Abstract
Reprogramming cells will play a fundamental role in shaping the future of cell therapies by developing new strategies to engineer cells for improved performance and higher-order physiological functions. Approaches in synthetic biology harness cells’ natural ability to sense diverse signals, integrate environmental inputs to make decisions, and execute complex behaviors based on the health of the organism or tissue. In this review, we highlight strategies in synthetic biology to reprogram cells, and discuss how recent approaches in the delivery of modified mRNA have created new opportunities to alter cell function in vivo. Finally, we discuss how combining concepts from synthetic biology and the delivery of mRNA in vivo could provide a platform for innovation to advance in vivo cellular reprogramming.
Introduction
There is an unmet need for medical technologies that can deliver or mediate therapeutic biomolecules and cellular treatments that are otherwise not possible with conventional pharmaceutical or surgical approaches. Cell therapy is a rapidly emerging field that involves the use of living cells to treat and potentially cure various diseases and medical conditions that were once considered untreatable [1–3].
The success of cell therapies lies in the ability of engineered cells to sense disease-associated biomarkers, perform complex computation to process this information, and produce a wide array of outputs to mitigate disease. A significant portion of initial efforts in synthetic biology was founded on assembling genetic parts to build synthetic gene circuits to respond to small molecules that diffuse through the cell membrane and activate synthetic circuits to implement new cellular functions [4–8]. The sophistication of sensing specific molecules within an environment has evolved with efforts to engineer receptors for improved target specificity that can be coupled to therapeutic responses, including activating cellular pathways, initiating a preprogrammed synthetic gene circuit, and participating in targeted payload delivery. Several cell types have been explored for the development of cell therapies, including bacteria, immune cells, and stem cells [9–11]. Engineering immune cells, specifically T cells, has become the focus of many studies due to the recent regulatory approval of chimeric antigen receptor (CAR) T cells, and the number of cancer patients who have been successfully treated with engineered CAR-T cells [12]. The premise of CAR-T-cell therapy is to couple the potency of a T cell with the specificity of an antibody to kill diseased cells [13]. However, the current high cost and complexity involved with manufacturing CAR-T cells ex vivo prohibits personalized T-cell therapy from being available to all cancer patients [14]. An exciting alternative is to target and reprogram circulating T cells for their direct reprogramming that would eliminate the need to perform costly manufacturing steps required to produce CAR-T cells ex vivo. The direct delivery of modified mRNA to T cells in vivo has been shown to enable T cells to translate new proteins, such as receptors, to target specific cell types for their destruction [15]. These early studies of delivering mRNA in vivo to alter the function of recipient cells strongly suggest that the delivery of mRNA in vivo may prove to be a new approach to advance cell therapies that can be used for treating many diseased and damaged tissues.
In recent years, the modification and optimization of mRNA for clinical applications has expanded the potential for in vivo reprogramming [16–18]. In 2020, the coronavirus disease 2019 (COVID-19) resulted in a global pandemic and catalyzed innovation in vaccine development with mRNA vaccines playing an essential role. Earlier studies by Kariko and Weissman demonstrated that base-modified mRNA prevents unintended immune responses [19] and increased protein production when delivered to immune cells compared with unmodified RNA [20,21]. With these discoveries, the concept of mRNA-based vaccines has been catapulted into the forefront of public awareness and has initiated additional research pursuits using mRNA in the clinic [22,23].
In this review, we highlight new developments in synthetic biology to reprogram cells using DNA- and RNA-based tools to emphasize the cellular reprogramming capabilities of synthetic biology, and we discuss recent studies that have successfully demonstrated the delivery of mRNA to cells in vivo. We propose that the success of delivering mRNA in vivo provides a roadmap for expanding synthetic biology devices to enhance the efficiency and precision of delivery, enable mRNA kinetics, and advance in vivo cellular reprogramming to address many challenges in medicine.
Reprogramming cells using synthetic biology
Boolean logic operations
A central goal of synthetic biologists is to reprogram cells with decision-making capabilities based on inputs from intrinsic and extrinsic cues for regulating target genes to produce a desired behavior. In this case, the processing of inputs has threshold states of ‘0’ or ‘1’ (‘OFF’ or ‘ON,’ respectively) [24] (Figure 1a). This Boolean logic can be used to distinguish between healthy and diseased cells, and upon sensing a target cell, produces a response that destines specific cells for destruction. Additionally, engineered cells can be programmed to produce therapeutic molecules to be delivered at controlled levels in response to the degree of disease.
Figure 1.
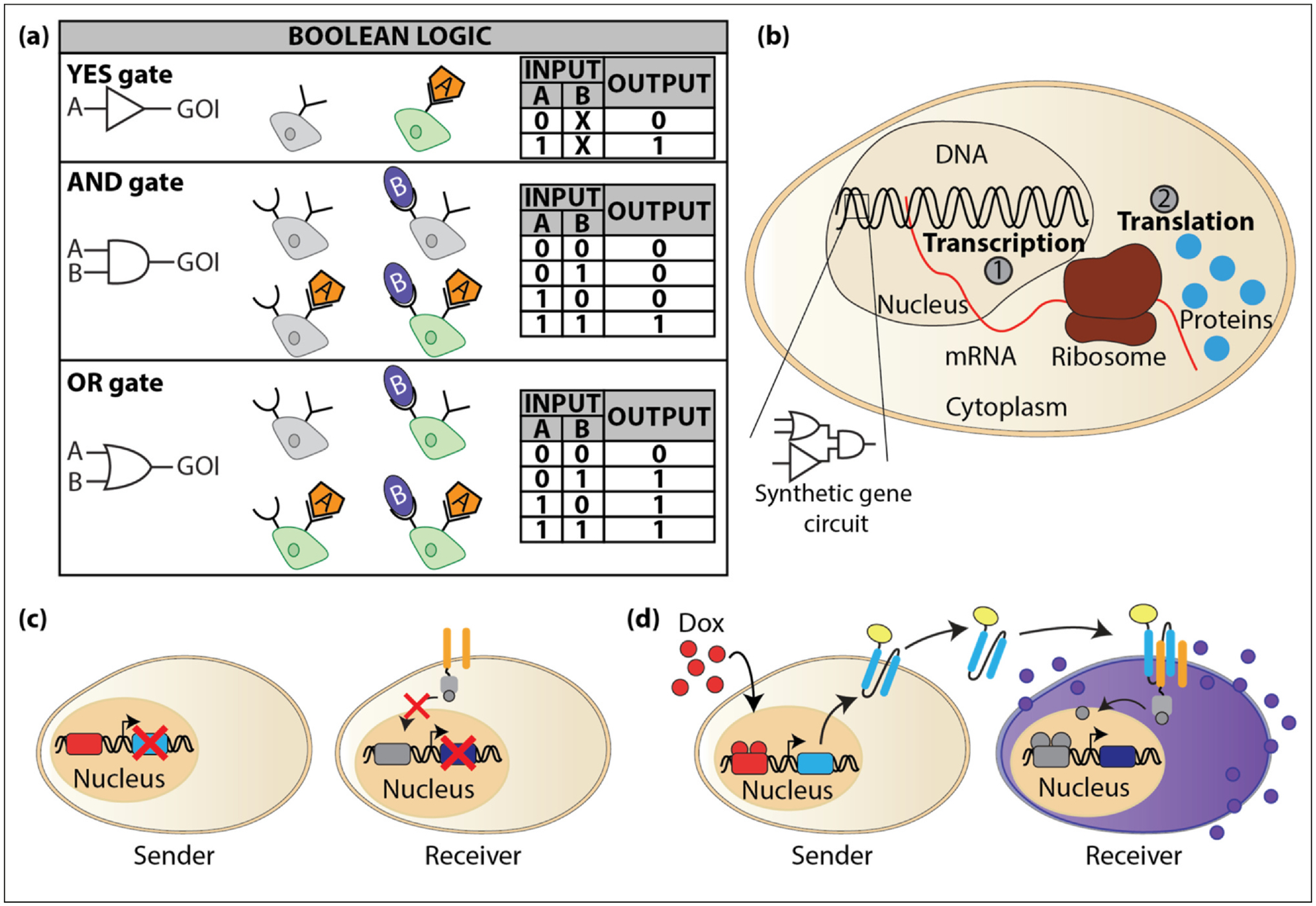
