Abstract
Freshwater macroinvertebrates are a diverse group and play key ecological roles, including accelerating nutrient cycling, filtering water, controlling primary producers, and providing food for predators. Their differences in tolerances and short generation times manifest in rapid community responses to change. Macroinvertebrate community composition is an indicator of water quality. In Europe, efforts to improve water quality following environmental legislation, primarily starting in the 1980s, may have driven a recovery of macroinvertebrate communities. Towards understanding temporal and spatial variation of these organisms, we compiled the TREAM dataset (Time seRies of European freshwAter Macroinvertebrates), consisting of macroinvertebrate community time series from 1,816 river and stream sites (mean length of 19.2 years and 14.9 sampling years) of 22 European countries sampled between 1968 and 2020. In total, the data include >93 million sampled individuals of 2,648 taxa from 959 genera and 212 families. These data can be used to test questions ranging from identifying drivers of the population dynamics of specific taxa to assessing the success of legislative and management restoration efforts.
Subject terms: Freshwater ecology, Community ecology, Biodiversity
Background & Summary
Freshwater ecosystems have been altered by a wide range of human impacts since the onset of the industrialization1,2, which have increased in intensity in Europe since World War II3. Legislation in Europe and elsewhere (e.g., CWA 19724, WFD 20005) has aimed to restore freshwater and riparian habitats, especially through improvements in wastewater treatment and reductions in airborne pollutants6. Other stressors such as climate change, land use change, physical alterations to waterways, including straightening, deepening, and impoundments, micropollutants, and invasive species, have either increased in intensity or continue to threaten freshwater ecosystems7. Moreover, despite some improvements, most freshwater ecosystems still fail to meet the European Water Framework Directive’s target of good ecological status8. Given freshwater biotas’ high biodiversity, importance for ecosystem functioning and provision of ecosystem services, monitoring the state of freshwater ecosystems remains an important task for managers and scientists.
Freshwater macroinvertebrates are commonly used as indicators of the status of freshwater ecosystems because of their taxon-specific sensitivity to environmental conditions. Long-term trends of freshwater communities have typically been examined at local to national scales9–11. To study large-scale patterns in biodiversity change, many efforts have been made to compile data on the abundance and occurrence of species into harmonized datasets (e.g., BioTIME12, Living Planet Index13, CESTES14). However, both freshwater biota and all invertebrates are often underrepresented in datasets of species abundance and occurrences. Currently, datasets focused on freshwater ecosystems include an earlier but smaller dataset of European macroinvertebrates15, macroinvertebrate communities in intermittent rivers16, abundance time-series data for fish17, and occurrence records for selected macroinvertebrate groups18. Freshwater insects (which constitute a large proportion of macroinvertebrates) are also represented by the InsectChange dataset19, but this dataset targeted community-level aggregated data and only the most publicly accessible time-series. In the United States, freshwater communities have been sampled across many sites by federal agencies, but most sites have been sampled only one to three times in the past three decades20. Hence, the TREAM (Time seRies of European freshwAter Macroinvertebrates) dataset, presented here, fills an important gap in data availability.
Here, we provide a new dataset of time-series abundance data for freshwater macroinvertebrates in Europe. We used our collaborator network to identify unpublished datasets and published but inaccessible datasets collected within different studies and in different countries. In total, data were sourced from 82 data co-authors including scientists, site managers, and representatives of regulatory agencies from 22 European countries. The TREAM dataset presented here differs from previously published datasets in its inclusion of longer time series (minimum of eight sampling years), of time series reporting all sampled members of the macroinvertebrate community (e.g. not time series of only Odonata or only Ephemeroptera, Plecoptera, and Trichoptera), and its larger number of time series (1,816) within one taxonomic group. Our dataset is outlined in detail below.
We have recently used our dataset to explore how trends in freshwater macroinvertebrate communities have changed through time1. The findings from this study indicate an overall increase in the abundance and richness of freshwater macroinvertebrates in Europe since 1968. However, the recovery has slowed since 2010. We further analysed site-level variation in trends in relation to environmental drivers, such as climate and land use1. Our dataset can be used for population-level and community-level analyses that address a wide range of questions about spatio-temporal patterns in taxonomic or functional taxon units21,22.
