Abstract
In flowering plants, the predominant sexual morph is hermaphroditism, and the emergence of unisexuality is poorly understood. Using Cucumis melo (melon) as a model system, we explore the mechanisms driving sexual forms. We identify a spontaneous mutant exhibiting a transition from bisexual to unisexual male flower, and identify the causal mutation as a Harbinger transposon impairing the expression of Ethylene Insensitive 2 (CmEIN2) gene. Genetics and transcriptomic analysis reveal a dual role of CmEIN2 in both sex determination and fruit shape formation. Upon expression of CmACS11, EIN2 is recruited to repress the expression of the carpel inhibitor, CmWIP1. Subsequently, EIN2 is recruited to mediate stamina inhibition. Following the sex determination phase, EIN2 promotes fruit shape elongation. Genome-wide analysis reveals that Harbinger transposon mobilization is triggered by environmental cues, and integrates preferentially in active chromatin, particularly within promoter regions. Characterization of a large collection of melon germplasm points to active transpositions in the wild, compared to cultivated accessions. Our study underscores the association between chromatin dynamics and the temporal aspects of mobile genetic element insertions, providing valuable insights into plant adaptation and crop genome evolution.
Subject terms: Flowering, Plant molecular biology, Plant hormones
In flowering plants, hermaphroditism is widespread. Here the authors identified a transposon insertion that triggers plant sexual transition. This study highlights the role of transposons in plant adaptation and evolution.
Introduction
Flowering plants exhibit a remarkable diversity of reproductive strategies, ranging from asexual to crossbreeding, each strategy influencing the genetic landscape and evolutionary trajectory of plant populations. One crucial aspect of this reproductive diversity lies in the development of unisexual flowers, a phenomenon intricately linked to sex determination mechanisms1. In dioecious species, plants bear either male or female flowers. In monoecious species, separate male and female flowers develop on the same individual. Other intermediary systems also exist. Andromonoecious plants bear both male and bisexual flowers on the same plants, and gynoecious and androecious plants bear only female and male flowers, respectively1,2.
Two models have been proposed to explain the transition to unisexuality. The first model, known as sex allocation theory, posits that ecological and physiological factors favor directing resources toward a single sexual function rather than both3,4. The second model relies on population genetic arguments, suggesting that dioecy becomes advantageous in the presence of inbreeding depression5,6. Consistent with these concepts, in monoecious species, the female function is prioritized when resources are ample, while the male function is expressed more often under adverse conditions, or when the female function is already fulfilled7. Male plants excel in pollen donation, potentially attributed to their capacity to produce a greater quantity of male flowers and pollen8. Besides, pollen development is highly sensitive to adverse temperatures9. Producing more male flowers could serve as a compensatory mechanism, mitigating the impact of the adverse conditions on male function.
In spite of our progress in decoding the genetic pathway leading to the development of unisexual flowers10–18, the molecular mechanisms integrating environmental cues driving the emergence of new sexual morphs are still obscure. The tapestry of evolutionary processes is woven with the intricate threads of genetic innovation. Among the most enigmatic contributors are transposable elements (TEs), ubiquitous mobile genetic elements that traverse the genomes of organisms across the tree of life19. Based on their mobility, TEs can be classified as DNA transposons, which use a cut-and-paste mechanism for their mobilization, and retro-transposons, which jump via a reverse-transcribed RNA intermediate. These two classes are further divided into TE superfamilies and families based on particular sequences, such as the presence of specific terminal repeats or conserved protein domains19. For instance, Harbinger transposons are DNA transposons which usually encode a DDE transposase and a SANT/Myb/trihelix domain-containing DNA-binding protein20. The role of transposons in adaptation and genome evolution is complex and multifaceted. Transposons can cause genetic instability by disrupting the functions of essential genes. Transposons can also contribute to genome evolution by creating advantageous genetic modifications21.
In the present work, we investigated how sexual morphs emerge in Cucurbitaceae, a large plant family that displays different sexual morphs. We focused our investigation on a monoecious species, Cucumis melo (melon). Male flowers are prevalent in melon, while female flowers develop on the initial nodes of the branches. During early developmental stages, flowers are initially bisexual. Subsequently, sex determination occurs through the developmental arrest of either stamen or carpel primordia2,18. This process is controlled by the monoecy pathway implicating the andromonoecious (M), androecious (A), and gynoecious (G) genes. The G gene codes for the transcription factor CmWIP1, a C2H2 zinc finger transcription factor, which functions as a master regulator in the process of sex determination in cucurbits16. The A and M genes encode for two aminocyclopropane-1-carboxylic acid synthase enzymes, CmACS7 and CmACS11, respectively14,15. CmACS11 expression in lateral branches suppresses the expression of the male-promoting gene CmWIP1. CmACS7 expression in carpel primordia of female flowers inhibits stamina development through a non-autonomous mechanism17.
Through characterization of a spontaneous sex transition mutant, we identified a Harbinger transposon, AndroPIF, inserted in the Ethylene Insensitive 2 (EIN2). We investigated the role of EIN2 in sex determination and fruit development, and the role of TEs in the emergence of sexual morphs in plants.
Results
A Harbinger transposon insertion in the EIN2 gene is responsible for androecy in CAM106 line
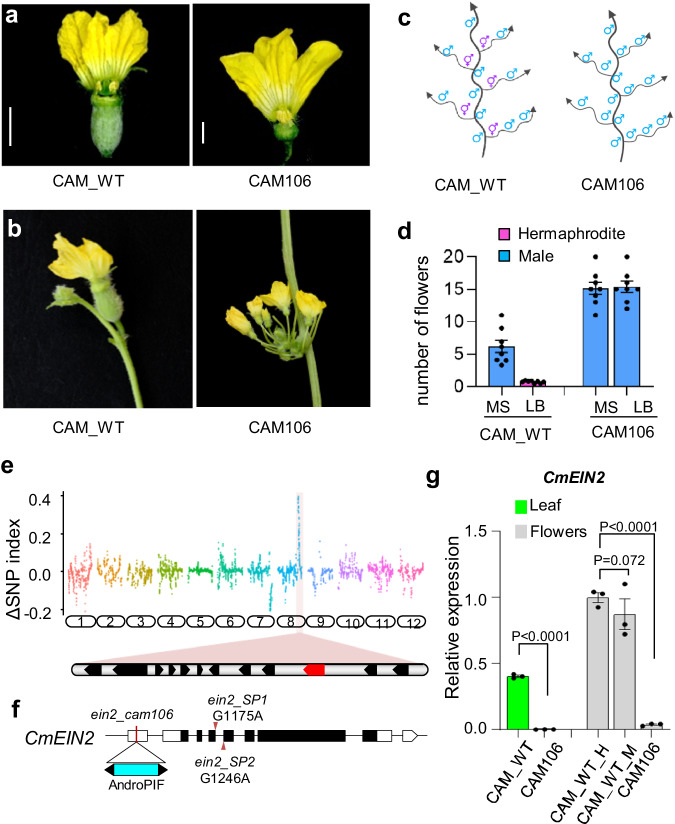
In andromonoecious melon, male flowers develop in inflorescences at every node of the main stem, and in distal nodes of the lateral branches. Hermaphrodite flowers develop at the first two nodes of lateral branches. During seed propagation of an andromonoecious Charentais line, hereafter named CAM_WT, we identified a spontaneous mutant line that is androecious, CAM106, where hermaphroditic flowers are converted to male unisexual flowers (Fig. 1a–c). Furthermore, male inflorescences develop in clusters of about 15 flowers, compared to 6 in the CAM_WT parental line (Fig. 1d). Androecy in melon can arise via the inactivation of CmACS11 or the melon CRABS CLAW gene (CmCRC)15,18. So, we sequenced the promoter and the coding regions of the two genes in CAM106 but found no mutations, pointing to a new locus controlling sex transition in cucurbits. To characterize the inheritance of such mutation, we crossed CAM106 to an andromonoecious line, CAM_Sister, and phenotyped 10 F1 hybrids and their F2 progenies. CAM_Sister, is genetically related to CAM_WT but not identical (see material and methods). We found all the F1 hybrids andromonoecious, indicating that the CAM106 mutation is recessive. Further phenotyping of 200 F2 plants showed 3:1 segregation of andromonoecy versus androecy, consistent with a single-locus recessive mutation leading to androecy (Supplementary Fig. S1a, b). We refer hereafter to this novel androecious mutation as a3. To identify the causal mutation in the CAM106 line, we sequenced bulked-genomic DNA from andromonoecious, and androecious segregant plants, and determined the delta-SNP index. We found a single genomic region on the right end of chromosome 8 completely associated with androecy (Fig. 1e). Fine mapping in a population of 1500 segregants plants, further delimited a3 to a single gene, CmEIN2, encoding the melon ortholog of the ethylene-insensitive protein 222 (Fig. 1f). Sequence analysis revealed an insertion of a DNA transposon of the P Instability Factor (PIF)/Harbinger superfamily8,20, henceforth termed AndroPIF at the 5’ UTR of CmEIN2 (Fig. 1f, Supplementary Figs. S1c and S2). Genotyping of plants harboring recombination events in the vicinity of a3, using a marker specific to the transposon insertion showed complete association of the transposon insertion in CmEIN2 and androecy (Supplementary Fig. S1d).
