAG monoclonal antibodies which recognize FLAG-tagged Luman and rabbit anti-Calnexin antibodies, followed by a mixture of Alexa488-tagged anti-rabbit and Alexa546-tagged anti-mouse antibodies. The cells were visualized in a fluorescent microscope by using either a 546-nm (A and C) or a 450- to 490-nm (B and D) filter.
The pattern of staining for Luman suggested that the protein was located in the endoplasmic reticulum (ER) and Golgi apparatus. To confirm this, transfected cells were probed simultaneously with mouse monoclonal anti-FLAG antibodies and rabbit polyclonal antibodies against Calnexin, an ER transmembrane protein (37). The anti-Calnexin bound to cytoplasmic structures in all cells (Fig. 1B and D) and in some cells we also saw intense staining of structures which are probably the Golgi apparatus. In cells expressing Luman the pattern of anti-Luman and anti-Calnexin staining could be superimposed (Fig. 1A and B). These results suggest that Luman is posttranslationally trapped in the ER and Golgi.
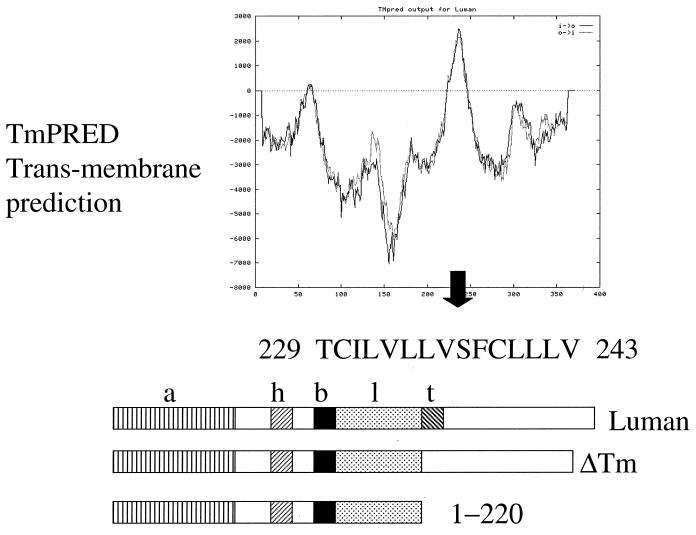
To determine if Luman contains potential transmembrane domains that may anchor it in the ER, we analyzed the primary structure of Luman by using TmPRED (K. Hofmann and W. Stoffel, http://www.ch.embnet.org/software/TmPRED-form.html) and PSORTII (K. Nakai and P. Horton, http://cookie.imcb.osaka-u.ac.jp/nakai/psort.html). Both programs detected a stretch of 15 hydrophobic amino acids that could serve as a transmembrane domain (Fig. 2). This putative transmembrane (Tm) domain lies downstream from all of the characterized functional domains of Luman, including those responsible for transcription activation, HCF binding, DNA binding, and protein dimerization (27, 28). To determine if deletion of the domain would change the location of Luman, we transfected cells with a plasmid coding for Luman in which amino acids 229 to 243, which make up the putative domain, had been deleted (Luman ΔTm, Fig. 2). The cells were then stained for Luman (Fig. 1C) as well as for Calnexin (Fig. 1D). In contrast to cells expressing full-length Luman (Fig. 1A), where most of the protein was associated with cytoplasmic structures, in cells expressing Luman ΔTm the protein was present largely in the nucleus (Fig. 1C). In these cells Luman and Calnexin did not colocalize (Fig. 1C and D). The latter was located only in the cytoplasm. In cells transfected with a plasmid expressing a mutant of Luman truncated just before the putative Tm domain (Luman 1–220), the protein was also located in the nucleus (not shown). However, in these cells relatively little Luman was present, suggesting that the truncated protein was unstable.
FIG. 2.
Putative transmembrane domain of Luman. The top figure shows the results of TmPRED analysis of the amino acid sequence of Luman. Portions of the protein with positive values (above the dotted line) have the potential of forming transmembrane domains. The portion of the Luman protein, residues 229 to 245, that display a high probability of forming a Tm domain are shown. The bottom figure is a schematic diagram of Luman and the ΔTm and 1–220 mutants showing their functional domains. Domains: a, activation; h, HCF binding; l, DNA binding; l, dimerization (leucine zipper); t, putative transmembrane.