Methods
Data call and criteria
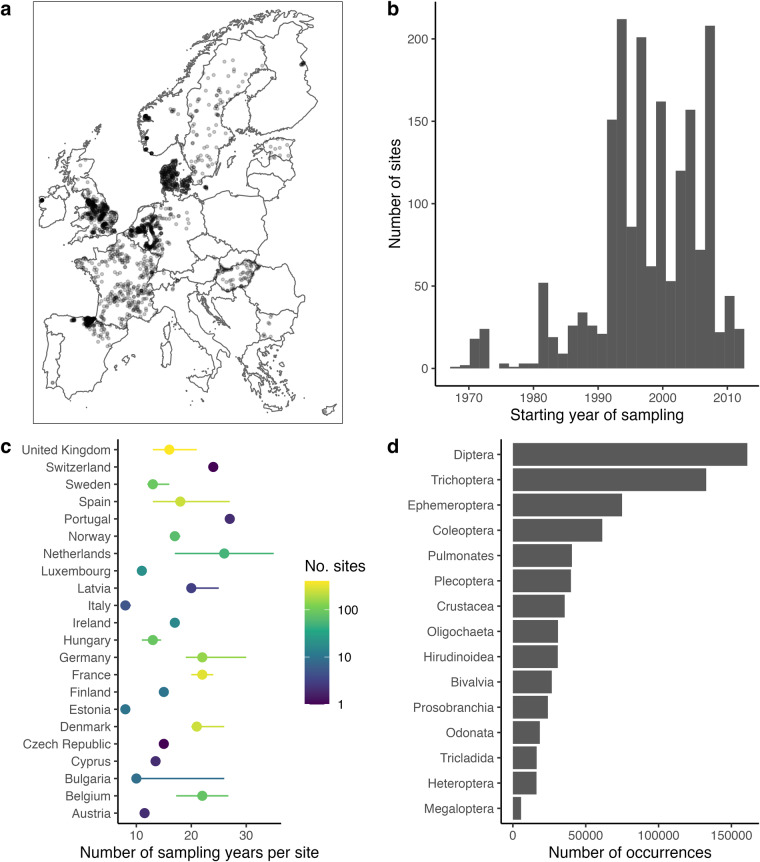
Data were sourced following a data call to water quality managers and freshwater ecologists in Europe from our professional networks. To be included in the TREAM dataset, data had to meet a number of criteria. First, we only included data that formed a time series (i.e., repeated sampling at the same location), with a minimum of eight sampling years, but sampling years did not have to be consecutive. Second, each time series represents whole sampled macroinvertebrate communities (i.e., all macroinvertebrates sampled were recorded). Third, we included only time series with counts or estimates of taxon abundance at the family, genus, or species level. Fourth, sampling location and method must be consistent over time within each time series, including the sampling method, the sampling effort, and the taxonomic resolution to which individual taxa were identified (see Usage notes regarding taxonomic resolution). Finally, all samples in a time series had to be collected within a period of three consecutive months of the year (e.g., June-August). Data were collected as part of 41 independent monitoring projects; project ID numbers and data provider names are listed in the file “TREAM_siteLevel.csv” within the deposited dataset23. Projects reported data from one or more sampling locations per country (Fig. 1; Austria: 2 time series, Belgium: 82, Bulgaria: 9, Cyprus: 2, Czechia: 1, Denmark: 248, Estonia: 10, Finland: 10, France: 307, Germany: 151, Hungary: 87, Ireland: 16, Latvia: 3, Luxembourg: 20, Netherlands: 51, Norway: 67, Portugal: 2, Spain: 245, Sweden: 91, Switzerland: 1, United Kingdom: 406).
Fig. 1.
Overview of dataset coverage including the distribution of sampling sites across 22 European countries (a), a histogram of the number of sampling sites per a given first year of sampling (b), the number of sampling years (median and interquartile range) per site within each country (c), and the top 15 most sampled orders and their occurrences across all surveys (d).
