Abstract
Cellular agriculture is an innovative technology for manufacturing sustainable agricultural products as an alternative to traditional agriculture. While most cellular agriculture is predominantly centered on the production of cultured meat, there is a growing demand for an understanding of the production techniques involved in dairy products within cellular agriculture. This review focuses on the current status of cellular agriculture in the dairy sector and technical challenges for cell-cultured milk production. Cellular agriculture technology in the dairy sector has been classified into fermentation-based and animal cell culture-based cellular agriculture. Currently, various companies synthesize milk components through precision fermentation technology. Nevertheless, several startup companies are pursuing animal cell-based technology, driven by public concerns regarding genetically modified organisms in precision fermentation technology. Hence, this review offers an up-to-date exploration of animal cell-based cellular agriculture to produce milk components, specifically emphasizing the structural, functional, and productive aspects of mammary epithelial cells, providing new information for industry and academia.
Keywords: Cell culture system, Cell-cultured milk, Mammary epithelial cells, Precision fermentation
Introduction
The dairy farming system has been directed toward enhancing efficiency of milk production through concentrated animal feeding operations, larger herds, advanced breeding technologies [1]. Over the past 80 years, milk yield in dairy farming has witnessed a 16.7-fold increase, from 53 million metric tons (Mt) in 1944 to 887 Mt in 2021 [2–4]. Furthermore, global milk production is forecasted to increase to 1,060 Mt by 2031 [3]. Genetic improvement has been a significant contributor to the increase in milk productivity. Specifically, three factors including transitioning from breeds such as Jersey and Guernsey to Holstein, widespread adoption of artificial insemination, and advancements in genetic evaluation procedures have played pivotal roles. These factors have collectively driven notable genetic changes in milk productivity [2].
Dairy intensification has been associated with adverse effects on the environment [4], animal welfare [5], human health [6], and rural livelihoods [1]. From an environmental standpoint, the intensification of dairy farming, encompassing enteric and manure storage, concentrated feed production, and farm crop cultivation, leads to greenhouse gas emissions, soil acidification, and eutrophication [7]. In addition, animals in dairy farms are raised in highly artificial environments to maximize milk yield, prompting concerns about animal welfare [8]. Nitrate contamination of soil, aquifers, and rivers through the accumulation of cattle urine in dairy farming is another major concern for human health, as exposure to such contaminated water is associated with colorectal cancer [9]. Thus, dairy farming and industries are focusing on producing milk and milk products in a sustainable rather than traditional manner [10].
Cellular agriculture is receiving attention as a new sustainable technology for agricultural food production that can incrementally positively affect the environment and society [11]. Cellular agriculture holds considerable promise over traditional agriculture, offering potential benefits in terms of environmental sustainability, economic value, enhanced animal welfare, and improved human health and well-being [12]. Several companies across the globe are focusing on the production of cellular agricultural products, such as cultured meat and cell-cultured milk, based on cellular agriculture technology [13]. Nevertheless, this technology has been predominantly applied for production of cultured meat [11, 14, 15]. Therefore, comprehensive studies are required to understand the production of sustainable cell-cultured milk. This review aimed to comprehensively identify the current status of cellular agriculture in the dairy sector and to understand the fundamental knowledge and challenges associated with cell culture-based dairy technology.
Cellular agriculture in the dairy sector
Cellular agriculture is a sustainable manufacturing technology that produces products such as meat components, milk components, and egg proteins using a cell culture system [16]. Various companies are currently trying to produce cell-cultured milk components using fermentation-based and animal cell culture-based technology (Fig. 1) [17]. However, prohibitive cost involved in the research, development, and production remain major obstacles [18]. Therefore, this section covers the techno-economic cost and technical state in current cell culture-based dairy field.
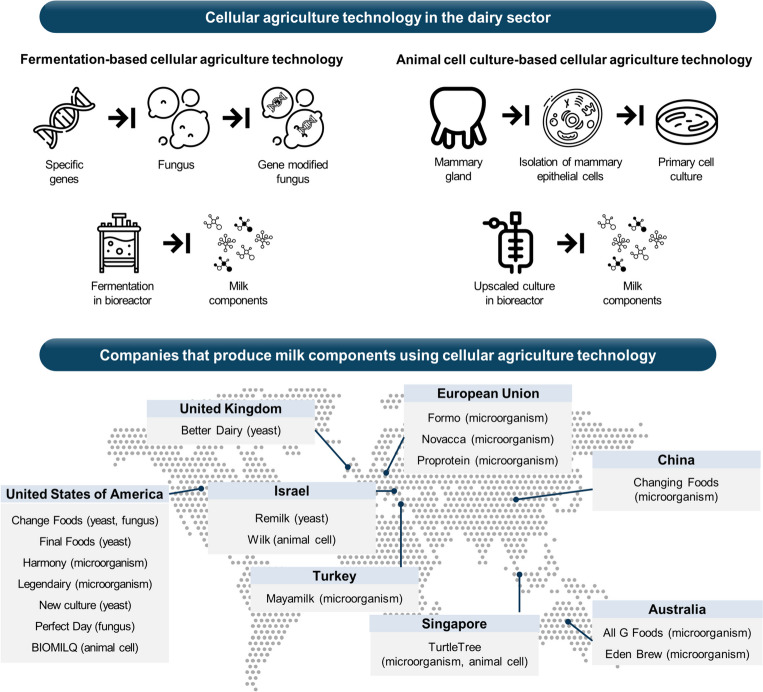
Fig. 1.
Cellular agriculture technology in the dairy sector and global startup companies to produce milk components. Various global companies are trying to synthesize milk components through fermentation- and animal cell culture-based technology
Techno-economic analysis
To assess the economic viability of cell-cultured milk components, a techno-economic analysis was conducted to estimate the cost-effectiveness of cell cultured milk components compared to whole milk. In the United States, a liter of whole milk typically costs around $1.00 USD [19]. One liter of milk contains about 26 g of casein (13 g of α-casein, 9.3 g of β-casein, and 3.3 g of κ-casein) and 40 g of fats [20]. Our previous research showed that MAC-T cells grown in a progesterone (P4)-based differentiation media were able to produce some key milk components [21]. Specifically, these cells synthesized 0.515 g of α-casein and 12.19 g of triglycerides per liter of media. However, it is important to consider that this media itself costs about $175 USD per liter [21]. Based on this cell-cultured milk technology, producing the same amounts of α-casein and triglycerides found in a liter of whole milk would be very expensive. The production for α-casein and triglycerides would likely cost around $4,417 USD and $574 USD, respectively. It is important to remember that this only considers the cell culture media. The actual production cost would be much higher because it excludes costs for labor, production facilities, separation and purifying the desired milk components, and other miscellaneous expenses. As a result, these projections indicate that the significant production expenses pose a significant obstacle in contemporary cellular agriculture. Therefore, it is important to develop cost-efficient methodologies to facilitate large-scale industrial production [18]. The primary focus of technical advancement involves enhancing cell lines, developing low-cost yet high-performance medium, optimizing bioreactors for efficiency, and refining down-stream processing methods for cost-effectiveness. These efforts are essential for realizing economically viable production of cell-cultured milk components.
Fermentation-based cellular agriculture
Fermentation-based cellular agriculture employs synthetic biology and genetic engineering to introduce specific genes into the DNA backbone of bacteria, yeast, or algae, to produce desired products [22]. Based on precision fermentation technology, various companies have developed and commercialized milk components, such as casein, whey protein, and lactoferrin (Table 1). However, most companies have chosen not to disclose details regarding the microorganisms or the techniques they utilized.
Table 1.
Companies that produce milk components using fermentation-based technology in cellular agriculture
| Company | Products | Microorganisms | Location | References |
|---|---|---|---|---|
| AII G Foods | Milk proteins | Undisclosed | Sydney, Australia | [22] |
| Better Dairy | Casein | Yeast | London, United Kingdom | [22, 23] |
| Change Foods | Casein | Bacteria, yeast, filamentous fungi | California, United States of America | [22, 23] |
| Changing Biotech | Undisclosed protein | Undisclosed | Shanghai, China | [23] |
| Eden Brew | Milk proteins | Undisclosed | Sydney, Australia | [22] |
| Final Foods | Whey proteins | Yeast | California, United States of America | [22] |
| Formo | Casein and whey protein | Undisclosed | Berline, Germany | [23] |
| Harmony | Human milk proteins | Undisclosed | Massachusetts, United States of America | [22] |
| Legendairy | Milk proteins | Undisclosed | Texas, United States of America | [22] |
| Mayamilk | Milk proteins | Undisclosed | Izmir, Turkey | [22] |
| New Culture | Casein | Yeast | California, United States of America | [23, 24] |
| Novacca | Milk proteins | Undisclosed | Denmark | [22] |
| Perfect Day | β-Lactoglobulin | Fungus | California, United States of America | [22–24] |
| Proprotein | Casein | Undisclosed | Tallinn, Estonia | [22] |
| Remilk | Casein and β-lactoglobulin | Yeast | Rehovot, Israel | [23, 24] |
| TurtleTree | Lactoferrin | Undisclosed | Singapore | [24] |
Fungi are known as the most suitable microbial hosts for precision fermentation because of their strong environmental adaptability [25]. From the metabolic engineering perspective, compared to bacteria, fungi possess better eukaryotic properties that allow them to express heterologous eukaryotic proteins, correcting protein folding and post-translation modifications [26, 27]. The filamentous fungus Trichoderma reesei is commonly utilized to synthesize recombinant food components because of its high protein productivity (up to 100 g/L); moreover, it is generally regarded as safe [22, 25]. Indeed, a prior study documented that T. reesei, employing precision fermentation technology, can produce β-lactoglobulin (BLG) at a level of 1 g/L, with the structural and functional properties of the recombinant BLG being consistent with bovine BLG [28].
