Abstract
CRISPR-Cas systems can be utilized as programmable-spectrum antimicrobials to combat bacterial infections. However, how CRISPR nucleases perform as antimicrobials across target sites and strains remains poorly explored. Here, we address this knowledge gap by systematically interrogating the use of CRISPR antimicrobials using multidrug-resistant and hypervirulent strains of Klebsiella pneumoniae as models. Comparing different Cas nucleases, DNA-targeting nucleases outperformed RNA-targeting nucleases based on the tested targets. Focusing on AsCas12a that exhibited robust targeting across different strains, we found that the elucidated modes of escape varied widely, restraining opportunities to enhance killing. We also encountered individual guide RNAs yielding different extents of clearance across strains, which were linked to an interplay between improper gRNA folding and strain-specific DNA repair and survival. To explore features that could improve targeting across strains, we performed a genome-wide screen in different K. pneumoniae strains that yielded guide design rules and trained an algorithm for predicting guide efficiency. Finally, we showed that Cas12a antimicrobials can be exploited to eliminate K. pneumoniae when encoded in phagemids delivered by T7-like phages. Altogether, our results highlight the importance of evaluating antimicrobial activity of CRISPR antimicrobials across relevant strains and define critical parameters for efficient CRISPR-based targeting.
Graphical Abstract
Graphical Abstract.
Introduction
The discovery and development of antimicrobial agents like antibiotics reshaped the medical field, allowing treatment for previously incurable infections and offering aid for surgeries, chemotherapy and organ transplants (1,2). However, the discovery pipeline for new antimicrobial agents has slowed down for both scientific and economic reasons, while the use and misuse of these agents has contributed to the rise of antibiotic resistance (2). The rise of antimicrobial resistance is recognized by the WHO and other global institutions as a major public health threat that calls for novel approaches (3,4).
Among the bacterial pathogens associated with antibiotic resistance, the ESKAPE group members Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp. are particularly concerning due to their ability to acquire and horizontally transfer multiple virulence and resistance genes (5,6). These bacteria are responsible for the majority of nosocomial infections with high mortality rates (3). Among these, Enterobacteriaceae that have acquired extended-spectrum β-lactamases and carbapenemases are considered of high priority (7). Indeed, the increasing occurrence of infections by these bacteria and lack of therapeutic options places an even greater urgency on the development of novel antimicrobials.
Among novel antimicrobial strategies, CRISPR-Cas systems have emerged as a promising means to specifically eliminate bacterial pathogens from a microbial community without affecting the surrounding bacteria (8). This property is advantageous compared to antibiotics, which generally kill multiple bacteria indiscriminately, thus altering the human microbiome and favoring the colonization of pathogenic bacteria (9,10). The programmability of CRISPR-Cas systems further offers advantages over lytic bacteriophages, which cannot be readily tailored to the genomic sequence of target bacteria. The specificity exhibited by CRISPR-Cas systems is granted by their natural role as adaptive immune systems (11,12). These systems store pathogen sequences (named spacers) flanked by palindromic recurrent sequences (named repeats) in a CRISPR array. Upon infection, the array is processed to generate an active guide RNA (gRNA) composed of a truncated spacer and repeat sequence. The gRNA directs a Cas nuclease to recognize a specific DNA or RNA sequence, thereby inducing an immune response (13–15).
There is a vast assortment of CRISPR-Cas systems, with known systems classified into two classes, seven types, and over 33 subtypes and variants (16). Each class comprises a multi-protein effector complex (class I) or a single-effector nuclease (class II) for immune defense. The type is generally identified based on the particular effector (e.g. Cas9 for type II systems), while the subtype captures different supporting cas genes or different phylogenies of the effector. Some systems act by targeting DNA (type I, II and most of type V), while others target RNA and have collateral activities (type III and VI), representing a broad range of activities that can be harnessed for different applications.
Within the diversity of CRISPR nucleases, the development of CRISPR antimicrobials has focused on type II Cas9 nucleases that introduce blunt cuts into the DNA, type VI Cas13 nucleases that drive cell dormancy through collateral RNA degradation and the type I multi-subunit complex Cascade that recruits Cas3 to nick and degrade a single DNA strand (10,17–23). Such nucleases have been used to either cure antibiotic-resistance plasmids or selectively kill cells by targeting chromosomal DNA or cellular RNA. In most studies, only one specific bacterial strain and one nuclease have been tested, with little guidance on which nuclease to select or how to design effective gRNAs. Delivery also remains a major challenge, with nuclease and gRNA delivery achieved mainly with temperate or lytic phages, conjugative plasmids or polymer-based and lipid nanoparticles (17,22,24–28). Phages have been the most widely utilized to-date, and the range of bacteria they can infect has been expanded through engineering of the tail fiber proteins to increase their efficiency across several strains (29–31). These advances set the stage to explore how to effectively develop and implement CRISPR antimicrobials, particularly in clinically-relevant pathogens in which new antimicrobial agents are needed.