Luman retains HCF in the cytoplasm.
We have shown that like HSV-1 VP16, Luman binds strongly to HCF (28). Both proteins, as well as their other viral and cellular homologues, have a common motif, D/EHXYS/A, for binding HCF. Mutations in the conserved residues of this motif reduce HCF binding both in vitro and in vivo.
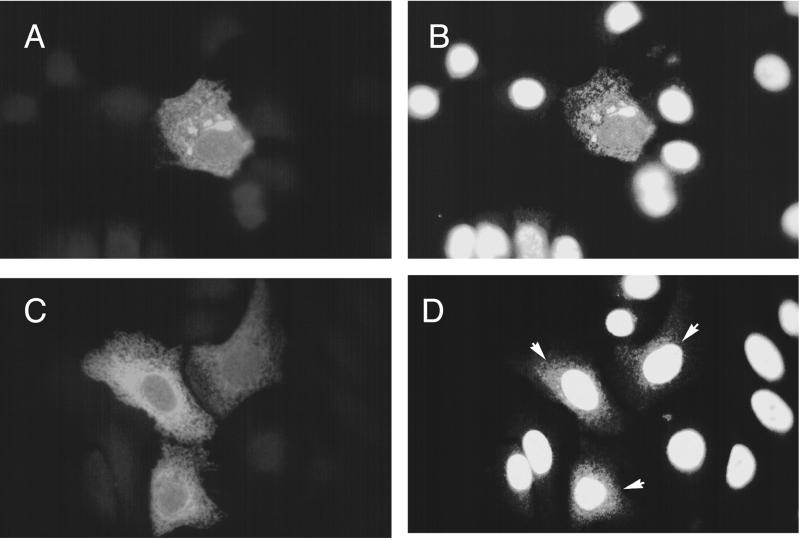
HCF is normally located in the nucleus of cells. To see if Luman anchored in the ER could alter the location of HCF, we simultaneously probed cells transfected with pcLuman with antibodies against HCF and the FLAG epitope. As expected, in cells not expressing Luman, HCF was located almost exclusively in the nucleus (Fig. 3B). However, in cells expressing Luman, HCF appeared to be retained in the cytoplasm with Luman (Fig. 3A and B). This retention of HCF by Luman was dependent on a functional HCF binding domain since Luman with a mutation in its HCF binding domain (Y81A) could not alter the nuclear localization of HCF (Fig. 3C and D). In these cells the mutant Luman remained in the cytoplasm but most of the HCF was nuclear.
FIG. 3.
Location of Luman and HCF in transfected cells. HeLa cells were transfected with either pcLuman (A and B) or pcLuman(Y81A) (C and D). Cells were stained 48 h later with a mixture of anti-FLAG monoclonal antibodies which recognize FLAG-tagged Luman and rabbit anti-HCF, followed by a mixture of Alexa488-tagged anti-rabbit and Alexa546-tagged anti-mouse antibodies. The cells were visualized in a fluorescent microscope by using either a 546-nm (A and C) or a 450- to 490-nm (B and D) filter.
Recently, Kristie et al. reported that while HCF is detected in the nucleus of most cells, it is located in the cytoplasm of neurons in the trigeminal ganglia of mice (24). Stimuli that reactivate latent HSV in the mouse model or postmortem changes caused the protein to move to the nucleus. To determine if Luman might be responsible for retaining HCF in the cytoplasm of neurons, we examined bovine trigeminal ganglia for the two proteins. Ganglia were scavenged from cattle submitted for postmortem examination after euthanasia. The ganglia were removed as soon as possible after death and frozen in liquid nitrogen.
Since antibodies against both Luman and HCF had been produced in rabbits, we were unable to probe individual sections of ganglia for both proteins. We did, however, examine consecutive 6-μm sections and were able to find the corresponding field in the sections (Fig. 4).
FIG. 4.
Location of Luman and HCF in trigeminal ganglia. Trigeminal ganglia were removed from a cow within 30 min of death, embedded, and frozen in liquid nitrogen. Consecutive 6-μm sections were either stained with hematoxylin-eosin (A) or for Luman (B) or HCF (C). Rabbit anti-Luman antibodies and Alexa546-tagged anti-rabbit antibodies were used to visualize Luman, while rabbit anti-HCF and Alexa488-tagged anti-rabbit antibodies were used to visualize HCF. Corresponding fields from the three sections were located and photographed by using the appropriate filters.