Precision fermentation has enabled the production of sustainable milk components, an emerging food trend in the fourth industrial revolution of the food industry [29]. However, the commercialization of genetically modified organisms (GMOs) used in precision fermentation has raised public concerns about food safety [30]. Accordingly, the production of food components using GMOs requires careful regulation, thorough safety evaluations, and consideration of consumer concerns [22]. Therefore, although precision fermentation as an innovative technology is anticipated to reduce the reliance on traditional dairy farming, overcoming GMO concerns remains a major challenge for fermentation-based cellular agriculture.
Animal cell culture-based cellular agriculture
Animal cell and tissue culture-based cellular agriculture involves tissue engineering to produce functional tissues using minimal cells or cell lines obtained from living animals [16]. Recent biotech startups have emerged, securing funds to pioneer the development of cell-cultured milk production (Table 2) [31]. As current animal cell culture-based cellular agriculture has technical difficulties in synthesizing whole milk, they mainly aim to produce a single component of milk using mammary epithelial cells (ECs) [32].
Table 2.
Companies that try to produce milk components using animal cell culture-based technology in cellular agriculture
| Company | Products | Animal cells | Location | References |
|---|---|---|---|---|
| BIOMILQ | Bovine and human milk components | Mammary epithelial cells | North Carolina, United States of America | [33] |
| Turtle Tree | Goat and human milk component | Mammary epithelial cells | Singapore | [33] |
| Wilk | Bovine and human milk components | Mammary epithelial cells | Rehovot, Israel | [23, 32] |
Milk components, such as casein, whey protein, and triglycerides, are primarily synthesized and secreted by ECs of the mammary gland [34, 35]. Thus, the primary step in the in vitro production of cell-cultured milk components is to obtain ECs. Companies such as BIOMILQ and Wilk isolate ECs from the milk-secreting parenchymal tissue of the mammary gland. In contrast, Turtle Tree isolates mesenchymal stem cells from mammary tissues, adipose tissues, and the umbilical cord, subsequently inducing differentiation into ECs [23, 32]. Despite the focus of the mentioned startup companies on the production of cell culture-based milk, technology related to animal cell culture for producing milk components is still in its early stages. Consequently, novel strategies are essential to surmount the technical barriers of animal cell culture-based cellular agriculture, necessitating a deeper understanding of milk biosynthesis in the mammary gland.
Current knowledge and technical challenges for producing cell-cultured milk
The mammary gland itself is a complicated bioreactor comprised of alveolar structure including various cell types. Technical challenges for producing cell-cultured milk include replicating the structure of milk-secreting mammary glands and reconstructing it within an in vitro environment. Cell-cultured milk can be produced through the intricate processes such as the structural interaction of cells and the regulation of milk synthesis-related hormones while cell-cultured meat is generally produced by culturing muscle cells or adipocytes [36]. Therefore, a detailed process of the milk synthesis and secretion in the mammary gland to produce cell-cultured milk would be described in this section. The short-term objective is to produce individual milk components using a two-dimensional (2D) culture of mammary cells, while the ultimate long-term goal is to achieve the production of whole milk through the three-dimensional (3D) culture of mammary glands [15]. To accomplish these objectives, the primary technical challenges will involve ensuring the sustainable resourcing of mammary cells, optimizing cell culture media, establishing a robust cell culture system, and down-stream processing of cell-cultured milk components (Fig. 2) [32].
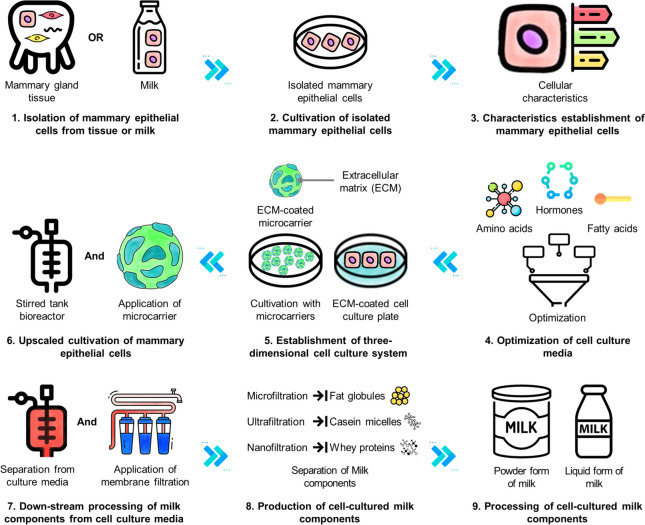
Fig. 2.
Principal processes and technical challenges for producing cell-cultured milk using animal cell culture-based technology. The production of cell-cultured milk, found on the cultivation of mammary epithelial cells (ECs), is through a series of sequential processes as follows: (1) isolation of mammary ECs from parenchymal tissues or milk, (2) cultivation of isolated mammary ECs for the establishment of a cell line, (3) evaluation of cellular characteristics, (4) optimization of cell culture media, (5) establishment of three-dimensional cell culture system using extracellular matrix, (6) upscale of cell cultivation using stirred tank bioreactor and microcarrier, (7) down-stream processing of cell culture media, (8) production of cell-cultured milk components, and (9) processing of cell-cultured milk components. The major technical challenges for the production of cell-cultured milk are resourcing the cell line (1–3), optimizing the cell culture media (4), establishing the cell culture system (5–6), and separating milk components (7–8). Comprehensive and detailed technical challenges for cell-cultured milk production are discussed in this review
Understanding functional and structural features of the mammary gland
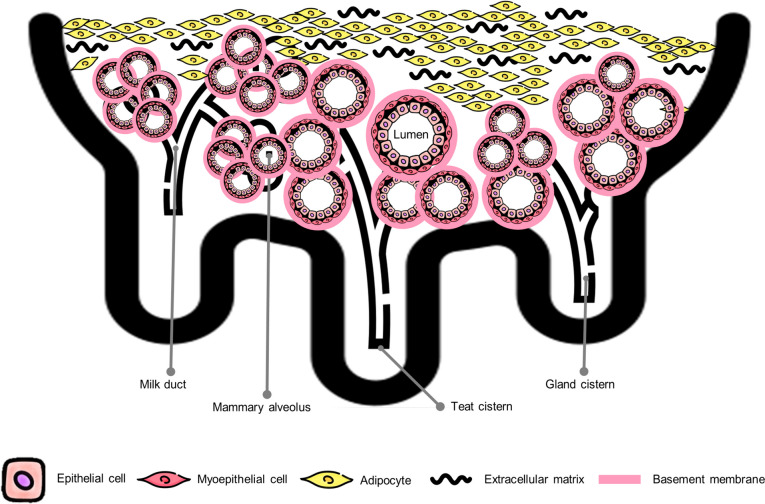
The mammary gland (breast) is a distinctive organ exclusive to mammals, characterized by an anatomical structure designed for the secretion of milk to nourish a newborn (Fig. 3) [37]. Herein, mammary alveolus is a fundamental constituent of mature mammary glands for milk production. Alveolar parenchyma comprises inner milk secretory ECs that surround the lumen, outer myoepithelial cells (MCs) that attach to the basal mammary epithelium, and the basement membrane (BM) that contacts the MCs. The stromal compartment comprises various stromal cells, including fibroblasts (FBs), adipocytes, endothelial cells, and extracellular matrix (ECM) [35, 38]. Thus, milk components are structurally synthesized by ECs, contracted by MCs, and secreted into the lumen [39].
Fig. 3.
Internal structure of mammary gland and mammary alveolus. The mammary gland is composed of lobes that comprise lobules containing 150–220 alveoli. Mammary alveoli are fundamental constituents that produce milk components. The mammary alveolus consists of parenchymal and stromal compartments based on the basement membrane. The parenchyma is constructed of inner milk secretory epithelial cells that surround a central lumen and outer myoepithelial cells that attach to the base of the mammary epithelium. In addition, the stroma is constituted of adipocytes and extracellular matrix
The functional capabilities of the mammary gland for milk synthesis and secretion occur primarily during physical development [40]. The mammary gland develops throughout the four growth stages (i.e., pre-puberty, post-puberty, pregnancy, and lactation), experiencing repeated apoptosis and growth in response to pregnancy cycles, parturition, lactation, and involution [40, 41]. In particular, the mature functional development of the mammary gland, which directly enables the synthesis and secretion of milk, occurs primarily during pregnancy and lactation [42], and is primarily regulated by the reproductive and metabolic hormones. Among the various hormones, prolactin (PRL) and P4 directly induce the alveologenesis and secretory differentiation through receptor activator of nuclear factor kappa-B ligand during pregnancy. In addition, 17β-estradiol (E2), cortisol (CORT), insulin (INS), and growth hormones support this development of mammary gland. After that, a decrease of P4 concentration in the presence of PRL, CORT, and INS triggers the secretory activation and the onset of milk production for the transition to lactation [43]. These features of the mammary gland are essential for the synthesis of milk components and are tightly regulated through the coordinated action of hormones within mammary cells.
Considering the structural and functional properties of the mammary gland as summarized above, the activation of mammary ECs or secretory differentiation through hormonal regulation are essential for producing cell-cultured milk components. To initiate this process in the laboratory, the first step involves isolating milk-secreting ECs and establishing their cellular characteristics.
