Significance
Myoblast fusion into multinuclear myotubes is a critical cellular process for the formation of skeletal muscle fibers. However, the mechanisms orchestrating cell fusion remain incompletely understood. In this study, using a cellular model of X-linked centronuclear myopathy, in which the phosphoinositide phosphatase MTM1 has been knockout, we unravel an unexpected pathway connecting MTM1 to cell-cell fusion. We demonstrate that MTM1 is the main enzyme in this cellular system responsible for synthesizing PI5P. Subsequently, PI5P is rapidly metabolized by the PI5P 4-kinase α into PI(4,5)P2 which accumulates at the plasma membrane thereby facilitating the formation of podosome-like protrusions, playing a crucial role in the spatiotemporal regulation of myoblast fusion.
Keywords: phosphoinositides, myoblast fusion, MTM1, podosomes
Abstract
Myogenesis is a multistep process that requires a spatiotemporal regulation of cell events resulting finally in myoblast fusion into multinucleated myotubes. Most major insights into the mechanisms underlying fusion seem to be conserved from insects to mammals and include the formation of podosome-like protrusions (PLPs) that exert a driving force toward the founder cell. However, the machinery that governs this process remains poorly understood. In this study, we demonstrate that MTM1 is the main enzyme responsible for the production of phosphatidylinositol 5-phosphate, which in turn fuels PI5P 4-kinase α to produce a minor and functional pool of phosphatidylinositol 4,5-bisphosphate that concentrates in PLPs containing the scaffolding protein Tks5, Dynamin-2, and the fusogenic protein Myomaker. Collectively, our data reveal a functional crosstalk between a PI-phosphatase and a PI-kinase in the regulation of PLP formation.
Phosphoinositides (PIs) are a minor class of phospholipids that play an essential role in diverse cellular functions highlighted by the direct involvement of their metabolizing enzymes in human pathologies (1, 2). However, there is still limited knowledge of the functions of low abundant PIs such as phosphatidylinositol 5-phosphate (PI5P) or phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2], the least characterized species (3). MTM1 is a lipid phosphatase that dephosphorylates phosphatidylinositol 3-phosphate (PI3P) into PI (4–6) and PI(3,5)P2 into PI5P (7, 8). Mutations in the myotubularin 1 (MTM1) gene are responsible for X-linked myotubular myopathy (XLMTM), the most severe form of centronuclear myopathy, which results in most cases from the lack of MTM1 (9). Interestingly, while MTM1 is ubiquitously expressed, only skeletal muscle is affected in XLMTM, suggesting the intervention of tissue-specific interactors/signaling pathways to mediate the impact of MTM1 on the disease. An Mtm1 knockout mouse model has been developed and it reproduces the human pathology (10). Data from studies on other animal models, like Drosophila and Zebrafish, support the muscular function of MTM1 in PI3P-dependent membrane/vesicular trafficking and the homeostasis of the endosomal system to regulate t-tubule organization, organelle positioning, excitation-contraction coupling, and cytoskeletal organization (11–13). However, our current knowledge about the role of MTM1 substrates and products during myogenesis is undefined as well as their roles in the etiology of the disease.
Results
MTM1 Is Required for Normal Myoblast Differentiation.
In order to gain insight into the function of MTM1, its importance in PI5P production from PI(3,5)P2, and the role of these lipids in the etiology of the disease, we used the C2C12 mouse myoblast cell line that is widely used to study myogenesis in vitro and that rapidly differentiates into myotubes under low-serum conditions (14–16). We succeeded to isolate an Mtm1-KO cell line that has retained its myotube formation capacity, using CRISPR–CRISPR-associated protein 9 (Cas9) technology via the ribonucleoprotein editing strategy. In KO cells, the presence of frameshift mutations leading to premature stop codons in the Mtm1 gene was identified by allelic sequencing (SI Appendix, Fig. S1 A and B) and confirmed by the absence of MTM1 on western blot; in parental WT cells, MTM1 expression normally increases steadily during differentiation (Fig. 1A). No indel mutations were observed in predictive off-targets (SI Appendix, Fig. S1C).
Fig. 1.
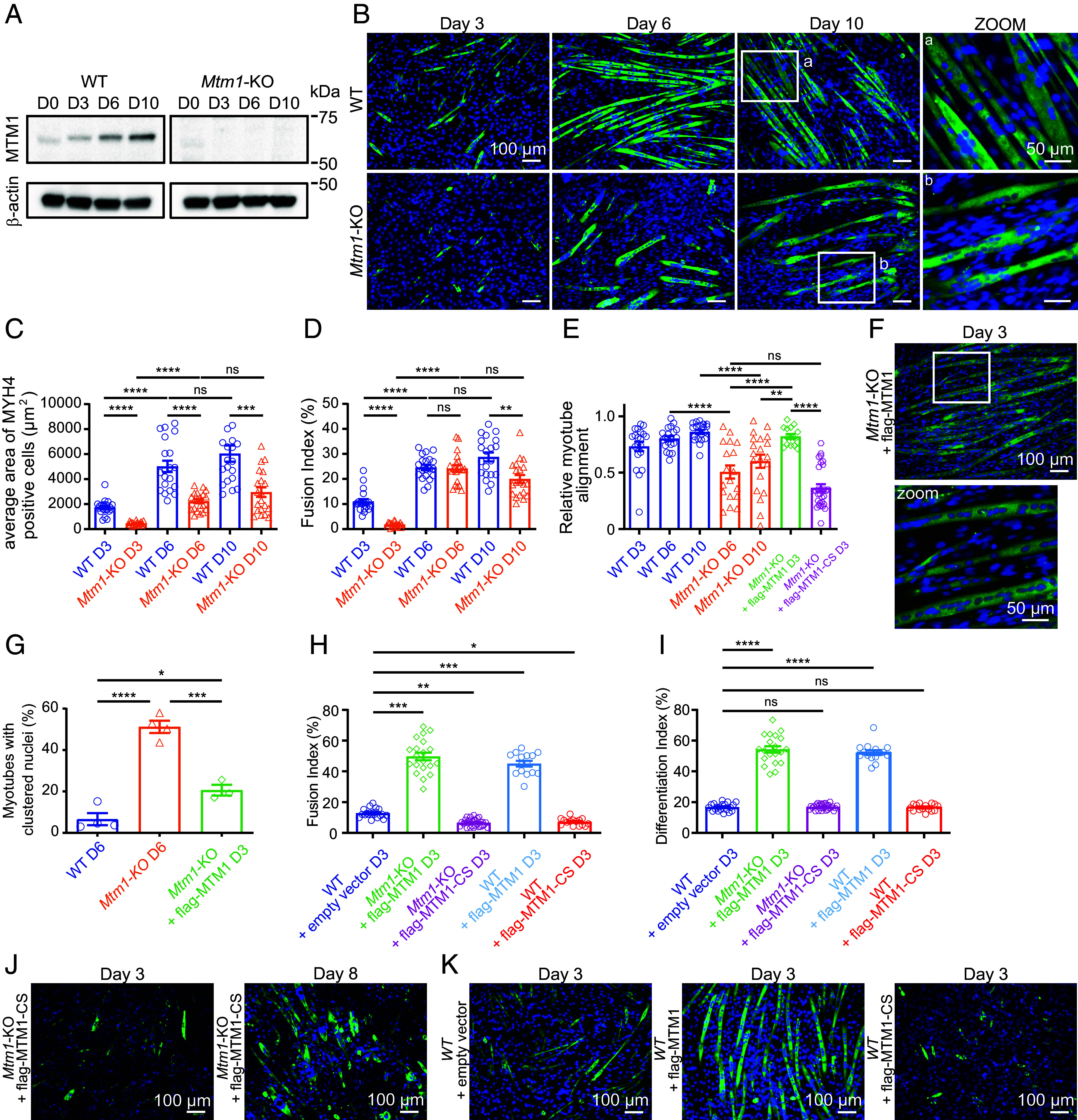
Phenotypic characterization of the Mtm1 knockout cell line. (A) Expression of MTM1 at the indicated time points of WT and Mtm1-KO C2C12 differentiation analyzed by western blot. Images are representative of six independent experiments. (B) MYH4 and DAPI staining of C2C12 WT and Mtm1-KO at the indicated time points. (Scale bar, 100 μm.) On the right, higher magnifications for the indicated areas shown as boxes. (Scale bar, 50 μm.) (C) Average area of MYH4 labeled WT and Mtm1-KO cells during differentiation. Data are represented as mean ± SEM, n = 4, five fields per independent experiment, each point represents one field of view, ns = not significant, ***P < 0.001, ****P < 0.0001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (D) Quantification of the fusion index for the WT and Mtm1-KO cells at the indicated time points. The fusion index represents the number of nuclei in MYH4 positive cells compared to the total number of nuclei. Data are represented as mean ± SEM, n = 4, five fields per independent experiment, each point represents one field of view, ns = not significant, **P < 0.01, ****P < 0.0001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (E) Relative myotube alignment from C2C12 WT, Mtm1-KO, Mtm1-KO expressing flag-MTM1, or flag-MTM1-CS for the indicated days of differentiation. Data are represented as mean ± SEM, n = 3 to 4, five fields per independent experiment, each point represents one field of view, ns = not significant, **P < 0.01, ****P < 0.0001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (F) MYH4 and DAPI staining of C2C12 Mtm1-KO expressing flag-MTM1 after 3 d of differentiation. (Scale bar, 100 μm.) The lower panel represent a higher magnification for the indicated area shown as a box. (Scale bar, 50 μm.) (G) Percentage of myotubes with clustered nuclei (myotube presenting more than five grouped nuclei) for the indicated days of differentiation of C2C12 WT, Mtm1-KO, and Mtm1-KO expressing flag-MTM1. Data are represented as mean ± SEM, n = 3 to 4, *P < 0.05, ***P < 0.001, and ****P < 0.0001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (H) Quantification of the fusion index for the indicated days of differentiation of C2C12 WT transduced with lentiviruses expressing an empty vector, Mtm1-KO cells expressing flag-MTM1 or flag-MTM1-CS, C2C12 WT cells expressing flag-MTM1 or flag-MTM1-CS. Data are represented as mean ± SEM, n = 3 to 4, five fields per independent experiment, each point represents one field of view, *P < 0.05, **P < 0.01, ***P < 0.001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (I) Quantification of the differentiation index for the indicated days of differentiation of C2C12 WT transduced with lentiviruses expressing an empty vector, Mtm1-KO cells expressing flag-MTM1 or flag-MTM1-CS, C2C12 WT cells expressing flag-MTM1 or flag-MTM1-CS. Data are represented as mean ± SEM, n = 3 to 4, five fields per independent experiment, each point represents one field of view, ns=not significant, ****P < 0.0001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (J) MYH4 and DAPI staining of C2C12 Mtm1-KO expressing flag-MTM1-CS after 3 d of differentiation (Left). MYH4 and DAPI staining of C2C12 Mtm1-KO expressing flag-MTM1-CS after 8 d of differentiation (Right). (Scale bar, 100 μm.) (K) MYH4 and DAPI staining of C2C12 WT expressing an empty vector (Left), expressing flag-MTM1 (Middle), expressing flag-MTM1-CS (Right), after 3 d of differentiation. (Scale bar, 100 μm.)