In the ganglia removed from adult animals within 30 min of death, we found that about a third to half of the neuronal cell bodies showed bright cytoplasmic fluorescence when stained for Luman. The pattern of staining was punctate and ranged from the entire visible cytoplasm (large arrow, Fig. 4B) to just the periphery of the cell (small arrow, Fig. 4B). When the adjacent section was probed for HCF, we saw that in every cell that stained for Luman, HCF could be detected in the cytoplasm as well (Fig. 4C). In these cells the pattern of cytoplasmic staining for Luman and HCF was the same. In addition, HCF could also be seen in neuronal nuclei, as well as the nuclei of accessory cells. When the removal of ganglia was delayed beyond 30 min, fewer cells were found to stain for Luman, and in almost all cases the staining was restricted to the periphery of the cell body. However, in all cells staining for Luman, HCF showed a similar cytoplasmic pattern of staining. The entire cytoplasm of all neuronal cell bodies stained for Calnexin (not shown). Like the cytoplasmic staining for Luman and HCF, the pattern of Calnexin staining was also punctate.
Luman protects cells from a productive infection by HSV-1.
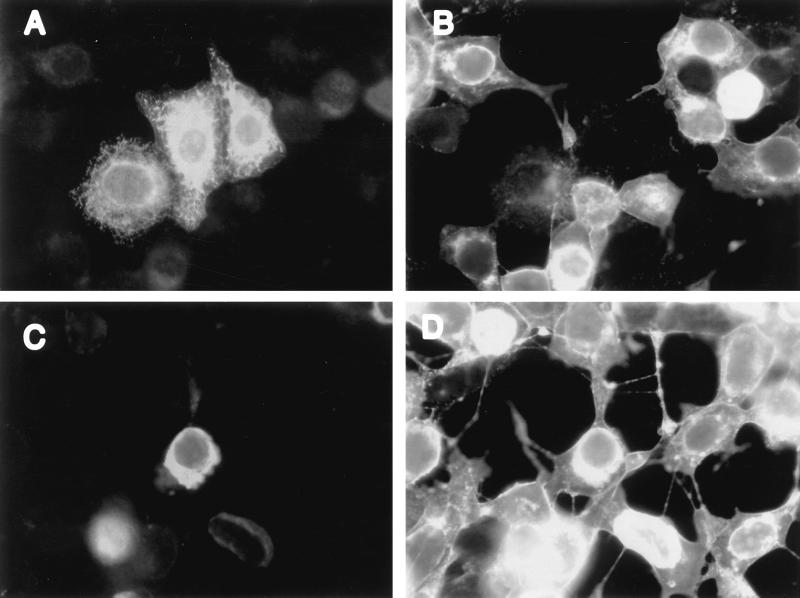
The VP16-HCF-mediated activation of IE gene expression is thought to substantially increase the efficiency with which the virus can initiate infection. Since our results suggested that in cells expressing Luman most of the HCF was sequestered in the cytoplasm, we hypothesized that these cells would be relatively insensitive to HSV-1 replication. To test this hypothesis, we transfected cells with pcLuman or pcLuman(Y81A). At 48 h after transfection the cells were infected with HSV-1. Another 20 h later the cells were fixed and probed with both polyclonal antibodies against Luman and monoclonal antibodies against HSV-1 gC. Every cell (5 to 10% of cells in the culture) that stained for Luman was devoid of any staining for gC (Fig. 5A and B) and did not show “cell rounding,” a characteristic sign of HSV cytopathic effect (CPE). Every cell that did not contain Luman showed strong staining for gC and HSV CPE. In contrast to cells expressing Luman, cells expressing Luman(Y81A), a mutant with a greatly reduced ability to bind HCF (28) and retain it in the cytoplasm (see Fig. 3B and C), showed brilliant staining for gC (Fig. 5C and D) and HSV CPE.
FIG. 5.
Luman protects cells from HSV in an HCF-dependent manner. HeLa cells were transfected with either pcLuman (A and B) or pcLuman(Y81A) (C and D). At 48 h after transfection cells were infected with HSV-1 (KOS) at a multiplicity of about 10 PFU/cell. Then, 20 h later, cells were fixed and stained with a mixture of anti-HSVgC monoclonal antibodies and rabbit anti-Luman antibodies, followed by a mixture of Alexa488-tagged anti-mouse and Alexa546-tagged anti-rabbit antibodies. The cells were visualized in a fluorescent microscope by using either a 546-nm (A and C) or a 450- to 490-nm (B and D) filter.