We aimed to take this next step using clinically-relevant strains of Klebsiella pneumoniae (Kp) as case studies. After comparing the activity of different nucleases in one Kp strain, we selected AsCas12a as one of the best antimicrobials. We then characterized how Kp escapes genome targeting, finding that mutational frequencies depend on the nuclease, gRNA or target and vary with the selected targeted chromosomal site. We next found that gRNAs could behave differently across multidrug-resistance (MDR) or hypervirulent (HV) Kp strains, which we linked to gRNA folding and double-stranded (ds)DNA repair. By combining a high-throughput screen with machine learning, we elucidated gRNA design rules specific for antimicrobial function in Kp and developed an algorithm that accurately predicts guide activity. Finally, by packaging CRISPR-encoding phagemids into replication-deficient phages having Kp-infecting tail fibers, we demonstrate that the CRISPR-Cas system can be delivered efficiently and exert its antimicrobial function. These findings lay the groundwork for the development of robust CRISPR antimicrobials across targeted strains.
Materials and methods
Strains
The Kp strains used in this study are Kp10031 (ATCC 10031), KPPR1 (ATCC 43816), CIP 52.145 (32), SB5442 and SB5961 (33), and NTUH-K2044 (34). Additionally, the Kp10031ΔrecA and SB5442ΔrecA strains were generated by lambda-Red recombineering (35). The recombineering protocol was adjusted by using the pKD46-hygR + flp plasmid to excise the kanR gene from the genome instead of the traditional pCP20 plasmid. The donor E. coli strain used in the library screen is E. coli MFDpir (36). Other strain information is reported in Supplementary Table S6. Cells were grown in LB medium at 37°C shaking at 220 rpm. The phage strain used in this study is T7Δ(11–12–17) (30), which is produced in an E. coli BW25113 strain encoding tail fibers on a plasmid.
Plasmids
Spacers were inserted into the gRNA backbone by digestion with BsmBI and ligation with Instant Sticky-end Ligase Master Mix (NEB, M0370S). The nuclease and the gRNA plasmid were modified with Q5 mutagenesis or Gibson assembly. Plasmids and oligos used in this study are listed in Supplementary Table S6.
Transformation-based targeting assays
Three biological replicates have been used for each targeting assay. Single colonies containing the nuclease plasmid were inoculated overnight in LB medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl in 1 l of dH2O) with chloramphenicol (Cm, 34 μg/ml) or hygromycin (Hyg, 100 μg/ml). The next day the samples were normalized based on their OD600, back-diluted 1:50 in fresh LB with Cm/Hyg and 0.7 mM EDTA (pH 8) and grown to an OD600 of ∼0.4. Cultures were placed on ice for at least 10 min before making them electrocompetent by pelleting and washing them twice with 10% glycerol. Then, 50 ng of the crRNA plasmids (having a kanR marker) were transformed into 40 μl of competent cells using the E. coli 1 program on the Gene Pulser Xcell Electroporator (Bio-rad). After 1 h of recovery in 500 μl of SOC medium (SOB medium: 20 g of Tryptone, 5 g of Yeast Extract, 0.5 g of NaCl, 800 ml of dH2O, and 10 ml of 250 mM KCl adjusted to pH 7. To SOB medium add 5 ml of 2 M MgCl2 and 20 ml of 1 M glucose), 5-fold dilutions of the cultures in 1× PBS (10× PBS: 80 g of NaCl, 2 g of KCl, 17.7 g of Na2HPO4·2H2O, 2.72 g of KH2PO4, fill up to 1 l with mqH2O, set pH to 7.4 and autoclave) were prepared, and 5-μl spot dilutions were plated on Cm/Hyg and kanamycin (Kan, 50 μg/ml) LB plates (prepared by mixing 1 l of LB medium with 18 g of Agar) and incubated at 37°C for 16–18 h. The colony number was counted and multiplied by the dilution row to get the number of transformants. The negative control was either a plasmid lacking a spacer or having a non-targeting spacer forming a hairpin.
Flow cytometry analysis
Cells expressing the nuclease and an empty gRNA plasmid were inoculated overnight in a selective LB medium and grown overnight at 37°C under shaking at 220 rpm. The cultures were normalized the next day after measuring the OD600, diluted to OD600 = 0.05, and grown until OD600 ∼ 0.6, after which 20 ul have been added to 180 ul of 1× PBS. A gate has been created on living cells and the mean fluorescence (FL1-H, 30 000 events) has been measured at the Novocyte flow cytometer (Agilent technologies). Three biological replicates have been used for each condition and GFP-positive colonies have been normalized with the correspondent cells not expressing GFP.
RT-qPCR
Three biological replicates of cells expressing the gRNA plasmid have been grown overnight at 37°C at 220 rpm. After measuring the OD600, the cells were normalized to reach the same concentration and back-diluted to OD600 0.05, after which they were grown until OD600 ∼ 0.4, placed on ice for 10 min, and pelleted by centrifugation at 3500 × g for 10 min. The cells have been snap-frozen in liquid nitrogen and placed at −80°C. The following day, total RNA was extracted using the Direct-zol RNA Miniprep Plus kit (Zymo Research, R2072), and DNase-treated with the Turbo DNA-free kit (Thermo Fisher Scientific, AM1907). Isolated RNAs (25 ng) have been used to run an RT-qPCR reaction (iTaq Universal SYBR Green One-Step Kit, Bio-Rad, 1725150). Two technical replicates have been used for each sample and recA and rho have been used as reference genes using already established primers (37). The primers used in the RT-qPCR reactions can be found in Supplementary Table S6.
