Abstract
Liver failure from chronic hepatitis C is the leading indication for liver transplantation in the United States. However, the pathogenesis of liver injury resulting from chronic hepatitis C virus (HCV) infection is not well understood. To examine the relationship between HCV replication in liver tissue and hepatocellular injury, a strand-specific in situ hybridization procedure was developed. The sensitivity and specificity of digoxigenin-labeled riboprobes were optimized by analyzing Northern blots and cell lines expressing HCV RNAs. For the current study, both genomic (sense) and replicative-intermediate (antisense) HCV RNAs were detected and quantified in 8 of 8 liver tissue specimens from infected patients versus 0 of 11 liver tissue specimens from noninfected controls. The distribution pattern for HCV replicative-intermediate RNA in liver was different from that for HCV genomic RNA. HCV genomic RNA was variably distributed throughout infected livers and was located primarily in the cytoplasm of hepatocytes, with some signal in fibroblasts and/or macrophages in the surrounding fibroconnective tissue. However, HCV replicative-intermediate RNA showed a more focal pattern of distribution and was exclusively localized in the cytoplasm of hepatocytes. There was no significant relationship between the distribution pattern for HCV genomic RNA and any indices of hepatocellular injury. However, a highly significant correlation was observed between the percentage of cells staining positive for replicative-intermediate RNA and the degree of hepatic inflammatory activity (P, < 0.0001). Furthermore, the ratio of cells staining positive for HCV replicative-intermediate versus genomic RNA correlated with the histological severity of liver injury (P, 0.0065), supporting the hypothesis that active replication of HCV in liver tissue may be a significant determinant of hepatocellular injury.
Hepatitis C virus (HCV) is an important cause of chronic liver disease leading to cirrhosis and end-stage liver disease in humans. Chronic hepatitis C is now recognized as a leading indication for orthotopic liver transplantation in the United States. HCV is a positive-strand RNA virus of approximately 9.4 kb with a single open reading frame encoding a polyprotein of 3,011 amino acids. HCV has been classified as a Hepacivirus within the family Flaviviridae, based on its genomic organization and genetic homology with pestiviruses and flaviviruses. By analogy with other members of the Flaviviridae, it is assumed that HCV replication requires the production of an antisense or replicative-intermediate RNA, from which progeny genomic strands are transcribed.
The diagnosis of HCV infection is established by detection of HCV-specific antibodies in patient serum by serological assays and by detection of HCV genomic (positive-strand) RNA in serum by molecular assays, such as reverse transcriptase (RT) PCR (RT-PCR) (10, 12). Detection of HCV replicative-intermediate RNA by RT-PCR has been controversial due to inherent limitations in achieving strand specificity (19). Recent studies with improved methods, however, appear to confirm the hypothesis that HCV replicates in human liver (20, 24). Several reports have described the detection of HCV RNA in liver tissue by the techniques of in situ hybridization and in situ PCR (2, 7–9, 13, 17, 18, 21, 22, 26, 28, 29, 31–34). However, several issues remain unresolved due to conflicting data. In some studies, only a subset of biopsies from patients with hepatitis C stained positive for HCV RNA, and only a small percentage of hepatocytes appeared to be infected (2, 23). In other studies, HCV genomes were found in over 90% of biopsies from infected patients (8, 9, 25, 34), suggesting that HCV infection may be more widely disseminated in human liver than previously thought. Previous studies have reported a wide range in the percentages of hepatocytes positive for HCV RNA in positive livers; the reports have also presented conflicting data on the correlations among hepatic RNA, serum RNA, and hepatocellular injury. Reports suggesting no relationship between the level of HCV RNA in the liver and the degree of hepatocellular injury have led to the hypothesis that HCV may not be a cytopathic virus (24, 25, 28). However, other reports have suggested a correlation between HCV RNA and/or HCV antigens in the liver and the degree of liver injury (1, 11, 13, 32).
To help resolve these important issues, the current report describes the development and optimization of a new quantitative and highly sensitive strand-specific in situ hybridization assay for detecting HCV RNAs in liver tissue. With this assay, different and distinct intrahepatic distribution patterns were observed for HCV genomic and replicative-intermediate RNAs, in terms of tissue distribution patterns and percentages of positive cells. The examination of HCV RNAs in infected tissues has led to the novel finding that the percentage of hepatocytes positive for replicative-intermediate RNA is significantly correlated with the index of liver injury, while the pattern of positivity for HCV genomic RNA shows no such correlation.
MATERIALS AND METHODS
Collection and processing of liver tissue.
Liver needle biopsy specimens were obtained from patients seen at the Hepatology Service of the University of Washington Medical Center under informed consent. Immediately after being released from the needle, the core biopsies were divided into three portions; the largest piece was processed by formalin fixation and paraffin embedding for histologic examination, the second piece was placed in guanidinium thiocyanate solution for HCV RNA testing by reverse transcription (RT)-PCR, and the remaining piece was snap-frozen in OCT medium (0.2% [wt/wt] polyvinyl alcohol, 4.3% [wt/wt] polyethylene glycol, 85.5% [wt/wt] nonreactive ingredients) and stored in a −70°C freezer for in situ hybridization.
Larger portions of explanted cirrhotic livers were collected from HCV-infected patients at the time of liver transplantation and processed in the manner described above. A human liver which was found to be unsuitable for organ transplantation due to excessive fat and which tested negative for HCV RNA by RT-PCR (12) was processed for use as negative control tissue. Ten HCV-negative needle biopsies examined in this study were obtained from HCV antibody-negative patients diagnosed with autoimmune cirrhosis, cryptogenic cirrhosis, or primary sclerosing cholangitis.
All liver biopsies and explants were evaluated by use of routine hematoxylin- and eosin-stained sections by pathologists who had no knowledge of the serum HCV infectivity status. Specific liver biopsies or explants were chosen for evaluation based upon the degree of inflammatory activity in the native livers or histologic evidence of recurrent hepatitis C infection in posttransplant biopsies. The degree of liver injury due to hepatitis C infection was assessed with the histologic activity index described by Knodell et al. (16). Components of liver injury that were assessed included portal inflammation, lobular degeneration and necrosis, piecemeal necrosis, and fibrosis. The hepatitis activity was deemed mild if the piecemeal necrosis and the lobular degeneration and necrosis were given scores of 1, moderate if they were given scores of 3, and severe if they received scores of 4. When there was a discrepancy between the scores for piecemeal necrosis and for lobular degeneration and necrosis, the higher score prevailed.
Generation of cDNA clones and riboprobes.
Portions of cDNAs of the human housekeeping genes for beta-actin and hypoxanthine-guanine phosphoribosyltransferase (HPRT) and HCV 5′ untranslated region (5′UTR) core and envelope 1 (E1) genes were amplified by RT-PCR with the primer pairs described in Table 1. E1 genes of both HCV genotypes 1a and 1b were incorporated because of the high genetic variability in this region (4). PCR products were purified with a PCR purification kit from QIAGEN Inc. (Chatsworth, Calif.) and cloned into plasmid pCRII (Invitrogen, San Diego, Calif.) according to the manufacturer's protocol to generate the recombinant plasmids illustrated in Fig. 1. Positive clones were selected by amplification with native primers to identify the correct inserts, and the orientation of the inserts was determined by examination of plasmid restriction enzyme digestion patterns.
TABLE 1.