Engineering cells. (a) Boolean logic can be programmed in cells using synthetic gene circuits to enable decision-making. Cells process inputs and when the logic input is true, the gene of interest (GOI) turns on (green). (b) DNA is transcribed into mRNA (red line) in the nucleus (1) and transported into the cytoplasm where it binds to a ribosome to be translated into protein (blue circles) (2). Transcriptional-based cellular reprogramming targets DNA located in the nucleus, whereas post-transcriptional-based reprogramming targets RNA in the cytoplasm. (c) An engineered communication system was built with engineered sender and receiver cells using coiled coils. In the absence of doxycycline (Dox), the coiled coils are not produced, and the receiver cell is in the off-state. (d) In the presence of Dox, the coiled coils are produced and bind to their cognate Epo receptors on the receiver cells. Upon binding, the STAT3 protein (gray ball) is released and translocates to the nucleus to activate the transcription of secreted alkaline phosphatase, which is secreted by the receiver cell (purple balls).
Epigenetic reprogramming
Epigenetics are stable alterations in gene expression that arise throughout the life of eukaryotic organisms without altering the DNA sequence. Epigenetic regulation can occur with the addition of methyl groups to cytosine or guanine nucleotides in the genomic DNA to silence genes [25], or when proteins bind to genomic DNA and alter the accessibility on chromatins for regulatory proteins to bind for activating or repressing gene expression [26]. Efforts to reprogram cells by targeting the genome directly are underway to create and optimize desired cell behavior. Specifically, synthetic transcription factors are designed to target specific sequences in the genome to either activate or repress gene expression. When transcription activator-like effectors and zinc finger proteins are fused to transcriptional activators or repressors, they can be custom-built to target specific locations in the genome to manipulate endogenous genes. Additionally, the clustered regulatory interspaced short palindromic repeat–Cas9 system enables programmable transcriptional regulation when the nuclease activity has been removed from the Cas9 protein (dCas9), and transcriptional activators or repressors are fused to dCas9 [27]. These synthetic transcription factors have been shown to be extremely versatile in a variety of applications in basic research and in biotechnology [28–31].
Transcriptional-based cellular reprogramming
Precise and sustainable control of gene expression is key to developing cell therapies. Enhancing cells’ natural functions, redirecting their natural abilities, and engineering them with new functions has improved the therapeutic efficacy of using synthetic biology approaches to reprogram cells [32–34]. Controlling gene expression at the transcriptional level remains a corner-stone of developing new gene regulatory parts and devices. In these devices, gene expression is typically controlled by transcription factors and their associated DNA-binding sequences that either prevent or enable RNA polymerase to transcribe downstream genes [35] (Figure 1b). A challenge with this approach can be transgene silencing, the loss of gene expression over time, especially when engineering primary cells and stem cells to be used for therapeutic applications. This is an active area of research to better understand the mechanisms associated with transgene silencing to develop strategies to mitigate this challenge [36–40]. Never-theless, controlling gene expression at the transcription level has proven to be a powerful approach to reprogramming cells, especially when cells can implement Boolean logic operations to autonomously respond to intracellular or environmental molecules that trigger regulated therapeutic responses. Much of this work continues to provide the foundation for innovative reprogramming approaches and capabilities.
Extracellular molecules also strongly influence cell behavior. Cells are exposed to hundreds of different signals in their environment, and receptors enable them to distinguish which molecules to respond to for directing an appropriate cellular response. Efforts in synthetic biology and receptor engineering have made it possible to repurpose natural cell receptors as custom environmental monitoring devices that can be linked to cell responses for defined cellular function. In general, synthetic receptors combine sensing capabilities to a desired extracellular molecule with the cell’s natural function or activate synthetic circuits to implement new cellular function. This is a key step in engineering cell-based therapies for customized sensing of a specific extracellular signal, responding with an appropriate therapeutic output, and communicating with appropriate cells to mitigate disease. These synthetic receptors have been extensively reviewed elsewhere [41,42].
Synthetic receptors can also be designed to reprogram how cells communicate with each other and within their external environments [43]. Recently, a synthetic mammalian communication platform was engineered using synthetic receptors and coiled-coil peptides [44] (Figure 1c). In this work, the domains in the erythropoietin receptor (EpoR) were functionalized to bind to coiled-coil peptides that can be secreted by mammalian cells and specifically bind to the engineered EpoRs, activating them and initiating the expression of a reporter gene. In this study, sender cells were engineered to secrete coiled-coil ligands and upon binding to their cognate engineered EpoRs expressed by neighboring cells, activate a reporter gene in the recipient cell (Figure 1d). This engineered cellular communication system can also produce 2-input Boolean logic operations and advances the capabilities to program complex biological functions to better understand relationships between extracellular inputs and intracellular responses.