Luman activates the IE110 promoter.
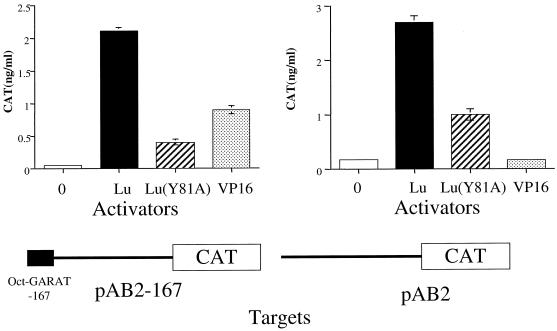
The IE110 protein is thought to be important for efficient reactivation of HSV from the latent state (25, 44, 48). To determine if Luman had the potential to initiate the replicative cascade by stimulating the synthesis of IE110, we determined if Luman could activate the IE110 promoter. Cells were transfected with pAB2-167, in which the basal IE110 promoter containing one octamer-GARAT element is linked to the reporter gene for CAT. We found that Luman could activate this promoter as efficiently as could VP16 (Fig. 6A). Also like VP16, reactivation of the IE110 promoter by Luman depended on the ability of the protein to bind HCF, as Luman (Y81A) was unable to stimulate the expression of CAT. We next examined the ability of VP16 and Luman to activate pAB2, which contains the minimal IE110 promoter from which all octamer-GARATs have been deleted. As expected, VP16 failed to activate this promoter, although Luman activated it to levels comparable to VP16 activation of the octamer-GARAT containing promoter (Fig. 6B). Incidently, pAB2 is almost completely inactive in HeLa cells. This suggests that these cells contain little Luman protein and is consistent with our inability to detect the protein in untransfected HeLa cells by immunofluorescence.
FIG. 6.
Luman activates the HSV IE110 promoter in an HCF-dependent and octamer-GARAT-independent manner. HeLa cells were transfected with target plasmid pAB2, which has the minimal IE110 promoter linked to coding sequences for CAT, or pAB2-167, which in addition has an IE110 octamer-GARAT motif. The cells were also transfected with pcDNA3-0 or pcDNA3 expressing Luman-Lu, Luman(Y81A)-Lu(Y81A), or VP16-VP16. At 48 h after transfection cells were lysed and assayed for CAT.
Luman activates the IE110 promoter through a CRE element.
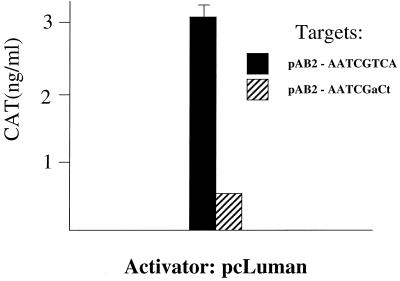
The IE110 promoter contains a putative CRE at position −68 relative to the start of transcription. In this site, AATCGTCA, the last five nucleotides (underlined) agree with the canonical CRE motif, TGACGTCA. Since Luman binds oligonucleotides containing the canonical CRE motif in vitro and activates promoters containing the sequence in vivo (27), we determined if Luman activated the IE110 promoter through its CRE element. We changed the GTCA in the motif in pAB2 to GaCt. Figure 7 shows that although Luman activated pAB2 in an HCF-dependent manner (Luman Y81A was unable to activate the promoter), it was unable to activate the promoter if two nucleotides in the putative CRE motif were changed.
FIG. 7.
Activation of the HSV IE110 promoter requires the putative CRE element. HeLa cells were transfected with target plasmid pAB2, which has the minimal IE110 promoter linked to coding sequences for CAT, or pAB2 in which two nucleotides of the CRE site have been altered (indicated). The cells were also transfected with pcLuman. At 48 h after transfection cells were lysed and assayed for CAT.
Luman activates the LAT promoter.