Library design and cloning
The library was designed to target a non-redundant collection of complete genomes of K. pneumoniae (38), by randomly choosing targets with a proper TTTV PAM shared by all the genomes in the collection. Guides matching at least 15 bp with the E. coli MG1655 genome were removed. An oligo pool (Twist Bioscience) containing 12 000 members (11 900 targets and 100 non-targeting guides) was constructed by Golden Gate cloning and transformed into E. coli MG1655. At least 106 clones were recovered (∼100× coverage), pooled in 10 ml LB with Kan, and stored as 1 ml aliquots in DMSO 10% at −80°C. The plasmid library was then extracted from 1 ml of pooled colonies using a miniprep kit (Macherey-Nagel) and electroporated into E. coli MFDpir. The transformed cells were then plated in LB with Kan and diaminopimelic acid (DAP) 0.3 uM, and incubated at 37°C obtaining at least 106 clones (∼100× coverage). Subsequently, the colonies were pooled in 10 ml LB with Kan and stored as 1 ml aliquots in DMSO 10% at −80°C. The plasmids used in the library screen can be found in Supplementary Table S6.
Guide library screen
The plasmid library was delivered by conjugation to three K. pneumoniae receptor strains (NTUH-K2044, KPPR1 and SB5442). Briefly, 108 donor cells were used to inoculate 50 ml of LB with Kan and DAP 0.3 uM, and cultivated at 37°C and 190 rpm until they reached the late-exponential phase (OD600 ≈ 1). Receptor cells were diluted 100X from an overnight culture and grown in LB until late exponential phase (OD600 ≈ 1), and 1:1 volume ratio of donor and receptor were mixed. For each sample, enough conjugations were performed to achieve at least 100× coverage of the library. The cells were plated and collected after 4 h of incubation at 37°C in LB agar with Kan, then pooled in 10 ml LB and stored as 1 ml aliquots in DMSO 10% at −80°C. To perform the assay, 1 ml of cells from −80°C was inoculated in 100 ml LB with Kan and grown to an OD600 of ∼0.2 at 37°C. Then, 1 μM of anhydrotetracycline (aTc) was added and cells were recovered at time zero, after 3 and 16 h. Finally, plasmids were extracted from 10 ml of these cultures and the donor strain culture (collected before conjugation) using a miniprep kit (Qiagen).
Guide library next-generation sequencing
To measure the relative abundance of guide RNAs in each sample, the samples were prepared for next-generation sequencing. Briefly, two nested PCR reactions were performed using Phusion High Fidelity Polymerase (Thermo Fisher Scientific). The first PCR was run for 25 cycles (10 ng plasmid DNA, 98°C for 10 s denaturation, 60°C for 30 s annealing, 72°C for 15 s extension), while in the second PCR the sample (5 μl) was amplified for 8 cycles under the same settings. The used primers are described in Supplementary Table S6. The first set amplified the sgRNA region adding a variable region to increase nucleotide diversity for optimal clustering. The second PCR added the indexes and flow cells' attachment sequences. The PCR primers are listed in Supplementary Table S6. After each PCR reaction, the samples were cleaned using AMPure beads (Beckman Coulter, A63881). Sequencing was then performed with Illumina sequencing primers using a NextSeq 500 benchtop sequencer.
Library screen analysis and machine learning
For analysis of the genome-wide screen, we considered the reads carrying a guide with a perfect match to a guide of the library. At 0 h, 11 722–11 742 guides (∼97%) were detected. After filtering guides for at least one count per million reads in at least two samples, the library sizes were normalized using the read counts for 86 remaining non-targeting guides with the trimmed mean of M-values (TMM) method in edgeR (version 3.38.1) (39,40). Differential abundance of 11 042 gRNAs between different time points (3 h–0 h and ON (overnight)–0 h) was assessed using the edgeR quasi-likelihood F test after fitting a generalized linear model.
The machine learning regression model was developed as previously described (41) with 738 features as predictors and the logFC values between ON and 0 h of gRNAs from the genome-wide screens in individual or both strains as targets using auto-sklearn version 0.14.6 (42) with all possible estimators included, all feature preprocessors excluded and parameters ‘ensemble_size’: 1, ‘resampling_strategy’: ‘cv’, ‘resampling_strategy_arguments’: {‘folds’: 10}, ‘per_run_time_limit’: 360 for individual dataset and 1000 for mixed datasets, and ‘time_left_for_this_task’: 3600 for individual dataset and 10 000 for mixed datasets. Features included dataset, gRNA GC content, three thermodynamic features (delta G of the hybridization of repeat-gRNA and target DNA, delta G of the homodimer of repeat-gRNA, and delta G of the monomer of repeat-gRNA), the number of consecutive nucleotides, the 732 one-hot-encoded single-nucleotide and dinucleotide features. Thermodynamic features were computed using the ViennaRNA Package (43): RNAduplex (version 2.4.14) for RNA:RNA hybrids; RNAduplex (version 2.1.9h) for DNA:RNA hybrids (44); RNAfold (version 2.4.14) for single RNA folding. 732 sequence features included the variable 4th position of the PAM, the 26 positions of the gRNA, and dinucleotide features from 5 nts upstream of the PAM to 5 nts downstream of the gRNA. When the data from only one screen were used to train, the dataset feature was removed. The optimal histogram-based gradient boosting model was evaluated using 10-fold cross-validation based on unique gRNA sequences and interpreted using TreeSHAP version 0.39.0 (45).