Dig-labeled riboprobes
| Clone | Insert (nucleotide [nt] location) | Size of riboprobe (nt) | Primers used for RT-PCR | Reference or source |
|---|---|---|---|---|
| pACS | Human beta-actin cDNA (408–1064)a | 657 | AT-1 and AT-4 | 27 |
| pCORE | Core gene of HCV genotype 1a (489–751)b | 263 | c104 and c186 | 30 |
| pCU | HCV 5′UTR and core gene (183–513)b | 331 | Ish1 and c186 (M&M) | 30 |
| pE1-1a | E1 gene of HCV genotype 1a (802–1639)b | 838 | JBE1_1S and JBE_1AS | 4 |
| pE1-1b | E1 gene of HCV genotype 1b (802–1639)b | 838 | JBE1_1S and JBE_1AS | 4 |
| pHR14 | Human HPRT cDNA (316–604)c | 289 | 5′HPRT and 3′HPRT | 37 |
| pSPT18-neo | Bacterial neomycin transferase gene (383–1114)d | 732 | Boehringer |
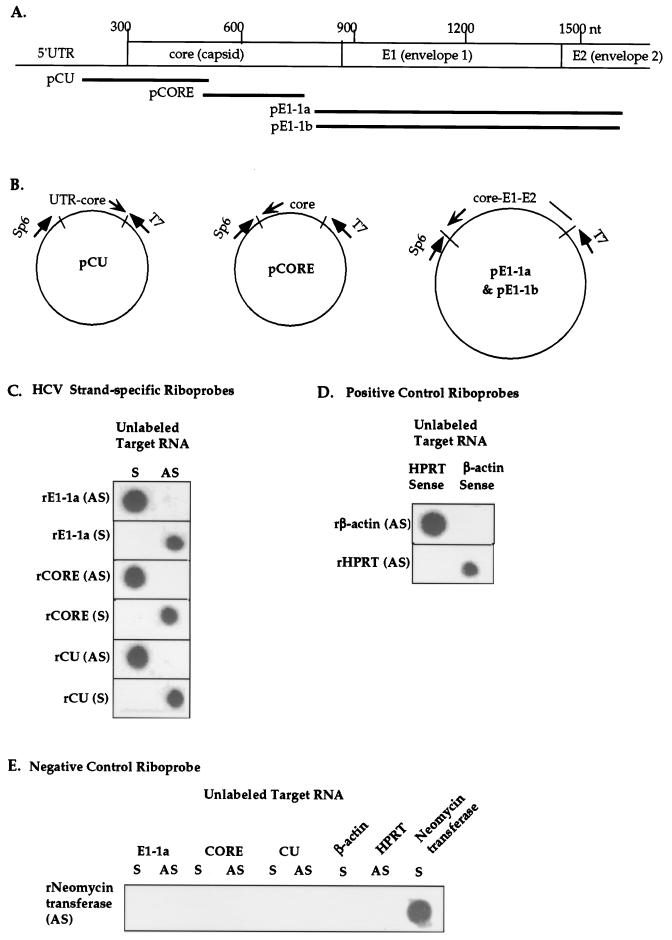
FIG. 1.
Characterization of Dig-labeled riboprobes specific for HCV sense and antisense RNAs. (A) Map location of riboprobes within the HCV genome (see Table 1 for nucleotide positions). Abbreviations: 1a, HCV genotype 1a; 1b, HCV genotype 1b; nt, nucleotide. (B) Recombinant plasmids containing subgenomic fragments of HCV genotype 1a or 1b were constructed for generation of riboprobes by in vitro transcription from bacteriophage T7 or Sp6 promoters. (C) Dot blot assay for assessing the strand specificity of HCV riboprobes. Unlabeled HCV genomic (sense [S]) and replicative-intermediate (antisense [AS]) target RNAs were synthesized by in vitro transcription from recombinant plasmid pE1-1a, pCORE, or pCU (B), treated with DNase I, and blotted onto nylon membranes. Unlabeled target RNAs were hybridized with Dig-labeled riboprobes synthesized from the genomic (S) or antigenomic (AS) strands of the corresponding recombinant DNA templates. For example, rE1-1a (AS), indicates the antigenomic riboprobe corresponding to the HCV genotype 1a E1 gene. (D) Specificity of positive control riboprobes specific for human beta-actin and HPRT sense RNAs. (E) Specificity of negative control riboprobe for Neo. Unlabeled sense (S) RNA probes and Dig-labeled antisense (AS) RNA probes were generated from recombinant DNA templates containing human and bacterial genes and were hybridized in the dot blot assay.
To generate riboprobes, cDNA inserts plus flanking bacteriophage promoter sequences (T7 or Sp6) were amplified by PCR with primers Csj7 (GACCATGATTACGCCAAAGC) and m13fII (GTAAAACGACGGCCAGTG). Digoxigenin-11-UTP (Dig)-labeled riboprobes were synthesized from purified PCR product templates by runoff transcription with T7 or Sp6 polymerase and with incorporation of Dig according to the manufacturer's protocol (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). The production of RNA and the subsequent removal of the DNA template were monitored by agarose gel electrophoresis and RNA denaturing gel electrophoresis. Briefly, 20 μl of in vitro transcription reaction mixture was incubated with 1 μl of DNase I (amplification grade) (Life Technologies, Gaithersburg, Md.) for 1 h or until the DNA template was invisible on agarose gels while a strong RNA band remained. DNase I was inactivated by incubation at 65°C for 10 min in the presence of 2.5 mM EDTA. Dig-labeled riboprobes were further broken down to an average size of 100 nucleotides by alkaline hydrolysis (6). The final riboprobe was precipitated and resuspended in 0.1% sodium dodecyl sulfate.
The pSTP18-neo plasmid containing the bacterial neomycin transferase gene (Neo) was purchased from Boehringer for use in generating negative control riboprobes. Plasmid DNA was cleaved with PvuII in preparation for in vitro transcription with T7 polymerase. The antisense transcript was processed by DNase digestion and alkaline hydrolysis.
Each newly made Dig-labeled riboprobe was evaluated against a known, standard Dig-labeled RNA according to the manufacturer's protocol (Boehringer). Typically, serial 10-fold dilutions of the Dig-labeled riboprobes were applied to nylon membranes along with serial 10-fold dilutions of standard Dig-labeled RNA. The concentrations of experimental riboprobes were estimated by comparing spot intensities of the standard control and the experimental dilutions. Probe concentrations were further optimized by Northern dot blot hybridization (see Results), and probe concentrations were adjusted to equivalent reactivity for all in situ hybridization experiments.
Generation of positive control cell lines.
To generate control cell lines expressing either HCV positive-strand (genomic) or HCV replicative-intermediate (antigenomic) RNA, DNA containing the HCV genotype 1a core plus E1 was amplified by PCR with primers Ish1 (CGACCGGTCCTTTTTGGA) and x(e2)19j (4). The Ish1 sequence is located 159 bp upstream of the start codon of the core gene, and the x(e2)19j sequence is located near the 3′ end of the hypervariable region of the E2 gene. The purified PCR product was subcloned into plasmid pCR2.1 (Invitrogen), and the colony PCR method was used to select a single clone containing the correct insert. HCV DNA inserts were subcloned into the eukaryotic expression vector pcDNA3 in both sense (pCE-S) and antisense (pCE-AS) orientations relative to the promoter-enhancer of human cytomegalovirus.
Huh7 cells (38), which are human hepatocellular carcinoma cells, were cultured in Dulbecco's modified Eagle medium containing high levels of glucose (0.45%) and supplemented with 10% fetal bovine serum, 50 U of penicillin per ml, and 50 μg of streptomycin per ml. Ten micrograms of purified pcDNA3, pCE-S, or pCE-AS DNA was transfected into 3 × 106 Huh7 cells by electroporation (electroporation system; BTX, San Diego, Calif.). Subsequently, transfected cell lines were selected by culturing in the presence of 600 μg of G418, a neomycin analog (Calbiochem, La Jolla, Calif.), per ml.
To prepare the cell lines for in situ hybridization analysis, approximately 8 × 104 cells were suspended in 50 μl of 1% bovine serum albumin (nuclease-free grade; Calbiochem) after being washed twice with 1× phosphate-buffered saline (PBS). Positively charged slides were precoated with 50 μl of 1% bovine serum albumin and centrifuged for 5 min at 800 rpm (Shandon Lipshaw, Pittsburgh, Pa.). Cells were air dried before fixation with formalin for 5 min at room temperature, after which they were washed and stored in 3× PBS at 4°C until in situ hybridization analysis.
In situ hybridization.
Frozen sections (6 μm) were initially placed on 50°C heating blocks for 2 min to improve adherence, fixed in 10% neutral buffered formalin for 5 min at room temperature, and sequentially washed in 3× PBS and 1× PBS for 5 min each. The tissue sections were treated with 0.2 N HCl for 2 min and proteinase K (1 μg/ml) for 2 to 3 min and rinsed with diethyl pyrocarbonate-treated deionized water. Slides were soaked in equilibration solution (50% formamide, 0.6 M NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 50 μg of heparin per ml, 10 mM dithiothreitol [DTT]) for 10 min at room temperature followed by prehybridization solution (50% formamide, 0.6 M NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 50 μg of heparin per ml, 10 mM DTT, 10% polyethylene glycol 8000, 1× Denhardt's solution) at 50°C for 1 h. The reagents were obtained from Novagen (Madison, Wis.).