Fig. 1. A Harbinger transposon insertion in EIN2 gene is responsible for androecy.
a Sexual phenotype of andromonoecious melon (CAM_WT), and androecious mutant (CAM106). Scale bar = 1 cm. b Lateral branches showing a hermaphrodite flower in CAM_WT, and a cluster of male flowers in CAM106. c Schematic of sexual morphs. d Number of hermaphroditic or male flowers per node on the main stem (MS), or at the 1st node of lateral branches (LB). Data are mean ± SE. (n = 8 biological replicates). e Cloning of the a3 locus. ∆ SNP index between andromonoecious and androecious bulks, calculated with a 1-Mb sliding window. Arrows indicate candidate genes on chromosome 8. f Structure of the CmEIN2 gene and the position of AndroPIF insertion. The ein2_SP1 and ein2_SP2 splicing mutations are shown by arrowhead. g Quantitative real-time PCR (qPCR) of CmEIN2 in leaf, hermaphrodite (H) and male (M) flowers of CAM_WT, and CAM106. P-values from an unpaired two-tailed Student’s t-test are indicated. Data are mean ± SE. (n = 3 biological replicates). Source data are provided as a Source Data file.
Transposable elements are exposed to epigenetic modifications, with consequences on the expression of nearby genes, especially through the genomic spreading of DNA methylation23. We examined the DNA methylation status of CAM_WT parental line and CAM106, using whole genome bisulfite sequencing. As expected, strong DNA methylation of AndroPIF was observed. However, weak methylation spreading was detected in AndroPIF flanking sequences (Supplementary Fig. S3). To test if AndroPIF insertion was interfering with CmEIN2 transcript accumulation, we measured the expression of CmEIN2, using quantitative real-time PCR. We found a weak to no expression of CmEIN2 in leaves and flowers of CAM106, indicating that a3 is likely a loss-of-function allele of CmEIN2 (Fig. 1g).
Induced mutations in EIN2 lead to androecy in melon
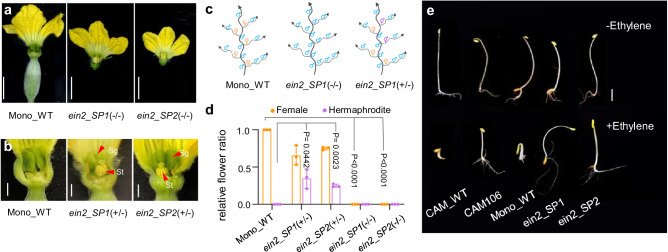
Since there is no efficient transformation protocol in cucurbits, to further validate the role of CmEIN2 in androecy, we used TILLING (Targeting Induced Local Lesions IN Genomes) collections from monoecious melon and screened for mutations in CmEIN2. Sixty-two induced mutations were identified, including two splicing, ein2-SP1 (G1175A) and ein2-SP2 (G1246A), and 17 missense mutations. As expected, missense mutations had no effect on the plant sexual phenotype (Supplementary Table 1), consistent with earlier reports on EIN2 mutagenesis in Arabidopsis22. We selected the two spicing mutants for further analysis. ein2-SP1 is altered in the splicing donor site of intron 4, while ein2-SP2 harbors a mutation in the splicing acceptor site of intron 4 (Supplementary Fig. S4a). Transcript analysis revealed introns 3 and 4 retention (30%), intron 4 retention (47%) or exon 4 skipping (23%) for ein2-SP1 mutant; similarly, ein2-SP2 mutation leads to intron 4 retention (27%) or to splice variant with a 4 bp deletion in exon 4 (73%). For both mutants, all the splicing forms lead to premature stop codons and thus are likely loss-of-function alleles (Supplementary Fig. S4b). We backcrossed the mutant lines harboring the splicing mutations to the wild type and followed the segregation of the mutations with the phenotypes. As predicted, plants homozygous for the splicing mutations were androecious, i.e., female flowers being transformed into male flowers (Fig. 2a, b; Supplementary Fig. S5). Interestingly, in plants heterozygous for ein2-SP1 and ein2-SP2 mutations, 20-40% of female flowers were transformed into bisexual flowers (Fig. 2c, d; Supplementary Fig. S5), highlighting also the role of CmEIN2 in stamen inhibition in female flowers. This phenotype was not observed in CAM106 mutant because CAM_WT parental line is andromonoecious developing male and hermaphrodite flowers (Fig. 1a).
Fig. 2. Induced mutations in CmEIN2 lead to androecy in melon.
a Sexual phenotype of a monoecious plant (Mono_WT), and ein2 splicing mutants (−/−). b Sexual phenotype of pistillate flowers of WT and ein2 heterozygous mutants (+/−). Sg, stigma; st, stamen. Scale bars in (a, b) = 0.5 cm. c Schematic of sexual morphs. d Relative ratio of female, or hermaphrodite flowers, per total carpel-bearing flowers. Data are mean ± SE. (n = 3 biological replicates). e Triple response phenotype of melon genotypes in absent (−Ethylene), and presence of ethylene (+Ethylene). The photography is taken 7 days after germination. Scale bars = 1 cm. Source data are provided as a Source Data file.
Since EIN2 is a key regulator of ethylene signaling24, we examined the ethylene response phenotype of 3 days old etiolated seedlings of CAM106-AndroPIF insertion mutant and ein2-SP1 and ein2-SP2 splicing mutants. Dark-grown melon seedlings treated with ethylene display a typical “triple response” consisting of inhibition of root development and hypocotyl elongation, radial swelling of the hypocotyl, and apical hook formation25. We found CAM106, ein2-SP1 and ein2-SP2 mutants to be insensitive to ethylene (Fig. 2e), supporting that the AndroPIF insertion and the splicing mutations are likely CmEIN2 loss-of-function alleles. Altogether, the characterization CmEIN2-AndroPIF and the CmEIN2-splicing mutants validate the role of CmEIN2 in the development of carpel-bearing flowers, both in andromonoecious and monoecious plants.