Production and testing of T7 transducing particles with different tail fibers
T7 transducing particles with different tail proteins were prepared as previously described (30). Briefly, ON culture of E. coli BW25113, harboring a plasmid encoding a T7 packaging signal and tail proteins were diluted 1:30 in LB supplemented with appropriate antibiotics and aerated at 37°C for an additional hour. The culture was then infected by ∼5 × 108 PFU of T7Δ(11–12-17) and aerated for ∼2 h at 37°C until lysis occurred. For the transduction assay, recipient cells in their exponential growth phase were mixed in a 1:1 ratio (v/v) with serial dilutions of the described T7 lysates. Cultures were incubated for 60 min at 37°C with shaking and plated on an LB-agar plate containing an appropriate antibiotic. Plates were incubated overnight at 37°C. The next day, colonies were counted, multiplied by 108 dilution factor, and normalized with the transductants numbers obtained in the indicated reference strain E. coli BW25113ΔtrxA to obtain TFU/ml. E. coli BW25113ΔtrxA lacks the host gene trxA that contributes to the T7 DNA polymerase (46), preventing lysis by WT T7 particles generated along with phagemid-harboring particles. The strain was used to normalize the number of transducing particles applied to each Kp strain as performed previously (30).
Targeting bacterial strains using CRISPR-harboring T7 particles
T7 lysates were prepared as described above except for the E. coli BW25113 which harbored two plasmids: (i) targeting plasmid with a T7 packaging signal, AsCpf1 Cas gene, and a guide RNA encoding sequence. (ii) tail-encoding plasmid with phiSG-JL2 tail genes, lacking a packaging signal. Transduction assays were also performed as described, using the T7 particle lysates with phiSG-JL2 tail carrying the targeting plasmid. TFU/ml was calculated relative to 108 transductants obtained for the non-targeting guide.
Data analysis and image visualization
The R and Python programming languages were used for analyzing the library screen data, while Microsoft Excel was used for the remaining data. GraphPad Prism was used to produce the scatter plots and pie charts. The graphs were then reformatted in Adobe Illustrator to generate the final figures with consistent sizing and formatting.
Results
Cas12a exhibits efficient antimicrobial activity in Klebsiella pneumoniae but yields variable escape mechanisms
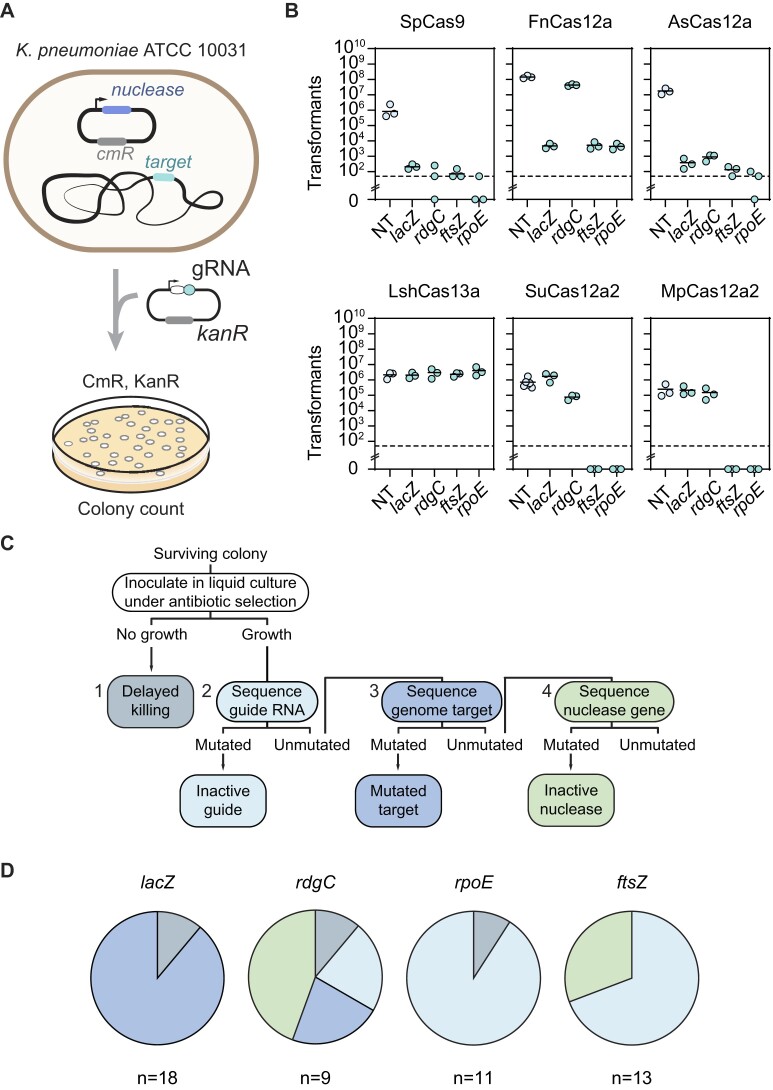
CRISPR-based antimicrobial activity relies on efficient cleavage of a target sequence located on the chromosome or on another essential replicon, or widespread collateral nucleic-acid degradation. In order to determine which Cas single-effector nucleases can maximize antimicrobial activity in Kp, we tested six distinct nucleases of varying types: the type II-A nuclease Cas9 from Streptococcus pyogenes (SpCas9), the type V-A nuclease Cas12a from Acidaminococcus species (AsCas12a) and from Francisella novicida (FnCas12a), the type V-A variant nuclease Cas12a2 from Sulfuricurvum sp. PC08-66 (SuCas12a2) and from a microbial community of Microcerotermes parvus (MpCas12a2) and the type VI-A nuclease from Leptotrichia shahii (LshCas13a). SpCas9 recognizes NGG PAM sequences and generates blunt dsDNA cuts, while AsCas12a and FnCas12a recognize T-rich PAM sequences and generate staggered dsDNA cuts (47–50). Target recognition by Cas12a nucleases elicits non-specific single-stranded DNA (ssDNA) cleavage, although this activity has not been associated with immune defense (51). In contrast, Cas12a2 and Cas13a target RNA flanked by a protospacer-flanking sequence (PFS), where target recognition elicits collateral cleavage activity against RNA, ssDNA, and dsDNA or just RNA, respectively, that leads to cell dormancy (52–55).