Approximately 15 μl of Dig-labeled riboprobes was applied to each slide at a final concentration of 2 to 4 ng/μl in hybridization buffer (50% formamide, 0.6 M NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 50 μg of heparin per ml, 10 mM DTT, 0.5 mg of carrier DNA per ml, 0.5 mg of tRNA per ml, 10% polyethylene glycol 8000, 1× Denhardt's solution). For analysis of HCV RNA, mixtures of core and genotype-specific E1 riboprobes (Fig. 1A) were used as HCV antisense or sense riboprobes. In all experiments, a mixture of HPRT and beta-actin antisense riboprobes was used as a positive control, and the antisense riboprobe of Neo served as a negative control. During the hybridization steps, tissue sections were covered with siliconized coverslips, sealed with rubber cement, and incubated at 50°C in a humidified chamber for 18 h.
After hybridization, sections were soaked in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to remove the coverslips, washed in 2× SSC at 50°C for 30 min, and treated with RNase (20 μg/ml in 2× SSC) at 37°C for 30 min to reduce nonspecific background. Subsequently, sections were washed once in 50% formamide–2× SSC for 30 min at 50 to 65°C and two times in 1× SSC at 50 to 65°C for 30 min. The wash temperature was optimized by titration for each probe.
Immunological detection.
Tissue sections were soaked in 2% blocking reagent in 100 mM Tris (pH 7.5)–150 mM NaCl for 30 min at room temperature, followed by incubation with anti-Dig-alkaline phosphatase conjugate (1:250 dilution) at 4°C overnight in a humidified chamber. Sections were washed twice with buffer 1 (100 mM Tris [pH 7.5], 150 mM NaCl) and once with 100 mM Tris buffer (pH 8.2) at room temperature. Vector red substrate (Vector Laboratories, Burlingame, Calif.) supplemented with 1.25 mM levamisole (Sigma, St. Louis, Mo.) was added for 30 minutes before the reaction was terminated by soaking the slides in buffer IV (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Slides were counterstained with 2% methyl green and dehydrated by successive washings with 95% ethanol, 100% ethanol, and xylene before permanent mounting. Slides were examined and photographed with bright-field microscopy.
For in situ experiments, the specificity of the positive signal was evaluated with the following controls. (i) Cells were stained with riboprobes against endogenous RNAs (HPRT and beta-actin) to evaluate the integrity of RNA in frozen sections, including specimens which were negative for HCV riboprobes, to confirm their RNA preservation. (ii) Cells were stained with the Neo riboprobe to establish the level of nonspecific background staining. (iii) All probes were omitted from the hybridization solution, allowing evaluation of any potential endogenous alkaline phosphatase activity in liver tissue. No endogenous alkaline phosphatase was detected by the in situ hybridization procedure as described (data not shown).
Statistical analysis.
To estimate the percentage of cells positive for either HCV genomic or replicative-intermediate RNA, we counted 50 to 120 cells per slide, distinguished by nuclear counterstaining, depending on the size of the liver tissue sample. The Pearson chi-square test was used to test the significance of differences between the percentages of positive cells in two inflammatory grades. Linearity was assessed for the percentage of cells positive for the HCV RNA signal and the severity of liver damage in three histopathologic classifications by use of the Mantel-Haenszel chi-square test. The statistical analyses were carried out by use of SPSS for Macintosh, version 6.1.1 (SPSS Inc., Chicago, Ill.).
RESULTS
Characterization of Dig-labeled riboprobes.
In order to develop an in situ hybridization system for the specific detection of HCV genomic and replicative-intermediate RNAs in human tissues, a series of control experiments were designed to optimize and characterize HCV riboprobes. Figure 1 presents a summary of such optimization experiments. A summary of HCV cDNA clones and their genetic locations, sizes of inserts, and primers used for amplification is listed in Table 1 and illustrated in Fig. 1A and B. Riboprobe synthesis is described in Materials and Methods.
For each riboprobe, Northern dot blot experiments were performed to determine positive reactivity as well as strand and genotype specificity. Representative Northern dot blot experiments are shown in Fig. 1C, D, and E. Unlabeled RNAs were synthesized from both strands of each clone and dotted to nylon membranes to serve as target RNA. The targets were probed with either sense or antisense Dig-labeled riboprobes synthesized from the same or different HCV genes or cellular control genes.
Three sets of sense and antisense Dig-labeled riboprobes were hybridized against the corresponding sense and antisense unlabeled RNA transcripts as described in the legend to Fig. 1C. Strong, highly specific reactivity was evident in each case. Positive control antisense riboprobes derived from beta-actin and HPRT cDNAs and the negative control riboprobe were hybridized against unlabeled transcripts from the same genes in Fig. 1D and E. Strong hybridization signals with high specificity were observed with these positive and negative control riboprobes. The weak reactivity between the Neo riboprobe and the HCV core antisense target was due to small amounts of homologous vector sequences present in both RNA constructs.
Establishment of cell lines expressing HCV sense and antisense RNAs.
To further evaluate the sensitivity and specificity of our in situ hybridization protocol, permanent cell lines expressing subgenomic regions of HCV sense and antisense RNAs were established. DNA containing the HCV 5′UTR, core, and E1 genes was amplified by PCR from plasmid pTET/HCV5"T73′AFL, obtained from Charles Rice (Washington University, St. Louis, Mo.), and subcloned into expression vector pcDNA3 as shown in Fig. 2A. Plasmid pcDNA3 contains enhancer-promoter sequences from the major immediate-early gene of human cytomegalovirus, which drives high-level transcription in human cell lines (3). Two plasmids, pCE-S and pCE-AS, were constructed to allow HCV RNA expression from either the positive (genomic) strand or the negative (antigenomic) strand of the HCV genome. Plasmids pCE-S and pCE-AS were transfected into Huh7 cell lines, and permanent transfectants (Huh7-hcvS and Huh7-hcvAS) were isolated after 3 weeks of selection in the presence of the neomycin analog G418.
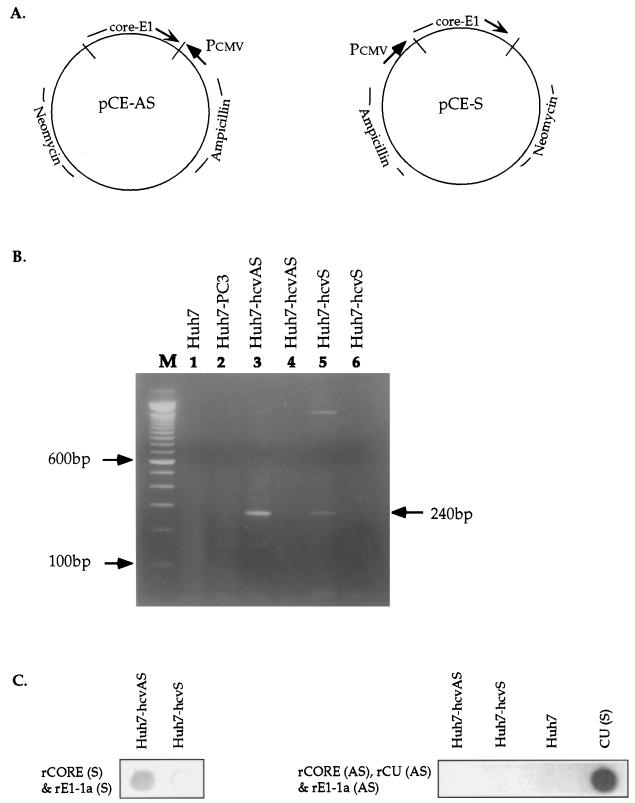
FIG. 2.