CmEIN2 is required both for carpel development and stamina inhibition
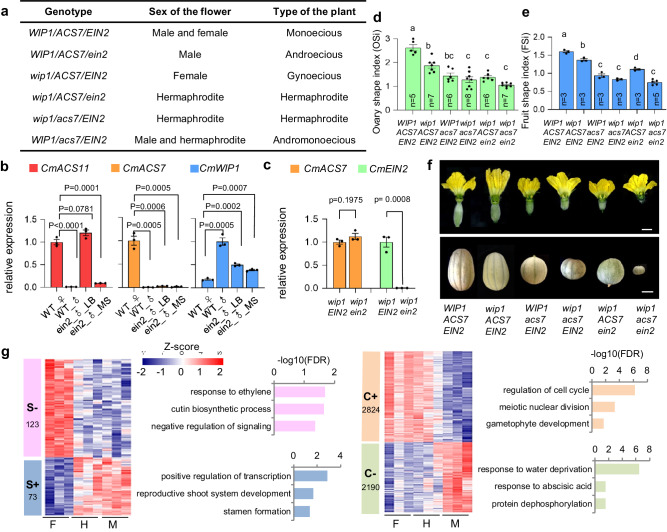
To integrate the function of CmEIN2 in the monoecy sex determination pathway, we generated mutant combinations with CmWIP1, CmACS7, and CmACS11, and examined the sex morphs of these double mutants in relation with the expression of sex genes (Fig. 3a). In melon, the inappropriate sexual organs are arrested at stage 6 after the initiation of both carpel and stamen primordia18. In monoecious plants, expression of CmACS11 in flower buds of lateral branches represses the expression of the male-promoting gene CmWIP1, leading to female flower development. Since loss of function of CmEIN2 leads to the development of male instead of female flowers in lateral branches, we first analyzed the expression of CmACS11, CmACS7, and CmWIP1. Compared to WT, the expression of CmACS11 was not compromised. In contrast, the repression of CmWIP1 expression was relieved, leading to male plants, and indirectly to repression of CmACS7 that relies on carpel development for its expression (Fig. 3a, b). To investigate the role of CmEIN2 in stamina inhibition, we crossed ein2 mutant to wip1 gynoecious line to generate ein2 and wip1 homozygotes double mutants, and analyzed the expression of the stamina inhibitor CmACS7. Compared to wip1 mutant, we found wip1 and ein2 homozygous double mutant fully hermaphroditic, despite the expression of the stamina inhibitor, CmACS7 (Fig. 3c).
Fig. 3. CmEIN2 is required both for carpel development and stamina inhibition.
a Sexual morphs of melon plants across combinations of alleles at different sex loci. b qPCR analysis of CmACS11, CmACS7 and CmWIP1 in Mono_WT female(♀) or male(♂) flowers, and in male flowers (♂) of CAM106(ein2) collected from main stems (MS), or lateral branches (LB). c qPCR analysis of CmACS7 and CmEIN2 in pistillate flowers of wip1,and wip1/ein2 double mutants. P-values from an unpaired two-tailed Student’s t-test are indicated in (b, c). Data in (b, c) are mean ± SE. (n = 3 biological replicates). d, e Ovary shape index(d) and fruit shape index(e) of different melon genotypes. Different letters indicate significant differences based on a one-way ANOVA with Tukey’s multiple comparisons test. Data in (d, e) are mean ± SE. n is indicated for number of biological replicates in (d, e). f Ovary and fruit phenotypes of the genotypes shown in (d and e). Scale bar in the flower panel = 1 cm. Scale bar in the fruit panel = 5 cm. g Gene-wise hierarchical clustering heat map of differentially expressed genes (DEGs, adjusted P value < 0.05) between female (F, wip1 mutant), hermaphrodite (H, wip1/ein2 double mutant), and males (M, ein2 mutant) flowers. DEGs (adjusted P value < 0.05) showing segregation into 4 clusters: stamen suppression (S−), stamen development (S+), carpel development (C+) and carpel suppression (C−). The z-score scale represents mean-subtracted regularized log-transformed read counts. GO terms specifying the biological process are shown at the right of the panels. Source data are provided as a Source Data file.
Ethylene has been associated with fruit shape development26. To investigate the role of CmEIN2 in fruit shape we measured the ovary shape index (OSi) and the fruit shape index (FSi). The OSi is determined as the ratio of the ovary length by the ovary width. FSi is determined as the ratio of the fruit length by the fruit diameter at mature stage. We found that OSi and FSi are significantly higher in Charentais Mono line (WT) compared to wip1/ein2 mutant. Conversely, the ovaries and fruits of wip1/ein2 mutants were flatter, phenocopying the one of acs716 mutant, while the double mutants showed an additive effect (Fig. 3d–f). Collectively, our findings highlight the dual function of EIN2 in both sex determination and fruit shape. In lateral branches, upon the expression of CmACS11, EIN2 is recruited to repress the expression of the carpel inhibitor, CmWIP1. Subsequent to the expression of CmACS7 in carpel primordia, EIN2 is recruited in the stamina inhibition processes. Following the sex determination phase, EIN2 promotes fruit shape elongation.
EIN2 signaling plays contrasting roles in carpel development and stamina inhibition
To gain insight into the gene network associated with CmEIN2 roles in stamina and carpel development, we examined the transcriptome of male, female, and hermaphrodite flower buds at stage 6. Male and female flower buds were collected from ein2 and wip1 loss-of-function mutants, respectively. Hermaphrodite flower buds were collected from wip1 and ein2 double mutant. Pairwise comparisons of the transcriptomes of male versus female, male versus hermaphrodite, and female versus hermaphrodite flowers, identified 8343, 5492 and 491 differentially expressed genes (DEG), respectively (Supplementary Fig. S6a, Supplementary Data 1). We grouped the differentially expressed genes by their expression patterns into 4 clusters: stamen suppression (S−), stamen development (S+), carpel suppression (C−), and carpel development (C+). Cluster S− and Cluster S+ denote genes that are upregulated, and genes that are downregulated, respectively, in female flowers compared to staminate flowers. Conversely, Cluster C− and Cluster C+ denote genes that are upregulated, and genes that are downregulated, respectively, in male flowers compared to pistillate flowers (Fig. 3g, Supplementary Fig. S6b). Gene Ontology (GO) term enrichment analysis revealed cluster S− enriched in GO terms of biological process related to response to ethylene and negative regulation of signaling (Fig. 3g). Conversely, cluster S+ is enriched in GO terms associated to stamen formation, reproductive shoot system development, and positive regulation of transcription (Fig. 3g, Supplementary Fig. S7). Consistent with this, RT-qPCR analysis showed the upregulation of ethylene responsive factor 1 (ERF1) and EIN3 Binding F-Box Protein (EBF1) associated with response to ethylene (GO: 0009723), and the downregulation of PISTILLATA (PI) and bHLH91 related to stamen development (GO: 0048443), in female, compared to staminate flowers (Supplementary Fig. S6c, d). Cluster C+ was found enriched in GO terms related to regulation of histone acetylation, gametophyte development, and mitotic cell cycle, while cluster C- is enriched in GO terms related to response to abscisic acid, water deprivation, and protein dephosphorylation (Fig. 3g, Supplementary Fig. S7). Alongside, RT-qPCR analysis validated the downregulation of the carpel marker genes, CRC and SEEDSTICK (STK) (GO: 0035065), and the upregulation of abscisic acid (ABA) associated genes, ABA RESPONSIVE ELEMENTS-BINDING FACTOR 3 (ABF3) and Cytochrome P450 (CYP707A10) (GO: 0009737) in male, compared to pistillate flowers (Supplementary Fig. S6e, f). In summary, our RNA-seq analysis reveals two contrasting indirect roles of CmEIN2 in carpel development, and in stamina inhibition. In developing carpel, through inhibition of the carpel repressor, CmWIP1 (Fig. 3b), EIN2 promotes the expression of genes associated with cell division and female gametophyte development, including the carpel identity gene, CRC (Fig. 3g, Supplementary Fig. S6e). In inhibited stamina, EIN2 promotes the expression of genes associated with negative regulation and signaling, and the repression of the stamina-promoting gene, PI (Fig. 3g, Supplementary Fig. S6c).