Resourcing of mammary epithelial cells
ECs can be isolated from the mammary gland tissues or milk of animals and humans, depending on the type of milk desired for production (Table 3). Isolating ECs from tissue is typically used in current research field due to its technical ease to apply. However, as the ECs can be obtained from the parenchymal tissue of the mammary gland after biopsy or slaughter, the procedures are uneconomical, time-consuming, and inconvenient. Furthermore, as mammary gland tissue comprises various cell populations such as ECs, MCs, FBs, adipocytes, and ECM [35], disassociating mammary gland tissue is essential to isolate ECs. Diverse enzymes such as collagenase, hyaluronidase, and trypsin have been used to isolate and purify ECs. In particular, trypsin has been widely used to remove FBs from the mammary gland tissue. However, according to a recent study, the combination of collagenase type 1 and hyaluronidase more effectively isolated ECs with better preservation of the physiological properties than trypsin [44]. Therefore, optimization of dissociation by applying multiple enzyme combinations would improve the physiological properties of isolated ECs, contributing to cell line resourcing.
Table 3.
Dissociation and sorting methods for the isolated mammary epithelial cells from the mammary gland and milk
| Species | Sources | Dissociation enzymes | Sorting methods | Markers | References |
|---|---|---|---|---|---|
| Buffalo | Mammary gland | Collagenase, hyaluronidase, and trypsin/EDTA | Selective trypsinization | CK18, vimentin | [45] |
| Caprine | Mammary gland | Collagenase type 1 | Collagen digestion | CK18, CK19, vimentin, α-SMA | [46] |
| Dairy cow | Mammary gland | Trypsin/EDTA | Selective trypsinization | CK18, vimentin | [47] |
| Dairy cow | Mammary gland | Trypsin/EDTA | Selective trypsinization | CK18 | [48] |
| Dairy cow | Mammary gland | Trypsin/EDTA | Selective trypsinization | CK18 | [49] |
| Dairy cow | Mammary gland | Trypsin/EDTA | Selective trypsinization | Pan-CK | [50] |
| Dairy cow | Milk | Not applicable | Centrifuge 1,850 × g, 10 min | Pan-CK | [51] |
| Dairy cow | Milk | Not applicable | Centrifuge 1,850 × g, 10 min | CK8, Pan-CK | [52, 53] |
| Goat | Mammary gland | Collagenase type 1, Trypsin/EDTA | Selective trypsinization | CK18 | [54] |
| Human | Mammary gland | Collagenase, hyaluronidase, Accumax | Collagen digestion | CK8, CK14, CK18 | [55] |
| Porcine | Mammary gland | Collagenase A, hyaluronidase | Collagen digestion | CK18, vimentin | [56] |
| Yak | Mammary gland | Trypsin/EDTA | Selective trypsinization | CK8, CK18, vimentin | [57] |
CK Cytokeratin, EDTA Ethylene-diamine-tetraacetic acid, α-SMA α-Smooth muscle actin
Recently, isolating ECs from milk is receiving attention as a novel strategy because it has several advantages, including non-invasiveness, repeatability, and less contamination by FBs. Notably, the cytoskeletal characteristics and milk productivity of primary bovine ECs extracted from milk were comparable to those of cells isolated from tissue [58]. In addition, human breast milk was a rich source of heterogeneous cell types such as milk-secreting ECs, MCs, progenitor cells, and multipotent mesenchymal stem cells [59, 60]. Therefore, isolating ECs from milk can be another effective alternative method for sustainable resourcing of ECs [52]. However, further study is required to demonstrate the suitability as an alternative to the tissue culture regarding gene expression and cellular functionality.
The cytoskeleton plays a crucial role in cellular integrity, structure, and function and expresses specific cytoskeletal proteins depending on the cell type [32]. Milk-producing ECs specifically express cytokeratin (CK) 8 [61]. ECs, MCs, and FBs selectively express CK18/19, α-smooth muscle actin (α-SMA), and vimentin as specific markers, respectively [62]. Accordingly, various cytoskeletal protein markers, including CK8, CK18, CK19, vimentin, and α-SMA, can be utilized to distinguish mammary cell line types. Taken together, isolating ECs from milk and evaluating reliable biomarker would contribute to the stable resourcing of the mammary cells.
Optimization of cell culture media for mammary epithelial cells
Optimal cultivation conditions for ECs can be established by imitating the in vivo circulatory system and physiological environment of the mammary alveolus. As all of the precursors for milk production are supplied from the blood [63], it plays an important role in providing hormones and nutrients for the growth, development, and lactation of the mammary gland [64]. Therefore, the most fundamental factor for the production of cell-cultured milk is to optimize the growth and differentiation media of ECs based on the levels of constituents in the blood during the development and lactation of the mammary gland [65].
Cell culture media are composed of a basal medium (comprising amino acids, vitamins, inorganic salts, glucose, among others), serum or serum alternatives (source of growth factors, hormones, and attachment factors), and several supplements [66]. Generally, Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham (DMEM/F12) with the addition of 10% fetal bovine serum (FBS) is used for cultivating ECs. In addition, antibiotics such as penicillin, streptomycin, gentamicin, and amphotericin B are added for aseptic cell culture. Therefore, the most fundamental growth media for the cultivation of ECs consist of DMEM/F12, 10% FBS, 1% penicillin/streptomycin, and 5 µg/mL amphotericin B (Table 4).
Table 4.
Culture conditions for the isolated mammary epithelial cells from mammary gland and milk
| Species | Basal media | Serum | Antibiotics | Hormones | Other additives | References |
|---|---|---|---|---|---|---|
| Buffalo | DMEM/F12 | FBS (10%) | Penicillin (100 U/mL), streptomycin (5 µg/mL), amphotericin (50 ng/mL) | INS (5 µg/mL), CORT (1 µg/mL), EGF (10 ng/mL), PRL (5 µg/mL) | Transferrin (1 µg/mL) | [45] |
| Caprine | DMEM/F12 | FBS (10%) | Penicillin (100 U/mL), streptomycin (100 µg/mL) | INS (10 µg/mL), CORT (5 µg/mL) | Sodium bicarbonate (2.2 mg/mL), sodium acetate (5 mmol/L), holo-transferrin (5 µg/mL), ethanolamine (0.5 mmol/L), | [46] |
| Dairy cow | DMEM/F12 | FBS (10%) | INS (5 µg/mL), P4 (5 µg/mL), CORT (1 µmol/L), EGF (10 ng/mL), E2 (5 µg/mL) | Transferrin (5 µg/mL) | [47] | |
| Dairy cow | DMEM/F12 | FCS (10%) | Penicillin (1%), streptomycin (1%) | INS (5 µg/mL), CORT (1 µg/mL), EGF (10 ng/mL), PRL (5 µg/mL) | Transferrin (5 µg/mL), glutamine (1%) | [48] |
| Dairy cow | DMEM/F12 | FBS (10%) | Penicillin/streptomycin (1%) | EGF (1%) | [49] | |
| Dairy cow | DMEM/F12 | FBS (20%) | Penicillin/streptomycin (50 IU/mL), amphotericin B (2.5 µg/mL) | INS (1 µg/mL) | [50] | |
| Dairy cow | DMEM/F12, INS (5 mg/mL) | FCS (10%) | Penicillin/streptomycin (100 µg/mL), gentamycin (100 µg/mL), amphotericin B (5 µg/mL) | Transferrin (5 mg/mL), sodium selenite (5 µg/mL) | [51] | |
| Dairy cow | DMEM/F12 | FBS (10%) | Amphotericin B (1.76 µg/mL) | INS (10 µg/mL), CORT (1 µg/mL), | Transferrin (5 µg/mL), sodium selenite (5 ng/mL) | [52, 53] |
| Goat | DMEM/F12 | FBS (10%) | Penicillin/streptomycin (100 IU/mL) | ITS (5 ng/mL), IGF-1 (10 ng/mL), EGF (10 ng/mL) | [54] | |
| Human | DMEM/F12 | FBS (5%) | INS (5 µg/mL), CORT (0.4 µg/mL), EGF (10 ng/mL) | Cholera toxin (8.4 ng/mL), adenine (24 µg/mL), Y-27632 (10 µmol/L) | [55] | |
| Porcine | DMEM/F12 | FBS (10%) | INS (0.5 µg/mL), CORT (1 µg/mL), PRL (0–2 µg/mL) | [56] | ||
| Yak | DMEM/F12 | FBS (10%) | Penicillin (100 IU/mL), streptomycin (5 µg/mL) | ITS (5 µg/ML), EGF (5 ng/mL), CORT (1 µg/mL), P4 (5 µg/mL) | [57] |
CORT Cortisol, DMEM/F12 Dulbecco’s Modified Eagle’s Medium/F-12 Nutrient Mixture Ham, E2 17β-estradiol, EGF Epidermal growth factor, FBS Fetal bovine serum, FCS Fetal calf serum, IGF-1 Insulin growth factor-1, INS Insulin, ITS Insulin-transferrin-selenium, P4 Progesterone, PRL Prolactin
Amino acids, fatty acids, glucose, vitamins, and minerals are key nutrients for the structural development of and milk component biosynthesis by ECs [67]. Reproductive and metabolic hormones such as PRL, P4, E2, CORT, and INS are also essential for the proliferation and differentiation of ECs [43]. Therefore, amino acids and hormones can promote the proliferation and differentiation of ECs [43, 67, 68]. The increase in the number of ECs through proliferation and differentiation enhanced the milk productivity in the development and lactation of mammary gland [69]. In addition, milk fat composed of triglycerides (98%), diglycerides (about 2%), cholesterol (less than 0.5%), phospholipids (about 1%), and free fatty acids (about 0.1%), are mainly biosynthesized by ECs from more than 400 different fatty acids [70]. The most abundant fatty acids in milk consist of long-chain fatty acids in the order of palmitic acid (C 16:0), oleic acid (18:1), stearic acid (18:0), and myristic acid (14:0) [71]. These long-chain (C18 and some C16) fatty acids are derived from the blood plasma lipid originating from the diet, while medium- and short-chain fatty acids are synthesized through de novo synthesis in ECs [72, 73]. Therefore, various additives, including hormones, amino acids, and fatty acids, can be supplemented to the cell culture media to promote the proliferation and differentiation of ECs, thereby enabling the production of cell-cultured milk components [74]. Finally, the optimal proliferation and differentiation media needs to be established based on the concentration of hormones, amino acids, and fatty acids in blood plasma during pregnancy and lactation (Table 5).