Data standardization
Following the data call, we standardized data by filtering subsets that did not meet our criteria listed above. For example, we excluded samples or whole time series for which sampling methods, effort, or primary level of taxonomic resolution changed during the time series (see Usage notes for a caveat regarding taxonomic resolution). When time series did not meet our inclusion criteria, including consistent sampling methods, for the minimum of eight sampling years, we excluded the whole time series. We only allowed one sampling event per year defined as sampling that occurred within a single day. For time series where multiple samples were provided within the same year, we excluded any sampling outside the most sampled period of three consecutive months. We excluded taxa that are not freshwater invertebrates, e.g., terrestrial taxa, vertebrates, and microinvertebrates that were not present in most of the time series, such as mites and copepods. Taxon names were harmonized according to freshwaterecology.info24.
Data description
The resulting TREAM dataset consists of 1,816 time series from 22 European countries. Time series had a mean of 14.9 (median: 14; range: 8–32) sampling years (not necessarily continuous, spanning 8–39 years). The most commonly sampled families by abundance were Chironomidae, Gammaridae, and Hydrobiidae, and by frequency of occurrence in site sampling years were Chironomidae, Baetidae, and Elmidae. Taxonomically, samples within time series were resolved to family level or finer (762 time series mainly at species level resolution, 537 at genus/mixed level, and 517 at family level). Samples were taken from the riverbed sediments and the majority of collections used forms of kick-net sampling, with either a fixed area or time-based sampling effort. Metadata including information on sampling effort and methods in terms of sampling approach and units are provided in the Knowledge Network for Biocomplexity repository23.
Environmental data
For each sampling location, we included stream characteristics (listed below), climate (cumulative precipitation and mean maximum temperature during the 12-month period before the sampling period), land cover (urban and agricultural land use), and dam impact score. To determine the sampling area of each site to extract these environmental data, we first assigned each sampling site to the corresponding stream segment of the Hydrography90m stream network25 using the v.net function in GRASS GIS26. The longest distance within the stream segment subcatchment was used as the distance threshold for the snapping function. We then computed the upstream catchment of each sampling site using the r.water.outlet function and stored the output as a raster GeoTIFF file. This raster file served as the basis for the subsequent environmental data extractions. While the point snapping and catchment delineation procedure were automated, we reviewed each resulting catchment. In cases where the site was assigned to the wrong segment, we manually corrected the stream segment assignment for a given sampling site and repeated the procedure. For stream characteristics, we extracted the topographical and topological attributes of each stream segment of the Hydrography90m dataset25. Specifically, we provide the extracted area of each site’s subcatchment, Strahler stream order, flow accumulation, elevation, and slope.
For climate data, we used the TERRA Climate monthly time series from 1967–2020 at 4-km resolution27 and overlaid the monthly precipitation and temperature raster layers with the upstream catchment of each sampling site using the r.univar function in GRASS GIS. In addition, we extracted the local temperature layers at each sampling site using the gdallocationinfo function. This resulted in monthly averaged upstream aggregated precipitation (in mm) and monthly averaged local maximum temperature (in °C). For each site and year, we then calculated the average of the monthly temperatures, and the sum of the monthly precipitation values for the year preceding the average month of sampling (e.g., for a site with most sampling occurring in May, year was defined as the preceding 12 months of May until April). The use of climatic data from one year prior to the start of the first time series allows the capture of the annual period of weather prior to the first sampling event, since population responses are typically lagged. We used Bayesian models in the program R v. 4.228 and package brms29 to calculate trends of change in precipitation and temperature over time (all sampling years of a given site/time series). Code for climate trends can be found at: https://github.com/Ewelti/EuroAquaticMacroInverts/blob/main/R/Initial_Biodiversity_FuncTrait_and_climate_calcs/climateTrends_brms.R.
To collect data on land cover, we used the ESA Land Cover CCI Product30 at 300-m spatial resolution to extract the percent coverage of the urban and crop land-cover categories from annual maps, available from 1992–2018. We first split each annual multi-category land cover raster layer into the single raster layers that hold only each single category. We then overlaid the urban and crop land cover category raster layers with the subcatchment raster layers and used the r.univar function to yield annual cover of the upstream urban and crop land cover within the catchment of each sampling site.