Transposons of the PIF/harbinger family are highly active transposons in melon
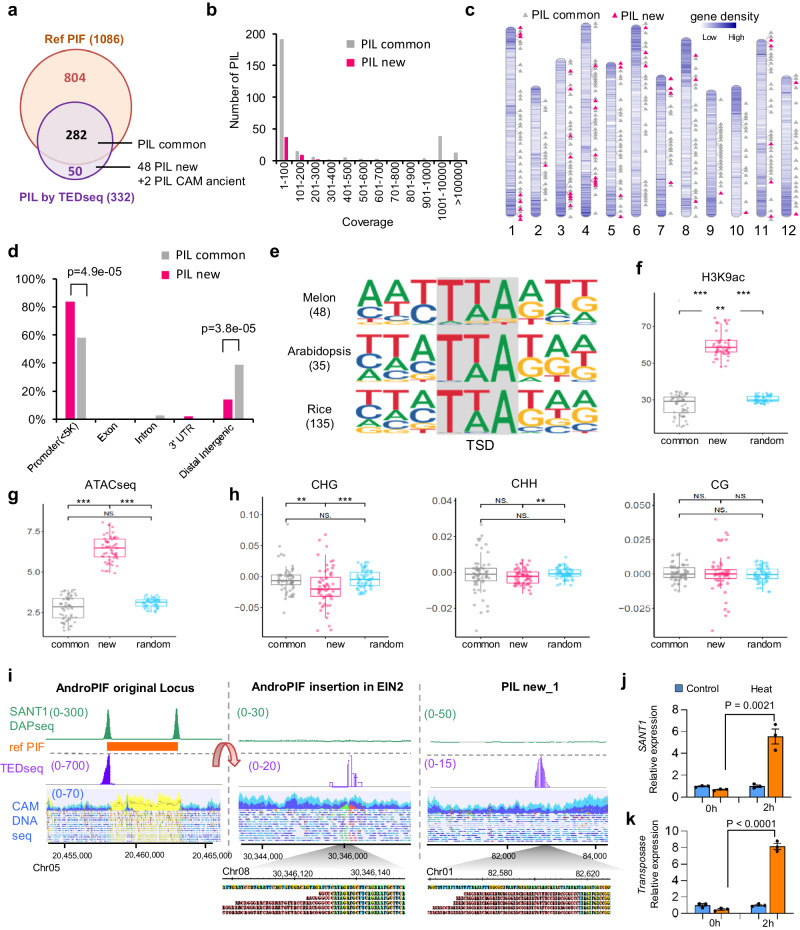
To determine if AndroPIF, and related PIF/Harbinger elements, are active transposons in the melon genome, we analyzed two biological replicates of pooled genomic DNA of 50 CAM_WT plants, including CAM106 control line. To identify AndroPIF insertions, we performed transposon display followed by illumina sequencing (TEDseq27; Supplementary Fig. S8). We detected an average of 34 read coverage per PIF/Harbinger insertion loci (PIL), in both replicates, indicating the high sensitivity of TEDseq (Supplementary Fig. S9a). In total, we found 332 PILs, out of which 282 were common between CAM and Mono_WT, and 50 were specific to CAM (Fig. 4a, b; Supplementary Data 2). Further analysis of the 50 CAM PILs identified two insertions as ancient ones, validated by broken-paired reads in the CAM_WT parental line, resulting in 48 new AndroPIF insertions. These new insertions were distributed over the 12 melon chomosomes, with preferential insertion bias in gene rich regions (Fig. 4c). Furthermore, 83% of novel insertions locate in promoter regions, compared to 58% of the common ones (Fig. 4d). Local sequence analysis of AndroPIFs insertion sites revealed the consensus motif ‘MWYTWARWK’ (Fig. 4e), similar to that previously reported for PIF transposons28,29. Open chromatin, H3K9ac histone mark, and low CHG/CHH DNA methylation are key features associated with active chromatin30. We observed a distinct pattern wherein active chromatin marks exhibit a positive correlation with the insertion of new PILs (Fig. 4f, h). In contrast, common AndroPIF insertions, indicative of ancient insertions, are consistently located in closed chromatin states, highlighting the temporal dynamics of chromatin from active to inactive, or reduced counter selection of insertions within inactive chromatin regions (Fig. 4g, h, Supplementary Fig. S9).
Fig. 4. AndroPIF transposon activity in melon genome.
a Venn plot of annotated PIF/Harbinger insertion Loci (PIL) in Mono genome (Ref PIL), and PIFs identified by TEDseq in poll of 50 CAM plants. b Sequence coverage of PILs identified by TEDseq. c Genomic position of PIL identified by TED-seq. PIL common, represent PILs that are common between CAM and Mono_WT. New PIL indicates new insertions observed in 50 plants descending from CAM parental line. d Distribution in percent of AndroPIF insertions in genes, and in intergenic regions. P-value from a Z-test are indicated and applied only to regions with non-zero values for both groups. e Target site insertion preference of new PILs, compared to mPing insertion sites, in Arabidopsis and rice28,29. f–h, Boxplots of H3K9ac (f), ATACseq(g), and DNA methylation(h) (CHG, CHH, and CG) levels in bin sizes of 100 bp in ±3 kb sequences flanking insertion sites of common PIFs, new PIFs, and random regions. For each boxplot, the lower and upper bounds of the box indicate the first (Q1) and third (Q3) quartiles, respectively, the center line indicates the median, and the whiskers extend to a maximum of 1.5 times the interquartile range (Q3–Q1). Number of bin is 60 for each box. Asterisks mark statistically significant differences (two-sided Wilcoxon rank-sum test, **P < 0.01, ***P < 0.001, NS. P > 0.05). i Genome browser view of SANT1 DAPseq peaks, TEDseq reads, and DNAseq, mapped on Mono genome. AndroPIF original locus in Mono genome, and in CAM_W (left), AndroPIF insertion in EIN2 in CAM106 line (middle), and a new PIL in CAM_W progeny plant (right). Color scheme in DNAseq track: blue and purple, pair-end reads; yellow, highly repetitive reads; green and red, discordant reads. Soft-clipped reads are shown as sequences with red background. J, k, qPCR of SANT1 (j) and Transposase (k) of AndroPIF in young leaf of CAM-WT under heat stress (42 °C/ 32 °C) and control (28 °C/22 °C) conditions. P-values from an unpaired two-tailed Student’s t-test are indicated. Data are mean ± SE. (n = 3 biological replicates). Source data are provided as a Source Data file.
PIF/harbinger SANT1 protein binds to PIF TIR sequences
AndroPIF, like most PIF/Harbinger transposons encodes two proteins: a nuclease and a DNA binding protein, SANT1 (Supplementary Fig. S2)20. Physical interaction between the two proteins is necessary for PIF/Harbinger transposition28,31. To further characterize AndroPIF, and related PIF/Harbinger transposon insertions in CAM melon line, we mapped genome wide DNA sequences binding to AndroPIF SANT1 protein, using amplified DNA affinity purification sequencing (ampDAP-seq)32. A total of 1521 SANT1 binding peaks were detected in the CMiso reference genome (Supplementary Data 3). Of this, 60,4% (n = 919) overlapped sequences annotated as PIF, 21.5% (n = 327) were linked to sequences annotated as non-TE, and 18% bound sequences annotated as TE, but not-PIF (Supplementary Fig. S10a, b). Sequence analysis of SANT1 insertion sites in melon genome revealed two consensus motifs. Motif 1 ‘TTYYTGTTTCTTGTT’ mapped at peak summits, while motif 2 ‘AGGGCCCGTTTGGTAACGTT’ was located at the flanking sides of the summits (Supplementary Fig. S10c–d). Further sequence annotation identified part of motif 2 as the TIR sequence ‘GGGCCCGTTTG’ of AndroPIF, pointing to the role of SANT1 in binding to the transposon extremities. Consistent with this, we systematically observed two SANT1 ampDAP-seq peaks (double peaks, Supplementary Fig. S10e, f) mapping to loci annotated as PIF, including the original copy of AndroPIF on chromosome 5, also captured by TEDseq (60,4% in Supplementary Fig. S10b). However, we found loci showing single peaks, corresponding to degenerate transposons that lost part of the TIR sequence, or random melon sequences that have similarities to TIR motif (18,5%, and 21.5% in Supplementary Fig. S10b). Several TEs have been shown to move following exposure to various environmental stressors33,34, we analyzed whether the expression of AndroPIF TRANSPOSASE and SANT1 genes are induced under heat stress conditions. CAM-WT plants were grown under control conditions (28 °C day/22 °C night), and shifted to heat stress conditions, for two hours at 42 °C. The expression of the TRANSPOSASE and SANT1 genes were then monitored by qPCR. We found the expression of both genes inducible by heat stress, pointing toward AndroPIF mobility triggered by abiotic factors (Fig. 4j, k).