Table 5.
Concentration of amino acids, hormones, and fatty acids in the bovine blood plasma
| Categories | Bioactive compounds | Concentration | References |
|---|---|---|---|
| Essential amino acids (µg/g of amino acids) | Arginine | 13.24−33 | [75, 76] |
| Isoleucine | 10.75−33.5 | ||
| Histidine | 4.65−42 | ||
| Leucine | 15.20−93.4 | ||
| Lysine | 9.21−74.7 | ||
| Methionine | 3.28−8.6 | ||
| Phenylalanine | 14.70−51.6 | ||
| Threonine | 7.03−66 | ||
| Tryptophan | 11.8−19.40 | ||
| Valine | 22.26−67.3 | ||
| Hormones (ng/mL in blood plasma) | INS | 0.35 (puberty), 0.416−0.625 (pregnancy−lactation), 0.25−0.5 (5 d before the onset of lactation) | [77, 78] |
| CORT | 9 (pregnancy), 3−5 (lactation), 5 (5 d before the onset of lactation) | [79–81] | |
| P4 | 0.5−3.5 (lactation), 4.5−6.5 (5 d before the onset of lactation) | [81, 82] | |
| E1 | 3.5 (pregnancy), 0.05 (lactation) | [83] | |
| E2 | 0.55 (pregnancy), 0.025 (lactation), 0.5−0.8 (5 d before the onset of lactation) | [83] | |
| PRL | 50 (5 d before the onset of lactation) | [78, 79, 84] | |
| Fatty acids (µg/g of fatty acids) | Myristic acid (14:0) | 7.7−10.2 | [85–87] |
| Palmitic acid (16:0) | 120−209 | [85–87] | |
| Palmitoleic acid (16:1) | 25.4−58 | [85, 86] | |
| Stearic acid (18:0) | 154.6−188 | [85–87] | |
| Oleic acid (18:1) | 86.5−149.6 | [85–87] | |
| Linoleic acid (18: 2n-6) | 280.2−376 | [85–87] | |
| Docosahexaenoic acid (n-3) | 10.3−34.2 | [85, 86] | |
| Arachidonic acid (n-6) | 414−421 | [85, 86] | |
| SFA | 402−405 | [85, 86] | |
| MUFA | 141−174 | [85, 86] | |
| PUFA | 425−456 | [85, 86] |
CORT Cortisol, E1 Estrone, E2 17β-estradiol, INS Insulin, MUFA Monounsaturated fatty acid, P4 Progesterone, PRL Prolactin, PUFA Polyunsaturated fatty acid, SFA Saturated fatty acid
Establishment of a cell culture system for mammary epithelial cells
ECs have typically been cultured using the 2D cell culture method to study the function of the mammary gland [49, 50]. Two-dimensional cultivation has the experimental advantage of promoting homogenous growth and proliferation of cells by supplying a consistent amount of nutrients and growth factors from cell culture media [88]. However, as 2D cell culture cannot completely mimic the structural shape of the tissues observed in the mammary gland, the bioactivities of the cells appear considerably different compared to those of tissues [89]. Furthermore, it has been reported that 2D-based cell culture is manual- and labor-intensive, demanding a significant amount of space and incurring a high manufacturing cost [90]. Consequently, 2D cell culture raises several strategic problems from the perspective of structural and productive cultivation of ECs for producing cell-cultured milk.
A 3D cell culture system is a potential approach for more effective production of cell-cultured milk. The first goal for 3D cell culture is to precisely mimic the structural and functional formation of mammary gland tissue. From the structural and functional perspectives of mammary alveolus, ECs are in contact with the thin and dense layers of a specialized ECM, termed the BM [89]. The BM, composed of a polymeric network of proteins, including laminins, collagen IV, heparin sulfate proteoglycan, and nidogen, has been reported to interact with ECs in the proliferation, differentiation, and metabolism processes [91–93]. Indeed, the culture of ECs with 3D collagen gels resulted in the maintenance of differentiation and synthesis of milk proteins, suggesting that ECM plays a key regulatory role in 3D cell culture [94, 95]. Therefore, the application of BM proteins is required for establishing a 3D cell culture of ECs.
Spheroids and scaffolds represent various 3D cell culture strategies. Additionally, microcarrier technology has emerged as suitable tool to apply BM proteins [96, 97]. Microcarriers are support matrices 100−300 μm diameter that enable the cultivation of anchorage-dependent adherent cells in a bioreactor system [98]. Several microcarriers are commercially synthesized using various materials, including glass, dextran, polystyrene, cellulose, collagen, gelatin, collagen, alginate, and chitosan [99]. These microcarriers can be coated with ECM proteins, such as laminin, collagen, and fibronectin, for the efficient adhesion of the cells. ECM proteins provide many RGD tripeptide (arginine-glycine-aspartate) motifs that can specifically bind to cell surface receptors [100]. Therefore, ECM proteins can enhance the cell attachment of microcarriers along with a high surface-to-volume ratio [101]. Indeed, a culture of 3 g/L Cytodex 1 (190 μm) and 3 (175 μm) microcarrier provides a surface area of 8.1–13.2 × 103 cm2 in 1L, which is equivalent to 108–176 of 75 cm2 cell culture flasks [101]. Various types of microcarriers have been mainly applied to cultivate human mesenchymal and pluripotent stem cells for cell therapy in clinical trials [101–103]. However, most studies related to ECs have primarily utilized microcarriers to establish 3D in vitro breast tumor models [104, 105]. Only one study reported the optimal cell adhesion, growth, and differentiation conditions on collagen-coated microcarriers (Cytodex 3) using bovine mammary epithelial cell line MAC-T [106]. Therefore, the optimization and application of ECs to ECM-coated microcarriers are required to overcome the structural and productive limitations of synthesizing cell-cultured milk.
To facilitate the upscale production of ECs, it is imperative to introduce a culture system that is more space-, labor-, and cost-efficient. A bioreactor stands out as a promising culture system for the large-scale cultivation of ECs [107]. Bioreactors have been extensively used for the industrial large-scale cultivation of mammalian cells under a controlled microenvironment [108]. Therefore, several bioreactor systems (stirred tank, wave, rotating wall, hollow fiber, and packed-bed), primarily developed for cultivating conventional mammalian cells, can potentially apply to the cultivation and scale-up of ECs [109]. Among the various bioreactor systems, a stirred tank bioreactor has many advantages for the commercial scale production, including ease of design, scale-up, in situ monitoring, and operation in different batch. While this bioreactor does have limitations in meeting the oxygen demand of large volumes (such as high-density cell cultures) and controlling excessive shear stress caused by the impeller, these challenges can be addressed through process optimization strategies. [110]. Considering the structural and functional characteristics of ECs and the application of microcarriers, a stirred tank bioreactor would be one of the most applicable systems for cell-cultured milk production. Stirred tank bioreactor is one of the most conventional bioreactors, which consist of a tank equipped with an impeller for efficient mixing and suspension [110]. The impeller, a core component of a stirred tank bioreactor, controls the culture environment, including pH, dissolved oxygen, temperature, nutrients, and metabolites through agitation [109, 111]. These bioreactors are simple and easy to monitor and control for large-scale cell cultivation [112]. Indeed, mammary epithelial stem cells inoculated at a 7.5 × 104 cells/mL in 125, 500, and 1,000 mL of stirred bioreactors resulted in the expansion of cell density of 3.38, 3.76, and 4.21 × 105 cells/mL, respectively, with the formation of aggregates (mammospheres) [113]. Moreover, stirred culture systems have the advantage of easy application of microcarrier technology that enhances the productivity of cells and their derivatives through an increase in the high surface-to-volume ratio [102]. Comprehensively, applying an ECM-coated microcarrier in a stirred tank bioreactor would be the most suitable cell culture system for the upscaled production of cell-cultured milk.
Down-stream processing of milk components from cell culture media
ECs cultured in a cell culture system secrete milk proteins and fat globules into the cell culture media. Concurrently, various cell culture additives including serum, hormones, and antibiotics used for the cellular proliferation and differentiation are contained in the cell culture media. However, only milk components should be separated and purified from the cell culture media. In these perspectives, membrane-based techniques can be effectively employed for isolating, purifying, and processing the milk components from the cell culture media.
Pressure-driven membrane separation process technology has widely been applied to produce high-value added dairy components in the dairy industry and to remove the hormones and antibiotics in the wastewater treatment industry [114, 115]. Membrane separation processes are classified as reverse osmosis (< 1 nm), nanofiltration (1–10 nm), ultrafiltration (10–100 nm), and microfiltration (100–10,000 nm) depending on the membrane pore size and molecular weight cutoff [114, 115]. One liter of milk contains 26 g casein micelles, 7 g whey proteins, and 40 g fat globules with sizes of 20–300 nm (average 110 nm), 3–6 nm, and 100–15,000 nm (average 3,400 nm), respectively [20, 116, 117]. Accordingly, fat globules, casein micelles, and whey proteins in the cell culture media can be separated using microfiltration, ultrafiltration, and nanofiltration [117]. In addition, since hormones and antibiotics have molecular weight of average 0.25–0.5 kDa, nanofiltration and reverse osmosis can be used for the removal [118, 119]. In detail, microfiltration, with a 1,400 nm pore size, is a standard method for separating fat globules and bacteria [120]. Microfiltration, with a 100–200 nm pore size, is employed to separate casein micelles from whey protein. Ultrafiltration and nanofiltration, featuring pore sizes of 1–100 nm and 2 nm, respectively, are used to concentrate whey protein [121]. Additionally, nanofiltration and reverse osmosis, which have a molecular weight cut off of 0.3–1 kDa and 0.1 kDa, respectively, are applied for removing the various types of hormones and antibiotics (Table 6) [114, 115].