We used the GRanD dataset31 to obtain the dams across the study area domain. First, we assigned each dam to the Hydrography90m network using the v.net function and using the longest distance within the stream segment subcatchment as the distance threshold. Afterwards, we visually checked the spatial assignment. We then computed the distance from each sampling site to the next upstream dam using the GRASS GIS function v.net.distance25. To create a proxy for dam impacts, we first extracted only dams within 100 km of a site that were connected to the site and upstream of the site. We then calculated a dam impact score for each site as:
where n represents the number of dams, i denotes a given dam, and d is the distance (km) of the dam from a site.
Data Records
Data are available from the Knowledge Network for Biocomplexity repository (https://knb.ecoinformatics.org/view/doi:10.5063/F1NG4P4R)23. Data are provided in three csv files listed below:
TREAM_siteLevel.csv
This file contains summary TREAM data where each row corresponds to a site (a site = location of one time series). Data include site characteristics (e.g. coordinates, number of sampling years, sampling season), data providers, and primary taxonomic resolution level, summary information on biodiversity across the time series (e.g. total taxa richness for all sampling years). The majority of available corresponding environmental data (i.e., dam impact score, mean values across sampling years of the mean temperature and precipitation in the 12 months prior to sampling, slope of temperature and precipitation over the sampled years, and percent upstream area of cropland and urban cover) and stream characteristic variables (i.e., area of subcatchment, Strahler stream order, flow accumulation, elevation, and slope) are included in this file in addition to sampling method information (e.g., sampling units, primary taxonomic resolution).
TREAM_siteYEARLevel.csv
This file contains TREAM data where each row corresponds to a site and a year of sampling. This file is the primary source for biodiversity summary metrics (e.g., taxa richness, functional redundancy, abundance of native taxa for each site and year) associated with this dataset. Environmental data that are available on an annual basis (i.e., mean temperature and precipitation in the 12 months prior to sampling, and percent upstream area of cropland and urban cover) are also provided in this file.
TREAM_alltaxa.csv
This file contains raw biodiversity data where each row corresponds to a taxon within a site and year of sampling. The abundance of each taxon per sampling unit (i.e., all sampling within one site and one time point) is included in this file (see TREAM_siteLevel.csv for sampling methods and effort information).
Technical Validation
Technical validation of the TREAM dataset was achieved through exclusion of time series data that did not match our inclusion criteria and data standardisation steps (outlined in Methods above). Any noted issues that did not adhere to the outlined standardisation within the datasets from the 41 independent projects included in this dataset were checked with data providers and corrected or removed when standardisation was not achievable (e.g., when collection methods changed over the course of the time series).
Usage Notes
Several key characteristics about these data should be noted by future data users. First, and most importantly, sampling methods and effort, and seasonality are standardised within individual time series but can vary across time series. This means that raw data are not directly comparable across the 41 independent projects included in the TREAM dataset.
Second, while we use the taxonomic backbone of freshwaterecology.info23 as it is a common tool used by European freshwater ecologists, we are aware that it does not capture all recent changes to taxonomic names.
Third, two pairs of time series overlap in sampling locations: 1) Site ID = 100000001 (SVD) & 100000309 (Bugey_SVD) refer to the same location; 2) Site ID = 100000002 (SVG) & 100000308 (Bugey_SVG) refer to the same location. The data from these sites were collected by the Institut national de la recherche agronomique (referent: Maxence Forcellini) between 1980 and 2014, and by Électricité de France (Referent: Anthony Maire) between 2000 and 2019.