Generation and analysis of human cell lines expressing HCV genomic and antigenomic RNAs. (A) Construction of expression plasmids pCE-AS and pCE-S. The entire core and E1 genes of HCV genotype 1a were aligned in either the sense orientation (pCE-S) or the antisense orientation (pCE-AS) relative to the human cytomegalovirus major immediate-early promoter regulatory region (Pcmv). Huh7 cells were transfected with plasmid pCE-S or pCE-AS, and stable transformants, designated Huh7-hcvS and Huh7-hcvAS, were selected as described in Materials and Methods. (B) Analysis of HCV RNA expression in cell lines Huh7-hcvS and Huh7-hcvAS by RT-PCR. Total RNA was extracted from cells, digested with DNase I prior to reverse transcription in the presence of a mixture of hexamer (pdN6) oligonucleotides (lanes 3 and 5), and then amplified with primers c256 and c186 (30). The arrow on the right denotes the appropriate expected band of 240 bp. In lanes 4 and 6, the reverse transcription step was eliminated prior to PCR amplification under conditions identical to those used for lanes 3 and 5, respectively. In lanes 1 and 2, RNA from negative control cell lines Huh7 and Huh7-PC3 was analyzed by the same method as in lanes 3 and 5. Lane M contains size markers. (C) Northern dot blot analysis of RNA from transfected or control cell lines with strand-specific HCV riboprobes. CU (S) designates a genomic-strand synthetic HCV RNA which was derived from the pCU clone and which served as a positive control for the antisense riboprobes. See the legend to Fig. 1 for an explanation of other designations.
To verify the expression of HCV RNA, total RNAs were extracted from Huh7, Huh7-PC3 (permanent pcDNA3-transfected cells), Huh7-hcvAS, and Huh7-hcvS cells and treated with DNase I prior to RT-PCR with primers c186 and c256 (30). The results of RT-PCR and control DNA PCR experiments are shown in Fig. 2B, indicating that subgenomic HCV RNAs were expressed in Huh7-hcvAS and Huh7-hcvS cell lines. Trace amounts of residual HCV DNA were detected in DNase I-treated Huh7-hcvAS RNA, as indicated by a weak positive signal (Fig. 2B, lane 4).
Northern dot blot analysis was performed to examine the expression of the positive- and negative-strand RNAs. Ten micrograms of RNA was isolated from Huh7-hcvAS and Huh7-hcvS cells, treated with DNase I, blotted to nylon membranes, and probed with strand-specific HCV riboprobes as described in the legend to Fig. 2C. Following hybridization with HCV sense riboprobes (Fig. 2C), total RNA from the Huh7-hcvAS cell line gave a positive hybridization signal, while RNA from the Huh7-hcvS cell line was negative. These results were consistent with the expectation that Huh7-hcvAS cells would produce only the antisense RNA of HCV genes. In a parallel experiment (Fig. 2C, right panel), HCV sense RNA was not detected in the Huh7-hcvS cell line by Northern dot blotting with antisense probes, although HCV sense RNA was detected in the cell line by strand-specific RT-PCR (Fig. 2B). Thus, Huh7-hcvS cells expressed small amounts of sense RNA detectable by RT-PCR but not by Northern dot blotting. The most likely explanation for this result is that the Huh7-hcvS cell line is not a clonal population, as was originally expected. In Fig. 2C, the positive control HCV sense RNA was efficiently detected by the HCV antisense riboprobes.
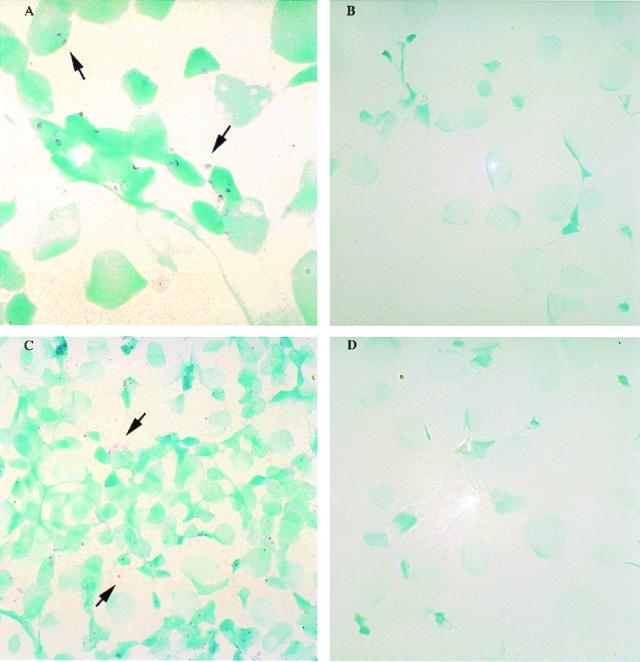
The expression of HCV RNAs in the cell lines was further examined by in situ hybridization. Huh7-hcvAS, Huh7-hcvS, and control Huh7 cells were harvested and each loaded on positively charged slides by cytospinning. Figure 3 shows in situ hybridization results obtained with HCV antisense riboprobes (Fig. 3A and B) and HCV sense riboprobes (Fig. 3C and D). The positive strand of HCV RNA was detected at low levels by antisense riboprobes in a subset of Huh7-hcvS cells (Fig. 3A) but not in Huh7 cells (Fig. 3B). Similarly, the negative strand of HCV RNA was appropriately detected by sense riboprobes in Huh7-hcvAS cells (Fig. 3C) but not in Huh7 cells (Fig. 3D). Importantly, sense and antisense riboprobes did not detect signals in Huh7-hcvS and Huh7-hcvAS cell lines, respectively (data not shown). The positive signals in Fig. 3A and C were located outside the nuclei, confirming that the signals resulted from in situ detection of cytoplasmic RNAs and not plasmid DNA. The results in Fig. 3 demonstrate (i) the establishment of positive control cell lines for in situ hybridization experiments, (ii) the specificity of the riboprobes for in situ experiments, and (iii) higher sensitivity of the in situ hybridization method than of Northern dot blot analysis for detecting HCV RNA transcripts.
FIG. 3.
In situ analysis of HCV genomic and replicative-intermediate RNAs in cell lines with Dig-labeled riboprobes. Cells in panels A and B were probed with HCV antisense riboprobes, while cells in panels C and D were probed with HCV sense riboprobes. (A) Huh7-hcvS cell lines, which express positive-strand (i.e., genomic polarity) HCV RNA. (C) Huh7-hcvAS cell lines, which express negative-strand (i.e. replicative-intermediate) HCV RNA. (B and D) Control Huh7 cell lines. Nuclear methyl green stain was used; magnifications, ×250 (A) and ×100 (B, C, and D). Arrows indicate the positive signal for HCV RNA.
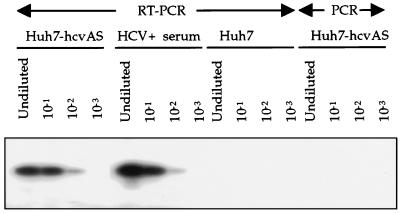
The cells expressing HCV antisense RNA (Huh7-hcvAS) appeared to be a particularly homogeneous population, since approximately 90% (95 of 107, 92 of 102, and 82 of 92 cells) of HCV-hcvAS cells showed a positive signal by in situ hybridization. The variance of signal intensity among cells was negligible. Therefore, experiments were performed to determine the sensitivity of our in situ hybridization assay. Total RNA was extracted from a known quantity of Huh7-hcvAS cells, converted to cDNA, and analyzed by serial dilution end-point PCR as described previously (12). The last specimen giving a positive RT-PCR result was used to calculate the HCV RNA titer. As a control, human serum containing known quantities of HCV RNA was spiked into solubilized Huh7 cells; the viral RNA was extracted and analyzed by RT-PCR in a parallel experiment (Fig. 4). Using this approach, we estimated that Huh7-hcvAS cells contained, on average, seven equivalent copies of HCV RNA per cell.
FIG. 4.
Determination of the level of HCV RNA expressed in Huh7-hcvAS cells. Total RNA was extracted from cells and a clinical serum sample and then digested with DNase I prior to reverse transcription in the presence of a mixture of hexamer (pdN6) oligonucleotides. Serial 10-fold dilution of cDNA was done before amplification with primers c256 and c186 (30). In the last four lanes, the reverse transcription step was eliminated prior to PCR amplification under conditions identical to those used for the first four lanes. PCR products were denatured and hybridized with 32P-labeled internal primer c104 before being applied to a 6% polyacrylamide gel.
In situ analysis of HCV genomic RNA in human liver tissue.