PIF/Harbinger display contrasted transposition in wild versus cultivated melons
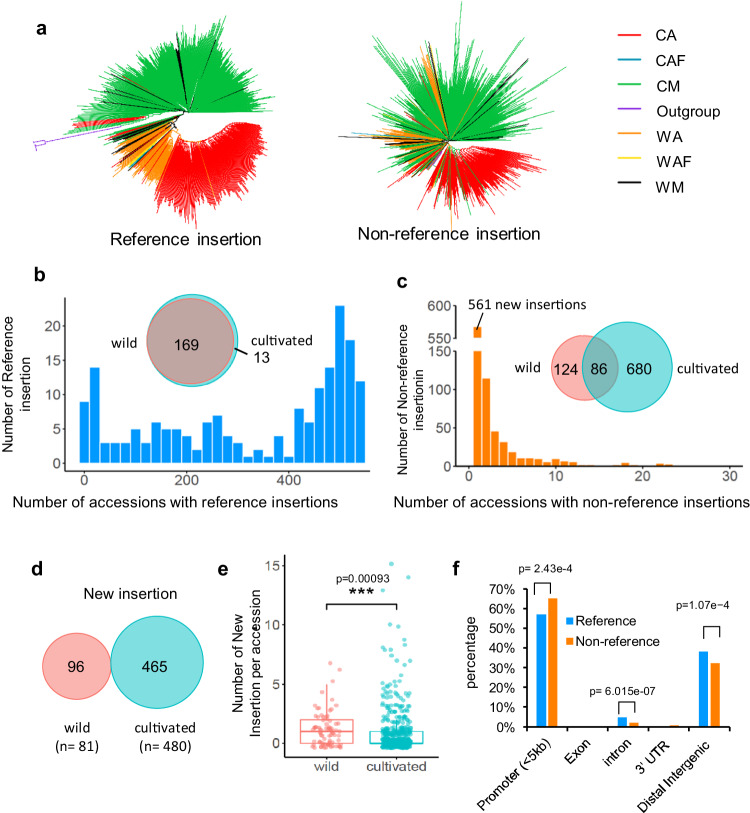
Crop domestication involved recurrent selection of desirable traits, leading to a severe loss in sequence diversity. We investigated whether sequence diversity due to PIF/Harbinger transposon insertions has been also constrained in cultivated compared to wild melon accessions. Positions of PIF/Harbinger polymorphic insertions in the genomes of 480 cultivated (group CM, CA, and CAF in Fig. 5a) and 81 wild (group WM, WAF, and WA in Fig. 5a) accessions35, collected from different geographic areas, was determined using short-read resequencing, and TEFLoN method36,37. Integration of PIF polymorphic insertion sites, SANT1 binding peak pairs, and search for TIR sequences resulted in the identification of 183 full-length AndroPIF copies (Supplementary Data 4, 5). The copies identified in the Mono-WT genome38 are denoted as reference insertions, while others are labeled as non-reference insertions (see Supplementary Fig. S11). Phylogenetic tree construction based on the presence/absence of reference AndroPIF elements exhibits a clustering structure akin to that generated by SNP polymorphism (Fig. 5a, Supplementary Data 6)35. In contrast, the tree generated based on non-reference insertions displays a mixed clustering pattern, suggesting that transposition of AndroPIF elements contributed to increased diversity in these populations. We found most PIF/Harbinger reference insertions (169 out of 182) shared between cultivated and wild accessions (Fig. 5b), indicating that they are ancestral. In contrast, out of the 894 non-reference-insertion, 561 are found in single accessions, pointing toward recent transposition events (Fig. 5c). We call this new category of PIF/Harbinger transposons “non-reference-new-insertions”. Distribution of non-reference-new-insertions in melon germplasms revealed active transpositions in wild melon accessions compared to melon breeding materials (Fig. 5d, e). As in CAM_WT, non-reference-new-insertions were overrepresented in promoter regions (65.14%), compared to reference insertion (57.09%), indicating that integration preference has shaped the genome landscape of AndroPIF insertions (Fig. 5f).
Fig. 5. PIF/Harbinger transposition in wild and cultivated melon accessions.
a Phylogenetic tree constructed using reference and non-reference PIF/Harbinger insertion sites identified using genomic re-sequencing data of 567 melon accessions, and 7 outgroup relatives (Zhao, 2019). AF, African; WAF, wild African; CAF, cultivated African; WM, wild melo; CM, cultivated melo; WA, wild agrestis; CA, cultivated agrestis. b, c Histogram and venn plots of reference (b) and non-reference (c) AndroPIF insertions identified in wild (WA and WM) and cultivated (CM and CA) accessions. d Venn plot of new insertions identified in wild and cultivated accessions. Number of accessions is indicated for each catalog. e Number of new-non-reference insertions per accession, in wild and cultivated accessions. P-values are calculated based on a two-sided Wilcoxon rank-sum test. For each boxplot, the lower and upper bounds of the box indicate the first (Q1) and third (Q3) quartiles, respectively, the center line indicates the median, and the whiskers extend to a maximum of 1.5 times the interquartile range (Q3–Q1). Number of accessions is indicated for each catalog in (d). f Distribution, in percents of AndroPIF insertions in genes, and intergenic regions. P-values from an Z-test are indicated and applied only to regions with non-zero values for both groups.
Discussion
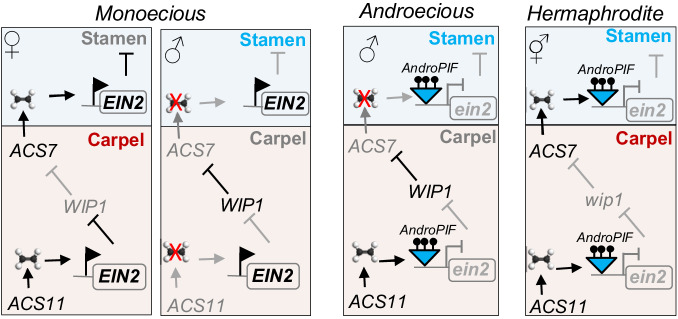
We investigated how new sex determination morphs could arise. During the propagation of an andromonoecious melon line, we encountered a spontaneous mutant, sexually converted from andromonoecy to androecy. This intriguing phenotype prompted us to study the causal mutation. We identified a PIF/Harbinger transposon as the origin of the sex transition, disrupting the expression of the Ethylene Insensitive 2 (CmEIN2) gene. Through genetic and expression analyses, we unveiled a dual function of CmEIN2 in sex determination and fruit shape. Upon the expression of CmACS11 in carpel primordia, of flowers developing on lateral branches, CmEIN2 is recruited to suppress the expression of the carpel inhibitor, CmWIP1. Subsequently, the expression of CmACS7, in developing carpel primordia, produce ethylene that is perceived in the stamina primordia, likely through a spatially differentially expressed ethylene receptors17. In stamina primordia, CmEIN2 is then recruited to mediate stamina inhibition, leading to female flower development. If CmEIN2 is inactivated, the repression of CmWIP1 expression is relieved leading to androecy. Inactivation of both CmEIN2 and CmWIP1 lead to hermaphrodite flower development (Fig. 6).
Fig. 6. Model of sex-determination pathway in melon integrating EIN2 function.