Fourth, although standardised taxonomic resolution within time series was a criterion for data inclusion, some datasets switch taxonomic resolution (e.g. from genus to species level) for a given taxon part-way through the time series. This is particularly the case for data from Denmark within the Baetidae, Brachycentridae, Chironomidae, Gammaridae, Oligochaeta and Simuliidae. We did not alter these names because they represent the original information provided for and published in Haase et al.1, and standardisation methods may vary depending on intended future use. In the time series provided, these issues could affect analyses of shifts in community composition, which could reflect a shift in identification level rather than compositional change. These issues do not affect analyses of total abundance and have little influence taxa richness or diversity, including their temporal trends, as they are typically substitutions of one unique taxon for another. As with all large datasets of ecological time series, data users should carefully check the data considering their intended use and when questions arise, contact data providers (listed in TREAM_siteLevel.csv).
Finally, since each record was assigned the Hydrography90m stream network25 subcatchment, users can directly interact with the hydrographr R-package which facilitates subsequent network and distance analyses using the data records32.
Acknowledgements
Nathalie Kaffenberger aided in initial data compilation. Funding for authors, data collection and processing was provided by the EU Horizon 2020 project eLTER PLUS (grant agreement no. 871128), German Federal Ministry of Education and Research (BMBF; 033W034A), German Research Foundation (DFG FZT 118, 202548816), the Collaborative Research Centre 1439 RESIST (DFG—SFB 1439/1 2021 –426547801), Czech Republic project no. GA23-05268S, the Leibniz Competition (J45/2018, P74/2018), the Spanish Ministerio de Economía, Industria y Competitividad - Agencia Estatal de Investigación and the European Regional Development Fund (MECODISPER project CTM 2017-89295-P), Ramón y Cajal contracts and the project funded by the Spanish Ministry of Science and Innovation (RYC2019-027446-I, RYC2020-029829-I, PID2020-115830GB-100), the Danish Environment Agency, the Norwegian Environment Agency, SOMINCOR – Lundin mining & FCT - Fundação para a Ciência e Tecnologia, Portugal, the Swedish University of Agricultural Sciences, the Swiss National Science Foundation (Grant PP00P3_179089), the EU LIFE programme (DIVAQUA project - LIFE18 NAT/ES/000121), and the UK Natural Environment Research Council (GLiTRS project -NE/V006886/1 and NE/R016429/1 as part of the UK-SCAPE programme), the Autonomous Province of Bolzano (Italy), Estonian Research Council (grant No PRG1266), Estonian national program ‘Humanitarian and natural science collections’. The Environment Agency of England, the Scottish Environmental Protection Agency and Natural Resources Wales provided publicly available data. The collection of data from the Rhône River in France was greatly aided by Marie-Claude Roger (INRAE Lyon), Jean-Claude Berger (INRAE AIX), and Pâquerette Dessaix (ARALEP). We are also grateful to the French Regional Environment Directorates (DREALs) for their collaboration in harmonising the long-term data series from the other French rivers. We thank the AWEL from the Canton of Zurich for providing access to macroinvertebrate data from the AWEL monitoring scheme. We acknowledge the Flanders Environment Agency, the Rhineland-Palatinate State Office for the Environment and the Bulgarian Executive Environment Agency for providing data. This manuscript is a contribution of the Alliance for Freshwater Life (www.allianceforfreshwaterlife.org). Any views expressed within this paper are those of the authors and do not necessarily represent the views of their respective employer organisations.