To assess the antimicrobial activity of each nuclease, the Kp strain ATCC 10031 (herein called Kp10031) was first transformed with a plasmid constitutively expressing the nuclease. The cells were then transformed with a plasmid encoding a gRNA targeting the bacterial genome or genomically-encoded transcript, and the number of antibiotic-resistant colonies was quantified (Figure 1A). Target sites were selected following PAM/PFS selection and guide length unique to each nuclease, which lent to non-overlapping and sometimes distant sites within a given gene. For SpCas9, the gRNA was expressed as a single-guide RNA (sgRNA) (47). As part of these experiments, SpCas9 and AsCas12a displayed strong antimicrobial activity, with a respective ∼103–104-fold and ∼104–105-fold reduction in transformants compared to a non-targeting gRNA control (Figure 1B). In contrast, Cas13a exhibited negligible antimicrobial activity for all targets, possibly due to insufficient nuclease or target expression as we reported previously (56). Finally, SuCas12a2 and MpCas12a2 exhibited a strong reduction in colony numbers for the two essential gene targets (>104-fold and >103-fold, respectively) but a negligible reduction for the two non-essential gene targets. Nucleases exhibiting strong reductions could not be further distinguished due to the limit-of-detection of the experimental setup. Ultimately, AsCas12a was selected for further use, as it has been widely characterized and employed for CRISPR technologies, and it yielded the strongest antimicrobial activity in Kp for both essential and non-essential gene targets. These findings demonstrate that CRISPR-Cas antimicrobials can be successfully applied to Kp10031, albeit with variable efficiencies depending on the adopted Cas nuclease and target.
Figure 1.
DNA-targeting nucleases outperform RNA-targeting nucleases as CRISPR antimicrobials for the selected targets, with AsCas12a exhibiting high antimicrobial activity against K. pneumoniae that drives infrequent but variable escape. (A) Experimental setup for the transformation-based targeting assay. (B) Drop in transformants following genome targeting with different Cas single-effector nucleases. Each dot represents a biological replicate obtained by preparing electrocompetent cells from a separate colony. ftsZ and rpoE: essential genes. rdgC and lacZ: non-essential genes. The dashed lines represent the limit-of-detection for colony numbers. The bars represent average values and are absent for samples in which at least one replicate had no colonies. (C) Flowchart of procedure to identify escape mutants. The mutations were screened following the indicated order until an escape mutation was identified. (D) Pie chart of the modes of escape from targeting for the different gRNAs. n indicates the number of screened colonies per condition.
While AsCas12a exhibited a consistently strong reduction in colony counts across the tested genomic targets, we still observed surviving colonies that presumably circumvented lethal attack by this nuclease. Such escape has been noted for other CRISPR antimicrobials and is normally attributed to mutations in the target region, the CRISPR nuclease, or the gRNA (8,17,19,22,57). To explore the modes of escape for AsCas12a in Kp, we picked colonies following the targeting of the same two non-essential genes (lacZ, rdgC) and two essential genes (rpoE, ftsZ) of the previous experiment. We then screened for escape mutations starting with the gRNA followed by the target and then the nuclease (Figure 1C). The screening was halted as soon as a disruptive mutation was detected.