Given the high sensitivity of our current assay, the next objective was to qualitatively assess HCV genomic RNAs in a panel of human liver tissues obtained from patients with active HCV infections and explanted liver tissues obtained from patients who had end-stage liver disease and who underwent liver transplantation at the University of Washington Medical Center (Table 2). Adjacent pieces of biopsies or explanted liver were submitted for histological analysis by hospital pathologists and also were tested for the presence of HCV genomic RNA by a highly sensitive RT-PCR assay (12). Altogether, 10 HCV RNA-negative biopsies, 6 HCV RNA-positive biopsies, one HCV RNA-negative explanted liver, and two HCV RNA-positive explanted livers were tested by in situ hybridization for genomic HCV RNA with antisense riboprobes and control riboprobes.
TABLE 2.
Clinical and histopathologic characteristics of liver specimens
| Specimen | Specimen type | Hepatitis C clinical status | Histopathology |
|---|---|---|---|
| A-1 | Posttransplant biopsy (3 mo) | Nondiseased | NSPAa |
| A-2 | Posttransplant biopsy (24 mo) | Nondiseased | NSPA |
| B-1 | Posttransplant biopsy (5 wk) | Nondiseased | NSPA |
| B-2 | Posttransplant biopsy (24 wk) | Mild acute hepatitis | Inflammation, necrosis |
| C | Nontransplant biopsy | Chronic hepatitis | Inflammation, necrosis, mild fibrosis |
| D | Liver explant | End-stage liver disease | Cirrhosis |
| E | Liver explant | End-stage liver disease | Cirrhosis |
NSPA, no specific pathological abnormality.
In all 19 cases, the qualitative results obtained for HCV genomic RNA by in situ hybridization were in perfect concordance with the RT-PCR results. Each HCV-infected tissue (n = 8) showed a positive signal with HCV antisense riboprobes, while each noninfected tissue (n = 11) was found negative by in situ hybridization. Among the HCV RNA-positive tissues, the percentage of cells containing genomic RNA ranged from 28 to 86%. Figure 5A illustrates the detection of HCV genomic RNA in a field of hepatocytes within cirrhotic nodules after staining with Vector red. Granular signals representing HCV genomic RNA showed a cytoplasmic localization. Overall, the abundance of the signal appeared to vary widely among cells, suggesting that different cells may harbor different quantities of viral genomic RNA. Within cirrhotic livers, the HCV genomic RNA signal varied from one nodule to another.
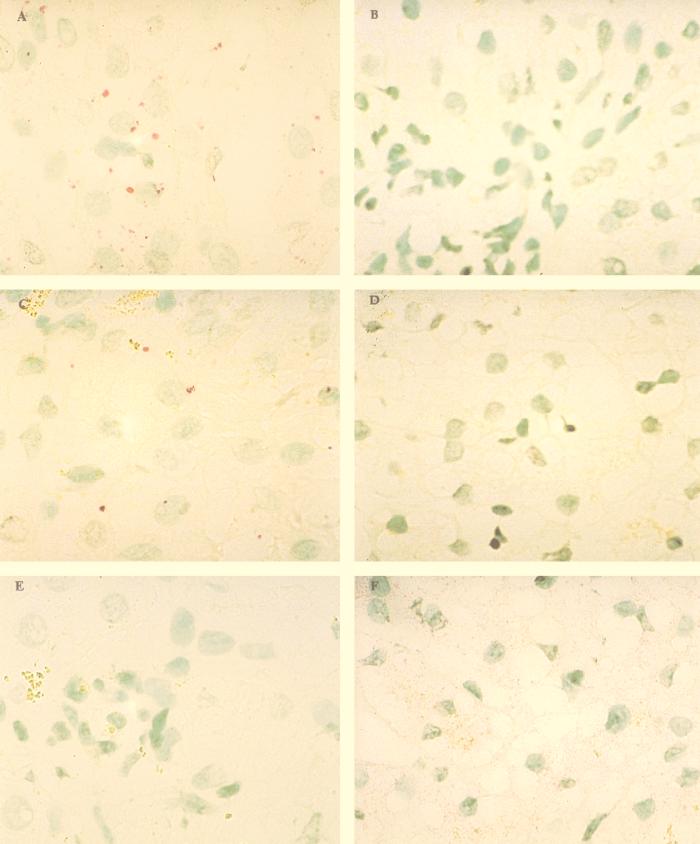
FIG. 5.
In situ analysis of HCV-infected, explanted cirrhotic liver tissue. (A, C, and E) HCV-infected liver tissue. (B, D, and F) Noninfected (normal) liver tissue. (A and B) Hybridization with antisense riboprobes. (C and D) Hybridization with sense riboprobes. (E) Hybridization with hybridization solution free of riboprobes. (F) Hybridization with human endogenous HPRT and beta-actin riboprobes. Nuclear methyl green stain was used; magnification, ×250.
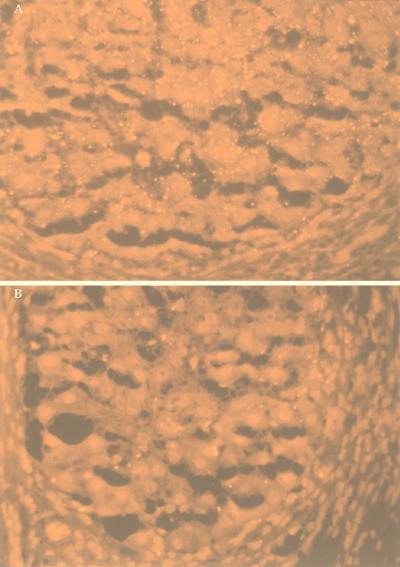
The fluorescent nature of the Vector red reaction product provides increased sensitivity when fluorescence microscopes are used. Figure 6A illustrates the detection of HCV genomic RNA in a cirrhotic liver viewed with a conventional Leitz fluorescence microscope with a rhodamine filter. The liver cells produced dark red autofluorescence, while the positive signal produced bright yellow fluorescence. As noted above, the HCV genomic signal was located primarily in the cytoplasm of hepatocytes, and the signal abundance varied from one nodule to another. However, no clear preferential localization within the hepatic lobule could be discerned. Most of the HCV genomic signal was located inside cirrhotic nodules, with some signal in fibroblasts and/or macrophages in the surrounding fibroconnective tissue. No convincing evidence for signal in the sinusoidal lining cells was identified, but this possibility cannot be excluded.
FIG. 6.
Fluorescent HCV genomic (A) and replicative-intermediate (B) signals in cirrhotic nodules of one explanted cirrhotic liver. Magnification, ∼×62.
In situ analysis of HCV replicative-intermediate RNA in human liver tissue.
The same 19 cases were analyzed for qualitative detection of HCV replicative-intermediate RNA with sense riboprobes. Again, in all 19 cases, the qualitative results obtained by in situ hybridization were in perfect concordance with the RT-PCR results. Figure 5C illustrates the detection of HCV replicative-intermediate RNA in a cirrhotic liver by in situ hybridization. The percentage of cells positive for replicative-intermediate RNA was uniformly lower than that observed for genomic RNA (4 to 25% versus 28 to 86%, respectively). Figure 6B shows the localization of the replicative-intermediate RNA signal exclusively inside cirrhotic nodules, a pattern observed among other cirrhotic nodules and other biopsies.
Relationship between HCV RNAs in serum and liver.
The virological characteristics of HCV in serum and liver specimens obtained during the same clinic visit are summarized in Table 3. In the five instances when serum specimens were available, HCV RNA titers ranged from 8,000 equivalents per ml to 500 million equivalents per ml. There was no relationship between levels of viral RNA in serum and the percentage of hepatocytes staining positive for either HCV genomic or HCV replicative-intermediate RNA. Specimen A-1, for example, was obtained 3 months after liver transplantation for end-stage hepatitis C; the liver specimen showed no specific pathological abnormality on examination (Table 2). The patient serum contained an unusually low titer of HCV RNA (8,000 copies/ml), considering that the patient was immunosuppressed. However, 73% of hepatocytes stained positive for HCV genomic RNA by in situ hybridization. In contrast, only 6% of hepatocytes stained positive for HCV replicative-intermediate RNA in this case. Specimen A-2, obtained from the same patient 21 months later, showed also no histological evidence of disease. However, the patient's serum contained an exceptionally high HCV load, 500 million equivalents per ml. On in situ examination, 86 and 11% of hepatocytes stained positive for HCV genomic and replicative-intermediate RNAs, respectively. It is noteworthy that there was no association between either the level of HCV genomic RNA in serum or the percentage of hepatocytes staining positive for HCV genomic RNA and histopathologic indices of liver disease, although the sample size was too small to allow us to make definite conclusions.