We next differentiated parental WT and KO cells for 10 d and assessed myotube formation [myosin heavy chain (MYH4)-positive cells]. As shown in Fig. 1B, myogenic differentiation in WT cells is visually apparent after 3 d, with aligned and elongated myotubes on day 6 that continue to grow until day 10. In contrast, in KO cells there is a clear delay in differentiation with formation of shorter non-spindle-shaped myotubes visible on day 6, with continued growth until day 10 but with adhesion defects of myotubes on the gelatin substratum, a phenotype already described in animal models (11, 17). There was no observed difference in cell spreading and migration speed between WT and Mtm1-KO myoblasts (SI Appendix, Fig. S1 D and E). Interestingly, we recently reported that valproic acid ameliorates the differentiation process and adhesion of our Mtm1-KO cells as in zebrafish and mouse XLMTM models (18). Importantly, knockdown of Mtm1 with shRNAs resulted in the same phenotypes as KO cells (SI Appendix, Fig. S1 F–H). Accordingly, average area of myotubes from KO cells was found reduced by ~50% at all stages of the differentiation process (Fig. 1C), with fusion indexes (percentage of nuclei in MYH4-positive cells containing more than 2 nuclei) in WT and KO cells of 11 ± 0.97% and 1.78 ± 0.19% after 3 d of differentiation, respectively (Fig. 1D). Fusion indexes were comparable on day 6, with a decrease in KO cells on day 10 that can be explained by myotube detachment from the substrate. This defect in the early phases of myoblast fusion is consistent with what has already been reported in primary cells from Mtm1−/y cells (17), where the authors observed a significant defect in myoblast fusion at 24 and 48 h of differentiation, but not at 72 h, which is reminiscent to the myofibers hypotrophy typically seen in patient’s muscles. While nuclei were found well aligned in WT myotubes (Fig. 1B, zoom a), nuclei were found mislocalized in KO myotubes (Fig. 1B, zoom b, Fig. 1G), a phenotype also reminiscent of the human XLMTM pathology. Moreover, quantification of alignment of myotubes, obtained by calculating the angle of deviation of myotubes from the principal axis (for more details, see Materials and Methods), identified the nonalignment of myotubes as a phenotype of Mtm1 KO (Fig. 1 B and E). To confirm that the defects in differentiation were a specific consequence of the loss of MTM1 rather than a nonspecific effect of CRISPR/Cas9 mutagenesis, we transduced KO cells with a lentivirus encoding flag-MTM1 and then allowed the cells to differentiate. Re-expression of MTM1 rescued the differentiation process, with the formation of well-structured and aligned myotubes (Fig. 1 F and E), a rescue in nuclei positioning (Fig. 1G), and an increase in the fusion index (Fig. 1H). On the contrary, re-expression of a catalytic-dead version of MTM1 (flag-MTM1-CS) failed to rescue the normal differentiation process (Fig. 1 E, H, and J). To confirm that MTM1 enzymatic function enhances the differentiation process, we overexpressed MTM1 or its catalytic-dead version in WT cells and allowed the cells to differentiate for 3 d. Similar to Mtm1-KO cells, overexpression of flag-MTM1 increased the fusion index in comparison to WT cells, whereas overexpression of flag-MTM1-CS had a negative effect on the formation of myotubes (Fig. 1 K and H). This result is consistent with the substrate-trapping function of MTM1-CS, which competes with endogenous MTM1 and acts as a dominant negative form (5, 19). Quantification of the differentiation index (percentage of nuclei in all MYH4-positive cells) revealed that MTM1-CS overexpression does not impact the myogenic program, consistent with observations in the Mtm1 knockout mouse model (Fig. 1I) (10). However, the overexpression of the catalytically active form of MTM1 increases the differentiation index. This may be attributed to an ectopic localization of MTM1 within the nucleus, where phosphoinositides have been demonstrated to regulate myogenic gene expression (20). All of these results validate the Mtm1-KO cell line as a cellular model and position MTM1 as a critical enzyme in myoblast differentiation as recently reported (21).
PI5P Level Is Dependent on MTM1 Activity in Muscle Cells and Unexpectedly Decreases during Myoblast Differentiation.
To evaluate the impact of Mtm1 KO on PIs metabolism, we quantified PIs during C2C12 differentiation by metabolically labeling cells with [32P] Phosphate. PI3P levels remained stable during WT cells differentiation, whereas it accumulates in KO myotubes (Fig. 2 A–C ), similar to what has been observed in muscle tissues from Mtm1-KO animal models (6, 12). Regarding the other in vitro substrate of MTM1, PI(3,5)P2, we measured an increase in its level by ~1.5-fold in KO myotubes (Fig. 2D), suggesting that PI(3,5)P2 is metabolized by endogenous MTM1 in skeletal muscle cells (7). However, one cannot exclude that one part of PI(3,5)P2 could come from the accumulation of PI3P and its subsequent phosphorylation by PIKfyve. Regarding PI(3,4)P2, a phosphoinositide unrelated to MTM1, no significant differences were observed between WT and Mtm1-KO cells, although its levels were slightly lower in Mtm1-KO cells (Fig. 2E). Because PI5P, the product of PI(3,5)P2 hydrolysis by MTM1, cannot be quantified by metabolic labeling, we used a specific radioactive mass assay. We measured a decrease of PI5P level by ~40% in Mtm1-KO myoblasts and ~70% in Mtm1-KO myotubes compared to WT cells (Fig. 2F) indicating that MTM1 is the major source for PI5P synthesis in this cell system. Unexpectedly, while an increase of PI5P level in myotubes derived from WT myoblasts was expected, due to the increase of MTM1 expression during differentiation (Fig. 1A), we measured a ~50% decrease in its level (Fig. 2F). We concluded that PI5P could be used as a substrate by phosphoinositide-metabolizing enzymes during myoblasts differentiation.
Fig. 2.