Post-transcriptional-based cellular reprogramming
RNA-based devices can be advantageous for cell therapies because they are delivered to the cytosol of cells where they control the translation of functional proteins, avoiding genomic integration and limiting epigenetic transgene silencing. Like transcription-based synthetic gene circuits, RNA-based genetic tools can sense inputs, process this information, and produce a controlled level of output in response to the degree of input. RNA is single-stranded and has the unique ability to base-pair with itself to form complex structures that enable the engineering of RNA-based tools to function by coupling the binding of a target molecule to alter its shape and facilitate translation [45,46]. This approach has been used to regulate the translation of mRNAs based on the presence or absence of RNA–protein interactions within the cytoplasm [47–52].
Toehold switches are RNA devices that are engineered to repress translation through base pairing around the translation start site of the mRNA of interest, creating a hairpin structure and preventing translation. Toeholds were initially designed to function in prokaryotes and have become a significant component in cell-free synthetic biology for diagnostics [53,54]. A eukaryotic version of toehold switches has recently been shown to function in mammalian cells [55]. These eToeholds contain internal ribosome entry site (IRES) sequences that can recruit translational machinery and initiate the translation of downstream genes. Gene expression is in the off-state in the absence of the cognate trigger RNA due to the inhibitory loops within the IRES structure (Figure 2a). Like the prokaryotic toeholds, once the trigger RNA anneals to a complementary sequence in the eToehold, the inhibitory loops are disrupted, and translation of the downstream mRNA is initiated (Figure 2b). This system is modular and can be designed to detect and respond to the presence of various intracellular RNA molecules, including viral RNA, that enables the translation of the mRNA linked to the toe-hold. eToehold devices can facilitate marking infected or diseased cells to be targeted by various therapies.
Figure 2.
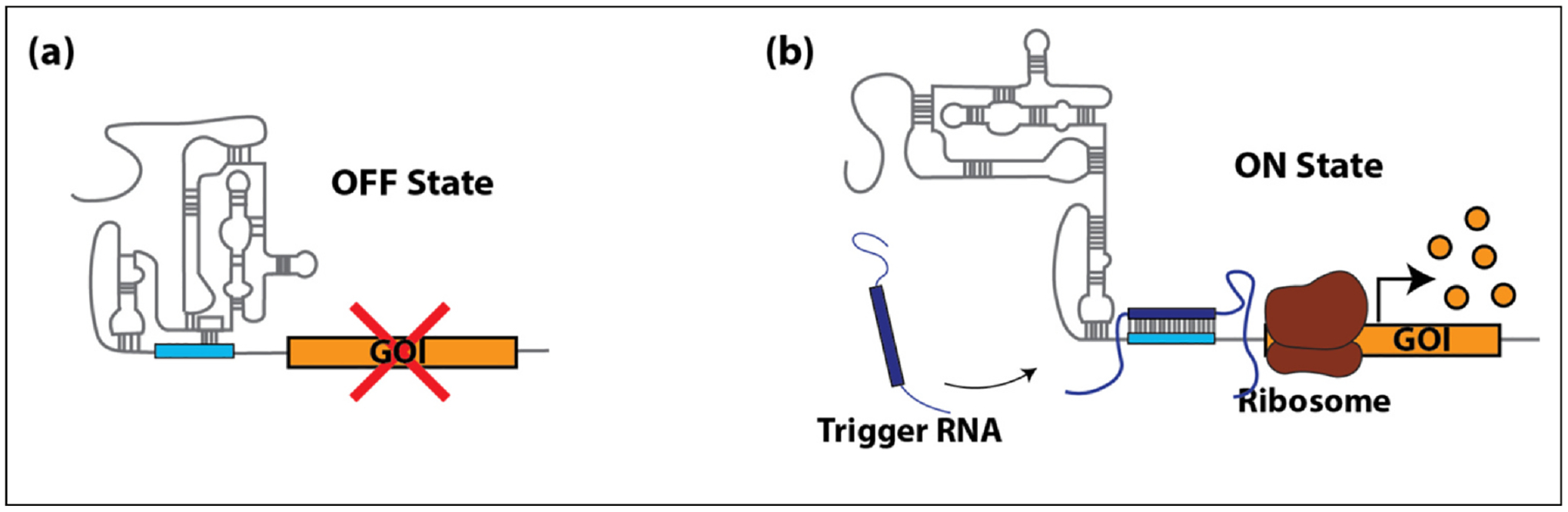
eToeholds. (a) Gene expression of the GOI is in the off-state in the absence of the trigger RNA due to inhibitory loops within the RNA structure. (b) In the presence of the trigger RNA (dark blue), it binds to the complementary RNA (teal) within the eToehold, causing disruption of the toehold structure to enable the assembly of ribosomes and translation of the downstream GOI.
Both an advantage and a disadvantage, mRNA therapeutics only allow for short-term expression of proteins because mRNA is usually degraded within a few hours after entering the cytosol. Using mRNA as a long-term, high-level expression therapeutic would therefore require repeated treatments. To mitigate this challenge, efforts are underway to prolong protein expression from mRNA molecules by building synthetic self-amplifying mRNA (saRNA). saRNAs are derived from positive-sense stranded viruses that can produce RNA-dependent RNA polymerase complexes that amplify synthetic transcripts in vivo at high levels [56]. Recently, an inducible saRNA switch was engineered to control mRNA kinetics in vivo [57]. This was accomplished by fusing a destabilization domain to the RNA-binding protein, L7Ae, that undergoes rapid degradation in the absence of the small molecule, trimethoprim (TMP). Adding TMP stabilizes the L7Ae protein that binds to structural motifs and suppresses translation of the downstream mRNA. This switch showed a significant increase and prolonged expression of the therapeutic protein compared with other in vivo approaches. Moreover, a recent study identifies multiple modified nucleotides that can be incorporated into saRNA that result in immune evasion and enhanced protein expression [18]. Circular mRNA is another approach for prolonged protein expression because it is more stable than linear RNA, less prone to degradation, and is less likely to cause an immune response in vivo [58–60].