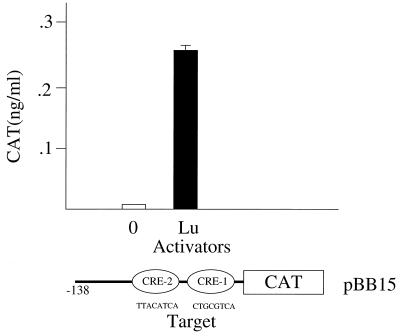
The HSV-1 LAT promoter also contains two putative CREs, CRE1 (26) and CRE2 (20). A mutation in CRE1 in the context of the viral genome reduces reactivation of virus by epinephrine iontophoresis (4), and in a rat pheochromocytoma cell line it confers cyclic AMP responsiveness to the LAT promoter (26). We examined the ability of Luman to activate either the entire LAT promoter represented by about 600 bp of DNA upstream from the start of transcription of LAT (pBB8) or a DNA fragment with just the proximal 143 bp (pBB15) which contains CRE1 and CRE2. Luman could activate both promoters about 20- to 30-fold. The results obtained with pBB15, which is inactive in HeLa cells in the absence of Luman, are shown in Fig. 8.
FIG. 8.
Luman activates the HSV LAT promoter. HeLa cells were transfected with the target plasmid pBB15, which contains 138 bp upstream from the start of transcription of LAT and either pcDNA-0 or pcLuman-Lu. At 48 h after transfection cells were lysed and assayed for CAT.
DISCUSSION
The cascade of herpesvirus gene expression that results in viral replication begins with the activation of IE genes by the virion-associated protein VP16. VP16 on its own is inefficient at associating with complexes formed on IE promoters and depends upon the cellular factor HCF for its activity. In this respect VP16 mimics the host bZIP protein Luman, which also requires HCF for activating transcription. Our objective is to explore interactions between Luman and HCF and to determine if they play a role in the biology of herpesviruses. The results presented here suggest that Luman may be involved in both the establishment of latent infections and the reactivation of latent virus. By sequestering HCF in the cytoplasm of sensory neurons, Luman may prevent the initiation of the replicative cascade leading to latent infection. Subsequent release of the Luman-HCF complex from the cytoplasm in response to external signals and its translocation to the nucleus may activate IE110 and LAT, key viral genes required for reactivation of the latent virus.
We found that ectopically expressed Luman in cultured epithelial cells, and possibly also in sensory neurons of the trigeminal ganglia, is retained in the ER. This is reminiscent of the sterol enhancer binding proteins (SREBPs) (5). These leucine-zipper-containing transcription activators are also sequestered in the ER and are released by proteolysis in response to reduced levels of cholesterol and sterols in the cellular environment. Subsequently, SREBPs are translocated to the nucleus, where they coordinately activate the expression of several genes involved in cholesterol and fatty acid biosynthesis. This strategy has probably evolved to allow a rapid response to low levels of substances that are critical for cellular survival.
Like the SREBPs, Luman is anchored to the ER by a hydrophobic domain. We found that deletion of this domain dramatically altered the location of Luman in the cell. Mutants of Luman that either lacked the transmembrane domain (Luman ΔTm) or lacked the domain as well as sequences downstream from it (Luman 1–220) accumulated in the nucleus. SREBP has two hydrophobic Tm domains that allow both the amino and carboxyl portions of the protein to protrude into the cytoplasm. The amino portion of the protein contains the activation domain, basic DNA binding domain, and leucine zipper, while the carboxyl terminus is thought to play a regulatory role. Luman appears to have a single transmembrane domain, suggesting that its proline-rich carboxyl terminus remains in the lumen of the ER. We speculate that as with the SREBPs the amino-terminal portion of Luman, which contains its domains for transcription activation dimerization and interactions with HCF and DNA, is released by proteolysis. The released product is probably unstable since mutants lacking the carboxyl portion of the protein accumulated in the cell at much lower levels than the wild-type protein (results not shown). The instability of the truncated protein may explain why, while we could visualize HCF in the nucleus of neurons of trigeminal ganglia, we were unable to follow the possible translocation of Luman from the cytoplasm to the nucleus in these cells. Instability of the truncated, and presumably active, form of Luman may be a way of ensuring that the activation of the genes that it regulates is transient.
We had previously shown that Luman strongly binds HCF both in vitro and in vivo. Here we demonstrate that Luman retained in the ER sequesters HCF. Since HCF is required for cell cycle progression, one consequence of this might be that cells expressing Luman are arrested in the G0 phase. This may explain why, despite several attempts, we have been unable to obtain cell lines expressing Luman, even when we have used tetracycline (14)- and Ecdysone (34)-inducible systems. It is possible that in the absence of inducers these systems allow the synthesis of small amounts of Luman which are enough to cause cell cycle arrest.