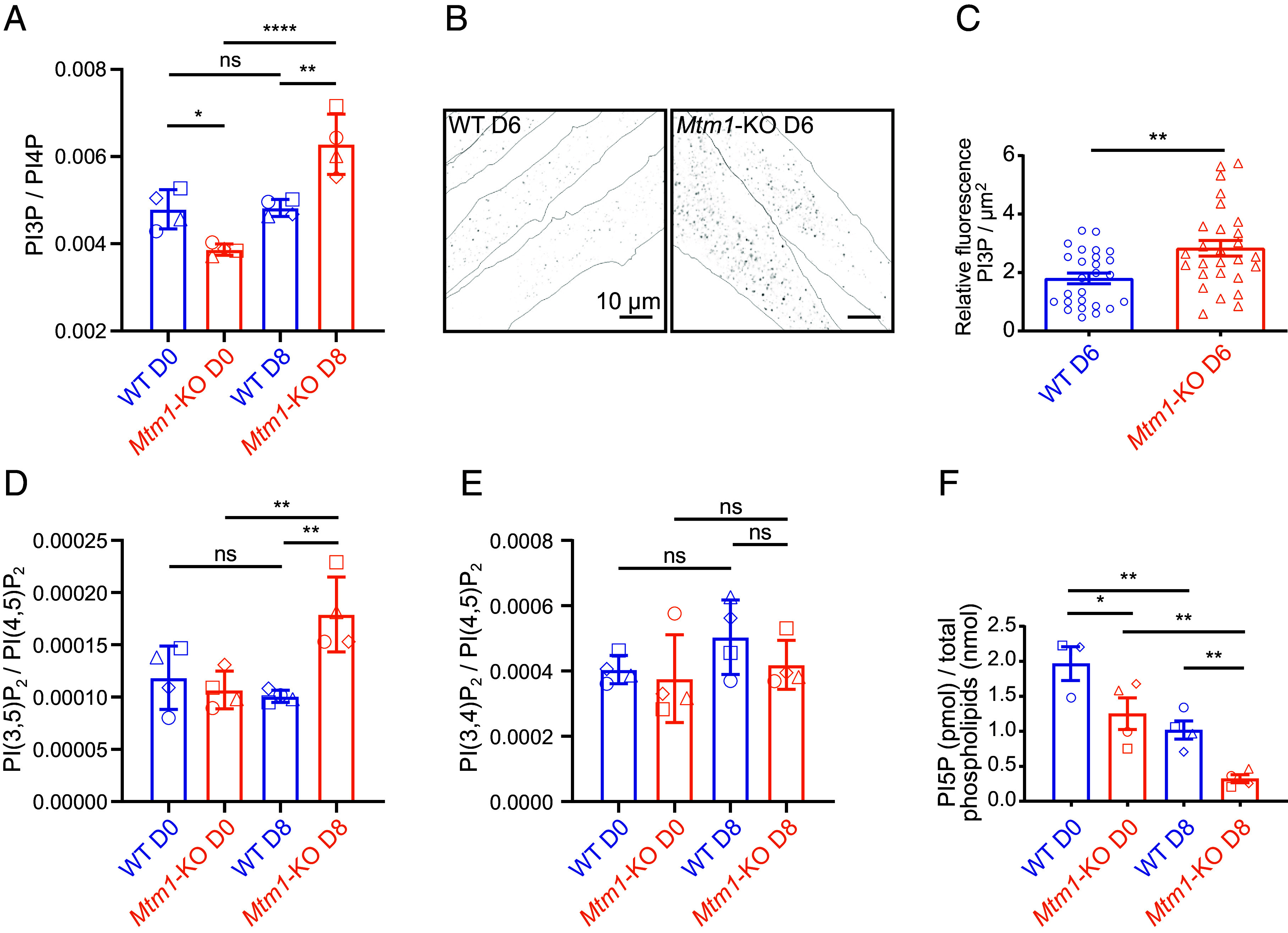
PI5P is mainly produced by MTM1 in skeletal muscle cells and is rapidly metabolized. (A) PI3P quantification in C2C12 WT and Mtm1-KO cells at the indicated days of differentiation quantified by HPLC following 32P metabolic labeling. Results are expressed as ratios to the most abundant PI-monophosphate PI4P. A unique symbol for each independent experiment is used. Data are represented as mean ± SEM, n = 4, ns = not significant, *P < 0.05, **P < 0.01, and ****P < 0.0001 according to one-way ANOVA test and Šídák’s multiple comparisons test. (B) Representative images of PI3P labeling (FYVEHRS probe) of C2C12 WT and Mtm1-KO myotubes on day 6 of differentiation. Cell contours are represented as dashed lines. (Scale bar, 10 μm.) (C) Quantification of the experiment described in (B). Data are represented as mean ± SEM, n = 3, each point represents one field of view, **P < 0.01 according to Student’s t test. (D) PI(3,5)P2 quantification as in (A). Results are expressed as ratios to the most abundant PI-bisphosphate PI(4,5)P2. A unique symbol for each independent experiment is used. Data are represented as mean ± SEM, n = 4, ns = not significant, **P < 0.01 according to one-way ANOVA test and Šídák’s multiple comparisons test. (E) PI(3,4)P2 quantification as in (A). Results are expressed as ratios to the most abundant PI-bisphosphate PI(4,5)P2. A unique symbol for each independent experiment is used. Data are represented as mean ± SEM, n = 4, ns = not significant according to one-way ANOVA test and Šídák’s multiple comparisons test. (F) PI5P quantification by mass assay in C2C12 WT and Mtm1-KO cells at the indicated days of differentiation. Data are represented as mean ± SEM, n = 4, *P < 0.05 and **P < 0.01 according to one-way ANOVA test and Šídák’s multiple comparisons test.
PI(4,5)P2, Tks5, Dynamin-2, and Myomaker Are Enriched in Podosome-Like Protrusions, the Myoblast Fusion-Promoting Sites.
The best characterized metabolic conversion mechanism of PI5P is mediated by PI5P 4-kinases (PI5P4Ks) to generate a minor pool of PI(4,5)P2, the major source being generated from PI4P by PI4P 5-kinases (22). However, it is largely accepted that their function would be to control PI5P levels in the cell (23). Interestingly, PI(4,5)P2 was shown to be locally enriched at the fusion site of Drosophila myoblast (24) in structures called podosome-like protrusions (PLPs), demonstrated to drive myoblast fusion by exerting an invasive force toward the opposing founder cell (25). Importantly, PLPs are essential to bring the cell membranes even closer together than the adhesion machinery and would be required to coordinate the action of cell fusogens (26–28). In this model, PI(4,5)P2 enrichment at the fusion site is thought to regulate fusion through the localization of activators of actin polymerization (24), which is consistent with the role of PI(4,5)P2 as a central regulator of actin cytoskeleton dynamics (29). More recently, those structures have been described to drive mammalian myoblast fusion (30). Remarkably, this study showed that the scaffolding proteins Tks5 (31) and Dynamin-2 (32) are also markers of those structures and are required for mammalian myoblast fusion. Therefore, we performed PI(4,5)P2 immunostaining using the established conditions reported by Hammond et al. that allow immunostaining of phosphoinositides at the plasma membrane (referred to as plasma membrane staining in the manuscript) or in endomembranes (referred to as endomembrane staining) (33). Immunofluorescence experiments on WT cells differentiated for 3 d revealed the presence of PI(4,5)P2 decorating all the plasma membrane, with PI(4,5)P2-enriched structures resembling PLPs (SI Appendix, Fig. S2A) (30). Using a fluorescent recombinant probe for PI(4,5)P2 (e.g., the PH domain from phospholipase Cδ1, PLCδ1) (34), we confirmed PI(4,5)P2 enrichment in those structures where α-actin accumulates (Fig. 3 A and B). Colabeling with Tks5 confirmed that these structures are indeed PLPs (Fig. 3 C and D). Time-lapse microscopy of WT C2C12 cells expressing Tks5-GFP through a lentiviral approach allowed us to observe the dynamics of structures resembling the labeling observed in fixed cells (Movie S1). However, excitation illumination hindered cell fusion in the imaged field areas. To address this issue, we transfected Tks5-GFP to minimize laser exposure and imaged cell fusion using confocal microscopy with a 15-min time-interval. Even though cell fusion remained low under fluorescent microscopy, Movie S2 shows a fusion event where Tks5 enrichment preceded cell fusion. Additionally, we demonstrated the enrichment of Tks5 and PI(4,5)P2 at the fusion site of fusing myocytes (Fig. 3E). We also identified dynamin-2 enrichment in PLPs (Fig. 3 F and G) and established that the skeletal muscle myosin MYH4 serves a marker for PLPs (Fig. 3 F–I and SI Appendix, Fig. S2 D and E). Importantly, confocal microscopy enabled us to illustrate its enrichment in fusing myocytes (SI Appendix, Fig. S2F). Thus, these results confirmed that PLPs are the fusion-promoting protrusion. Since the fusogenic protein Myomaker is known to be essential for the fusion process (35), and the formation of the hemifusion stalk (36), it was tempting to speculate that there may be a correlation between PLPs and this fusion factor. This factor has been suggested to converge to the invasive protrusions of Zebrafish myoblast, initiating myoblast fusion, as demonstrated for Myomerger (27). Therefore, we colabeled Myomaker with an antibody that we first validated (SI Appendix, Fig. S3 A and B) along with PI(4,5)P2 and MYH4. We observed a strong enrichment of Myomaker in the plasma membrane enriched with PI(4,5)P2 (Fig. 3 H and I and SI Appendix, Fig. S3C), while MYH4 was localized in the cytosol (Fig. 3I and SI Appendix, Fig. S3C). These findings further emphasize the role of PLPs in mammalian myoblast fusion. Moreover, colabeling of PI(4,5)P2 with cadherin, a plasma membrane marker, revealed that membrane folding does not account for the excess PI(4,5)P2 staining in PLPs (Fig. 3J). Importantly, imaging and quantification of PLPs using Tks5 and PI(4,5)P2 colabeling in WT and Mtm1-KO cells after 3 d of differentiation revealed a defect in their formation in the absence of MTM1 (Fig. 3K and SI Appendix, Fig. S3 D and E).
Fig. 3.