Delivery of modified mRNA in vivo
The ability to manipulate genes and pathways in vivo has largely been dependent on the generation of genetically engineered mouse models or the transfer of transgenes using a variety of vectors that include viruses, plasmids, and nanoparticles. Lipid nanoparticles (LNPs) have been an essential component in the COVID-19 vaccine for delivering mRNA in vivo and are the most clinically advanced delivery vehicle to date. LNPs are therefore promising vehicles to deliver a variety of therapeutics, including mRNA in vivo. LNPs are small fat droplets composed of cholesterol and lipids that stabilize the particle’s structure, and the positively charged lipids bind to the negatively charged mRNA (Figure 3a). Once LNPs enter the cell, they are taken up by the endosome, which breaks open the LNP releasing the mRNA into the cytosol to be translated (Figure 3b). Tremendous progress has been made using LNPs for delivering mRNA in vivo.
Figure 3.
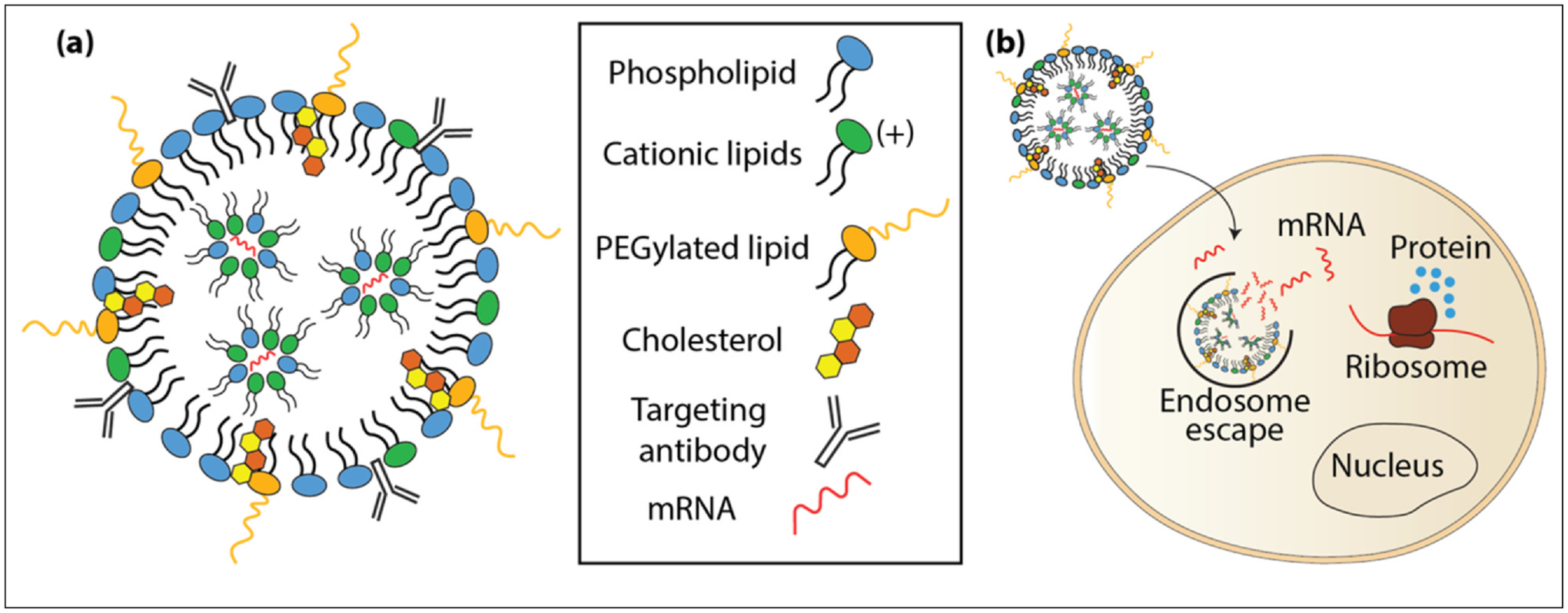
LNPs used to deliver modified mRNA. (a) LNPs are fat droplets composed of a mixture of lipids and cholesterol. They can be designed to target specific cells by functionalizing them with targeting antibodies. (b) Once inside the cell, the LNPs enter the endosome where they are broken apart and the mRNA escapes into the cytosol where it is translated into functional protein.
Recent studies have demonstrated the potential for the targeted delivery of mRNA using LNPs to reprogram cells in vivo to direct cell behavior and improve clinical outcomes in animal models. In many chronic heart diseases and injuries, cardiac fibroblasts become activated and secrete excessive extracellular matrix, resulting in fibrosis that stiffens the myocardium and eventually leads to the loss in the ability to pump blood properly [61]. To address this, a recent study demonstrated that T cells could be reprogrammed in vivo to target and remove activated fibroblasts [62]. In this study, mRNA encoding a CAR designed against the fibroblast activation protein (FAP) was packaged into LNPs decorated with antibodies for CD5, a marker for T cells (Figure 4a). Once the mRNA is delivered to the cytosol of T cells, it is translated into functional receptors against FAP, and these new CAR-T cells target and kill the activated fibroblasts. Using this in vivo platform demonstrated that cardiac function was improved in mice with cardiac injuries.
Figure 4.