We observed that Luman-expressing cells were resistant to productive infection by HSV, at least as assessed by the lack of gC in these cells after infection. Since gC is made late in infection, we do not know at which stage of the replicative cycle the infection was interrupted or the mechanism by which Luman blocked HSV replication. Interaction between Luman and HCF was required for protection against HSV since the Luman (Y81A) mutant that does not bind HCF and does not retain it in the cytoplasm, failed to protect cells. This suggests that cells expressing Luman were protected from HSV replication by a mechanism that relied on retention of HCF in the cytoplasm. A simple explanation for how Luman might protect cells could be that Luman prevents HCF from entering the nucleus, thereby blocking VP16-mediated IE gene expression. However, our observations suggest that the mechanism might be more complex. In transfected cells enough Luman-HCF makes it to the nucleus to activate the IE110-ICP0 promoter in a VP16-independent manner. In addition, our preliminary results (unpublished) suggest that although Luman-expressing HSV-infected cells fail to make detectable levels of the structural proteins gC, gB, and gD, they do synthesize the IE proteins ICP0, ICP4, and ICP27.
Kristie and others found that, in contrast to other cell types, in neurons of the trigeminal ganglia HCF is located largely in the cytoplasm. It is translocated to the nucleus after death of the animal and in response to other stimuli that lead to reactivation of HSV in the mouse model (24). These authors suggested that translocation of HCF to the nucleus may lead to the activation of IE gene expression and reactivation. Since latently infected neurons would not be expected to contain VP16, activation of IE genes by HCF would likely be by a VP16-independent mechanism. Kristie et al. suggest that this may involve GA binding protein (GABP). The IE110 promoter has binding sites for GABP, and T. M. Kristie (unpublished observations) has indicated that activation by GABP may require HCF.
Our observations of sections of bovine trigeminal ganglia support the results of our experiments with transfected cells in culture and may explain two of Kristie's observations. First, these results provide a mechanism for the sequestering of HCF in the neuronal cytoplasm. We found Luman and HCF in neuronal cytoplasm in a pattern similar to that observed by Kristie for HCF. In addition, in sections of ganglia the ER marker Calnexin had the same punctate pattern, suggesting that as in transfected cells Luman and HCF were associated with the ER. In transfected cells we showed that the sequestering of HCF by Luman in the ER correlated with resistance to HSV replication. Similar association of the two proteins in neurons could also suppress replication and lead to latency, if the initiation of viral replication at this site relied on an HCF-dependent process. Second, our observations suggest a mechanism for the initiation of the replicative cascade once both HCF and Luman are translocated to the nucleus. We showed that Luman could efficiently activate the promoters of IE110 and LAT, two genes that appear to be critical for reactivation. Our observations do not rule out the possibility that other neuronal factors, such as GABP (24), also mediate activation. They simply suggest that Luman may also play a role in reactivation from latency. Recently, however, Davido and Leib (8) showed that the region of the IE110 promoter that lies between −420 and −70 and includes the GABP-responsive sites is dispensable for reactivation from latency. The Luman-responsive CREs lie downstream from this dispensable region. Those authors discounted the role of the CRE because it did not bind CREB and was nonfunctional in undifferentiated neuroblastoma and pheochromocytoma cells (PC12). However, our preliminary data suggest that the IE110 CRE is relatively specific for Luman, and we have been unable to find Luman in undifferentiated PC12 cells. It is possible that Luman is expressed only after differentiation of these cells into neurons, and we are exploring this possibility. Interestingly, Su et al. (49) have shown that HSV causes a long-term “quiescent” infection in differentiated PC12 cells. We have not examined differentiated PC12 cells for Luman expression, but it is tempting to speculate that Luman synthesis in response to differentiation causes these cells to become resistant to virus infection.
We hypothesize that, as with SREBP, the retention of Luman in the ER provides a means of responding rapidly to conditions that might be detrimental to neurons. Release of the complex from the ER and translocation to the nucleus leads to the coordinate expression of genes that can correct these conditions. Furthermore, rapid degradation of Luman released from the ER ensures that its activation of downstream genes was transient. By requiring HCF for the initiation of its replicative cascade, HSV exploits this pathway both for the establishment of latency and reactivation from it. In the absence of free nuclear HCF in differentiated neurons of the trigeminal ganglia, the virus is unable to replicate and becomes latent. Release of Luman-HCF and translocation to the nucleus activates IE expression in the absence of viral proteins and leads to reactivation.