The modes of escape varied with the employed gRNA and target (Figure 1D and Supplementary Figure S1A). For the essential targets, escape was dominated by mutations that inactivate the gRNA, including mutations in the repeat or the first nucleotide of the guide for the ftsZ target, and mutations within the repeat or deletion of the entire gRNA for the rpoE target (Supplementary Figure S1A). The rdgC and ftsZ targets also yielded escape through disruption of the Ascas12a gene via premature stop codons, small deletions, or insertion of endogenous transposable elements, paralleling prior studies (8,22) (Supplementary Table S1). For the non-essential lacZ target, escape was dominated by deletions that removed the target, including deletions exceeding 20 kb. For the other non-essential rdgC target, the escape mutants fell into all three categories. Finally, some of the colonies did not grow when inoculated into a liquid culture, possibly due to delayed killing or the presence of mutations that prevent further growth in liquid culture with antibiotic selection. Altogether, escape mutants follow different mechanisms in a target-dependent manner and may be difficult to completely avoid. However, Cas12a displayed strong antimicrobial activity in Kp, with mutational escape occurring at a low frequency (<0.01%) in our experimental setup. Furthermore, prior work did not observe escape mutants when testing phage-delivered CRISPR antimicrobials (17,74), even though such mutants were observed as part of plasmid transformation. Therefore, the escape observed when testing individual gRNAs via plasmid transformation may not be a major practical concern for downstream applications of CRISPR antimicrobials.
Guides can yield strain-dependent antimicrobial activity
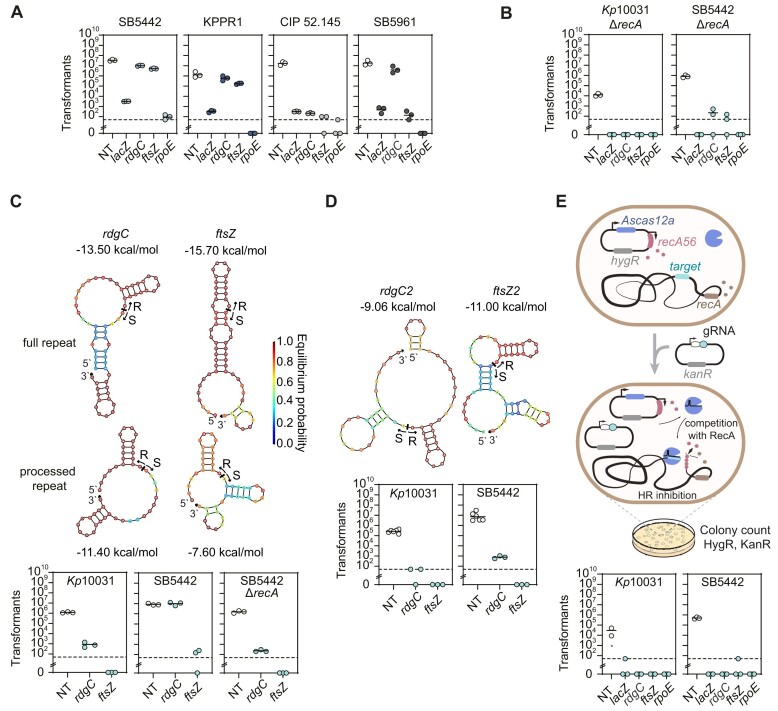
Studies on CRISPR antimicrobials commonly investigate the targeting activity of antimicrobials on single bacterial strains and show a lack of targeting for the surrounding strains (17,19,21,57). However, a pathogenic species typically comprises numerous strains, which could not only differ in antibiotic resistance or virulence but also possess genotypic and phenotypic differences that impact CRISPR antimicrobial activity. To begin exploring this possibility, we asked how the same AsCas12a gRNAs function in different MDR (SB5442 and ATCC 43816) and HV (SB5961 and CIP 52.145) Klebsiella strains.
After establishing selection with hygromycin and confirming homology of the four tested gRNAs to each respective target, we applied the same transformation-based targeting assay to characterize each gRNA’s antimicrobial activity. Surprisingly, the ftsZ and rdgC guides behaved differently across strains, displaying comparable activity to Kp10031 (∼105-fold reduction) in CIP 52.145, poor activity (<10-fold reduction) in SB5442 and ATCC 43816 and variable activity (∼105-fold reduction for ftsZ and < 10-fold reduction for rdgC) in SB5961 (Figure 2A). The strain-specific impact of these gRNAs could not be explained by nuclease expression as measured by flow cytometry analysis of a transcriptional reporter, since the expression did not correlate with the observed transformation fold-reductions (Supplementary Figure S2A). Likewise, the strain-specific impact could not be explained by gRNA expression as measured by RT-qPCR, since the transformation fold-reduction was higher in Kp10031 than in other strains with equal or greater gRNA expression (i.e. SB5442, SB5961 and KPPR1) (Figure 2A and Supplementary Figure S2B).
Figure 2.