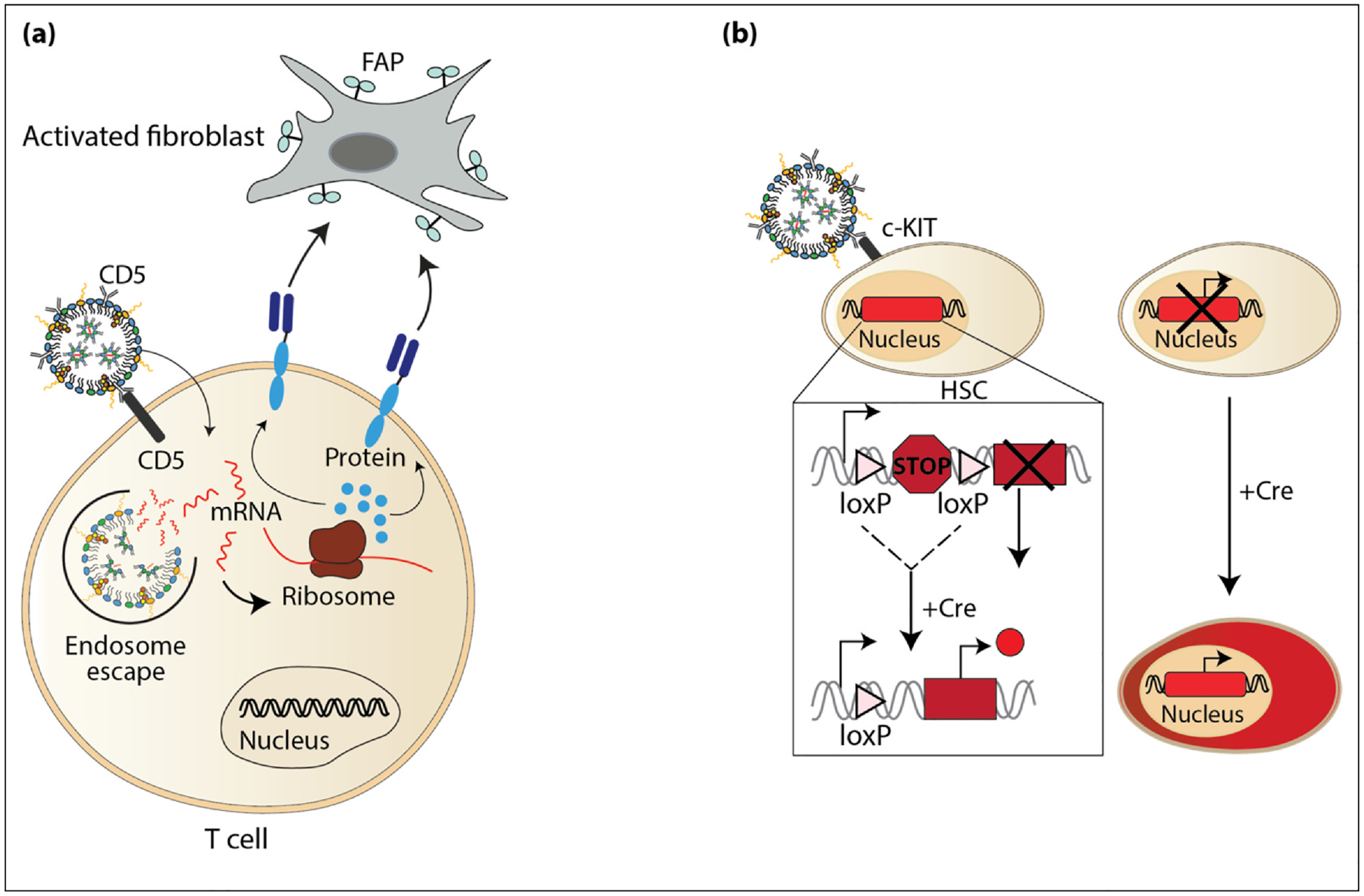
Delivery of mRNA in vivo. (a) LNPs functionalized with antibodies for CD5, a marker for T cells, loaded with mRNA encoding a CAR designed against the FAP. Once the mRNA is released from the endosome, it is translated into a functional CAR and the newly engineered CAR-T cells target and kill activated fibroblasts. (b) LNPs functionalized with antibodies against c-KIT, a marker for mouse HSCs, were filled with mRNA encoding the enzyme Cre recombinase. HSCs in the transgenic mouse model harboring a loxP-flanked STOP cassette that prevents the expression of tdTomato were targeted. In the absence of Cre recombinase, the cells do not express tdTomato and are not red. However, when Cre is delivered to the cells, it causes a homologous recombination event at the loxP sites and removes the STOP cassette and turns on the expression of tdTomato turning the cells that turn red.
Another demonstration of modifying cells in vivo is the recent study targeting hematopoietic stem cells (HSCs). HSCs reside in the bone marrow and have the potential to become all cells in the blood system throughout an individual’s lifetime [9]. HSCs are a therapeutically relevant cell population to target because diseased HSCs can lead to leukemia, lymphoma, cardiac failure, immunodeficiencies, autoimmune diseases, bleeding disorders, and others [63]. Recently, it was shown that LNP–mRNA technology could be used to target HSCs in vivo for the stable gene editing of HSCs. In this study, the outside of LNPs was functionalized with antibodies against the mouse HSC marker, c-KIT, and filled with Cre recombinase mRNA to target HSCs in the Ai9 mouse line [64]. This mouse line has a loxP-flanked STOP cassette preventing the transcription of tdTomato in the absence of Cre recombinase (Figure 4b) [65]. After intravenous administration of c-KIT/LNP-Cre, Cre mRNA was delivered to HSCs and the expression of tdTomato was observed in peripheral blood cells for up to 4 months, demonstrating the delivery and translation of Cre mRNA in HSCs. Importantly, long-term HSCs are a critical population that replenishes the hematopoietic system and must persist for the lifespan of the organism [66,67]. Remarkably, in this study, the long-term HSCs were also reprogrammed and tdTomato expression was observed in all peripheral blood lineages, indicating that the edits made in progenitor cells are maintained throughout differentiation. This represents a promising modality to target HSCs in vivo for reprogramming stem cells with enhanced capabilities and open the possibilities for future cell therapies to treat hematologic diseases.
Conclusions and outlook
A powerful feature of using cells as therapeutic devices is their inherent compatibility, and their natural ability to sense, integrate, and respond to dynamic environments in vivo. Synthetic biology approaches offer platform technologies to augment these features to reprogram cells that can discriminate between cell states, produce a regulated dose of therapeutic biomolecules, and function to deliver payloads in a spatial and temporal fashion. Synthetic receptors can be built to sense specific molecules and cells within the environment and be coupled to synthetic gene circuits to facilitate tailored cell behavior. The recent success of mRNA-based COVID-19 vaccines shows promise for delivering mRNA to cells in vivo. In particular, the demonstration that mRNA for chimeric antigen receptors can be delivered to recipient T cells in vivo and translated into functional receptors to redirect immune cells for targeting specific cell types opens the door for a new wave of innovation in cell therapies and in vivo reprogramming.
Here, we highlight recent advances in synthetic biology to reprogram cells at the transcription and translation levels to emphasize the programmability of cells for sensing intracellular and extracellular molecules to activate native pathways, or synthetic circuits for implementing new cellular function. We envision the possibility of loading programmable synthetic gene circuits with multistep functions in vivo that can be used to enhance information processing to produce appropriately graded outputs based on the level of therapeutic need to mitigate disease. For example, eToeholds were shown to detect the presence of intracellular viral RNA and, if infected, produced a reporter protein. Instead of producing a reporter protein, eToeholds could produce mRNA for a unique transmembrane peptide to mark the infected cell for CAR-T-cell destruction (Figure 5). This approach can be coupled to the recent multiplexing of functionalizing LNPs for diverse targeting and reprogramming of T cells [68]. Similar approaches could be used with other cell types for tissue regeneration to augment the healing process by recruiting specific cells to locations of injury and targeting progenitor cells to enhance their differentiation.