ACKNOWLEDGMENTS
We are grateful to Peter O'Hare and Sylvie LaBoissiere of the Marie Curie Institute for discussions, reagents, and advice on immunofluorescence; to Ian Shirley and Leah Heasman for assistance with the bovine ganglia; and to Kevin Snyder for technical assistance.
R.L. is a postdoctoral fellow of the Saskatchewan Health Services Utilization and Research Commission, and this work was funded by an operating grant to V.M. from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.apRhys C M, Ciufo D M, O'Neill E A, Kelly T J, Hayward G S. Overlapping octamer and TAATGARAT motifs in the VF65-response elements in herpes simplex virus immediate-early promoters represent independent binding sites for cellular nuclear factor III. J Virol. 1989;63:2798–2812. doi: 10.1128/jvi.63.6.2798-2812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batchelor A H, O'Hare P. Localization of cis-acting sequence requirements in the promoter of the latency-associated transcript of herpes simplex virus type 1 required for cell-type-specific activity. J Virol. 1992;66:3573–3582. doi: 10.1128/jvi.66.6.3573-3582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom D C, Hill J M, Devi-Rao G, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom D C, Stevens J G, Hill J M, Tran R K. Mutagenesis of a cAMP response element within the latency-associated transcript promoter of HSV-1 reduces adrenergic reactivation. Virology. 1997;236:202–207. doi: 10.1006/viro.1997.8723. [DOI] [PubMed] [Google Scholar]
- 5.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 6.Chen C A, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 7.Chen S H, Kramer M K, Schaffer P S, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davido D J, Leib D A. Analysis of the basal and inducible activities of the ICP0 promoter of herpes simplex virus type 1. J Gen Virol. 1998;79:2093–2098. doi: 10.1099/0022-1317-79-9-2093. [DOI] [PubMed] [Google Scholar]
- 9.Devi-rao G B, Bloom D C, Stevens J G, Wagner E K. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J Virol. 1994;68:1271–1282. doi: 10.1128/jvi.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drolet B S, Perng G C, Villosis R J, Slanina S M, Nesburn A B, Wechsler S L. Expression of the first 811 nucleotides of the herpes simplex virus type 1 latency-associated transcript (LAT) partially restores wild-type spontaneous reactivation to a LAT-null mutant. Virology. 1999;253:96–106. doi: 10.1006/viro.1998.9492. [DOI] [PubMed] [Google Scholar]
- 11.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber D A, Schaffer P A, Knipe D M. The LAT-associated function reduces productive cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71:5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerster T, Roeder R G. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 16.Halford W P, Gebhardt B M, Carr D J. Mechanisms of herpes simplex virus type 1 reactivation. J Virol. 1996;70:5051–5060. doi: 10.1128/jvi.70.8.5051-5060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill J M, Garza H H J, Su Y H, Meegalla R, Hanna L A, Loutsch J M, Thompson H W, Varnell E D, Bloom D C, Block T M. A 437-base-pair deletion at the beginning of the latency-associated transcript promoter significantly reduced adrenergically induced herpes simplex virus type 1 ocular reactivation in latently infected rabbits. J Virol. 1997;71:6555–6559. doi: 10.1128/jvi.71.9.6555-6559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones C. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv Virus Res. 1998;51:81–133. doi: 10.1016/s0065-3527(08)60784-8. [DOI] [PubMed] [Google Scholar]
- 19.Katan M, Haigh A, Verrijzer C P, van der Vliet P C, O'Hare P. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res. 1990;18:6871–6880. doi: 10.1093/nar/18.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenny J J, Krebs F C, Hartle H T, Gartner A E, Leiden C B, J M, Hoeffler J P, Weber P C, Wigdahl B. Identification of a second ATF/CREB like element in the herpes simplex virus type 1 (HSV-1) latency associated transcript (LAT) promoter. Virology. 1994;171:607–610. doi: 10.1006/viro.1994.1180. [DOI] [PubMed] [Google Scholar]