Table 6.
Membrane types based on milk components in membrane separation process technology
| Membrane type | Pore size, nm | Molecular weight cut off (pressure) | Separation component (Size distribution, nm) |
Reference |
|---|---|---|---|---|
| Microfiltration |
100–10,000 100–200 |
> 200 kDa (low, below 2 bar, 0.2 Mpa) |
Fat globules (100–15,000) Casein micelles (20–300) |
[117–119, 122–124] |
| Ultrafiltration | 1–100 | 1–200 kDa (medium, 1–10 bar, 0.1–1 Mpa) |
Casein micelles (20–300) Whey proteins (3–6) |
|
| Nanofiltration | 1–10 | 0.3–1 kDa (medium to high, 5–40 bar, 0.5–4 MPa) |
Whey proteins (3–6) Hormones and antibiotics (0.25–0.5 kDa) |
|
| Reverse osmosis | < 1 | 0.1 kDa (high, 10–100 bar, 1–10 MPa) |
Lactose and water (0.35 kDa) Hormones and antibiotics (0.25–0.5 kDa) |
Two types of tubular ceramic membranes (TCMs) and spiral-wound membranes (SWMs) are typically applied in the separation of milk components using microfiltration, ultrafiltration, and nanofiltration [117]. TCM is widely used for membranes because of its narrow pore distribution, high hydraulic performance, and high thermal stability [125, 126]. However, compared with SWM, TCM has high transmembrane pressure (∆pTM), which increases the flux value (L/m2·h) and membrane fouling, resulting in high energy consumption and low separation efficiency for milk protein fractionation [127]. Thus, recent studies have focused on optimizing the efficiency of milk protein separation using SWM to improve membrane fouling [128, 129]. A 0.3-µm pore size SWM can achieve a whey protein separation ratio of up to 97% from skim milk, surpassing the 95% ratio achieved with the 0.1-µm pore size TCM [128]. Furthermore, although pore size did not affect the flux value in 0.1- and 0.2-µm pore size SWM, the 0.1-µm pore size SWM was more suitable for milk protein separation because of a high loss of protein in the 0.2-µm SWM [129]. Therefore, establishing an optimal process of SWM based on the pore size can increase the separation efficiency of milk components from cell culture media. In summary, the milk components present within cell culture media can be effectively separated into fat globules, casein micelles, and whey proteins by implementing an optimal process that integrates microfiltration, ultrafiltration, and nanofiltration, using SWM.
Conclusions
Cellular agriculture in the dairy sector may provide a wide range of opportunities for the sustainable production of dairy products, such as milk components or cell-cultured milk. Cellular agriculture in the dairy sector is categorized into fermentation-based and animal cell culture-based cellular agriculture. While several companies predominantly focus on producing milk components through fermentation-based technology, precision fermentation technology still faces the challenges with respect to GMO concerns. Several startup companies are attempting to produce milk components by cultivating ECs in animal cell culture-based technology. However, the technology is still in its early stages of development. This review summarized the structural and functional attributes of the mammary gland and discussed the technologies for cell-cultured milk production. The major technologies were (1) resourcing of mammary EC line, (2) optimizing the cell culture media, (3) establishing the cell culture system, and (4) down-stream processing of milk component. Additionally, future developments and areas of further research include several key areas. Firstly, there is a need to efficiently cultivate milk-derived mammary ECs, achieved through the identification of reliable biomarkers and the use of optimal proliferation and differentiation media. Secondly, the application of 3D cell culture techniques, bioreactors, and membrane separation systems holds promise for scaling up the production of cell-cultured milk. Lastly, it is imperative to reduce the cost of cell-cultured milk compared to traditional milk, ensuring consumer accessibility to the product. In conclusion, this review presents current insights and challenges regarding cell culture-based dairy production and offers implications for ongoing efforts required to produce commercially significant quantities of cell-cultured milk.
Acknowledgements
None.
Abbreviations
- 2D
Two-dimensional
- 3D
Three-dimensional
- BLG
β-lactoglobulin
- BM
Basement membrane
- CK
Cytokeratin
- CORT
Cortisol
- DMEM/F12
Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham
- E1
Estrone
- E2
17β-Estradiol
- ECM
Extracellular matrix
- ECs
Epithelial cells
- EDTA
Ethylene-diamine-tetraacetic acid
- EGF
Epidermal growth factor
- FBS
Fetal bovine serum
- FBs
Fibroblasts
- FCS
Fetal calf serum
- GMO
Genetically modified organism
- IGF-1
Insulin growth factor-1
- INS
Insulin
- ITS
Insulin-transferrin-selenium
- MCs
Myoepithelial cells
- Mt
Million metric tons
- MUFA
Monounsaturated fatty acid
- P4
Progesterone
- PRL
Prolactin
- PUFA
Polyunsaturated fatty acid
- SFA
Saturated fatty acid
- α-SMA
α-Smooth muscle actin
- SWM
Spiral-wound membrane
- TCM
Tubular ceramic membrane
Authors’ contributions
HCK and SGH conceptualized the review. HCK, HSJ, and VK wrote the manuscript. HCK, HSJ, and VK collected the data. VK corrected the language. HCK and SGH revised and finalized the manuscript. SGH supervised the review. All authors read and approved the final manuscript.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2022R1A2C1008327).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Clay N, Garnett T, Lorimer J. Dairy intensification: Drivers, impacts and alternatives. Ambio. 2020;49:35–48. 10.1007/s13280-019-01177-y. [DOI] [PMC free article] [PubMed]
- 2.Capper JL, Cady RA, Bauman DE. The environmental impact of dairy production: 1944 compared with 2007. J Anim Sci. 2009;87(6):2160–2167. doi: 10.2527/jas.2009-1781. [DOI] [PubMed] [Google Scholar]
- 3.OECD/FAO. OECD-FAO Agricultural Outlook 2021–2030. OECD Publishing. 2021. 10.1787/19428846-en.
- 4.Salou T, Le Mouël C, van der Werf HM. Environmental impacts of dairy system intensification: the functional unit matters! J Clean Prod. 2017;140:445–454. doi: 10.1016/j.jclepro.2016.05.019. [DOI] [Google Scholar]
- 5.Tucker CB, Mench JA, von Keyserlingk MAG. Animal welfare: an integral component of sustainability. In: Kebreab E, editor. Sustainable animal agriculture. Wallingford: CAB International; 2013. p. 42–52. 10.1079/9781780640426.0042.
- 6.Westhoek H, Lesschen JP, Rood T, Wagner S, De Marco A, Murphy-Bokern D, et al. Food choices, health and environment: Effects of cutting Europe's meat and dairy intake. Glob Environ Change. 2014;26:196–205. doi: 10.1016/j.gloenvcha.2014.02.004. [DOI] [Google Scholar]
- 7.Bava L, Sandrucci A, Zucali M, Guerci M, Tamburini A. How can farming intensification affect the environmental impact of milk production? J Dairy Sci. 2014;97(7):4579–4593. doi: 10.3168/jds.2013-7530. [DOI] [PubMed] [Google Scholar]
- 8.Godfray HCJ, Garnett T. Food security and sustainable intensification. Philos Trans R Soc B-Biol Sci. 2014;369(1639):20120273. doi: 10.1098/rstb.2012.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joy M. The environmental and human health impacts of dairy intensification: a Canterbury case study. Vetscript. 2019;32(8):32–36. [Google Scholar]
- 10.Feil AA, Schreiber D, Haetinger C, Haberkamp ÂM, Kist JI, Rempel C, et al. Sustainability in the dairy industry: a systematic literature review. Environ Sci Pollut Res. 2020;27:33527–33542. doi: 10.1007/s11356-020-09316-9. [DOI] [PubMed] [Google Scholar]
- 11.Mattick CS. Cellular agriculture: the coming revolution in food production. Bull Atom Scient. 2018;74(1):32–35. doi: 10.1080/00963402.2017.1413059. [DOI] [Google Scholar]
- 12.Kadim IT, Mahgoub O, Baqir S, Faye B, Purchas R. Cultured meat from muscle stem cells: A review of challenges and prospects. J Integr Agric. 2015;14(2):222–233. doi: 10.1016/S2095-3119(14)60881-9. [DOI] [Google Scholar]
- 13.Mouat MJ, Prince R. Cultured meat and cowless milk: On making markets for animal-free food. J Cult Econ. 2018;11(4):315–329. doi: 10.1080/17530350.2018.1452277. [DOI] [Google Scholar]
- 14.Nyika J, Mackolil J, Workie E, Adhav C, Ramadas S. Cellular agriculture research progress and prospects: Insights from bibliometric analysis. Curr Res Biotechnol. 2021;3:215–224. doi: 10.1016/j.crbiot.2021.07.001. [DOI] [Google Scholar]
- 15.Hong TK, Shin D-M, Choi J, Do JT, Han SG. Current issues and technical advances in cultured meat production: a review. Food Sci of Anim Resour. 2021;41(3):355. doi: 10.5851/kosfa.2021.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens N, Di Silvio L, Dunsford I, Ellis M, Glencross A, Sexton A. Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci Technol. 2018;78:155–166. doi: 10.1016/j.tifs.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens N, Ellis M. Cellular agriculture in the UK: a review. Wellcome Open Res. 2020;5:12. 10.12688/wellcomeopenres.15685.2. [DOI] [PMC free article] [PubMed]
- 18.Bhat ZF, Kumar S, Fayaz H. In vitro meat production: Challenges and benefits over conventional meat production. J Integr Agric. 2015;14(2):241–248. doi: 10.1016/S2095-3119(14)60887-X. [DOI] [Google Scholar]
- 19.Liu Y, Rabinowitz AN. The impact of the COVID-19 pandemic on retail dairy prices. Agribusiness. 2021;37(1):108–121. doi: 10.1002/agr.21687. [DOI] [Google Scholar]
- 20.Pereira PC. Milk nutritional composition and its role in human health. Nutrition. 2014;30(6):619–627. doi: 10.1016/j.nut.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Kwon HC, Jung HS, Kim DH, Han JH, Han SG. The role of progesterone in Elf5 activation and milk component synthesis for cell-cultured milk production in MAC-T cells. Animals. 2024;14(4):642. doi: 10.3390/ani14040642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Augustin MA, Hartley CJ, Maloney G, Tyndall S. Innovation in precision fermentation for food ingredients. Crit Rev Food Sci Nutr. 2023:1–21. 10.1080/10408398.2023.2166014. [DOI] [PubMed]
- 23.Waltz E. Cow-less milk: the rising tide of animal-free dairy attracts big players. Nat Biotechnol. 2022;40:1534–1536. doi: 10.1038/s41587-022-01548-z. [DOI] [PubMed] [Google Scholar]
- 24.Linder T. Beyond agriculture-how microorganisms can revolutionize global food production. ACS Food Sci Technol. 2023;3(7):1144–1152. doi: 10.1021/acsfoodscitech.3c00099. [DOI] [Google Scholar]
- 25.Chai KF, Ng KR, Samarasiri M, Chen WN. Precision fermentation to advance fungal food fermentations. Curr Opin Food Sci. 2022;47:100881. 10.1016/j.cofs.2022.100881.