TABLE 3.
Virological characteristics of hepatitis C in serum and liver specimens
| Specimen | HCV genotype | Serum viral loada | % of hepatocytes staining positive for HCV RNA
|
R/G ratiob | |
|---|---|---|---|---|---|
| Genomic | Replicative intermediate | ||||
| A-1 | 1b | 8 × 103 | 73 | 6 | 0.08 |
| A-2 | 1b | 500 × 106 | 85 | 11 | 0.13 |
| B-1 | 1a | 100 × 106 | 28 | 4 | 0.14 |
| B-2 | 1a | 288 × 106 | 31 | 12 | 0.39 |
| C | 1a | 37 × 106 | 60 | 21 | 0.35 |
| D | 1a | NA | 47 | 21 | 0.45 |
| E | 1a | NA | 71 | 25 | 0.35 |
Expressed as HCV RNA equivalents per milliliter. NA, not assessed.
Percentage of hepatocytes staining positive for HCV replicative-intermediate RNA (R) divided by percentage of hepatocytes staining positive for HCV genomic RNA (G).
Specimens B-1 and B-2 were obtained from a second patient at 5 and 24 weeks after liver transplantation, respectively. Both serum specimens had high HCV RNA titers (100 million and 288 million equivalents per ml, respectively), and 28 to 31% of hepatocytes stained positive for HCV genomic RNA by in situ hybridization (Table 3). The B-1 liver biopsy specimen showed no histological evidence of hepatitis, while the B-2 specimen showed mild to moderate hepatitis (Table 2). Of interest, the percentage of cells staining positive for HCV replicative-intermediate RNA was increased threefold in specimen B-2 compared to specimen B-1, and the serum HCV RNA titer was increased to the same degree. However, the percentage of hepatocytes staining positive for HCV genomic RNA did not change appreciably in these specimens.
Specimen C was obtained from a nontransplant patient with chronic hepatitis C. The patient's serum HCV RNA titer was relatively high for a nontransplant patient (37 million equivalents per ml). The liver specimen from patient C showed a higher percentage of cells staining positive for HCV replicative-intermediate RNA than any of the posttransplant specimens (21 versus 4 to 12%, respectively), a result which may reflect a longer duration of disease in this case. Similarly high percentages of cells staining positive for HCV replicative-intermediate RNA were seen in specimens D and E (21 and 25% of cells, respectively), both of which came from cirrhotic livers of patients with end-stage hepatitis C.
The percentage of cells positive for HCV replicative-intermediate RNA correlates with liver injury.
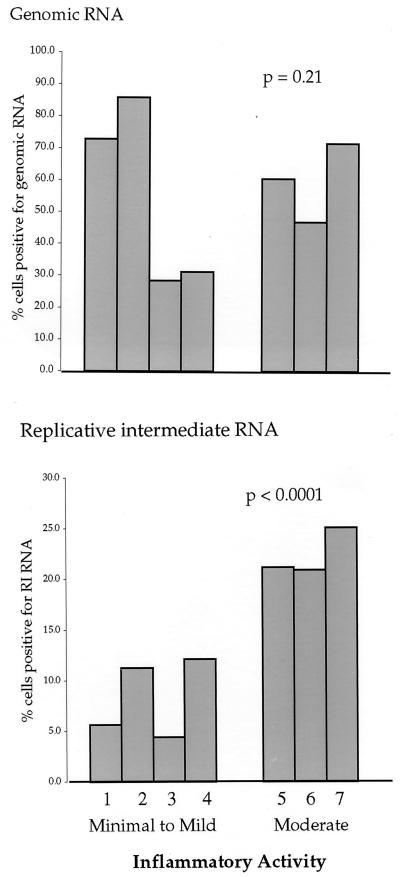
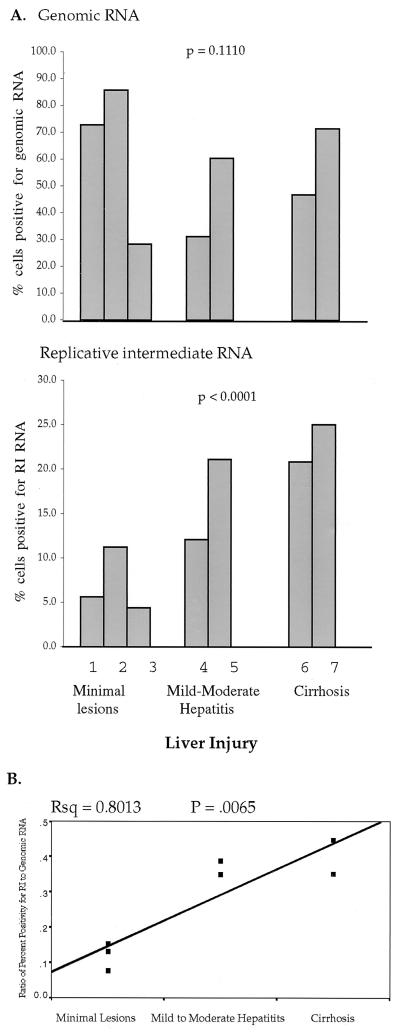
The relationship between the percentage of hepatocytes staining positive for HCV RNA and indices of liver disease is summarized in Fig. 7 and 8. Figure 7 illustrates a highly significant correlation between the percentage of cells staining positive for replicative-intermediate RNA and hepatic inflammatory activity (χ2, 26.82; P, < 0.0001). Specimens with higher inflammation scores had a significantly higher percentage of cells staining positive for HCV replicative-intermediate RNA than specimens with lower inflammation scores (one-tailed P, < 0.0001). On the other hand, the percentage of cells staining positive for HCV genomic RNA did not correlate with liver inflammation (χ2, 1.54; P, 0.21). The percentage of cells staining positive for HCV replicative-intermediate RNA was also significantly correlated with the histopathologic classification of liver injury (Fig. 8A) (χ2, 24.97; P, < 0.0001). Specimens from patients with cirrhosis had higher percentages of positivity for replicative-intermediate RNA than specimens from patients with minimal lesions. However, the percentage of cells staining positive for HCV genomic RNA did not correlate with liver injury (χ2, 2.54; P, 0.111). Figure 8B illustrates linear regression analysis of the significant relationship between the ratio of percent positivity for replicative-intermediate RNA/genomic RNA and liver injury (R2, 0.80; P, 0.0065). In summary, the percentage of hepatocytes staining positive for HCV replicative-intermediate RNA but not HCV genomic RNA showed significant correlation with indices of liver disease.
FIG. 7.
Association between percentage of hepatocytes positive for HCV genomic RNA or replicative-intermediate (RI) RNA and inflammatory activity in seven liver specimens. The data were analyzed by a chi-square test.
FIG. 8.
Percentage of hepatocytes positive for HCV RNA in seven liver specimens and correlation with the status of liver injury. (A) Correlation between percentage of cells positive for genomic or replicative-intermediate (RI) RNA and the histopathologic assessment of liver injury. (B) Linear regression model expressing the ratio of percentage of replicative-intermediate RNA-positive cells to that of genomic RNA-positive cells. Rsq, squared multiple correlation coefficient.
DISCUSSION
Since the discovery of HCV over a decade ago, major progress has been made in understanding this important viral infection. However, the basic mechanisms of HCV pathogenesis remain to be elucidated, and two schools of thought predominate. The “immunopathogenic” model argues that the disease is largely mediated by host immune responses, while the “pathogenic virus” model argues that HCV is capable of cytopathic replication, in at least some instances. Studies which assess HCV RNA in liver specimens have attempted to provide insight into these two models; however, results to date have been conflicting, possibly due to technical limitations and also because of variability in the quality of liver biopsy specimens. The present study addressed many limitations of in situ technology in the following manner. (i) Sensitivity and specificity of riboprobes were well characterized, and genotype-specific probes were used in all cases. To maximize sensitivity, riboprobes incorporating the core and the E1 regions of the HCV genome were used. The 5′UTR was not used for riboprobe production because, although mostly conserved, it has been shown to form a complex secondary structure such that some portions of the RNA sequence hybridize with both positive- and negative-strand RNAs (15, 35). (ii) Liver cell lines expressing HCV subgenomic RNAs were used as controls. (iii) All tissues were snap-frozen at the time of biopsy and were systematically evaluated for integrity of endogenous mRNAs (beta-actin plus HPRT). Only high-quality tissues were accepted for study. (iv) Negative control riboprobes were used in all cases to evaluate and control for background staining. Rigorous optimization of probe and hybridization parameters resulted in an assay with exquisite sensitivity, calculated to be less than 10 HCV RNA copies per cell, without any evidence of nonspecific staining in negative control cell lines or noninfected human liver.