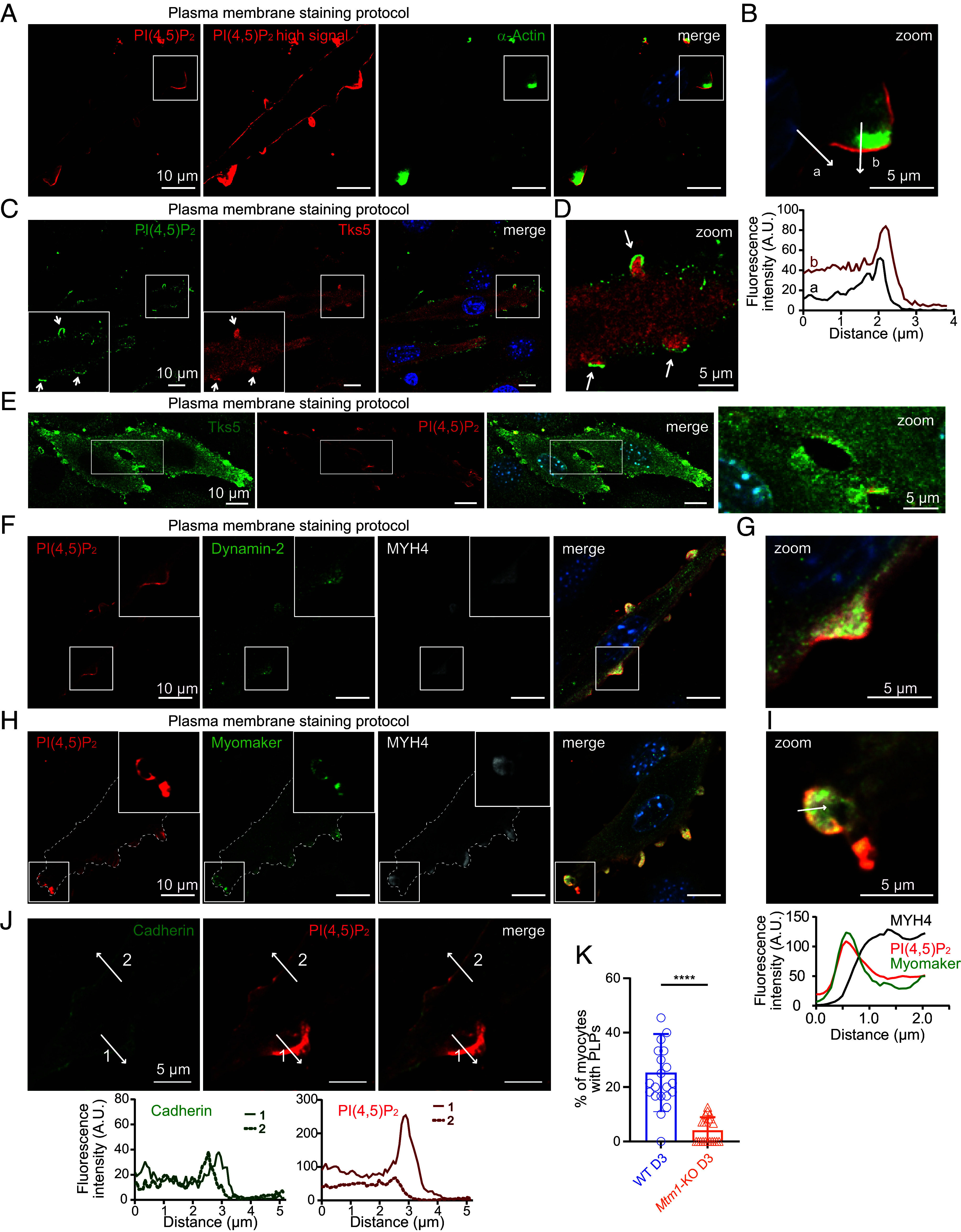
PI(4,5)P2 is highly enriched in podosome-like protrusions. (A) Representative confocal images of C2C12 WT after 3 d of differentiation and labeled with a PI(4,5)P2 probe (PH domain of PLCδ1), α-actin, and DAPI. (Scale bar, 10 μm.) (B) Magnification of the boxed area shown on the left. Respective line scans are shown below the zoom panel. (Scale bar, 5 μm.) (C) Representative confocal images of C2C12 WT on day 3 of differentiation labeled with a PI(4,5)P2 antibody and for Tks5 and DAPI. Arrows indicate podosome-like protrusions. (Scale bar, 10 μm.) (D) Magnification of the boxed area shown in (C). Arrows indicate podosome-like protrusions. (Scale bar, 5 μm.) (E) Confocal images of fusing C2C12 WT on day 3 of differentiation labeled for Tks5, a PI(4,5)P2 probe, and DAPI. (Scale bar, 10 μm.) A magnification of the boxed area depicting the fusion site is shown on the right. (Scale bar, 5 μm.) (F) Representative confocal images of C2C12 WT after 3 d of differentiation labeled with a PI(4,5)P2 probe and for Dynamin-2, Myosin Heavy Chain 4 (MYH4) and DAPI. (Scale bar, 10 μm.) (G) Magnification of the boxed area shown in (F). (Scale bar, 5 μm.) (H) Representative confocal images of C2C12 WT after 3 d of differentiation and labeled with a PI(4,5)P2 probe, Myomaker, MYH4, and DAPI. Cell contour is represented as dashed lines. (Scale bar, 10 μm.) (I) Magnification of the boxed area shown on the left. A line scan is shown on the bottom. (Scale bar, 5 μm.) (J) Representative confocal images of C2C12 WT after 3 d of differentiation and labeled for Cadherin and with the PI(4,5)P2 probe. Respective line scans are shown on the bottom. (Scale bar, 5 μm.) (K) Quantification of podosome-like protrusions [positives for PI(4,5)P2 and Tks5 enrichments] in WT and Mtm1-KO myocytes at the indicated time points of differentiation. Data are represented as mean ± SEM, n = 4, five fields per independent experiment, each point represents one field of view, ****P < 0.001 according to unpaired t test.
PI5P4Kα and γ Are Enriched in Podosome-Like Protrusions.
We next tested whether PI(4,5)P2 enrichment was dependent on PI5P4K activity. This family encompasses three genes in mammals: PIP4K2A, PIP4K2B, and PIP4K2C that encode the enzymes PI5P4Kα, PI5P4Kβ, and PI5P4Kγ, respectively, with very different in vitro PI5P 4-kinase activities. PI5P4Kα is at least 100-fold more active than PI5P4Kβ and 1,000-fold more active than PI5P4Kγ (37). However, recent data in Drosophila suggest that all mammalian PI5P4K isoforms have in vivo lipid-kinase activity since the phenotype observed in the absence of the sole Drosophila PI5P4K can be compensated by each mammalian isoform (38). Moreover, individual PI5P4Ks have been shown to localize in different cellular compartments and to homo and hetero-dimerize (39). Notably, PI5P4Kβ, which is localized in the nucleus through a nuclear localization sequence (39), has been shown to regulate nuclear PI5P and the expression of myogenic genes during myoblast differentiation (20). Concerning PI5P4Kα and PI5P4Kγ, they have been shown to hetero-dimerize in vitro (37) and to localize mainly in the cytosol including the Golgi and undefined endomembrane compartments (39). We thus focused on PI5P4Kα and PI5P4Kγ and localized endogenous PI5P4Ks using antibodies that we first validated (SI Appendix, Fig. S4 A, Right) and that didn’t exhibit cross-reactivity with one another (SI Appendix, Fig. S4 D–F). PI5P4Kα and PI5P4Kγ were found to localize as cytoplasmic puncta that accumulate in MYH4-enriched PLPs (Fig. 4 A and B). Immunofluorescence of PI(4,5)P2-enriched PLPs confirmed those observations (Fig. 4 C and D). Particularly, we observed that PI5P4Kα was strongly enriched in all PLPs, while PI5P4Kγ accumulated in larger PLPs (Fig. 4 E and F). These findings further support the hypothesis that PI5P4Ks, and most probably PI5P4Kα, are responsible for producing this localized pool of PI(4,5)P2 from PI5P. In contrast, PI5P4Kβ localized homogeneously as cytosolic puncta (SI Appendix, Fig. S5 A and B).
Fig. 4.
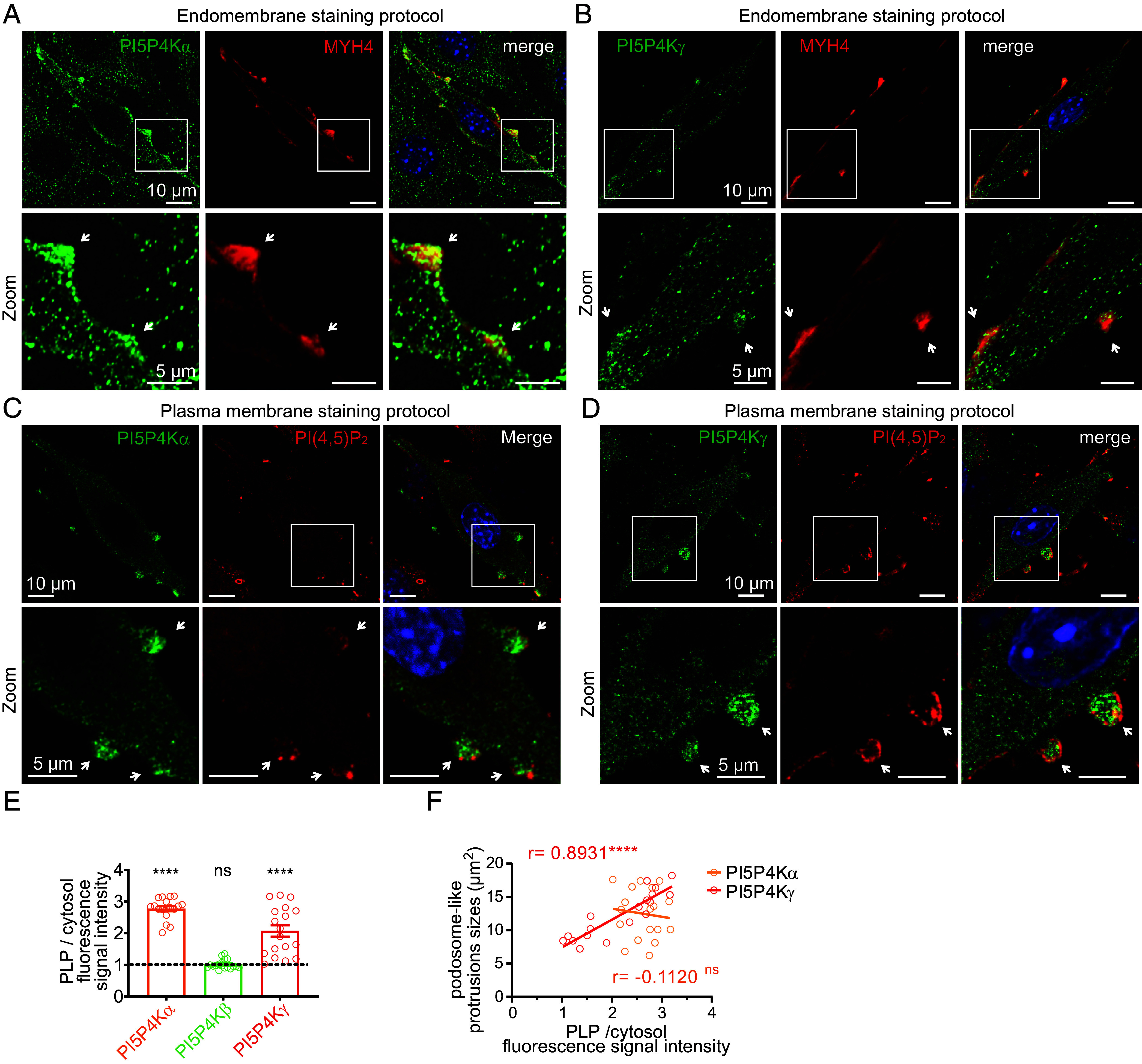
PI5P4Kα and γ localize to PI(4,5)P2-enriched podosome-like protrusions. (A) Top: Representative confocal images of C2C12 WT after 3 d of differentiation and labeled for PI5P4Kα, MYH4 and DAPI. (Scale bar, 10 μm.) Bottom: higher magnifications of areas shown as boxes on the top. The arrows show the strong enrichment of PI5P4Kα in MYH4-positive PLPs. (Scale bar, 5 μm.) (B) Top: Representative confocal images of C2C12 WT after 3 d of differentiation and labeled for PI5P4Kγ, MYH4 and DAPI. (Scale bar, 10 μm.) Bottom: higher magnifications of areas shown as boxes on the top. The arrows indicate the enrichment of PI5P4Kγ in MYH4-positive PLPs. (Scale bar, 5 μm.) (C) Top: Representative confocal images of C2C12 WT after 3 d of differentiation and labeled for PI5P4Kα, PI(4,5)P2 (PH domain of PLCδ1), and DAPI. (Scale bar, 10 μm.) Bottom: higher magnifications of areas shown as boxes on the top. (Scale bar, 5 μm.) (D) Top: Representative confocal images of C2C12 WT at 3 d of differentiation labeled for PI5P4K2γ, PI(4,5)P2 (PH domain of PLCδ1), and DAPI. (Scale bar, 10 μm.) Bottom: magnifications of areas shown as boxes on the top. (Scale bar, 5 μm.) (E) Quantification of the fluorescence ratio of the indicated proteins in podosome-like protrusions (PLPs) versus cytosol. Data are represented as mean ± SEM, n = 3, each point represents the quantification of one field of view, ns = not significant, ****P < 0.0001 according to One-sample t test (two tailed). (F) Correlation plot between the size of PLPs and the fluorescence ratio of the indicated proteins in podosome-like protrusions versus cytosol, n = 3, r = Pearson correlation coefficient, ns = not significant, ****P < 0.0001, according to simple linear regression analysis.