Author contributions
P.H. initiated the project and put out the data call. E.A.R.W. and J.S.S. performed data cleaning. E.A.R.W. and D.E.B. designed and conducted analyses. S.D. and J.R.G. provided environmental data with assistance from G.Amatulli, J.K. and L.Shen. E.A.R.W., D.E.B., S.D. and J.R.G. wrote the first draft with assistance from P.H., and all authors contributed to additional versions, especially J.S.S. Authors who contributed macroinvertebrate data were F.A., M.Á.-C., D.G.A., G.A., I.A.J., T.A., I.A., I.B., J.B.O., C.L.B., L.B., N.B., R.B., M.C.-A., N.J.B., Z.C., T.D., E.d.E., A.D., G.D., E.D., K.A.E., J.E., T.E.E., V.E., M.J.F., M.Ferréol, M.Floury, M.Forcellini, M.A.F., R.F., N.F., J.-F.F., G.G., P.G., M.A.S.G., A.H., P.H., K.-L.H., T.C.J., R.K.J., J.I.J., L.K., A.L., P.L., L.L. M.-H.L., A.W.L., A.Maire., J.A.M.A., B.G.M., A.Millán, D.M., T.M., J.F.M., D.O., R.P., F.J.P.S., F.P., M.P., P.P., J.J.R., M.R., D.S.-F., L.Sandin, R.B.Schäfer, A.S.-K., A.Scotti, A.Skuja, S.S., M.S., R.Stubbington, H.T., V.G.T., I.T., Y.U., G.H.v., R.V., E.V., G.Várbíró, G.Velle., P.F.M.V., R.C.M.V., Y.V., and P.W.-L.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Code availability
Code used to conduct analyses for the corresponding publication analysing these data2 is available at: https://github.com/Ewelti/EuroAquaticMacroInverts. Code to calculate site and year level biodiversity metrics is available at: https://github.com/Ewelti/EuroAquaticMacroInverts/blob/main/R/Initial_Biodiversity_FuncTrait_and_climate_calcs/TaxaDiversity_calculation.R.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Haase P, et al. The recovery of European freshwater biodiversity has come to a halt. Nature. 2023;620:582–588. doi: 10.1038/s41586-023-06400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vörösmarty CJ, et al. Global threats to human water security and river biodiversity. Nature. 2010;467:555–561. doi: 10.1038/nature09440. [DOI] [PubMed] [Google Scholar]
- 3.Steffen W, et al. Planetary boundaries: guiding human development on a changing planet. Science. 2015;347:1259855. doi: 10.1126/science.1259855. [DOI] [PubMed] [Google Scholar]
- 4.United States. Federal Water Pollution Control Act Amendments. Public Law 92–500 (1972).
- 5.European Union. Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 establishing a framework for community action in the field of water policy. Community, European vol. Directive 2000/60/EC 1–72 (2000).
- 6.European Environment Agency. Air quality in Europe — 2018 report., (2018).
- 7.Birk S, et al. Impacts of multiple stressors on freshwater biota across spatial scales and ecosystems. Nat Ecol Evol. 2020;4:1060–1068. doi: 10.1038/s41559-020-1216-4. [DOI] [PubMed] [Google Scholar]
- 8.Whelan MJ, et al. Is water quality in British rivers “better than at any time since the end of the Industrial Revolution”? Sci Total Environ. 2022;843:157014. doi: 10.1016/j.scitotenv.2022.157014. [DOI] [PubMed] [Google Scholar]
- 9.Vaughan IP, Gotelli NJ. Water quality improvements offset the climatic debt for stream macroinvertebrates over twenty years. Nat Commun. 2019;10:1956. doi: 10.1038/s41467-019-09736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Klink R, et al. Meta-analysis reveals declines in terrestrial but increases in freshwater insect abundances. Science. 2020;368:417–420. doi: 10.1126/science.aax9931. [DOI] [PubMed] [Google Scholar]
- 11.Bowler DE, et al. Winners and losers over 35 years of dragonfly and damselfly distributional change in Germany. Divers and Distrib. 2021;27:1353–1366. doi: 10.1111/ddi.13274. [DOI] [Google Scholar]
- 12.Dornelas M, et al. BioTIME: A database of biodiversity time series for the Anthropocene. Glob Ecol Biogeogr. 2018;27:760–786. doi: 10.1111/geb.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoological Society of London and the World Wildlife Fund. Living Planet Index database. 2022 (2022).