The model integrates ACS11, WIP1, and ACS7 sex-determination genes. Pink box indicates carpel primordia and blue box indicates stamen primordia. Spatial expression of ACS11, ACS7, or WIP1 is restricted to carpel primordia. EIN2 is expressed both in the carpel and stamina primordia. The chemical symbol indicates ethylene gas. Red cross indicates no ethylene produced by ACS7 or ACS11. Ethylene produced by ACS7 in the carpel is perceived in the stamina primordia likely through a spatially differentially expressed ethylene receptors17. Monoecious plants develop male and female flowers. AndroPIF insertion in EIN2 leads to Androecy. Ein2/wip1 double mutant is hermaphrodite. EIN2-mediated repression of WIP1 depends on the expression of ACS11. EIN2-mediated repression of stamen requires the expression of ACS7.
Transposons are powerful drivers of genome evolution and plasticity and contribute to genetic variability, potentially under stress conditions34. We thus tested whether PIF/Harbinger mobility is induced by adverse conditions. Structurally, PIF/Harbinger transposons are predicted to encode two proteins, a transposase and a DNA-binding protein. We found the expression of both genes activated by heat stress, pointing toward triggering of PIF/Harbinger mobility under adverse conditions. Similar stress-responsive transcription or movement has been reported in several TEs33,39. In rice, the DNA transposon mPing can be activated in response to cold and salt stress27; in tobacco, Tnt1 element can be induced by different biotic and abiotic stress34.
Given the sensitivity of TEs to environmental threats, our findings suggest that androecy may arise as a result of transposition triggered by stress into CmEIN2. Ethylene is a key feminizing hormone in cucurbits40,41. Consistent with this scenario, we found CmEIN2 required for the transcriptional repression of the male-promoting gene CmWIP1, leading to female flower development (Fig. 6). Under adverse conditions, fruit set could be costly, inactivation of CmEIN2 relieves CmWIP1 from the repression, and consequently leading to androecy. By producing only male flowers, androecious plants can have an advantage in promoting outcrossing.
The role of transposon mobilization in shaping sexual reproduction presented here could be a general mechanism. Besides AndroPIF transposon leading to androecy, we previously showed that the DNA transposon, Gyno-hAT is at the origin of male to female sex transition, resulting in gynoecious plants16. In species harboring sex chromosomes, TEs also tend to accumulate in genomic regions containing sex genes, underscoring their significant role in the emergence and development of sex chromosomes42.
In addition to promoting female flower development, ethylene is required for inhibition of stamina in pistillate flowers (Fig. 6). We introduced ein2 mutation in monoecious genetic background. We found plants heterozygous for ein2 mutation having 20–40% of female flowers converted to bisexual, pointing to the quantitative role of ethylene in sex determination (Fig. 2d). Furthermore, ethylene is a key regulator for fruit elongation in melon. Consistent with this, fruits developing on plant harboring ein2 mutation are significantly affected in both ovary and fruit shape indexes. In summary, our work proposes a model in which CmEIN2 play a central role both in the control of unisexual flowers in monoecious plants as well as the emergence of male and female unisexual plants.
The transcriptome analysis of flower buds collected from female (wip1/EIN2), hermaphrodite (wip1/ein2), and male (ein2) plants, revealed 4 clusters of gene expression patterns. We found S- cluster enriched in GO terms such as ethylene response genes, while S+ cluster contains several TFs, such as PI, implicated in stamen development. We also found C+ cluster enriched in GO terms related to female gametophyte development, and mitotic cell cycle, while C- cluster enriched in GO terms related to abscisic acid, water deprivation, and protein dephosphorylation. These results corroborate that ethylene signaling negatively regulates stamen development, while simultaneously exerting a positive effect on carpel development.
Annotating PIF/Harbinger elements in plant genomes presents several challenges due to their high diversity in sequences, sizes, and organizations. Beside in silico annotation, we used SANT1 ampDAPseq analysis as a tool to identify and annotate PIF/Harbinger transposons in melon genomes. In loci displaying ampDAPseq double peaks, we systematically found either a new or an ancient PIF element. However, in loci showing single ampDAPseq peaks, the accuracy of the method is weak, as the binding could be due to degenerate transposon or random sequences that has similarities to TIR sequences. Nevertheless, when we combined transposon annotation with SANT1 ampDAPseq analysis, the approach proved highly efficient in identifying new transpositions as well as ancient and degenerate transpositions.
Following the annotation of PIF/Harbinger elements, we classified them into three categories, refence PILs, non-reference PILs and new PILs. We found most reference and non-reference PILs reflect the genetic relationship between cultivated and wild melon accessions, while new PILs corresponded to recent insertions. Interestingly, we found the new PILs more frequent in wild melon accessions compared to breeding materials, pointing toward their roles in plant adaptations. The movement of transposons in wild and cultivated plants is of particular interest because it can provide new genetic variation for plant breeding43–45. The majority of novel insertions are located in promoter regions. It will be interesting to explore their influence on the modulation of gene expressions and, as a result, on plant adaptation.
Our work reveals a central role of PIF/Harbinger class of transposons in the emergence of sexual morphs in flowering plants. As PIF/Harbinger are present in organisms spanning the tree of life19, our study will likely inspire future investigations beyond cucurbits.
Methods
Plant material
The CAM106 spontaneous mutant was discovered during the propagation of the andromonoecious line CAM_WT (Cucumis melo L. var. cantalupensis). To identify the causal mutation, we crossed CAM106 to CAM_Sister, an andromonoecious EMS-mutagenized mutant line derived from CAM_WT, and phenotype the F2 segregant plants. The plants were cultivated in a greenhouse with a daytime temperature of 27 °C, a nighttime temperature of 21 °C, and a photoperiod of 16 hours of light. The plants were assessed for flower sex type and fruit shape.
Bulk segregant analysis
Genomic DNA was extracted from CAM106 X CAM_Sister F2 population. For bulk segregation analysis, equal amounts of genomic DNA from 10 individual plants with either andromonoecious or androecious phenotypes were pooled to create the andromonoecious and androecious pools. Subsequently, DNA libraries were prepared, using the Next Ultra DNA Library Prep Kit, and 150 bp paired-end reads sequenced on HiSeq 2000 Illumina platform. Reads were aligned to the reference Charentais Mono melon genome CMiso1.138 (here referred to as Mono_WT). The SNP-index across all loci was calculated, and the delta (Δ) SNP-index was determined by subtracting the SNP-indices of the two bulks at each locus (SNP-index_mutant – SNP-index_WT).
Triple response assay
Melon seeds were germinated in a 10-litter wet box in the dark for 7 days at 27 °C, either with or without 50 μM ethylene. Ethylene solution at 50 μM concentration was prepared by diluting 1 M ethephon (Sigma) in 5 mM Na2HPO3 buffer, as described previously25.
Sexual morph phenotyping and epistasis analysis
Monoecious melons carry only male flowers in the main stem, female flowers develop on the first nodes (L1) of lateral branches. The following flowers on L2 and L3 nodes of the lateral branches can be female depending upon plant age and environment. Melon also exhibits other sex morphs, andromonoecious, gynoecious, androecious and hermaphrodite. Androecious plants bear only male flowers. Gynoecious and hermaphrodite plants develop only female and hermaphrodite flowers, respectively. Andromonoecious plants develop separate male and hermaphrodite flowers. To position EIN2 function in the monoecy pathway we generated double mutants, through crossing of the single mutant and genotyping F2 segregant plants. The loss-of-function alleles leading to gynoecy (wip1), androecy (acs11) or andromomonoecy (acs7) used in the epistasis analysis are described in Boualem et al14., Martin et al16., and Boualem et al15., respectively14–16. Single, and double homozygous plants for wip1, acs11, acs7, and ein2 mutations were phenotyped for sexual transition of the flowers on the male stem and lateral branches.