PI5P4Kα Plays a Major Role in Myoblast Differentiation.
We then analyzed the expression of PI5P4Kα and PI5P4Kγ by immunoblot, which showed that both proteins increased gradually during C2C12 differentiation, with no difference in Mtm1-KO cells (SI Appendix, Fig. S5C). This suggests that these proteins may play a role in myogenesis but also in mature muscle cells, as already demonstrated for PI5P4Kα in myotubes, in which it regulates the PI5P level to control insulin-stimulated glucose uptake (40). We then knock down either Pip4k2a or Pip4k2c in C2C12 cells using lentiviruses expressing shRNA. We achieved efficient knockdown with Pip4k2a shRNA #1 and #2 (84.3% and 94.2% of protein extinction, respectively), while shRNA #3 led to a more modest reduction (42.8% of extinction) (SI Appendix, Fig. S4 A and B). Knockdown of Pip4k2a with shRNAs #1 and #2 resulted in a significant impairment of myoblast fusion (Fig. 5A) and with a high proportion of MYH4-positive cells with 1 nucleus (Fig. 5B). Regarding Pip4k2a shRNA #3, myoblast fusion still occurred, indicating that substantial knockdown levels are required to inhibit the fusion process. Immunoblot analysis of Myogenin expression in cells depleted for Pip4k2a with the most effective shRNA (shRNA #2) during differentiation indicated that the myogenic program was unaffected (Fig. 5C). Re-expression of PI5P4Kα in knockdown cells for Pip4k2a (shRNA #2) using a lentivirus that is resistant to shRNA successfully restored the fusion process (Fig. 5 A and B), thereby confirming that this phenotype is specifically due to Pip4k2a knockdown. Importantly, re-expression of a kinase-dead version of PI5P4Kα failed to rescue the fusion process, indicating the essential role of its catalytic activity (Fig. 5 A and B). Concerning PI5P4Kγ, robust knockdown was achieved with Pip4k2c shRNAs #1 and #2 (60% and 93.6% of protein extinction, respectively) (SI Appendix, Fig. S4 A and C). This also led to the inhibition of myoblast fusion and a high proportion of MYH4-positive cells containing 1 nucleus and re-expression of PI5P4Kγ, with either the WT or kinase-dead version, rescued the fusion defect (Fig. 5 A and B). This validates the observed phenotype and suggests a potential role for this isoform in the regulation of PI5P4Kα activity. Similar to Pip4k2a knockdown, immunoblot analysis of Myogenin expression during differentiation in cells depleted for Pip4k2c using the most effective shRNA (shRNA #2) revealed no impact on the myogenic program (Fig. 5C). Consistent with the phenotypes observed during the myoblast differentiation, and their enrichment in PLPs, the knockdown of Pip4k2a and Pip4k2c reduced the number of PLPs in myoblasts after 3 d of differentiation (Fig. 5D). To further investigate the role of PI5P4Kα in myogenesis, we used lentiviruses to overexpress the lipid kinase in C2C12 myoblasts and allowed the cells to differentiate for 3 d. The results showed that overexpression of flag-PI5P4Kα accelerated differentiation, with a fusion index of 15.08% compared to a fusion index of 12.33% observed with vector-control lentiviruses (Fig. 5E). Importantly, PI5P4Kα overexpression caused a marked increase in fusion with a robust increase in the appearance of myotubes with several nuclei in the cultures infected with PI5P4Kα (Fig. 5F), further suggesting that PI5P4Kα likely plays a role in myoblast fusion.
Fig. 5.
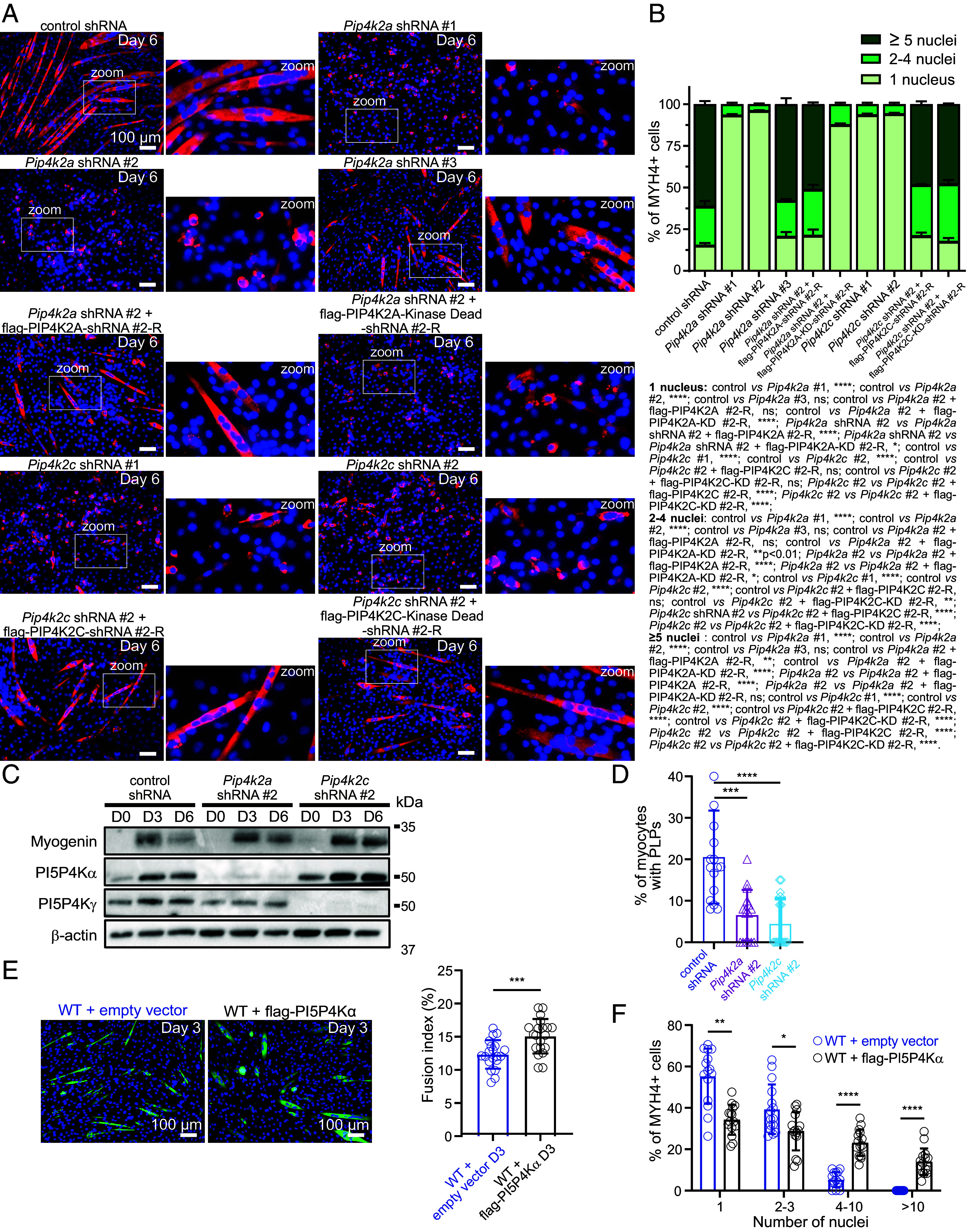
PI5P4Kα play a major role in myoblast differentiation. (A) MYH4 and DAPI staining of differentiated C2C12 cells expressing control shRNA, Pip4k2a shRNAs, Pip4k2c shRNAs and re-expressing or not a shRNA-resistant PI5P4K (-R) or a shRNA-resistant kinase-dead PI5P4K, after 6 d of differentiation. Higher magnifications of the boxed areas are shown on the right of each image. (Scale bar, 100 μm.) (B) Percentage of MYH4 positive cells containing 1, 2 to 4, or ≥5 nuclei from the experiment described in (A). Data are represented as mean ± SEM, n = 3, five fields per independent experiment, ns = not significant, *P < 0.05, **P < 0.01, ****P < 0.0001, according to Two-way ANOVA and Šídák’s multiple comparisons test. (C) Expression of PI5P4Kα, PI5P4Kγ, Myogenin, and actin, at the indicated time points of C2C12 cells expressing control shRNA, Pip4k2a shRNA #2, Pip4k2c shRNA #2 analyzed by western blot. Images are representative of three independent experiments. (D) Quantification of podosome-like protrusions [positives for PI(4,5)P2 and Tks5 enrichment] in WT myocytes expressing control shRNA, Pip4k2a shRNA #2, or Pip4k2c shRNA #2 at the indicated time points of differentiation. Data are represented as mean ± SEM, n = 3, five fields per independent experiment, each point represents one field of view, ***P < 0.001 according to unpaired t test. (E) Representative images of C2C12 WT cells transduced with lentiviruses expressing an empty vector or flag-PI5P4Kα after 3 d of differentiation and labeled for MYH4 and DAPI. (Scale bar, 100 μm.) Quantification of the fusion index for the experiment shown on the right. Data are represented as mean ± SEM, n = 4, five fields per independent experiment, each point represents one field of view. ***P < 0.001 according to unpaired t test. (F) Percentage of MYH4+ cells that contained the indicated number of nuclei of the experiment described in (E). Data are represented as mean ± SEM, n = 3, five fields per independent experiment, each point represents one field of view, *P < 0.05, **P < 0.01, according to Mann–Whitney U test.