Figure 5.
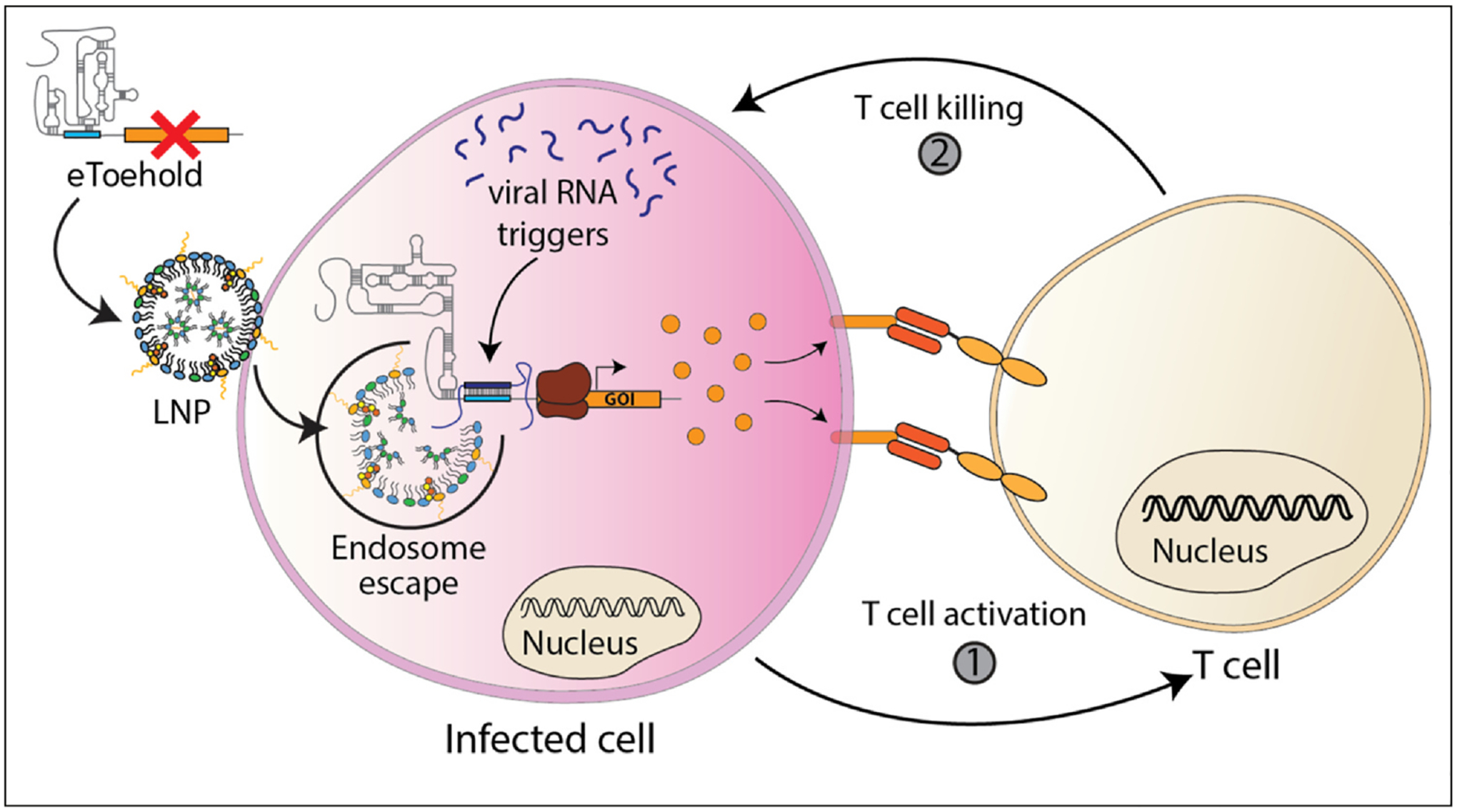
eToeholds for detecting infected cells. eToeholds can be delivered by LNPs or other delivery mechanisms and upon detecting viral RNA in cells, it activates the translation of transmembrane proteins that function as T-cell beacons (orange) for T-cell destruction. (1) Upon T binding to the infected cell, it is activated and (2) the T cell is signaled to kill the infected cell.
LNPs have shown tremendous success in vaccine development, however, challenges remain with low targeting specificity and toxicity, suggesting a need for improved delivery approaches [69,70]. Indeed, cells can be programmed for targeting molecules and cellular surface markers, however, nucleated cells come with their own set of safety risks. More recently, efforts have been made to utilize nonnucleated extracellular vesicles, red blood cells, and platelets as delivery vehicles that have shown promising early preliminary results for delivering payloads to potentially enhance targeted delivery and improve cell-based therapeutic approaches [71–74]. While in vivo reprogramming is still in its infancy, we envision that synthetic biology approaches will provide a platform for innovation that will offer next-generation cell therapy technologies.
Acknowledgements
This work was supported by the National Institute of Health, USA Trailblazer Award 1R21EB025413-01, the National Institute of Health, USA Director New Innovator Award 1DP2CA250006-01, and the National Institutes of Health, USA Award 1R01EB033851-01.
Footnotes
CRedit authorship contribution statement
Farhana Islam: Writing – original draft, Writing – review & editing. Mitchell Lewis: Writing – original draft, Writing – review & editing. James Craig: Writing – original draft, Writing – review & editing. Peyton Leyendecker: Writing – original draft. Tara L. Deans: Conceptualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
Nothing declared.