Quantitative RT-PCR
Total RNA was extracted from frozen leaves or flowers. First-strand cDNA was synthesized from 2 pg of total RNA with the Superscript® III reverse transcriptase (Invitrogen). Primer design was performed using the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Primer sequences used are listed in Supplementary Table 2. Polymerase chain reactions were performed with the Bio-Rad CFX96 Real-time PCR apparatus, with qPCR MasterMix Plus for SYBR® Green I (Eurogentec, France). Gene expression is normalized to the expression levels of housekeeping genes: CmActin2. The qRT-PCR results were analyzed using a ΔΔCt methodology.
RNA-seq assay
Flower buds at developmental stage 6 were sampled, and total RNA was extracted using the NucleoSpin RNA Plus kit (Macherey Nagel). The quality and concentration of the RNA were determined using Agilent RNA chips (Bioanalyser 2100). Three biological replicates for female (wip1), hermaphrodite (wip1/ein2), and male (ein2) flowers were used for library preparation. RNA-seq was performed on the Illumina NovaSeq 6000 with 150 bp paired-end reads. The reads were aligned to the reference genome CMiso1.1 by STAR (v2.7.5c)46, and the mapped reads were assigned to genes with featureCount (v2.0.3)47. Differentially expressed gene calling was conducted using DiCoExpress48, with a p-value adjusted <0.05. GO enrichments were completed by ClusterProfiler49 with customized GO annotation.
Whole-genome bisulfite sequencing
Genomic DNA of young leaves of CAM_WT and CAM106 were extracted with CTAB. The gDNA was used to generate the libraries and treated by bisulfite. Sequencing was done on Illumina NovaSeq 6000 with 150 bp paired-end reads. The reads were analyzed by MethylStar pipeline50. The differentially methylated sites (DMRs) were identified by methylkit51 and DMRcaller52.
TE display sequencing
TEDseq was performed on a DNA pool of 50 CAM_WT seedlings, and CAM-106 DNA, used a control. Genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN). Sequencing libraries were prepared from 1 μg of pooled DNA using the NEBNext® Ultra™ II DNA Library Prep Kit. PCR enrichment was carried out for 20 cycles, using Illumina P7 Primer with index, and P5 primers PIF-P1 or PIF-P2 (Supplementary Table 2; Supplementary fig. S8). Nested PCR enrichment was then carried out for 12 cycles, using PIF-P3 or PIF-P4. This process was independently repeated twice. Pair-end sequencing was carried out using the Illumina Miseq, generating 150 bp reads. To identify new TE insertions, soft-clipped and discordant reads that partially mapped to the reference sequence of AndroPIF were extracted. The reads were subsequently remapped to the CMisoV1.1 reference genome using Bowtie2 v2.3.5 with the arguments ‘very-sensitive-local’53. Putative insertions were retained if they were supported by more than one set of 3 soft-clipped and discordant reads in at least one repeat at the same locus. TE target site analysis was performed with a Rscript, using packages ‘dplyr’, ‘readxl’, ‘knitr’, ‘ggplot2’, ‘ggseqlogo’ and ‘Biostrings’. For each identified region, reads overlapping putative site and TE sequence were retrieved and trimmed with Cutadapt54, and were aligned with Bowtie2 on CMisoV1.1 genome. The exact positions of the insertion sites were calculated by strand specificity and CICAR flag. A sequence of 6 nucleotides flanking the insertion site were extracted to calculate consensus matrix and motifs. The level of epigenetic marks around the TE insertion sites were calculated by Deeptools3.5.5 using the data described previously17,55,56 (accession number: PRJNA383830).
DNA affinity purification sequencing
AndroPIF SANT1coding sequence was isolated from leaf RNA by RT-PCR and cloned into pIX-HALO by Gateway recombination according to the manufacturer’s protocol (Invitrogen). The primers used in cloning of SANT1 are listed in the Supplementary Table 2. Expression of HALO-SANT1 fusion protein and enrichment of DNA targets were performed as described previously32. The DNA used for DNA library preparations was extracted from the Charentais Mono line (WT). AmpDAP-seq was employed, utilizing amplification to eliminate DNA methylation. Library preparations followed the manufacturer’s protocol for the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (Illumina). The final enriched DNA library was sequenced on a NextSeq500 as 2×75 nt reads. The control HALO protein AmpDAP-seq data were generated previously18 (accession numbers: SRX15631305, SRX15631306).
Transposable elements insertion detection in melon germplasm
The genomic sequences of the 567 C. melo accessions, and the 7 related species (Supplementary Data 6) were retrieved from previous study35. The genome wide identification of AndroPIF related elements was performed using the TEFLoN method and the McClintock pipeline36.
TEs were subsequently annotated manually, using SANT1 ampDAPseq peaks, and the search of TIR sequences ‘GGGCCCGTTTG’, allowing 2 bp mismatches. CMisoV1.1 genome was used a reference genome for the mapping of the germplasm sequences.
The gff3 results from McClintock pipeline were converted to vcf format with the code present(REF0/0)/deletion(1/1) for reference insertion, and absence(0/0)/insertion(1/1) for non-reference insertion. For phylogenetic tree building, Euclidean distance was computed for bitwise matrix and the Neighbor Joining method was used for clustering. R packages ‘vcfR’, ‘poppr’, ‘ggtree’, and ‘dplyr’ were used for the analysis.
Heat stress assays
Germinated seeds were grown for 3 weeks in the greenhousse at 27 °C daytime, 21 °C nighttime, and a photoperiod of 16 hours of light. For the heat stress assay, the plants were transferred to a growth chamber with a temperature set at 42 °C. Total RNA samples were extracted at 0 hours, and 2 hours after the initiation of the heat stress.
Statistical analyses
All experiments were carried out in at least three biological replicates. Differences in mean for RT-qPCR data were assessed using an unpaired two-sided Student’s t-test. For genomic distribution in percent of TE insertions in promoter, genes, or in intergenic regions, a two-sided Wilcoxon rank-sum test was performed and applied only to regions with non-zero values for both groups. For each boxplot, the lower and upper bounds of the box indicate the first (Q1) and third (Q3) quartiles, respectively, the center line indicates the median, and the whiskers extend to a maximum of 1.5 times the interquartile range (Q3–Q1). All the specific statistic methods were described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
The authors thank Olivier Martin for the critical reading of the manuscript. We thank Vincent Rittener for technical assistance, and IPS2 glasshouse staffs and the research facilities provided by GAFL and the experimental unit, for plant handling. We thank the IPS2 bioinformatics team for providing help and computing resources. This work was supported by European Research Council(grant ERC-NectarGland Project, No.101095736 to A.Be.), Agence National de la Recherche (grant NECTAR, No. ANR-19-CE20-0023 to A.Be.), Inititiative d’Excellence Paris-Saclay (grant Lidex-3P, No. ANR-11-IDEX-0003-02 to A.Be. and A.Bo.).
Author contributions
The conceptualization of the project was undertaken by A.Be. and A.Bo. The methodology. involved the efforts of H.H., S.Z., F.A.C., F.T., C.P., H.S.P., C.T., F.S., Q.C., and F.M. The investigation was conducted by A.Be., H.H., S.Z., F.A.C., M.V., A.Bo., C.D., and L.Q. The visualization was handled by H.H., S.Z., and M.V. Funding was acquired by A.Be., and project administration was led by A.Be. Supervision of the project was provided by A.Be., A.Bo., and C.D. The original draft of the writing was done by A.Be. and S.Z., and the review and editing were carried out by A.Be., S.Z., A.Bo., L.Q., and C.D.
Peer review
Peer review information
Nature Communications thanks Pierre Bourguet and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive database under the accession number PRJNA1085522, including RNA-Seq, Bisulfite-Seq, TEDseq, and AmpDAP-seq. Previously published data used in this study are SRX15631305, SRX15631306, and PRJNA383830. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hsin-Ya Huang, Siqi Zhang, Fadi Abou Choucha.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-49250-9.