PI5P Is Likely Converted into PI(4,5)P2 by PI5P4Kα on Cytosolic Vesicles.
In order to demonstrate that PI5P4Ks are responsible for the transformation of PI5P produced from MTM1 into PI(4,5)P2, we measured PI5P levels in 6 d differentiated WT cells depleted for either PI5P4Kα or PI5P4Kγ. A ~fivefold increase in PI5P levels was observed when Pip4k2a was knocked down (as compared to control cells), whereas a ~twofold increase was observed when PI5P4Kγ was knocked down, showing that PI5P4Kα is responsible for the conversion of a large amount of PI5P into PI(4,5)P2 (Fig. 6A). Importantly, PIP2 levels were unchanged, showing that PI5P4Kα and γ are responsible for the synthesis of a quantitatively minor pool of PI(4,5)P2 (Fig. 6B). In contrast, knock down of Pip4k2a or Pip4k2c in Mtm1-KO C2C12 cells (SI Appendix, Fig. S5 D–G) had no effect on PI5P levels, demonstrating that the pool of PI5P produced by MTM1 is metabolized into PI(4,5)P2 mainly by PI5P4Kα (Fig. 6A). Because there is currently no specific probe to visualize endogenous PI5P, we reasoned that overexpressing PI5P4Kα together with a PI(4,5)P2 probe would define the localization of PI5P. We thus overexpressed mCherry-PH (PLCδ1) alone (as a control) or cooverexpressed mCherry-PH (PLCδ1) and GFP-PI5P4Kα [converting PI5P into PI(4,5)P2], in WT C2C12 cells, differentiated the cells for 3 d, and labeled them for active β1 integrin as a ventral plasma membrane marker. As expected and previously reported, blocking PI(4,5)P2 accessibility with a PI(4,5)P2 probe inhibits myoblast fusion and PLP formation (24, 41). Besides the expected labeling of PI(4,5)P2 at the plasma membrane in both conditions, we observed approximately a fourfold increase in the labeling of cytosolic vesicles upon overexpression of GFP-PI5P4Kα (Fig. 6 C and D and SI Appendix, Fig. S5H). These PI(4,5)P2-rich vesicles, whose origin remains to be determined, colocalized with GFP-PI5P4Kα (Fig. 6D), suggesting that PI5P is primarily localized on vesicles, mirroring the localization pattern of endogenous PI5P4Kα, which is found to be enriched in PLPs (Fig. 4 A and C). It is worth noting that PI(4,5)P2 signals in the ventral region of cells were much lower compared to those at the equatorial planes. This discrepancy could potentially be attributed to strong interactions of PI(4,5)P2 with proteins from the adhesion machinery, which could hinder its accessibility for the PI(4,5)P2 probe. Interestingly, we observed a significant enrichment of flag-MTM1 in PLPs (Fig. 6 F and G), suggesting that MTM1 may convert PI(3,5)P2 into PI5P in proximity to the plasma membrane. Subsequently, PI5P would be transformed into PI(4,5)P2 by PI5P4Kα, fueling the plasma membrane with PI(4,5)P2 to promote PLP formation.
Fig. 6.
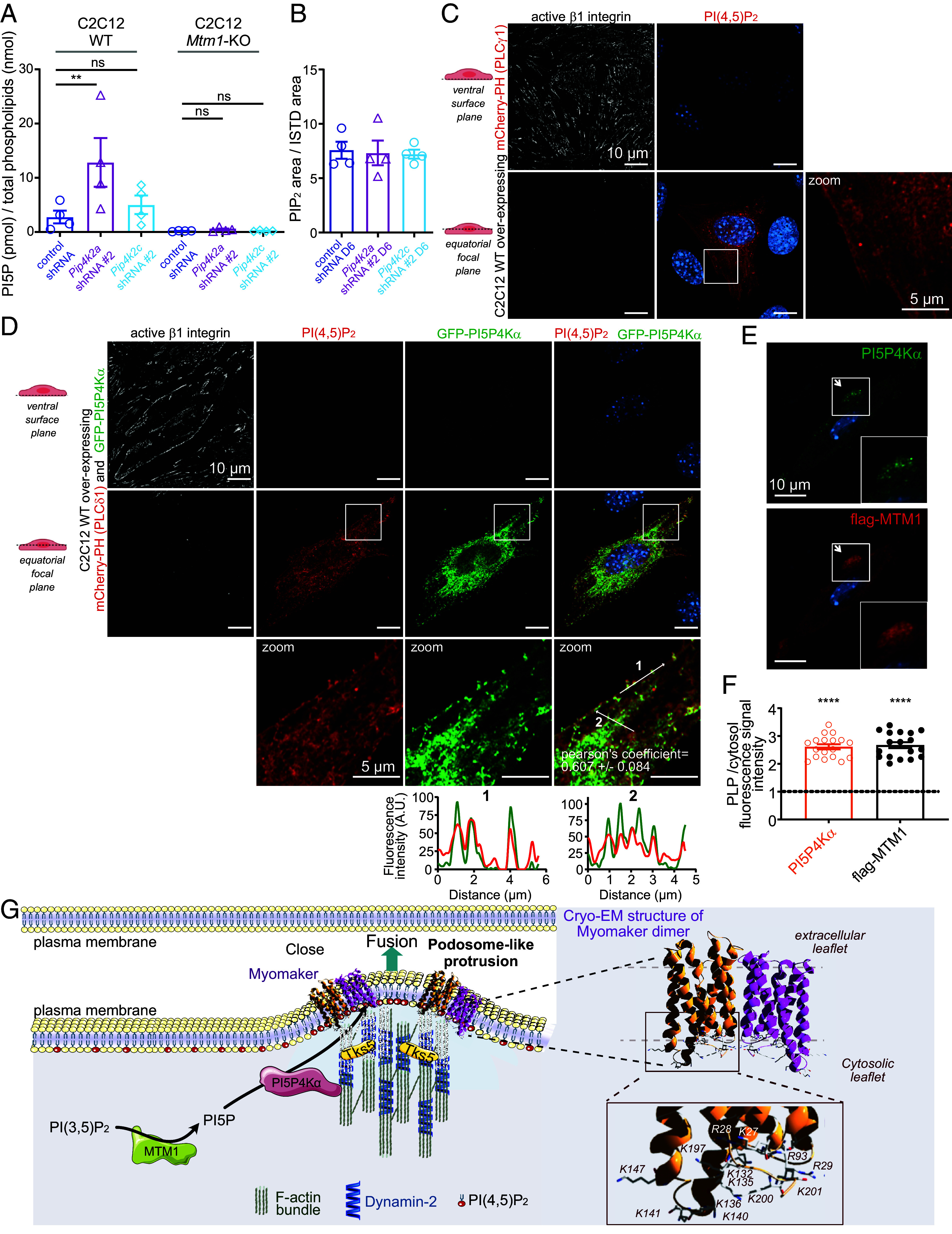
PI5P4Kα is responsible for the conversion of PI5P produced by MTM1 into a minor PI(4,5)P2 pool on cytosolic vesicles. (A) PI5P quantification by mass assay of 6 d differentiated WT and Mtm1-KO C2C12 cells expressing control shRNA, Pip4k2a shRNA #2, or Pip4k2c shRNA #2. Data are represented as mean ± SEM, n = 4, ns = not significant, **P < 0.01 according to one-way ANOVA test and Šídák’s multiple comparisons test. (B) PIP2 quantification by mass spectrometry of 6 d differentiated C2C12 cells expressing control shRNA, Pip4k2a shRNA #2, or Pip4k2c shRNA #2. Data are represented as mean ± SEM, n = 4, no statistical significance, according to one-way ANOVA test and Šídák’s multiple comparisons test. (C) Representative confocal images of C2C12 WT cells overexpressing mCherry-PH (PLCδ1), differentiated for 3 d, and labeled for active β1 integrin and DAPI. (Scale bar, 10 μm.) Magnification of the boxed area shown on the right. (Scale bar, 5 μm.) (D) Representative confocal images of C2C12 WT cells overexpressing mCherry-PH (PLCδ1) and GFP-PI5P4Kα, differentiated for 3 d, and labeled for active β1 integrin and DAPI. (Scale bar, 10 μm.) Magnification of the boxed areas shown on the bottom. (Scale bar, 5 μm.) Respective line scans are shown below. Colocalization between mCherry-PH (PLCδ1) and GFP-PI5P4Kα was quantified by means of Pearson’s correlation coefficient using the Fiji JaCoP Plugin, n = 3 (10 cells per experiment). r = 0.607 ± 0.084. (E) Representative confocal images of C2C12 WT cells differentiated for 3 d and overexpressing flag-MTM1, and labeled for PI5P4Kα, flag, and DAPI. (Scale bar, 10 μm.) Higher magnifications of the boxed areas are shown as Insets. (F) Quantification of the fluorescence ratio of the indicated proteins in podosome-like protrusions (PLPs) versus cytosol. Data are represented as mean ± SEM, n = 3, each point represents the quantification of one field of view, ****P < 0.0001 according to One-sample t test (two tailed). (G) Hypothetical model for the production of PI(4,5)P2 in podosome-like protrusions via MTM1 through PI5P4Kα. Right: Ribbon diagram representation of Myomaker dimer (PDB: 8T04). The side chains of all basic residues are shown in stick form.