With the optimized in situ hybridization assay, HCV genomic and replicative-intermediate RNAs were detected in 100% of liver tissue specimens from infected patients versus 0% of specimens from noninfected controls. A granular nature of the positive signal was seen for either HCV RNA or endogenous cellular mRNAs (beta-actin and HPRT) in both cell lines and native liver tissue. For HCV genomic RNA in infected liver tissue, we noticed that some cells had a few dots of signal while other cells had multiple dots aggregating to form complex shapes. On the other hand, endogenous HPRT and beta-actin mRNAs generated more consistent and diffuse patterns of positive signal and varied less in abundance among the same types of cells. HCV genomic RNA signal granules were located mostly in the cytoplasm of hepatocytes; the abundance of the signal varied widely among different cells, in agreement with previous reports (7–9). In the current report, the percentage of hepatocytes positive for genomic RNA ranged from 28 to 86%, while the percentage of hepatocytes positive for replicative-intermediate RNA ranged from 4 to 25%. The range of replicative-intermediate RNA signal abundance was more limited than that of genomic RNA; however, the focal intensity of replicative-intermediate RNA granules was equal to or even greater than that of the genomic signal (data not shown). The signal distribution patterns were quite distinct: HCV genomic RNA was distributed throughout the entire tissue, including nonhepatic fibroconnective tissue, while the HCV replicative-intermediate RNA was arranged in a more restricted pattern resembling foci and was found only in hepatocytes. To explain this phenomenon, we propose that HCV replication occurs in a limited number of infected hepatocytes, possibly due to local interference by endogenous antiviral mediators at the molecular level.
Previous studies have provided conflicting results on the relationship between HCV RNA in liver and indices of hepatocellular injury (1, 5, 9, 10, 18, 27, 36). In a recent study with a well-characterized in situ hybridization assay, Agnello and colleagues convincingly demonstrated that HCV RNA molecules are more widespread in human liver than previously appreciated (1). In their study, levels of HCV genomes and antigenomes were lowest in biopsies with minimal disease activity. In this study, we also found low levels of HCV antigenomes in biopsies with minimal lesions but found widespread distribution of HCV genomes in the same biopsies. The cases with minimal liver lesions differed from those in the study of Agnello et al. in at least two ways: in our study, HCV infection of the liver was relatively recent (during the first year posttransplant), and the patients were immunosuppressed. However, it is noteworthy that in both studies, low levels of HCV antigenomes were associated with minimal histological lesions.
In this study, the finding that the percentage of cells staining positive for HCV replicative-intermediate RNA (but not HCV genomic RNA) correlated with hepatic inflammation and histopathologic assessment of liver disease is novel, as it has not been previously reported. Using strand-specific semiquantitative RT-PCR, Negro et al. concluded that the amount of HCV replicative-intermediate RNA in infected liver is not correlated with the degree of liver damage (24, 25). Our study differs in that we used in situ hybridization to examine the distribution of HCV RNAs in tissue. Preliminary results obtained with computerized image analysis show that there is no significant difference in the absolute amount of replicative-intermediate RNA per 100 hepatocytes analyzed among biopsies with various degrees of damage (M. Chang et al., unpublished data). This information seems to agree with the observation of Negro et al. that the measurement of replicative-intermediate RNA is not a good predictor of liver injury. However, our quantitative in situ method allows evaluation of the percentage of positive cells as well as viral RNA signal abundance per positive cell. In this study, we found that two parameters increased in direct proportion to the degree of inflammatory activity: (i) the percentage of hepatocytes harboring replicative-intermediate RNA and (ii) the ratio of replicative-intermediate to genomic HCV RNA. These observations suggest that HCV-associated cytopathic damage and immune system-mediated inflammatory activity in hepatocytes may be induced not simply by the presence of virions but rather by the process of viral replication.
For the disease-free cases, in which we observed high levels of HCV genomes but low levels of HCV antigenomes, it is reasonable to speculate that the accumulation of nonreplicating HCV genomes may represent some form of viral latency or inactivity in the infected host. Such a model would help explain earlier reports that HCV genomic RNA levels in liver do not correlate with liver injury (24, 25, 28). We and others have previously reported that the levels of HCV nonstructural antigen in liver correlate with the degree of liver injury (11, 14, 32, 36); such reports seem consistent with our present findings and the hypothesis that active HCV replication in the liver is important in the pathogenesis of hepatocellular injury.
In conclusion, the present study describes a reliable method for high-sensitivity in situ detection of HCV genomic and replicative-intermediate RNAs in liver tissue specimens. This method provides a useful tool for investigating viral replicative functions in the host liver and in model systems and has potential diagnostic value. We found a significant correlation between HCV replication and liver injury which needs to be verified with a larger number of liver biopsy specimens from diseased and nondiseased patients.
ACKNOWLEDGMENTS
We deeply appreciate Vaughn Fierke (Histology Laboratory, University of Washington Medical Center) for preparing frozen sections of clinical samples and the advice of Shu-Kuang Lee (Department of Biostatistics) on statistical analysis. We thank members of the Viral Hepatitis Laboratory, Department of Laboratory Medicine, University of Washington Medical Center, including Jean-Baptiste Nousbaum, for critical review of the manuscript; Steve Polyak for assistance with transfected cell lines; Dan Sullivan for providing the pCORE clone; Minjun Chung, Maureen Guajardo, and Ka Wing Ng for technical assistance; and Jane Ninh for assistance in manuscript preparation.
The work was supported in part by NIH grants AI 39049-03 and AI/OK 41320-02.