Data Availability
No data was used for the research described in the article.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•• of special interest
•• of outstanding interest
- 1.Fischbach MA, Bluestone JA, Lim WA: Cell-based therapeutics: the next pillar of medicine. Sci Transl Med 2013, 5:179ps177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee S, Khalil AS, Wong WW: Recent progress of gene circuit designs in immune cell therapies. Cell Syst 2022, 13:864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie M, Fussenegger M: Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat Rev Mol Cell Biol 2018, 19:507–525. [DOI] [PubMed] [Google Scholar]
- 4.Deans TL, Cantor CR, Collins JJ: A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell 2007, 130:363–372. [DOI] [PubMed] [Google Scholar]
- 5.Gardner TS, Cantor CR, Collins JJ: Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403:339–342. [DOI] [PubMed] [Google Scholar]
- 6.Khalil AS, Collins JJ: Synthetic biology: applications come of age. Nat Rev Genet 2010, 11:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guet CC, Elowitz MB, Hsing W, Leibler S: Combinatorial synthesis of genetic networks. Science 2002, 296:1466–1470. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald IC, Deans TL: Tools and applications in synthetic biology. Adv Drug Deliv Rev 2016, 105:20–34. [DOI] [PubMed] [Google Scholar]
- 9.Bush LM, Healy CP, Javdan SB, Emmons JC, Deans TL: Biological cells as therapeutic delivery vehicles. Trends Pharmacol Sci 2021, 42:106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent RL, Gurbatri CR, Li F, Vardoshvili A, Coker C, Im J, Ballister ER, Rouanne M, Savage T, de Los Santos-Alexis K, et al. : Probiotic-guided CAR-T cells for solid tumor targeting. Science 2023, 382:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoang DM, Pham PT, Bach TQ, Ngo ATL, Nguyen QT, Phan TTK, Nguyen GH, Le PTT, Hoang VT, Forsyth NR, et al. : Stem cell-based therapy for human diseases. Signal Transduct Target Ther 2022, 7:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullard A: FDA approves first CAR T therapy. Nat Rev Drug Discov 2017, 16:669. [DOI] [PubMed] [Google Scholar]
- 13.••.Baker DJ, Arany Z, Baur JA, Epstein JA, June CH: CAR T therapy beyond cancer: the evolution of a living drug. Nature 2023, 619:707–715. [DOI] [PubMed] [Google Scholar]; A comprehensive review for using CAR T cells as living drugs.
- 14.Irvine DJ, Maus MV, Mooney DJ, Wong WW: The future of engineered immune cell therapies. Science 2022, 378:853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parayath NN, Stephan MT: In situ programming of CAR T cells. Annu Rev Biomed Eng 2021, 23:385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin H, Song CQ, Suresh S, Wu Q, Walsh S, Rhym LH, Mintzer E, Bolukbasi MF, Zhu LJ, Kauffman K, et al. : Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol 2017, 35:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhary N, Weissman D, Whitehead KA: mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov 2021, 20:817–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.•.McGee JE, Kirsch JR, Kenney D, Chavez E, Shih TY, Douam F, Wong WW, Grinstaff MW: Complete substitution with modified nucleotides suppresses the early interferon response and increases the potency of self-amplifying RNA. bioRxiv; 2023.. [Google Scholar]; A study showing multiple modified nucleotides that can be incorporated into saRNA to improve immune tolerance and translation rate.
- 19.Kariko K, Buckstein M, Ni H, Weissman D: Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23:165–175. [DOI] [PubMed] [Google Scholar]
- 20.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Kariko K: Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 2010, 38:5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D: Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 2008, 16:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain S, Venkataraman A, Wechsler ME, Peppas NA: Messenger RNA-based vaccines: past, present, and future directions in the context of the COVID-19 pandemic. Adv Drug Deliv Rev 2021, 179:114000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin U, Kariko K, Tureci O: mRNA-based therapeutics — developing a new class of drugs. Nat Rev Drug Discov 2014, 13:759–780. [DOI] [PubMed] [Google Scholar]
- 24.Siuti P, Yazbek J, Lu TK: Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol 2013, 31:448–452. [DOI] [PubMed] [Google Scholar]
- 25.Jurkowski TP, Ravichandran M, Stepper P: Synthetic epigenetics-towards intelligent control of epigenetic states and cell identity. Clin Epigenetics 2015, 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goell JH, Hilton IB: CRISPR/Cas-based epigenome editing: advances, applications, and clinical utility. Trends Biotechnol 2021, 39:678–691. [DOI] [PubMed] [Google Scholar]
- 27.Thakore PI, Black JB, Hilton IB, Gersbach CA: Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods 2016, 13:127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF 3rd: Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth Biol 2014, 3:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farzadfard F, Perli SD, Lu TK: Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2013, 2:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ: A synthetic biology framework for programming eukaryotic transcription functions. Cell 2012, 150:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon JB, Vankara A, Ettyreddy AR, Bohning JD, Gersbach CA: Myogenic progenitor cell lineage specification by CRISPR/Cas9-based transcriptional activators. Stem Cell Rep 2020, 14:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auslander S, Fussenegger M: From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol 2013, 31:155–168. [DOI] [PubMed] [Google Scholar]
- 33.Chin JW: Programming and engineering biological networks. Curr Opin Struct Biol 2006, 16:551–556. [DOI] [PubMed] [Google Scholar]
- 34.Ruder WC, Lu T, Collins JJ: Synthetic biology moving into the clinic. Science 2011, 333:1248–1252. [DOI] [PubMed] [Google Scholar]
- 35.Deans TL: Parallel networks: synthetic biology and artificial intelligence. ACM J Emerg Technol Comput Syst (JETC) 2014, 11:21. [Google Scholar]
- 36.••.Cabera A, Edelstein HI, Glykofrydis F, Love KS, Palacios S, Tycko J, Zhang M, Lensch S, Shields CE, Livingston M, et al. : The sound of silence: transgene silencing in mammalian cell engineering. Cell Syst 2022, 13:950–973. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review on transgene silencing.
- 37.Fitzgerald M, Livingston M, Gibbs C, Deans TL: Rosa26 docking sites for investigating genetic circuit silencing in stem cells. Synth Biol 2020, 5:ysaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calero-Nieto FJ, Bert AG, Cockerill PN: Transcription-dependent silencing of inducible convergent transgenes in transgenic mice. Epigenetics Chromatin 2010, 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godecke N, Zha L, Spencer S, Behme S, Riemer P, Rehli M, Hauser H, Wirth D: Controlled re-activation of epigenetically silenced Tet promoter-driven transgene expression by targeted demethylation. Nucleic Acids Res 2017, 45:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kues WA, Schwinzer R, Wirth D, Verhoeyen E, Lemme E, Herrmann D, Barg-Kues B, Hauser H, Wonigeit K, Niemann H: Epigenetic silencing and tissue independent expression of a novel tetracycline inducible system in double-transgenic pigs. FASEB J 2006, 20:1200–1202. [DOI] [PubMed] [Google Scholar]
- 41.Manhas J, Edelstein HI, Leonard JN, Morsut L: The evolution of synthetic receptor systems. Nat Chem Biol 2022, 18:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javdan SB, Deans TL: Design and development of engineered receptors for cell and tissue engineering. Curr Opin Syst Biol 2021, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim WA: The emerging era of cell engineering: harnessing the modularity of cells to program complex biological function. Science 2022, 378:848–852. [DOI] [PubMed] [Google Scholar]
- 44.••.Makri Pistikou AM, Cremers GAO, Nathalia BL, Meuleman TJ, Bogels BWA, Eijkens BV, de Dreu A, Bezembinder MTH, Stassen O, Bouten CCV, et al. : Engineering a scalable and orthogonal platform for synthetic communication in mammalian cells. Nat Commun 2023, 14:7001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper introduces an orthogonal communication platform using synthetic receptors that respond to coiled coils.