AsCas12a guides can exhibit variable behavior across strains that can be countered by blocking the homologous recombination pathway or opening the predicted secondary structure. (A) Transformation-based targeting assay in different Klebsiella pneumoniae strains with the same four guides used for K. pneumoniae ATCC 10031. (B) Targeting assay in two Klebsiella strains with the recA gene deleted. (C) Folding predictions and targeting assay with gRNAs with processed repeats against the rdgC and ftsZ genes. (D) Folding predictions and targeting assay with gRNA guides with open secondary structure and against the rdgC and ftsZ genes. (E) Targeting assay with two Klebsiella strains expressing the dominant negative inhibitor of the RecA protein (RecA56). In all the experiments, the dashed lines represent the limit of detection of colony numbers. *: The sample yielded a lawn that was uncountable. ftsZ and rpoE: essential genes. rdgC and lacZ: non-essential genes. Each dot represents a biological replicate obtained by preparing electrocompetent cells from a separate colony. The bars represent average values and are absent for samples in which at least one replicate had no colonies.
Another possibility is that variable survival could be linked to how well the cells repair dsDNA cleavage by AsCas12a via homology-directed repair (HDR) (57,58). Such a repair mechanism is mediated by the RecA protein, so disrupting the encoding recA gene and applying CRISPR targeting should be cytotoxic. Therefore, we knocked out recA in Kp10031 as well as in the MDR strain SB5442 in which the ftsZ and rdgC guides had yielded negligible fold-reduction. Performing the transformation-based targeting assay in the SB5442 ΔrecA strain, we observed restored targeting activity for all the guides, with ∼104-fold reduction in colony numbers (Figure 2B). We also observed > 102-fold reduction for strain Kp10031 ΔrecA. The colony number decreased also for the non-targeting control, indicating reduced fitness of this deletion mutant in general. Thus, we inferred that the guides are active in the different Kp strains, although some guides yield variable antimicrobial activity that is RecA-dependent.
While exploring different potential explanations for the strain-specific activity, we noticed hairpins within the rdgC- and ftsZ-targeting gRNAs, with the rdgC guide forming an internal hairpin and the ftsZ guide pairing with the repeat (Figure 2C). We hypothesized that the hairpins were hindering different steps leading up to DNA cleavage, such as gRNA binding or processing by AsCas12a (59,60). Strain-specific differences, such as the ability to recognize and repair the resulting dsDNA breaks, could then account for the differential antimicrobial activity of these attenuated gRNAs.
To initially test this hypothesis, we encoded the two guides with a processed repeat, where the processed version removes the hairpin predicted in the ftsZ gRNA but not the one predicted in the rdgC gRNA. We then repeated the transformation assay in Kp10031 and the wild-type and SB5442 ΔrecA strains (Figure 2C). The processed ftsZ-targeting gRNA yielded strong antimicrobial activity (∼104–105-fold transformation reduction) in all three strains, in line with the folded secondary structure inhibiting gRNA processing and consequently targeting activity. As expected, the processed rdgC-targeting gRNA exhibited the same trend as the unprocessed gRNA given the persistence of the predicted internal hairpin (Figure 2C). Building on this finding, we designed guides directed to the same two genes but with predicted lower folding free energies that should adopt a structure less prone to internal hairpin formation (Figure 2D). As part of the transformation assay, both gRNAs promoted robust antimicrobial activity in both strains (>103 and >104-fold decrease in colony counts in Kp10031 and SB5442, respectively). These results suggest that poor gRNA folding can reduce targeting activity, which can lead to strain-dependent survival based on variable capacities to repair and survive dsDNA breaks.
As antimicrobial activity depends both on targeting efficiency and DNA break repair, we explored ways to block repair without altering the targeted bacterial genome. Following prior efforts to inhibit RecA-mediated repair (57), we expressed a dominant-negative inhibitor of RecA, RecA56, that blocks HDR by competing with the binding of the RecA protein to ssDNA (61). Expressing RecA56 as part of the targeting assay decreased colony counts for all tested targeting gRNAs, but also to a lesser extent for the non-targeting gRNA (Figure 2E). The reduction for the non-targeting gRNA was more evident for Kp10031 and mirrored the toxicity observed when knocking out recA (Figure 2B). We alternatively tried to express the Gam protein from Mu phage known to hinder HDR (57). However, its constitutive expression proved cytotoxic since we could not obtain viable colonies. We deduce that altering the HDR pathway can help boost antimicrobial activity but has repercussions on cell growth. Future work could focus on transiently expressing the RecA inhibitor to enhance the potency of CRISPR antimicrobials without severely impacting the viability of non-target cells. Altogether, these findings indicate that gRNAs can yield different outcomes in highly similar strains, although these differences can be overcome through proper gRNA design or inhibiting dsDNA break repair.