REFERENCES
- 1.Agnello V, Abel G, Knight G B, Muchmore E. Detection of widespread hepatocyte infection in chronic hepatitis C. Hepatology. 1998;28:573–584. doi: 10.1002/hep.510280240. [DOI] [PubMed] [Google Scholar]
- 2.Blight K, Trowbridge R, Rowland R, Gowans E. Detection of hepatitis C virus RNA by in situ hybridization. Liver. 1992;12:286–289. doi: 10.1111/j.1600-0676.1992.tb01062.x. [DOI] [PubMed] [Google Scholar]
- 3.Boshart M, Weber F, Jahn G, Dorsch Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 4.Bukh J, Purcell R H, Miller R H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina Selby R, Barr P J, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox K H, DeLeon D V, Angerer L M, Angerer R C. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 7.Felgar R E, Montone K T, Furth E E. A rapid method for the detection of hepatitis C virus RNA by in situ hybridization. Modern Pathol. 1996;9:696–702. [PubMed] [Google Scholar]
- 8.Gastaldi M, Massacrier A, Planells R, Robaglia Schlupp A, Portal Bartolomei I, Bourliere M, Quilici F, Fiteni J, Mazzella E, Cau P. Detection by in situ hybridization of hepatitis C virus positive and negative RNA strands using digoxigenin-labeled cRNA probes in human liver cells. J Hepatol. 1995;23:509–518. doi: 10.1016/0168-8278(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 9.Gosalvez J, Rodriguez Inigo E, Ramiro Diaz J L, Bartolome J, Tomas J F, Oliva H, Carreno V. Relative quantification and mapping of hepatitis C virus by in situ hybridization and digital image analysis. Hepatology. 1998;27:1428–1434. doi: 10.1002/hep.510270534. [DOI] [PubMed] [Google Scholar]
- 10.Gretch D R. Diagnostic tests for hepatitis C. Hepatology. 1997;26:S43–S47. doi: 10.1002/hep.510260708. [DOI] [PubMed] [Google Scholar]
- 11.Gretch D R, Bacchi C E, Corey L, dela Rosa C, Lesniewski R R, Kowdley K, Gown A, Frank I, Perkins J D, Carithers R L., Jr Persistent hepatitis C virus infection after liver transplantation: clinical and virological features. Hepatology. 1995;22:1–9. [PubMed] [Google Scholar]
- 12.Gretch D R, Wilson J J, Carithers R L, Jr, de la Rosa C, Han J H, Corey L. Detection of hepatitis C virus RNA: comparison of one-stage polymerase chain reaction (PCR) with nested-set PCR. J Clin Microbiol. 1993;31:289–291. doi: 10.1128/jcm.31.2.289-291.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haruna Y, Hayashi N, Hiramatsu N, Takehara T, Hagiwara H, Sasaki Y, Kasahara A, Fusamoto H, Kamada T. Detection of hepatitis C virus RNA in liver tissues by an in situ hybridization technique. J Hepatol. 1993;18:96–100. doi: 10.1016/s0168-8278(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu N, Hayashi N, Haruna Y, Kasahara A, Fusamoto H, Mori C, Fuke I, Okayama H, Kamada T. Immunohistochemical detection of hepatitis C virus-infected hepatocytes in chronic liver disease with monoclonal antibodies to core, envelope and NS3 regions of the hepatitis C virus genome. Hepatology. 1992;16:306–311. doi: 10.1002/hep.1840160205. [DOI] [PubMed] [Google Scholar]
- 15.Honda M, Beard M R, Ping L H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knodell R G, Ishak K G, Black W C, Chen T S, Craig R, Kaplowitz N, Kiernan T W, Wollman J. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1:431–435. doi: 10.1002/hep.1840010511. [DOI] [PubMed] [Google Scholar]
- 17.Komminoth P, Adams V, Long A A, Roth J, Saremaslani P, Flury R, Schmid M, Heitz P U. Evaluation of methods for hepatitis C virus detection in archival liver biopsies. Comparison of histology, immunohistochemistry, in-situ hybridization, reverse transcriptase polymerase chain reaction (RT-PCR) and in-situ RT-PCR. Pathol Res Pract. 1994;190:1017–1025. doi: 10.1016/s0344-0338(11)80896-4. [DOI] [PubMed] [Google Scholar]
- 18.Lamas E, Baccarini P, Housset C, Kremsdorf D, Bréchot C. Detection of hepatitis C virus (HCV) RNA sequences in liver tissue by in situ hybridization. J Hepatol. 1992;16:219–223. doi: 10.1016/s0168-8278(05)80119-9. [DOI] [PubMed] [Google Scholar]
- 19.Lanford R E, Chavez D, Chisari F V, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol. 1995;69:8079–8083. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford R E, Sureau C, Jacob J R, White R, Fuerst T R. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 21.Lau G K, Fang J W, Wu P C, Davis G L, Lau J Y. Detection of hepatitis C virus genome in formalin-fixed paraffin-embedded liver tissue by in situ reverse transcription polymerase chain reaction. J Med Virol. 1994;44:406–409. doi: 10.1002/jmv.1890440417. [DOI] [PubMed] [Google Scholar]
- 22.Lau J Y, Davis G L. Detection of hepatitis C virus RNA genome in liver tissue by nonisotopic in situ hybridization. J Med Virol. 1994;42:268–271. doi: 10.1002/jmv.1890420313. [DOI] [PubMed] [Google Scholar]
- 23.Lau J Y, Krawczynski K, Negro F, González Peralta R P. In situ detection of hepatitis C virus—a critical appraisal. J Hepatol. 1996;24:43–51. [PubMed] [Google Scholar]
- 24.Negro F, Giostra E, Krawczynski K, Quadri R, Rubbia-Brandt L, Mentha G, Colucci G, Perrin L, Hadengue A. Detection of intrahepatic hepatitis C virus replication by strand-specific semiquantitative RT-PCR: preliminary application to the liver transplantation model. J Hepatol. 1998;29:1–11. doi: 10.1016/s0168-8278(98)80172-4. [DOI] [PubMed] [Google Scholar]
- 25.Negro F, Krawczynski K, Quadri R, Rubbia-Brandt L, Mondelli M, Zarski J P, Hadengue A. Detection of genomic- and minus-strand of hepatitis C virus RNA in the liver of chronic hepatitis C patients by strand-specific semiquantitative reverse-transcriptase polymerase chain reaction. Hepatology. 1999;29:536–542. doi: 10.1002/hep.510290223. [DOI] [PubMed] [Google Scholar]
- 26.Negro F, Pacchioni D, Shimizu Y, Miller R H, Bussolati G, Purcell R H, Bonino F. Detection of intrahepatic replication of hepatitis C virus RNA by in situ hybridization and comparison with histopathology. Proc Natl Acad Sci USA. 1992;89:2247–2251. doi: 10.1073/pnas.89.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng S Y, Gunning P, Eddy R, Ponte P, Leavitt J, Shows T, Kedes L. Evolution of the functional human beta-actin gene and its multipseudogene family: conservation of noncoding regions and chromosomal dispersion of pseudogenes. Mol Cell Biol. 1985;5:2720–2732. doi: 10.1128/mcb.5.10.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nouri Aria K T, Sallie R, Sangar D, Alexander G J, Smith H, Byrne J, Portmann B, Eddleston A L, Williams R. Detection of genomic and intermediate replicative strands of hepatitis C virus in liver tissue by in situ hybridization. J Clin Investig. 1993;91:2226–2234. doi: 10.1172/JCI116449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuovo G J, Lidonnici K, MacConnell P, Lane B. Intracellular localization of polymerase chain reaction (PCR)-amplified hepatitis C cDNA. Am J Surg Pathol. 1993;17:683–690. doi: 10.1097/00000478-199307000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y, et al. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 31.Saito K, Sullivan D, Haruna Y, Theise N D, Thung S N, Gerber M A. Detection of hepatitis C virus RNA sequences in hepatocellular carcinoma and its precursors by microdissection polymerase chain reaction. Arch Pathol Lab Med. 1997;121:400–403. [PubMed] [Google Scholar]
- 32.Sansonno D, Cornacchiulo V, Iacobelli A R, Gatti P, Di Stasi M, Dammacco F. Demonstration and distribution of HCV RNA sequences by in situ hybridization and HCV-related proteins by immunohistochemistry in the liver tissue of patients with chronic HCV infection. Pathobiology. 1995;63:239–248. doi: 10.1159/000163956. [DOI] [PubMed] [Google Scholar]
- 33.Sansonno D, Cornacchiulo V, Racanelli V, Dammacco F. In situ simultaneous detection of hepatitis C virus RNA and hepatitis C virus-related antigens in hepatocellular carcinoma. Cancer. 1997;80:22–33. [PubMed] [Google Scholar]
- 34.Tanaka Y, Enomoto N, Kojima S, Tang L, Goto M, Marumo F, Sato C. Detection of hepatitis C virus RNA in the liver by in situ hybridization. Liver. 1993;13:203–208. doi: 10.1111/j.1600-0676.1993.tb00631.x. [DOI] [PubMed] [Google Scholar]
- 35.Tang S, Collier A J, Elliott R M. Alterations to both the primary and predicted secondary structure of stem-loop IIIc of the hepatitis C virus 1b 5′ untranslated region (5′UTR) lead to mutants severely defective in translation which cannot be complemented in trans by the wild-type 5′UTR sequence. J Virol. 1999;73:2359–2364. doi: 10.1128/jvi.73.3.2359-2364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsumi M, Urashima S, Takada A, Date T, Tanaka Y. Detection of antigens related to hepatitis C virus RNA encoding the NS5 region in the livers of patients with chronic type C hepatitis. Hepatology. 1994;19:265–272. [PubMed] [Google Scholar]
- 37.Van Voorhis W C, Barrett L K, Sweeney Y T, Kuo C C, Patton D L. Repeated Chlamydia trachomatis infection of Macaca nemestrina fallopian tubes produces a Th1-like cytokine response associated with fibrosis and scarring. Infect Immun. 1997;65:2175–2182. doi: 10.1128/iai.65.6.2175-2182.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoo B J, Selby M J, Choe J, Suh B S, Choi S H, Joh J S, Nuovo G J, Lee H S, Houghton M, Han J H. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long-term culture persistently infected with HCV. J Virol. 1995;69:32–38. doi: 10.1128/jvi.69.1.32-38.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]