- 21.Kosz-Vnenchak M, Jacobson J, Coen D M, Knipe D M. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67:5383–5393. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 23.Kristie T M, Roizman B. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc Natl Acad Sci USA. 1987;84:71–75. doi: 10.1073/pnas.84.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristie T M, Vogel J L, Sears A E. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci USA. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leib D A, Nadeau K C, Rundle S A, Schaffer P A. The promoter of the latency associated transcripts of herpes simplex virus type 1 contain a functional cAMP response element: role of the latency associated transcripts and cAMP in reactivation of viral latency. Proc Natl Acad Sci USA. 1991;88:48–52. doi: 10.1073/pnas.88.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, Luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsden H S, Campbell M E, Haarr L, Frame M C, Parris D S, Murphy M, Hope R G, Muller M T, Preston C M. The 65,000-Mr DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987;61:2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKnight J L, Kristie T M, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misra V, Blumenthal R M, Babiuk L A. Proteins specified by bovine herpesvirus-1 (infectious rhinotracheitis virus) J Virol. 1981;40:367–378. doi: 10.1128/jvi.40.2.367-378.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misra V, Bratanich A C, Carpenter D, O'Hare P. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV alpha gene trans-inducing factor. J Virol. 1994;68:4898–4909. doi: 10.1128/jvi.68.8.4898-4909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misra V, Walker S, Yang P, Hayes S, Ohare P. Conformational alteration of Oct 1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Mol Cell Biol. 1996;16:4404–4413. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.No D, Yao T P, Evans R M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Hare P, Goding C R. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 36.O'Rourke D, O'Hare P. Mutually exclusive binding of two cellular factors within a critical promoter region of the gene for the IE110k protein of herpes simplex virus. J Virol. 1993;67:7201–7214. doi: 10.1128/jvi.67.12.7201-7214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou W J, Cameron P H, Thomas D Y, Bergeron J J. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 38.Perng G C, Dunkel E C, Geary P A, Slanina S M, Ghiasi H, Kaiwar R, Nesburn A B, Wechsler S L. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol. 1994;68:8045–8055. doi: 10.1128/jvi.68.12.8045-8055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perng G C, Slanina S M, Yukht A, Drolet B S, Keleher W J, Ghiasi H, Nesburn A B, Wechsler S L. A herpes simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. J Virol. 1999;73:920–929. doi: 10.1128/jvi.73.2.920-929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston C M, Cordingley M G, Stow N D. Analysis of DNA sequences which regulate the transcription of a herpes simplex virus immediate early gene. J Virol. 1984;50:708–716. doi: 10.1128/jvi.50.3.708-716.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preston C M, Frame M C, Campbell M E. A complex formed between cell components and a HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell. 1988;52:425–434. doi: 10.1016/s0092-8674(88)80035-7. [DOI] [PubMed] [Google Scholar]
- 42.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Field's virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 2221–2230. [Google Scholar]
- 43.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Field's virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 2231–2296. [Google Scholar]
- 44.Russell J, Stow E C, Stow N D, Preston C M. Herpes simplex virus genes involved in latency in vitro. J Gen Virol. 1987;68:3009–3019. doi: 10.1099/0022-1317-68-12-3009. [DOI] [PubMed] [Google Scholar]
- 45.Sawtell N M, Thompson R L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner I, Spivack J G, Deshmane S L, Ace C I, Preston C M, Fraser N W. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. J Virol. 1990;64:1630–1638. doi: 10.1128/jvi.64.4.1630-1638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stern S, Tanaka M, Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 48.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 49.Su Y H, Meegalla R L, Chowhan R, Cubitt C, Oakes J E, Lausch R N, Fraser N W, Block T M. Human corneal cells and other fibroblasts can stimulate the appearance of herpes simplex virus from quiescently infected PC12 cells. J Virol. 1999;73:4171–4180. doi: 10.1128/jvi.73.5.4171-4180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Leary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Triezenberg S J, LaMarco K L, McKnight S L. Evidence of DNA:protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 52.Wada I, Rindress D, Cameron P H, Ou W J, Doherty J J D, Louvard D, Bell A W, Dignard D, Thomas D Y, Bergeron J J. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- 53.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 54.Xiao P, Capone J P. A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein-DNA complex assembly with Oct-1. Mol Cell Biol. 1990;10:4974–4977. doi: 10.1128/mcb.10.9.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]