- 26.Lyu X, Lee J, Chen WN. Potential natural food preservatives and their sustainable production in yeast: terpenoids and polyphenols. J Agric Food Chem. 2019;67(16):4397–4417. doi: 10.1021/acs.jafc.8b07141. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Ma L, Su P, Huang L, Gao W. Cytochrome P450s in plant terpenoid biosynthesis: discovery, characterization and metabolic engineering. Crit Rev Biotechnol. 2023;43(1):1–21. doi: 10.1080/07388551.2021.2003292. [DOI] [PubMed] [Google Scholar]
- 28.Aro N, Ercili-Cura D, Andberg M, Silventoinen P, Lille M, Hosia W, et al. Production of bovine beta-lactoglobulin and hen egg ovalbumin by Trichoderma reesei using precision fermentation technology and testing of their techno-functional properties. Food Res Int. 2023;163:112131. 10.1016/j.foodres.2022.112131. [DOI] [PubMed]
- 29.Hassoun A, Bekhit AE-D, Jambrak AR, Regenstein JM, Chemat F, Morton JD, et al. The fourth industrial revolution in the food industry—part II: Emerging food trends. Crit Rev Food Sci Nutr. 2022:1–31. 10.1080/10408398.2022.2106472. [DOI] [PubMed]
- 30.Banovic M, Grunert KG. Consumer acceptance of precision fermentation technology: A cross-cultural study. Innov Food Sci Emerg Technol. 2023;88:103435. 10.1016/j.ifset.2023.103435.
- 31.Cohen M, Cassidy T. Milk from Mars. The challenges of regulating lab-produced (human) milk. Food & Drug Law J. 2022;77:6. [Google Scholar]
- 32.Gan J, Cao C, Stahl B, Zhao X, Yan J. Advances and challenges for obtaining human milk oligosaccharides: Extraction from natural sources and synthesis by intentional design. Trends Food Sci Technol. 2023;141:104203. 10.1016/j.tifs.2023.104203.
- 33.Eibl R, Senn Y, Gubser G, Jossen V, van den Bos C, Eibl D. Cellular agriculture: Opportunities and challenges. Annu Rev Food Sci Technol. 2021;12:51–73. doi: 10.1146/annurev-food-063020-123940. [DOI] [PubMed] [Google Scholar]
- 34.Weaver S, Hernandez L. Autocrine-paracrine regulation of the mammary gland. J Dairy Sci. 2016;99(1):842–853. doi: 10.3168/jds.2015-9828. [DOI] [PubMed] [Google Scholar]
- 35.Jaswal S, Jena MK, Anand V, Jaswal A, Kancharla S, Kolli P, et al. Critical review on physiological and molecular features during bovine mammary gland development: recent advances. Cells. 2022;11(20):3325. doi: 10.3390/cells11203325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Post MJ, Levenberg S, Kaplan DL, Genovese N, Fu J, Bryant CJ, et al. Scientific, sustainability and regulatory challenges of cultured meat. Nat Food. 2020;1(7):403–15. 10.1038/s43016-020-0112-z.
- 37.Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev -Dev Biol. 2012;1(4):533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biswas SK, Banerjee S, Baker GW, Kuo C-Y, Chowdhury I. The mammary gland: basic structure and molecular signaling during development. Int J Mol Sci. 2022;23(7):3883. doi: 10.3390/ijms23073883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McManaman JL, Neville MC. Mammary physiology and milk secretion. Adv Drug Deliv Rev. 2003;55(5):629–641. doi: 10.3390/ijms23073883. [DOI] [PubMed] [Google Scholar]
- 40.Yart L, Lollivier V, Marnet P-G, Dessauge F. Role of ovarian secretions in mammary gland development and function in ruminants. Animal. 2014;8(1):72–85. doi: 10.1017/S1751731113001638. [DOI] [PubMed] [Google Scholar]
- 41.Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142(6):1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- 42.Hassiotou F, Geddes D. Anatomy of the human mammary gland: Current status of knowledge. Clin Anat. 2013;26(1):29–48. doi: 10.1002/ca.22165. [DOI] [PubMed] [Google Scholar]
- 43.Hannan FM, Elajnaf T, Vandenberg LN, Kennedy SH, Thakker RV. Hormonal regulation of mammary gland development and lactation. Nat Rev Endocrinol. 2023;19(1):46–61. doi: 10.1038/s41574-022-00742-y. [DOI] [PubMed] [Google Scholar]
- 44.Hu Z, Chen Y, Gao M, Chi X, He Y, Zhang C, et al. Novel strategy for primary epithelial cell isolation: Combination of hyaluronidase and collagenase I. Cell Prolif. 2023;56(1):e13320. 10.1111/cpr.13320. [DOI] [PMC free article] [PubMed]
- 45.Anand V, Dogra N, Singh S, Kumar SN, Jena MK, Malakar D, et al. Establishment and characterization of a buffalo (Bubalus bubalis) mammary epithelial cell line. PLoS ONE. 2012;7(7):e40469. 10.1371/journal.pone.0040469. [DOI] [PMC free article] [PubMed]
- 46.Pantschenko A, Woodcock-Mitchell J, Bushmich S, Yang T. Establishment and characterization of a caprine mammary epithelial cell line (CMEC) In Vitro Cell Dev Biol -Anim. 2000;36:26–37. doi: 10.1290/1071-2690(2000)036<0026:EACOAC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 47.Hu H, Wang J, Bu D, Wei H, Zhou L, Li F, et al. In vitro culture and characterization of a mammary epithelial cell line from Chinese Holstein dairy cow. PLoS ONE. 2009;4(11):e7636. 10.1371/journal.pone.0007636. [DOI] [PMC free article] [PubMed]
- 48.Zhao K, Liu HY, Zhou MM, Liu JX. Establishment and characterization of a lactating bovine mammary epithelial cell model for the study of milk synthesis. Cell Biol Int. 2010;34(7):717–721. doi: 10.1042/CBI20100023. [DOI] [PubMed] [Google Scholar]
- 49.Lu C, Yang R, Liu B, Li Z, Shen B, Yan S, et al. Establishment of two types of mammary epithelial cell lines from Chinese Holstein dairy cow. J Anim Vet Adv. 2012;11(8):1166–1172. doi: 10.3923/javaa.2012.1166.1172. [DOI] [Google Scholar]
- 50.Jedrzejczak M, Szatkowska I. Bovine mammary epithelial cell cultures for the study of mammary gland functions. In Vitro Cell Dev Biol -Anim. 2014;50:389–398. doi: 10.1007/s11626-013-9711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danowski K, Gross JJ, Meyer H, Kliem H. Effects of induced energy deficiency on lactoferrin concentration in milk and the lactoferrin reaction of primary bovine mammary epithelial cells in vitro. J Anim Physiol Anim Nutr. 2013;97(4):647–655. doi: 10.1111/j.1439-0396.2012.01305.x. [DOI] [PubMed] [Google Scholar]
- 52.Sorg D, Fandrey E, Frölich K, Meyer H, Kliem H. Mammary immunity of White Park and Highland cattle compared with Brown Swiss and Red Holstein. Anim Genet Resour. 2013;52:91–104. 10.1017/S2078633612000781.