- 45.Schmidt CM, Smolke CD: RNA switches for synthetic biology. Cold Spring Harb Perspect Biol 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dykstra PB, Kaplan M, Smolke CD: Engineering synthetic RNA devices for cell control. Nat Rev Genet 2022, 23:215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endo K, Hayashi K, Inoue T, Saito H: A versatile cis-acting inverter module for synthetic translational switches. Nat Commun 2013, 4:2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawasaki S, Fujita Y, Nagaike T, Tomita K, Saito H: Synthetic mRNA devices that detect endogenous proteins and distinguish mammalian cells. Nucleic Acids Res 2017, 45:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, Furushima R, Inoue T: Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat Chem Biol 2010, 6:71–78. [DOI] [PubMed] [Google Scholar]
- 50.Ono H, Kawasaki S, Saito H: Orthogonal protein-responsive mRNA switches for mammalian synthetic biology. ACS Synth Biol 2020, 9:169–174. [DOI] [PubMed] [Google Scholar]
- 51.Wroblewska L, Kitada T, Endo K, Siciliano V, Stillo B, Saito H, Weiss R: Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat Biotechnol 2015, 33:839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.•.Ono H, Saito H: Sensing intracellular signatures with synthetic mRNAs. RNA Biol 2023, 20:588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review on using RNA to sense intracellular molecules.
- 53.Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ: Paper-based synthetic gene networks. Cell 2014, 159:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green AA, Silver PA, Collins JJ, Yin P: Toehold switches: de-novo-designed regulators of gene expression. Cell 2014, 159:925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.••.Zhao EM, Mao AS, de Puig H, Zhang K, Tippens ND, Tan X, Ran FA, Han I, Nguyen PQ, Chory EJ, et al. : RNA-responsive elements for eukaryotic translational control. Nat Biotechnol 2022, 40:539–545. [DOI] [PubMed] [Google Scholar]; This paper demonstrates a mammalian toehold switch that is capable of sensing and responding to viral infections and discriminating cell states.
- 56.Bloom K, van den Berg F, Arbuthnot P: Self-amplifying RNA vaccines for infectious diseases. Gene Ther 2021, 28:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.•.Mc Cafferty S, De Temmerman J, Kitada T, Becraft JR, Weiss R, Irvine DJ, Devreese M, De Baere S, Combes F, Sanders NN: In vivo validation of a reversible small molecule-based switch for synthetic self-amplifying mRNA regulation. Mol Ther 2021, 29:1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates the use of synthetic self-amplifying RNA that can control mRNA kinetics in vivo.
- 58.Wesselhoeft RA, Kowalski PS, Anderson DG: Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun 2018, 9:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Unti MJ, Jaffrey SR: Highly efficient cellular expression of circular mRNA enables prolonged protein expression. Cell Chem Biol 2024, 31:163–176 e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loan Young T, Chang Wang K, James Varley A, Li B: Clinical delivery of circular RNA: lessons learned from RNA drug development. Adv Drug Deliv Rev 2023, 197:114826. [DOI] [PubMed] [Google Scholar]
- 61.Henderson NC, Rieder F, Wynn TA: Fibrosis: from mechanisms to medicines. Nature 2020, 587:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.••.Rurik JG, Tombacz I, Yadegari A, Mendez Fernandez PO, Shewale SV, Li L, Kimura T, Soliman OY, Papp TE, Tam YK, et al. : CAR T cells produced in vivo to treat cardiac injury. Science 2022, 375:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses LNPs to deliver mRNA for producing CAR T cells in vivo.
- 63.Chivu-Economescu M, Rubach M: Hematopoietic stem cells therapies. Curr Stem Cell Res Ther 2017, 12:124–133. [DOI] [PubMed] [Google Scholar]
- 64.••.Breda L, Papp TE, Triebwasser MP, Yadegari A, Fedorky MT, Tanaka N, Abdulmalik O, Pavani G, Wang Y, Grupp SA, et al. : In vivo hematopoietic stem cell modification by mRNA delivery. Science 2023, 381:436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses LNPs to deliver mRNA to HSCs to edit blood progenitor cells in vivo.
- 65.Ferrari S, Naldini L: A step toward stem cell engineering in vivo. Science 2023, 381:378–379. [DOI] [PubMed] [Google Scholar]
- 66.Bush LM, Healy CP, Marvin JE, Deans TL: High-throughput enrichment and isolation of megakaryocyte progenitor cells from the mouse bone marrow. Sci Rep 2021, 11:8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang G, Liu F: Long-term expansion of human hematopoietic stem cells. Cell Regen 2023, 12:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.••.Su FY, Zhao QH, Dahotre SN, Gamboa L, Bawage SS, Silva Trenkle AD, Zamat A, Phuengkham H, Ahmed R, Santangelo PJ, et al. : In vivo mRNA delivery to virus-specific T cells by light-induced ligand exchange of MHC class I antigen-presenting nanoparticles. Sci Adv 2022, 8:eabm7950. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper designs nanoparticles for targeting multiple T cell epitopes by making recombinant peptide MHC class I complexes.
- 69.Kumah EA, Fopa RD, Harati S, Boadu P, Zohoori FV, Pak T: Human and environmental impacts of nanoparticles: a scoping review of the current literature. BMC Public Health 2023, 23:1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park K: Facing the truth about nanotechnology in drug delivery. ACS Nano 2013, 7:7442–7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stranford DM, Simons LM, Berman KE, Cheng L, DiBiase BN, Hung ME, Lucks JB, Hultquist JF, Leonard JN: Genetically encoding multiple functionalities into extracellular vesicles for the targeted delivery of biologics to T cells. Nat Biomed Eng 2023,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Javdan SB, Islam F, Deans TL: Reprogramming megakaryocytes to engineer platelets as delivery vehicles. bioRxiv; 2023. [Google Scholar]
- 73.Chen M, Leng Y, He C, Li X, Zhao L, Qu Y, Wu Y: Red blood cells: a potential delivery system. J Nanobiotechnol 2023, 21:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO: Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007, 9:654–659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.