References
- 1.Barrett SC. The evolution of plant sexual diversity. Nat. Rev. Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- 2.Ming R, Bendahmane A, Renner SS. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- 3.Charnov EL, Bull JJ, Maynard Smith J. Why be an hermaphrodite? Nature. 1976;263:125–126. doi: 10.1038/263125a0. [DOI] [Google Scholar]
- 4.Pannell JR, Jordan CY. Evolutionary transitions between hermaphroditism and dioecy in animals and plants. Annu. Rev. Ecol. Evol. Syst. 2022;53:183–201. doi: 10.1146/annurev-ecolsys-102320-085812. [DOI] [Google Scholar]
- 5.Charlesworth D, Charlesworth B. Population genetics of partial male-sterility and the evolution of monoecy and dioecy. Heredity. 1978;41:137–153. doi: 10.1038/hdy.1978.83. [DOI] [Google Scholar]
- 6.Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. Am. Nat. 1978;112:975–997. doi: 10.1086/283342. [DOI] [Google Scholar]
- 7.Dai C, Galloway LF. Male flowers are better fathers than hermaphroditic flowers in andromonoecious Passiflora incarnata. N. Phytol. 2012;193:787–796. doi: 10.1111/j.1469-8137.2011.03966.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang SQ. Flower dimorphism and the maintenance of andromonoecy in Sagittaria guyanensis ssp. lappula (Alismataceae) N. Phytol. 2003;157:357–364. doi: 10.1046/j.1469-8137.2003.00676.x. [DOI] [PubMed] [Google Scholar]
- 9.Hedhly A. Sensitivity of flowering plant gametophytes to temperature fluctuations. Environ. Exp. Bot. 2011;74:9–16. doi: 10.1016/j.envexpbot.2011.03.016. [DOI] [Google Scholar]
- 10.Akagi T, et al. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant cell. 2018;30:780–795. doi: 10.1105/tpc.17.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akagi T, Henry IM, Tao R, Comai L. A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science. 2014;346:646–650. doi: 10.1126/science.1257225. [DOI] [PubMed] [Google Scholar]
- 12.Müller NA, et al. A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants. 2020;6:630–637. doi: 10.1038/s41477-020-0672-9. [DOI] [PubMed] [Google Scholar]
- 13.Harkess A, et al. Sex determination by two Y-linked genes in garden asparagus. Plant cell. 2020;32:1790–1796. doi: 10.1105/tpc.19.00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boualem A, et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science. 2008;321:836–838. doi: 10.1126/science.1159023. [DOI] [PubMed] [Google Scholar]
- 15.Boualem A, et al. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science. 2015;350:688–691. doi: 10.1126/science.aac8370. [DOI] [PubMed] [Google Scholar]
- 16.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 17.Rashid D, et al. Ethylene produced in carpel primordia controls CmHB40 expression to inhibit stamen development. Nat. Plants. 2023;9:1675–1687. doi: 10.1038/s41477-023-01511-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, et al. The control of carpel determinacy pathway leads to sex determination in cucurbits. Science. 2022;378:543–549. doi: 10.1126/science.add4250. [DOI] [PubMed] [Google Scholar]
- 19.Wells JN, Feschotte C. A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 2020;54:539–561. doi: 10.1146/annurev-genet-040620-022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapitonov VV, Jurka J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004;23:311–324. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- 21.Fedoroff NV. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 22.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 23.Cui X, Cao X. Epigenetic regulation and functional exaptation of transposable elements in higher plants. Curr. Opin. Plant Biol. 2014;21:83–88. doi: 10.1016/j.pbi.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Li W, et al. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell. 2015;163:670–683. doi: 10.1016/j.cell.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 25.Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boualem A, et al. Ethylene plays a dual role in sex determination and fruit shape in cucurbits. Curr. Biol. 2022;32:2390–2401.e2394. doi: 10.1016/j.cub.2022.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Va D, et al. Transposon display identifies individual transposable elements in high copy number lines. Plant J. 1998;13:121–129. doi: 10.1046/j.1365-313X.1998.00004.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang G, Zhang F, Hancock CN, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2007;104:10962–10967. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 30.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019;20:207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 31.Sinzelle L, et al. Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc. Natl Acad. Sci. USA. 2008;105:4715–4720. doi: 10.1073/pnas.0707746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartlett A, et al. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 2017;12:1659–1672. doi: 10.1038/nprot.2017.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarevitch I, et al. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015;11:e1004915. doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidl MF, Thomma BP. Transposable elements direct the coevolution between plants and microbes. Trends Genet. 2017;33:842–851. doi: 10.1016/j.tig.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhao G, et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019;51:1607–1615. doi: 10.1038/s41588-019-0522-8. [DOI] [PubMed] [Google Scholar]
- 36.Nelson MG, Linheiro RS, Bergman CM. McClintock: an integrated pipeline for detecting transposable element insertions in whole-genome shotgun sequencing data. G3-Genes Genom. Genet. 2017;7:2763–2778. doi: 10.1534/g3.117.043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adrion JR, Song MJ, Schrider DR, Hahn MW, Schaack S. Genome-wide estimates of transposable element insertion and deletion rates in Drosophila melanogaster. Genome Biol. Evol. 2017;9:1329–1340. doi: 10.1093/gbe/evx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pichot C, et al. Cantaloupe melon genome reveals 3D chromatin features and structural relationship with the ancestral Cucurbitaceae karyotype. Iscience. 2022;25:103696. doi: 10.1016/j.isci.2021.103696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietzenuk B, et al. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements. Genome Biol. 2016;17:1–15. doi: 10.1186/s13059-016-1072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Switzenberg JA, Little HA, Hammar SA, Grumet R. Floral primordia-targeted ACS (1-aminocyclopropane-1-carboxylate synthase) expression in transgenic Cucumis melo implicates fine tuning of ethylene production mediating unisexual flower development. Planta. 2014;240:797–808. doi: 10.1007/s00425-014-2118-y. [DOI] [PubMed] [Google Scholar]
- 41.Martínez C, Jamilena M. To be a male or a female flower, a question of ethylene in cucurbits. Curr. Opin. Plant Biol. 2021;59:101981. doi: 10.1016/j.pbi.2020.101981. [DOI] [PubMed] [Google Scholar]
- 42.Hobza R, et al. Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Res. 2015;23:561–570. doi: 10.1007/s10577-015-9496-2. [DOI] [PubMed] [Google Scholar]
- 43.Castanera R, Morales-Díaz N, Gupta S, Purugganan M, Casacuberta JM. Transposons are important contributors to gene expression variability under selection in rice populations. Elife. 2023;12:RP86324. doi: 10.7554/eLife.86324.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domínguez M, et al. The impact of transposable elements on tomato diversity. Nat. Commun. 2020;11:4058. doi: 10.1038/s41467-020-17874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carpentier M-C, et al. Retrotranspositional landscape of Asian rice revealed by 3000 genomes. Nat. Commun. 2019;10:24. doi: 10.1038/s41467-018-07974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 48.Lambert I, Paysant-Le Roux C, Colella S, Martin-Magniette M-L. DiCoExpress: a tool to process multifactorial RNAseq experiments from quality controls to co-expression analysis through differential analysis based on contrasts inside GLM models. Plant Methods. 2020;16:1–10. doi: 10.1186/s13007-020-00611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahryary Y, Hazarika RR, Johannes F. MethylStar: A fast and robust pre-processing pipeline for bulk or single-cell whole-genome bisulfite sequencing data. BMC Genom. 2020;21:1–8. doi: 10.1186/s12864-020-06886-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:1–9. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Catoni M, Tsang JM, Greco AP, Zabet NR. DMRcaller: a versatile R/Bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Res. 2018;46:e114. doi: 10.1093/nar/gky602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 55.Latrasse D, et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics Chromatin. 2017;10:1–17. doi: 10.1186/s13072-017-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
Sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive database under the accession number PRJNA1085522, including RNA-Seq, Bisulfite-Seq, TEDseq, and AmpDAP-seq. Previously published data used in this study are SRX15631305, SRX15631306, and PRJNA383830. Source data are provided with this paper.