- 53.Hillreiner M, Müller NI, Koch HM, Schmautz C, Küster B, Pfaffl MW, et al. Establishment of a 3D cell culture model of primary bovine mammary epithelial cells extracted from fresh milk. In Vitro Cell Dev Biol -Anim. 2017;53:706–720. doi: 10.1007/s11626-017-0169-7. [DOI] [PubMed] [Google Scholar]
- 54.Tong H-L, Li Q-Z, Gao X-J, Yin D-Y. Establishment and characterization of a lactating dairy goat mammary gland epithelial cell line. In Vitro Cell Dev Biol -Anim. 2012;48:149–155. doi: 10.1007/s11626-012-9481-4. [DOI] [PubMed] [Google Scholar]
- 55.Jin L, Qu Y, Gomez LJ, Chung S, Han B, Gao B, et al. Characterization of primary human mammary epithelial cells isolated and propagated by conditional reprogrammed cell culture. Oncotarget. 2018;9(14):11503. doi: 10.18632/oncotarget.23817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dahanayaka S, Rezaei R, Porter W, Johnson G, Burghardt R, Bazer F, et al. Isolation and characterization of porcine mammary epithelial cells. J Anim Sci. 2015;93(11):5186–5193. doi: 10.2527/jas.2015-9250. [DOI] [PubMed] [Google Scholar]
- 57.Fu M, Chen Y, Xiong X, Lan D, Li J. Establishment of mammary gland model in vitro: culture and evaluation of a yak mammary epithelial cell line. PLoS ONE. 2014;9(12):e113669. 10.1371/journal.pone.0113669. [DOI] [PMC free article] [PubMed]
- 58.Sorg D, Potzel A, Beck M, Meyer H, Viturro E, Kliem H. Effects of cell culture techniques on gene expression and cholesterol efflux in primary bovine mammary epithelial cells derived from milk and tissue. In Vitro Cell Dev Biol -Anim. 2012;48:550–553. doi: 10.1007/s11626-012-9544-6. [DOI] [PubMed] [Google Scholar]
- 59.Patki S, Kadam S, Chandra V, Bhonde R. Human breast milk is a rich source of multipotent mesenchymal stem cells. Hum cell. 2010;23:35–40. doi: 10.1111/j.1749-0774.2010.00083.x. [DOI] [PubMed] [Google Scholar]
- 60.Witkowska-Zimny M, Kaminska-El-Hassan E. Cells of human breast milk. Cell Mol Biol Lett. 2017;22(1):11. 10.1186/s11658-017-0042-4. [DOI] [PMC free article] [PubMed]
- 61.Lemay DG, Hovey RC, Hartono SR, Hinde K, Smilowitz JT, Ventimiglia F, et al. Sequencing the transcriptome of milk production: milk trumps mammary tissue. BMC Genomics. 2013;14:872. 10.1186/1471-2164-14-872. [DOI] [PMC free article] [PubMed]
- 62.Ontsouka EC, Bertschi JS, Huang X, Lüthi M, Müller S, Albrecht C. Can widely used cell type markers predict the suitability of immortalized or primary mammary epithelial cell models? Biol Res. 2016;49:1. 10.1186/s40659-015-0063-2. [DOI] [PMC free article] [PubMed]
- 63.Pandey Y, Taluja J, Vaish R, Pandey A, Gupta N, Kumar D. Gross anatomical structure of the mammary gland in cow. J Entomol Zool Stud. 2018;6(4):728–733. [Google Scholar]
- 64.Cai J, Wang D, Liu J. Regulation of fluid flow through the mammary gland of dairy cows and its effect on milk production: a systematic review. J Sci Food Agric. 2018;98(4):1261–1270. doi: 10.1002/jsfa.8605. [DOI] [PubMed] [Google Scholar]
- 65.Arora M. Cell culture media: a review. Mater Methods. 2013;3(175):24. [Google Scholar]
- 66.Yao T, Asayama Y. Animal-cell culture media: History, characteristics, and current issues. Reprod Med Biol. 2017;16(2):99–117. doi: 10.1002/rmb2.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rezaei R, Wu Z, Hou Y, Bazer FW, Wu G. Amino acids and mammary gland development: nutritional implications for milk production and neonatal growth. J Anim Sci Biotechnol. 2016;7:20. 10.1186/s40104-016-0078-8. [DOI] [PMC free article] [PubMed]
- 68.Akers RM. Lactation and the mammary gland. Ames: Blackwell publishing company; 2016. pp. 165–198. [Google Scholar]
- 69.Capuco AV, Choudhary RK. Symposium review: Determinants of milk production: Understanding population dynamics in the bovine mammary epithelium. J Dairy Sci. 2020;103(3):2928–2940. doi: 10.3168/jds.2019-17241. [DOI] [PubMed] [Google Scholar]
- 70.Lindmark MH. Fatty acids in bovine milk fat. Food Nutr Res. 2008;52(1):1821. doi: 10.3402/fnr.v52i0.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markiewicz-Kęszycka M, Czyżak-Runowska G, Lipińska P, Wójtowski J. Fatty acid profile of milk-a review. J Vet Res. 2013;57(2):135–139. doi: 10.2478/bvip-2013-0026. [DOI] [Google Scholar]
- 72.Mohammad MA, Sunehag AL, Haymond MW. De novo synthesis of milk triglycerides in humans. Am J Physiol -Endocrinol Metab. 2014;306(7):E838–E847. doi: 10.1152/ajpendo.00605.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez-Bermúdez R, Fouz R, Rico M, Camino F, Souza TK, Miranda M, et al. Factors affecting fatty acid composition of Holstein cow’s milk. Animals. 2023;13(4):574. doi: 10.3390/ani13040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee DY, Lee SY, Jung JW, Kim JH, Oh DH, Kim HW, et al. Review of technology and materials for the development of cultured meat. Crit Rev Food Sci Nutr. 2023;63(27):8591–8615. doi: 10.1080/10408398.2022.2063249. [DOI] [PubMed] [Google Scholar]
- 75.Duarte RT, Carvalho Simões MC, Sgarbieri VC. Bovine blood components: fractionation, composition, and nutritive value. J Agric Food Chem. 1999;47(1):231–236. doi: 10.1021/jf9806255. [DOI] [PubMed] [Google Scholar]
- 76.Rius A, Appuhamy J, Cyriac J, Kirovski D, Becvar O, Escobar J, et al. Regulation of protein synthesis in mammary glands of lactating dairy cows by starch and amino acids. J Dairy Sci. 2010;93(7):3114–3127. doi: 10.3168/jds.2009-2743. [DOI] [PubMed] [Google Scholar]
- 77.Brickell J, McGowan M, Wathes D. Effect of management factors and blood metabolites during the rearing period on growth in dairy heifers on UK farms. Domest Anim Endocrinol. 2009;36(2):67–81. doi: 10.1016/j.domaniend.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 78.Bionaz M, Loor JJ. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform Biol Insights. 2011;5:BBI–S7003. doi: 10.4137/BBI.S7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukherjee J, Mallick S, Chaudhury M, Prakash B, Dang A. Infradian rhythmicity in milk leukocyte activity together with plasma cortisol and prolactin levels throughout the lactation period in high-yielding crossbred cows. Biol Rhythm Res. 2015;46(6):909–917. doi: 10.1080/09291016.2015.1066544. [DOI] [Google Scholar]
- 80.Fustini M, Galeati G, Gabai G, Mammi L, Bucci D, Baratta M, et al. Overstocking dairy cows during the dry period affects dehydroepiandrosterone and cortisol secretion. J Dairy Sci. 2017;100(1):620–628. doi: 10.3168/jds.2016-11293. [DOI] [PubMed] [Google Scholar]
- 81.Hiew M, Megahed A, Horstman L, Constable P. Clinical utility of plasma progesterone and blood and plasma glucose concentrations in predicting parturition in Holstein cows. J Dairy Sci. 2020;103(6):5575–5590. doi: 10.3168/jds.2019-17800. [DOI] [PubMed] [Google Scholar]
- 82.Kamada H. Effects of selenium-rich yeast supplementation on the plasma progesterone levels of postpartum dairy cows. Asian Australasian J Animal Sci. 2017;30(3):347. doi: 10.5713/ajas.16.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Erb R, Chew B, Keller H. Relative concentration of extrogen and progestrone in milk and blood, and excretion of estrogene in urine. J Anim Sci. 1977;45(3):617–626. doi: 10.2527/jas1977.453617x. [DOI] [PubMed] [Google Scholar]
- 84.Accorsi P, Govoni N, Gaiani R, Pezzi C, Seren E, Tamanini C. Leptin, GH, PRL, insulin and metabolic parameters throughout the dry period and lactation in dairy cows. Reprod Domest Anim. 2005;40(3):217–223. doi: 10.1111/j.1439-0531.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 85.Zachut M, Arieli A, Lehrer H, Livshitz L, Yakoby S, Moallem U. Effects of increased supplementation of n-3 fatty acids to transition dairy cows on performance and fatty acid profile in plasma, adipose tissue, and milk fat. J Dairy Sci. 2010;93(12):5877–5889. doi: 10.3168/jds.2010-3427. [DOI] [PubMed] [Google Scholar]
- 86.Szczechowiak J, Szkudelska K, Szumacher-Strabel M, Sadkowski S, Gwozdz K, El-Sherbiny M, et al. Blood hormones, metabolic parameters and fatty acid proportion in dairy cows fed condensed tannins and oils blend. Ann Anim Sci. 2018;18(1):155. doi: 10.1515/aoas-2017-0039. [DOI] [Google Scholar]
- 87.Contreras G, O’boyle N, Herdt T, Sordillo L. Lipomobilization in periparturient dairy cows influences the composition of plasma nonesterified fatty acids and leukocyte phospholipid fatty acids. J Dairy Sci. 2010;93(6):2508–2516. doi: 10.3168/jds.2009-2876. [DOI] [PubMed] [Google Scholar]
- 88.Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32(4):266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 89.Vidi PA, Bissell MJ, Lelièvre SA. Three-dimensional culture of human breast epithelial cells: the how and the why. Methods Mol. 2013;945:193–219. doi: 10.1007/978-1-62703-125-7_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abraham E, Gupta S, Jung S, McAfee E. Bioreactor for scale-up: process control. In: Biswanathan S, Hematti P, editors. Mesenchymal Stromal Cells. UK: Elsevier; 2017. p.139–178. 10.1016/B978-0-12-802826-1.00006-4.