Discussion
Our results reported here identify PI5P4Kα/γ, members of the lipid kinases family implicated in metabolism, immune function, and growth control (23), as important regulators of myoblast differentiation. We show, by biochemical experiments combined with studies in genome-engineered cells, that MTM1 is a master enzyme for the production of PI5P from PI(3,5)P2 in skeletal muscle cells, highlighting an unconsidered role of PI(3,5)P2 and PI5P compared to PI3P in the pathogenesis of XLMTM. Moreover, we uncover a direct role of MTM1 activity in the early phase of myogenesis, showing that MTM1 may participate in different regulatory mechanisms during myogenesis as compared to its function in mature skeletal muscle (42). We reveal an unexpected mechanism whereby PI5P is rapidly transformed into PI(4,5)P2 by PI5P4Kα to drive PI(4,5)P2 accumulation in PLPs where it localizes with α-actin, Tks5, and Dynamin-2, the molecular markers of PLPs (30), as well as the skeletal muscle myosin MYH4 and the fusogen Myomaker. This allows the generation of a localized area of close cell–cell contact to create a favorable environment for membranes to fuse (Fig. 6F). Therefore, the pathway unraveled here identifies PI5P4Kα as responsible for the synthesis of a minor yet functional pool of PI(4,5)P2, as also shown in B cells (43), while most of previous studies suspected that it mainly acts by controlling PI5P levels (23). Concerning PI5P4Kγ, we found it accumulating during PLP formation. Given the evidence that knockdown of either pip4k2a or pip4k2c negatively affects PLP formation, and that PI5P4Kα activity is essential for myocyte fusion, while PI5P4Kγ activity is not, our main hypothesis is that PI5P4Kγ could play a role in localizing PI5P4Kα to the site of PI(4,5)P2 generation (37) and that its accumulation may impair PI5P4Kα activity by competing for substrate binding. The major pool of PI(4,5)P2, generated from PI4P by PI4P 5-kinases (22), and decorating the entire plasma membrane would, in turn, play a major role in cytoskeleton dynamics, focal adhesion assembly (44), second messenger production (IP3 and DAG), as well as in cell-cell contacts through cadherin stabilization (45). Based on our results, we propose a model in which the production of PI(4,5)P2 from PI5P within vesicles is as a mechanism for myocytes to efficiently supply a local amount of PI(4,5)P2 for PLP formation. This would imply exocytosis of these vesicles to specific regions of the plasma membrane, suggesting the involvement of the actin and/or tubulin cytoskeleton (46). Interestingly, this hypothesis aligns with the crucial role of the actin cytoskeleton in the targeted exocytosis of prefusion vesicles during myoblast fusion in Drosophila (47) and is consistent with previous electron microscopy studies revealing the accumulation of vesicles at localized points of cell-cell apposition in fusing cells (48, 49). In line with the recognized role of PI(4,5)P2 in vesicle exocytosis and membrane fusion (50), these vesicles will subsequently fuel the plasma membrane with PI(4,5)P2. This, in turn, could recruit proteins capable of inducing curvature in the plasma membrane through curvature-sensing domains. Interesting candidates are I-BAR domain-containing proteins, well-documented for generating negative membrane curvature on PI(4,5)P2-rich membranes, linking direct membrane deformation to actin polymerization, and inhibiting the lateral diffusion of phosphoinositide molecules (51). Moreover, it was proposed that exocytosis of these vesicles may release fusogenic materials to trigger fusion (52). The analysis of the recent cryo-EM structure of the fusogen Myomaker (53) revealed clusters of positively charged amino acid residues (14 arginine and lysine) facing the cytosolic leaflet, compatible with its binding with phosphoinositides and that could be responsible for Myomaker clustering (Fig. 6F). Interestingly, Syntaxin, a plasma membrane protein crucial for vesicle fusion with target membranes, has been demonstrated to interact with and be clustered by PI(4,5)P2 through a polybasic region, thus strengthening this hypothesis (54). Moreover, Myomaker could also promote PI(4,5)P2 clustering through electrostatic interactions, thereby facilitating the recruitment of other effectors (55–57). In line with the phenotype observed when Pip4k2a was knocked down in C2C12, knockdown of the Zebrafish PI5P4Kα has been shown to lead to severe morphological abnormalities features, most likely due to defects in midbody curvature, which are largely dependent on structurally and functionally intact muscles (58). However, mouse knockout for PI5P4Kα (59) or PI5P4Kγ (60) appear normal in regard to growth and viability. Nevertheless, it is conceivable that most tissues can compensate developmental events when there is deletion of a single isoform of PI5P4K as suggested by the facts that phenotypes are only observed or worsen when multiple isoforms are targeted (59) and that compensation of the Drosophila PI5P4K deletion phenotype is observed with any of the three mammalian PI5P4K isoforms (38). Finally, because we didn’t succeed to identify the vesicular compartments where PI5P is produced by MTM1 and transformed into PI(4,5)P2 by PI5P4Kα, further investigations are needed to understand the origin of the different actors implicated in this molecular pathway. With better tools being made available, such as a specific PI5P probe and MTM1 antibodies working on immunofluorescence, we hope to answer precisely these questions to understand the spatiotemporal regulation of MTM1 functions particularly during PLP formation. Collectively, these findings provide a key missing link in the understanding of muscle cell fusion, potentially explaining the hypotrophy and abnormal shape of myofibers observed in XLMTM patients (17, 61), that we also observed in our Mtm1-KO cell model. Notably, the absence of MTM1 leads to impaired PLP formation, causing delayed myoblast fusion during early differentiation. However, at high cell confluency, fusion still occurs but in a disorganized manner, resulting in the formation of shorter, non-spindle-shaped myotubes with clustered nuclei. Given Myomaker’s essential role in myoblast fusion, this suggests that its trafficking to the plasma membrane may operate independently of MTM1, or alternatively, compensatory mechanisms within the cells may facilitate Myomaker localization to the plasma membrane. Consequently, the defect in PLP formation resulting from the absence of MTM1 may result in a uniform distribution of Myomaker at the plasma membrane of myocytes, rather than its enrichment in PLPs. This uniform distribution likely underlies the dysregulation of the spatiotemporal fusion process, ultimately leading to the observed phenotypes. Taken together, our study reveals a phosphoinositide conversion mechanism involving MTM1 and PI5P4Kα that generates a functional pool of PI(4,5)P2, which accumulates in PLPs and plays a crucial role in the precise regulation of myoblast fusion.
Materials and Methods
Detailed information on cell culture; antibodies (SI Appendix, Table S1); plasmids (SI Appendix, Table S2); main reagents; CRISPR-Mediated Genome Editing in C2C12 Cells; lentivirus production and transduction; Western Blotting; protein purification; microscopy and image analysis; phospholipid extraction and analysis; and PIP2 measurement by mass spectrometry is provided in SI Appendix.
Statistics and Reproducibility.
All data were represented as the mean ± SEM. Statistical analysis was performed as described for each experiment in the figure legends using GraphPad Prism 9. In all tests and all statistical analyzed datasets, the levels of significance were defined as *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. For comparisons between two experimental groups, after a normality test, data were analyzed by Mann–Whitney U test or unpaired t test. For comparisons between more than two experimental groups, data were analyzed by one-way ANOVA with a post hoc test (Šídák’s multiple comparisons test). One-sample t test was used for comparisons with control group values that had been set to 1 for normalization purposes. For each experiment, data are representative of at least three independent replications, with similar results obtained.
Supplementary Material
Appendix 01 (PDF)
Time-lapse microscopy of Tks5-GFP in differentiated C2C12 myoblasts. Day 3 differentiated C2C12 myoblasts transduced with lentivirus encoding Tks5-GFP were imaged with videomicroscopy using a 10-min time-interval. Arrows indicate contact-site that look-like podosome-like protrusions enriched in Tks5-GFP.
Time-lapse confocal microscopy of Tks5-GFP during C2C12 fusion. Day 3 differentiated C2C12 myoblasts transfected with a plasmid encoding Tks5-GFP were imaged with an LSM900 Zeiss microscope (Zen software, x20 objective) equipped with a ibidi stage Top incubator, using a 15-min time-interval and a single Z-plane. An arrow indicates the presence of a podosome-like protrusion enriched in Tks5-GFP before fusion.
Acknowledgments
We thank Jennifer Doudna and Romain Rouet (University of California, Berkeley/HHMI) for sharing with us the plasmid pMJ915, their protocol for Cas9 purification and their advices. We thank Jonathan Clarke (University of Cambridge) for the gift of the PI5P4Kα, β, and γ antibodies and the pEGFP vectors of PI5P4Kα, β, and γ. We thank Damien Ramel (UMR1297, Toulouse, France) for helpful discussions and Jim Dowling (Department of Molecular Genetics, University of Toronto, Canada) for critical reading of the manuscript. We thank the imaging, cytometry, We-Met, and Metatoul-Lipidomics facilities of the INSERM UMR1297 and the Integrated Screening Platform of Toulouse (Virginie Nahoum, PICT, IBiSA, Toulouse) for providing access to their equipments. This work was initiated by a grant from AFM-Téléthon (#20186 to J.V.) and supported by Inserm, grants from E-Rare JTC 2017 TREAT-MTMs (to B.P.), AFM-Téléthon (#23101 to J.V. and K.H.), and ANR (NEWPI, ANR-22-CE44-0026 to J.V.). M.P. is a recipient of a PhD fellowship from AFM-Téléthon (#22115). We thank the four anonymous reviewers for their insightful comments.