- 14.Jeliazkov A, et al. A global database for metacommunity ecology, integrating species, traits, environment and space. Sci Data. 2020;7:6. doi: 10.1038/s41597-019-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Statzner B, Bonada N, Dolédec S. Conservation of taxonomic and biological trait diversity of European stream macroinvertebrate communities: a case for a collective public database. Biodivers Conserv. 2007;16:3609–3632. doi: 10.1007/s10531-007-9150-1. [DOI] [Google Scholar]
- 16.Leigh C, et al. IRBAS: An online database to collate, analyze, and synthesize data on the biodiversity and ecology of intermittent rivers worldwide. Ecol Evol. 2017;7:815–823. doi: 10.1002/ece3.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comte L, et al. RivFishTIME: A global database of fish time-series to study global change ecology in riverine systems. Glob Ecol Biogeogr. 2021;30:38–50. doi: 10.1111/geb.13210. [DOI] [Google Scholar]
- 18.Grigoropoulou A, et al. The global EPTO database: Worldwide occurrences of aquatic insects. Glob Ecol Biogeogr. 2023;32:642–655. doi: 10.1111/geb.13648. [DOI] [Google Scholar]
- 19.van Klink R, et al. InsectChange: a global database of temporal changes in insect and arachnid assemblages. Ecology. 2021;102:e03354. doi: 10.1002/ecy.3354. [DOI] [PubMed] [Google Scholar]
- 20.Rumschlag SL, et al. Density declines, richness increases, and composition shifts in stream macroinvertebrates. Sci Adv. 2023;9:eadf4896. doi: 10.1126/sciadv.adf4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinclair JS, et al. Multi-decadal improvements in the ecological quality of European rivers are not consistently reflected in biodiversity metrics. Nat Ecol Evol. 2024;8:430–441. doi: 10.1038/s41559-023-02305-4. [DOI] [PubMed] [Google Scholar]
- 22.Sexton, A. N. et al. Inland navigation and land use interact to impact European freshwater biodiversity. Nat Ecol Evol 1–11, 10.1038/s41559-024-02414-8 (2024). [DOI] [PubMed]
- 23.Welti E, Haase P. 2023. TREAM: Time series of freshwater macroinvertebrate abundances and site characteristics of European streams and rivers. Knowledge Network for Biocomplexity. [DOI] [PubMed]
- 24.Schmidt-Kloiber A, Hering D. – An online tool that unifies, standardises and codifies more than 20,000 European freshwater organisms and their ecological preferences. Ecol Indic. 2015;53:271–282. doi: 10.1016/j.ecolind.2015.02.007. [DOI] [Google Scholar]
- 25.Amatulli G, et al. Hydrography90m: a new high-resolution global hydrographic dataset. Earth Syst Sci Data. 2022;14:4525–4550. doi: 10.5194/essd-14-4525-2022. [DOI] [Google Scholar]
- 26.Neteler, M. & Mitasova, H. Open Source GIS: A GRASS GIS Approach. (Springer, New York, NY, 2007).
- 27.Abatzoglou JT, Dobrowski SZ, Parks SA, Hegewisch KC. TerraClimate, a high-resolution global dataset of monthly climate and climatic water balance from 1958–2015. Sci Data. 2018;5:170191. doi: 10.1038/sdata.2017.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2023).
- 29.Bürkner, P.-C. Bayesian item response modeling in R with brms and Stan. J. Stat. Soft. 100 (2021).
- 30.European Space Agency. Land Cover CCI Product User Guide Version 2. Tech. Rep. (2017).
- 31.Lehner B, 2019. Global reservoir and dam database, Version 1 (GRanDv1). Palisades, NY: NASA Socioeconomic Data and Applications Center (SEDAC) [DOI]
- 32.Schürz M, et al. hydrographr: An R package for scalable hydrographic data processing. Methods in Ecology and Evolution. 2023;14:2953–2963. doi: 10.1111/2041-210X.14226. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Welti E, Haase P. 2023. TREAM: Time series of freshwater macroinvertebrate abundances and site characteristics of European streams and rivers. Knowledge Network for Biocomplexity. [DOI] [PubMed]
- Lehner B, 2019. Global reservoir and dam database, Version 1 (GRanDv1). Palisades, NY: NASA Socioeconomic Data and Applications Center (SEDAC) [DOI]
Data Availability Statement
Code used to conduct analyses for the corresponding publication analysing these data2 is available at: https://github.com/Ewelti/EuroAquaticMacroInverts. Code to calculate site and year level biodiversity metrics is available at: https://github.com/Ewelti/EuroAquaticMacroInverts/blob/main/R/Initial_Biodiversity_FuncTrait_and_climate_calcs/TaxaDiversity_calculation.R.