- 91.Kruegel J, Miosge N. Basement membrane components are key players in specialized extracellular matrices. Cell Mol Life Sci. 2010;67:2879–2895. doi: 10.1007/s00018-010-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finot L, Chanat E, Dessauge F. Mammary gland 3D cell culture systems in farm animals. Vet Res. 2021;52(1):78. doi: 10.1186/s13567-021-00947-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kass L, Erler JT, Dembo M, Weaver VM. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. Int J Biochem Cell Biol. 2007;39(11):1987–1994. doi: 10.1016/j.biocel.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Emerman JT, Pitelka DR. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 1977;13(5):316–328. doi: 10.1007/BF02616178. [DOI] [PubMed] [Google Scholar]
- 95.Hall HG, Farson DA, Bissell MJ. Lumen formation by epithelial cell lines in response to collagen overlay: a morphogenetic model in culture. Proc Natl Acad Sci. 1982;79(15):4672–4676. doi: 10.1073/pnas.79.15.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang L, Abdalla AM, Xiao L, Yang G. Biopolymer-based microcarriers for three-dimensional cell culture and engineered tissue formation. Int J Mol Sci. 2020;21(5):1895. doi: 10.3390/ijms21051895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Justice BA, Badr NA, Felder RA. 3D cell culture opens new dimensions in cell-based assays. Drug Discov Today. 2009;14(1–2):102–107. doi: 10.1016/j.drudis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 98.YekrangSafakar A, Acun A, Choi JW, Song E, Zorlutuna P, Park K. Hollow microcarriers for large-scale expansion of anchorage-dependent cells in a stirred bioreactor. Biotechnol Bioeng. 2018;115(7):1717–1728. doi: 10.1002/bit.26601. [DOI] [PubMed] [Google Scholar]
- 99.Koh B, Sulaiman N, Fauzi MB, Law JX, Ng MH, Idrus RBH, et al. Three dimensional microcarrier system in mesenchymal stem cell culture: a systematic review. Cell Biosci. 2020;10(1):75. 10.1186/s13578-020-00438-8. [DOI] [PMC free article] [PubMed]
- 100.Derakhti S, Safiabadi-Tali SH, Amoabediny G, Sheikhpour M. Attachment and detachment strategies in microcarrier-based cell culture technology: A comprehensive review. Mater Sci Eng C. 2019;103:109782. 10.1016/j.msec.2019.109782. [DOI] [PubMed]
- 101.Chen AK-L, Reuveny S, Oh SKW. Application of human mesenchymal and pluripotent stem cell microcarrier cultures in cellular therapy: achievements and future direction. Biotechnol Adv. 2013;31(7):1032–1046. doi: 10.1016/j.biotechadv.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 102.Badenes SM, Fernandes TG, Rodrigues CA, Diogo MM, Cabral JM. Microcarrier-based platforms for in vitro expansion and differentiation of human pluripotent stem cells in bioreactor culture systems. J Biotechnol. 2016;234:71–82. doi: 10.1016/j.jbiotec.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 103.Couto PS, Rotondi M, Bersenev A, Hewitt C, Nienow A, Verter F, et al. Expansion of human mesenchymal stem/stromal cells (hMSCs) in bioreactors using microcarriers: lessons learnt and what the future holds. Biotechnol Adv. 2020;45:107636. 10.1016/j.biotechadv.2020.107636. [DOI] [PubMed]
- 104.Yang C, Burg KL. Designing a tunable 3D heterocellular breast cancer tissue test system. J Tissue Eng Regen Med. 2015;9(3):310–314. doi: 10.1002/term.1660. [DOI] [PubMed] [Google Scholar]
- 105.Becker JL, Blanchard DK. Characterization of primary breast carcinomas grown in three-dimensional cultures. J Surg Res. 2007;142(2):256–262. doi: 10.1016/j.jss.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 106.Roper AM. Growth of a bovine mammary epithelial cell line (Mac-T) on Cytodex 3 microcarriers. 1993. https://escholarship.mcgill.ca/concern/theses/b2773z07d. Accessed 8 Jan 2024.
- 107.Moritz MS, Verbruggen SE, Post MJ. Alternatives for large-scale production of cultured beef: A review. J Integr Agric. 2015;14(2):208–216. doi: 10.1016/S2095-3119(14)60889-3. [DOI] [Google Scholar]
- 108.Stephenson M, Grayson W. Recent advances in bioreactors for cell-based therapies. F1000Research. 2018;7:517–525. doi: 10.12688/f1000research.12533.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kropp C, Massai D, Zweigerdt R. Progress and challenges in large-scale expansion of human pluripotent stem cells. Process Biochem. 2017;59:244–254. doi: 10.1016/j.procbio.2016.09.032. [DOI] [Google Scholar]
- 110.Kumar A, Starly B. Large scale industrialized cell expansion: producing the critical raw material for biofabrication processes. Biofabrication. 2015;7(4):044103. 10.1088/1758-5090/7/4/044103. [DOI] [PubMed]
- 111.Yang ST. Bioprocessing for value-added products from renewable resources: new technologies and applications. Amsterdam: Elsevier; 2011. pp. 491–507. [Google Scholar]
- 112.Warnock JN, Al-Rubeai M. Bioreactor systems for the production of biopharmaceuticals from animal cells. Biotechnol Appl Biochem. 2006;45(1):1–12. doi: 10.1042/BA20050233. [DOI] [PubMed] [Google Scholar]
- 113.Youn BS, Sen A, Kallos MS, Behie LA, Girgis-Gabardo A, Kurpios N, et al. Large-scale expansion of mammary epithelial stem cell aggregates in suspension bioreactors. Biotechnol Prog. 2005;21(3):984–993. doi: 10.1021/bp050059f. [DOI] [PubMed] [Google Scholar]
- 114.Benoit S, Chamberland J, Doyen A, Margni M, Bouchard C, Pouliot Y. Integrating pressure-driven membrane separation processes to improve eco-efficiency in cheese manufacture: a preliminary case study. Membranes. 2020;10(10):287. doi: 10.3390/membranes10100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shojaee Nasirabadi P, Saljoughi E, Mousavi SM. Membrane processes used for removal of pharmaceuticals, hormones, endocrine disruptors and their metabolites from wastewaters: A review. Desalination Water Treat. 2016;57(51):24146–24175. doi: 10.1080/19443994.2016.1140081. [DOI] [Google Scholar]
- 116.Korhonen H, Pihlanto A. Technological options for the production of health-promoting proteins and peptides derived from milk and colostrum. Curr Pharm Design. 2007;13(8):829–843. doi: 10.2174/138161207780363112. [DOI] [PubMed] [Google Scholar]
- 117.Carter B, Cheng N, Kapoor R, Meletharayil G, Drake M. Invited review: Microfiltration-derived casein and whey proteins from milk. J Dairy Sci. 2021;104(3):2465–2479. doi: 10.3168/jds.2020-18811. [DOI] [PubMed] [Google Scholar]
- 118.Koyuncu I, Arikan OA, Wiesner MR, Rice C. Removal of hormones and antibiotics by nanofiltration membranes. J Membr Sci. 2008;309(1–2):94–101. doi: 10.1016/j.memsci.2007.10.010. [DOI] [Google Scholar]
- 119.Yang L, Xia C, Jiang J, Chen X, Zhou Y, Yuan C, et al. Removal of antibiotics and estrogens by nanofiltration and reverse osmosis membranes. J Hazard Mater. 2024;461:132628. 10.1016/j.jhazmat.2023.132628. [DOI] [PubMed]
- 120.Reig M, Vecino X, Cortina JL. Use of membrane technologies in dairy industry: An overview. Foods. 2021;10(11):2768. doi: 10.3390/foods10112768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kowalik-Klimczak A. The possibilities of using membrane filtration in the dairy industry. J Mach Constr Maintenance. 2017;2:99–108. [Google Scholar]
- 122.Brans G, Schroën C, Van der Sman R, Boom R. Membrane fractionation of milk: state of the art and challenges. J Membr Sci. 2004;243(1–2):263–272. doi: 10.1016/j.memsci.2004.06.029. [DOI] [Google Scholar]
- 123.Kumar P, Sharma N, Ranjan R, Kumar S, Bhat Z, Jeong DK. Perspective of membrane technology in dairy industry: A review. Asian Australasian J Anim Sci. 2013;26(9):1347. doi: 10.5713/ajas.2013.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pouliot Y. Membrane processes in dairy technology—From a simple idea to worldwide panacea. Int Dairy J. 2008;18(7):735–740. doi: 10.1016/j.idairyj.2008.03.005. [DOI] [Google Scholar]
- 125.Zulewska J, Newbold M, Barbano D. Efficiency of serum protein removal from skim milk with ceramic and polymeric membranes at 50 °C. J Dairy Sci. 2009;92(4):1361–77. 10.3168/jds.2008-1757. [DOI] [PubMed]
- 126.Fernández García L, Riera Rodríguez FA. Microfiltration of milk with third generation ceramic membranes. Chem Eng Commun. 2015;202(11):1455–1462. doi: 10.1080/00986445.2014.950731. [DOI] [Google Scholar]
- 127.Schopf R, Schmidt F, Linner J, Kulozik U. Comparative assessment of tubular ceramic, spiral wound, and hollow fiber membrane microfiltration module systems for milk protein fractionation. Foods. 2021;10(4):692. doi: 10.3390/foods10040692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beckman S, Zulewska J, Newbold M, Barbano D. Production efficiency of micellar casein concentrate using polymeric spiral-wound microfiltration membranes. J Dairy Sci. 2010;93(10):4506–4517. doi: 10.3168/jds.2010-3261. [DOI] [PubMed] [Google Scholar]
- 129.Mercier-Bouchard D, Benoit S, Doyen A, Britten M, Pouliot Y. Process efficiency of casein separation from milk using polymeric spiral-wound microfiltration membranes. J Dairy Sci. 2017;100(11):8838–8848. doi: 10.3168/jds.2017-13015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.