Author contributions
M.M. and J.V. designed research; M.M., A.O.K., G.C., M.P., A.M., R.F.-F., and J.V. performed research; M.M., A.O.K., G.C., R.F.-F., B.P., K.H., and J.V. analyzed data; B.P., K.H., and J.V. acquired financial support; and J.V. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. E.H.C. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Viaud J., et al. , Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie 125, 250–258 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Posor Y., Jang W., Haucke V., Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 23, 797–816 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasegawa J., Strunk B. S., Weisman L. S., PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct. Funct. 42, 49–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondeau F., et al. , Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet 9, 2223–2229 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Taylor G. S., Maehama T., Dixon J. E., Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. U.S.A. 97, 8910–8915 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amoasii L., et al. , Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle in vivo. J. Cell Sci. 126, 1806–1819 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Tronchere H., et al. , Production of phosphatidylinositol 5-phosphate by the phosphoinositide 3-phosphatase myotubularin in mammalian cells. J. Biol. Chem. 279, 7304–7312 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Schaletzky J., et al. , Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr. Biol. 13, 504–509 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Tasfaout H., Cowling B. S., Laporte J., Centronuclear myopathies under attack: A plethora of therapeutic targets. J. Neuromuscul. Dis. 5, 387–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buj-Bello A., et al. , The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 99, 15060–15065 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribeiro I., Yuan L., Tanentzapf G., Dowling J. J., Kiger A., Phosphoinositide regulation of integrin trafficking required for muscle attachment and maintenance. PLoS Genet. 7, e1001295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowling J. J., et al. , Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 5, e1000372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Qusairi L., et al. , T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl. Acad. Sci. U.S.A. 106, 18763–18768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaffe D., Saxel O., Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270, 725–727 (1977). [DOI] [PubMed] [Google Scholar]
- 15.Burattini S., et al. , C2C12 murine myoblasts as a model of skeletal muscle development: Morpho-functional characterization. Eur. J. Histochem. 48, 223–233 (2004). [PubMed] [Google Scholar]
- 16.Hindi S. M., Tajrishi M. M., Kumar A., Signaling mechanisms in mammalian myoblast fusion. Sci. Signal 6, re2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lionello V. M., et al. , Amphiphysin 2 modulation rescues myotubular myopathy and prevents focal adhesion defects in mice. Sci. Transl. Med. 11, eaav1866 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Volpatti J. R., et al. , X-linked myotubular myopathy is associated with epigenetic alterations and is ameliorated by HDAC inhibition. Acta Neuropathol. 144, 537–563 (2022), 10.1007/s00401-022-02468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begley M. J., et al. , Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. 103, 927–932 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stijf-Bultsma Y., et al. , The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell 58, 453–467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samso P., et al. , Antagonistic control of active surface integrins by myotubularin and phosphatidylinositol 3-kinase C2beta in a myotubular myopathy model. Proc. Natl. Acad. Sci. U.S.A. 119, e2202236119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan X., Thapa N., Choi S., Anderson R. A., Emerging roles of PtdIns(4,5)P2–beyond the plasma membrane. J. Cell Sci. 128, 4047–4056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghu P., Emerging cell biological functions of phosphatidylinositol 5 phosphate 4 kinase. Curr. Opin. Cell Biol. 71, 15–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bothe I., Deng S., Baylies M., PI(4,5)P2 regulates myoblast fusion through Arp2/3 regulator localization at the fusion site. Development 141, 2289–2301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D. M., Chen E. H., Drosophila myoblast fusion: Invasion and resistance for the ultimate union. Annu. Rev. Genet. 53, 67–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez J. M., Podbilewicz B., The hallmarks of cell-cell fusion. Development 144, 4481–4495 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Luo Z., et al. , The cellular architecture and molecular determinants of the zebrafish fusogenic synapse. Dev. Cell 57, 1582–1597.e6 (2022), 10.1016/j.devcel.2022.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millay D. P., Regulation of the myoblast fusion reaction for muscle development, regeneration, and adaptations. Exp. Cell Res. 415, 113134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senju Y., Lappalainen P., Regulation of actin dynamics by PI(4,5)P2 in cell migration and endocytosis. Curr. Opin. Cell Biol. 56, 7–13 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Chuang M. C., et al. , Tks5 and Dynamin-2 enhance actin bundle rigidity in invadosomes to promote myoblast fusion. J. Cell Biol. 218, 1670–1685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudlik G., et al. , Advances in understanding TKS4 and TKS5: Molecular scaffolds regulating cellular processes from podosome and invadopodium formation to differentiation and tissue homeostasis. Int. J. Mol. Sci. 21, 8117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R., et al. , Dynamin regulates the dynamics and mechanical strength of the actin cytoskeleton as a multifilament actin-bundling protein. Nat. Cell Biol. 22, 674–688 (2020), 10.1038/s41556-020-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond G. R., Schiavo G., Irvine R. F., Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem. J. 422, 23–35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemmon M. A., Ferguson K. M., O’Brien R., Sigler P. B., Schlessinger J., Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 92, 10472–10476 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millay D. P., et al. , Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leikina E., et al. , Myomaker and myomerger work independently to control distinct steps of membrane remodeling during myoblast fusion. Dev. Cell 46, 767–780.e767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke J. H., Irvine R. F., Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 454, 49–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathre S., et al. , Functional analysis of the biochemical activity of mammalian phosphatidylinositol 5 phosphate 4-kinase enzymes. Biosci. Rep. 39, BSR20182210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulley S. J., Clarke J. H., Droubi A., Giudici M. L., Irvine R. F., Exploring phosphatidylinositol 5-phosphate 4-kinase function. Adv. Biol. Regul. 57, 193–202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grainger D. L., Tavelis C., Ryan A. J., Hinchliffe K. A., Involvement of phosphatidylinositol 5-phosphate in insulin-stimulated glucose uptake in the L6 myotube model of skeletal muscle. Pflugers Arch. 462, 723–732 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Bach A. S., et al. , ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways. Mol. Biol. Cell 21, 2412–2424 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joubert R., et al. , Site-specific Mtm1 mutagenesis by an AAV-Cre vector reveals that myotubularin is essential in adult muscle. Hum. Mol. Genet. 22, 1856–1866 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Bulley S. J., et al. , In B cells, phosphatidylinositol 5-phosphate 4-kinase-alpha synthesizes PI(4,5)P2 to impact mTORC2 and Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 10571–10576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katan M., Cockcroft S., Phosphatidylinositol(4,5)bisphosphate: Diverse functions at the plasma membrane. Essays Biochem. 64, 513–531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charrasse S., Comunale F., De Rossi S., Echard A., Gauthier-Rouviere C., Rab35 regulates cadherin-mediated adherens junction formation and myoblast fusion. Mol. Biol. Cell 24, 234–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng J., Feng S., Wu B., Guo W., Polarized exocytosis. Cold Spring Harb. Perspect Biol. 9, a027870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S., et al. , A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev. Cell 12, 571–586 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Engel L. C., Egar M. W., Przybylski R. J., Morphological characterization of actively fusing L6 myoblasts. Eur. J. Cell Biol. 39, 360–365 (1986). [PubMed] [Google Scholar]
- 49.Kalderon N., Gilula N. B., Membrane events involved in myoblast fusion. J. Cell Biol. 81, 411–425 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin T. F., Role of PI(4,5)P(2) in vesicle exocytosis and membrane fusion. Subcell Biochem. 59, 111–130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao H., Pykalainen A., Lappalainen P., I-BAR domain proteins: Linking actin and plasma membrane dynamics. Curr. Opin. Cell Biol. 23, 14–21 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Doberstein S. K., Fetter R. D., Mehta A. Y., Goodman C. S., Genetic analysis of myoblast fusion: Blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 136, 1249–1261 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long T., et al. , Cryo-EM structures of Myomaker reveal a molecular basis for myoblast fusion. Nat. Struct. Mol. Biol. 30, 1746–1754 (2023), 10.1038/s41594-023-01110-8. [DOI] [PubMed] [Google Scholar]
- 54.van den Bogaart G., et al. , Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honigmann A., et al. , Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 20, 679–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.James D. J., Khodthong C., Kowalchyk J. A., Martin T. F., Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J. Cell Biol. 182, 355–366 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picas L., et al. , BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 5, 5647 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Elouarrat D., et al. , Role of phosphatidylinositol 5-phosphate 4-kinase alpha in zebrafish development. Int. J. Biochem. Cell Biol. 45, 1293–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Emerling B. M., et al. , Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 155, 844–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shim H., et al. , Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc. Natl. Acad. Sci. U.S.A. 113, 7596–7601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawlor M. W., et al. , Skeletal muscle pathology in X-linked myotubular myopathy: Review with cross-species comparisons. J. Neuropathol. Exp. Neurol. 75, 102–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Time-lapse microscopy of Tks5-GFP in differentiated C2C12 myoblasts. Day 3 differentiated C2C12 myoblasts transduced with lentivirus encoding Tks5-GFP were imaged with videomicroscopy using a 10-min time-interval. Arrows indicate contact-site that look-like podosome-like protrusions enriched in Tks5-GFP.
Time-lapse confocal microscopy of Tks5-GFP during C2C12 fusion. Day 3 differentiated C2C12 myoblasts transfected with a plasmid encoding Tks5-GFP were imaged with an LSM900 Zeiss microscope (Zen software, x20 objective) equipped with a ibidi stage Top incubator, using a 15-min time-interval and a single Z-plane. An arrow indicates the presence of a podosome-like protrusion enriched in Tks5-GFP before fusion.
Data Availability Statement
All study data are included in the article and/or supporting information.