A guide library screen provides design rules for efficient antimicrobial activity
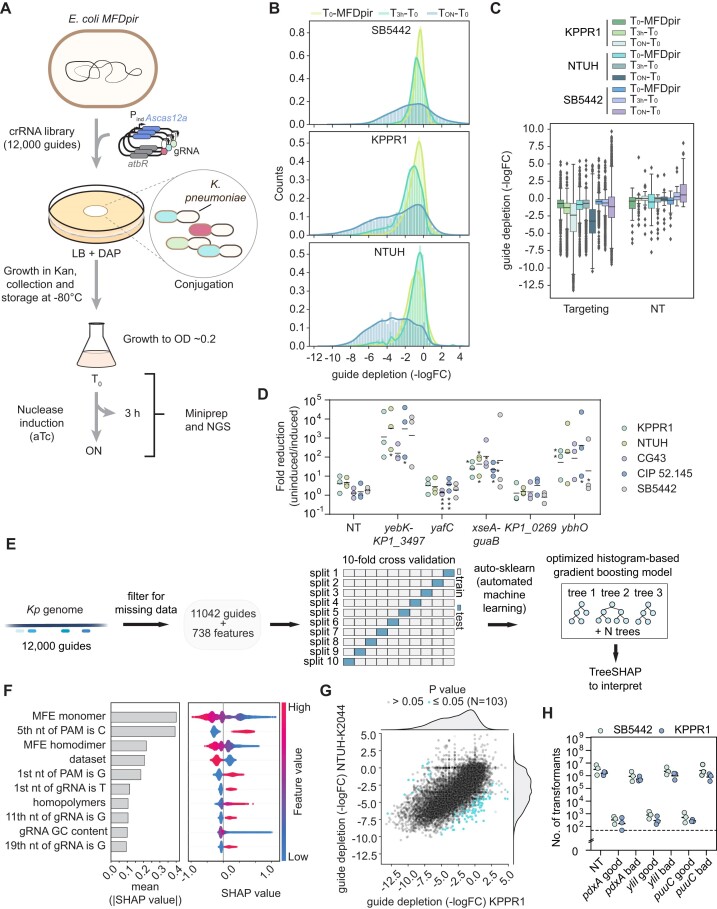
Given the relevance of guide design for consistent antimicrobial activity across strains, we next evaluated gRNA features associated with efficient clearance of Kp. Genome-wide guide screens have proven incredibly informative and formed the basis of design tools (62–64). However, they remain to be explored for CRISPR antimicrobial design, particularly across multiple strains.
To apply such a screen, we first designed 11900 guides with sequences matching the coding and non-coding chromosomal regions of two hypervirulent Kp strains (NTUH-K2044, KPPR1) and the MDR strain SB5442. Next, we transformed these three strains with a conjugative plasmid encoding the constitutively expressed gRNA library and inducibly expressed AsCas12a (Figure 3A). We induced nuclease expression for three hours or overnight and examined the extent of guide depletion as compared to non-targeting control guides within the library at each time point. We measured only minor changes in relative guide abundance across the library after three hours of induction and stronger guide depletion after overnight induction (−2.4, −3.3 and −1.2 logFC for KPPR1, NTUH-K2044 and SB5442 respectively). Therefore, we compared overnight induction to before induction for further analysis (Figure 3B, C). Strain SB5442 exhibited wider variability in the logFC of non-targeting guides as well as apparent enrichment of many targeting guides (Figure 3B, C, Supplementary Figure S3A) that could confound subsequent analyses, so we decided to focus only on the results from the KPPR1 and NTUH-K2044 strains.
Figure 3.
A library screen confirms differential depletion for some AsCas12a guides across strains and sets guide design rules for efficient antimicrobial activity. (A) Schematic of the library screen setup. (B) Depletion of guides following nuclease induction for 3 h or overnight. (C) Box plots of the guide depletion distribution before and after aTc nuclease induction for the targeting and NT guides. MFDpir: E. coli donor strain. KPPR1, SB5442 and NTUH-K2044 (indicated as NTUH): different Kp strains. T0, T3h, TON: time 0, 3 h, overnight after induction. (D) Library verification with five different gRNAs. After transforming the library into E. coli, five colonies were randomly selected, transformed into different Kp strains and induced to determine the reduction in colony numbers. Subsequently, they were sequenced to determine the expressed gRNA sequence and are reported in the initial assigned order. KP1_0269 indicates a gene encoding a putative protein, while xseA-guaB and yebK-KP1_3497 are placed in intergenic regions. The gRNA sequences can be found in Supplementary Table S1. *: small colonies on plates. (E) Depiction of the method used for developing the machine learning algorithm. (F) Contribution of different features to guide depletion from the library using the mixed dataset for training the algorithm. The left barplot depicts the absolute weight of each feature to the depletion values, while the right beeswarm plot shows how each datapoint impacts depletion according to each feature. The PAM nucleotides are counted from 3′ to 5′ starting from the −1 position before the gRNA sequence. (G) Correlation between guide depletion in the KPPR1 and NTUH-K2044 strains. (H) Validation of algorithm prediction of high and low-efficiency guides in KPPR1 and SB5442. The dashed line represents the limit of detection of the assay. Each dot represents a biological replicate obtained by preparing electrocompetent cells from a separate colony.
For the most depleted guides, targets were present in intergenic regions as well as in genes representing a broad range of classes, suggesting that the entire genome is available for designing guides as antimicrobials. To validate the screen, we tested five randomly selected gRNAs constructs from the library in several Kp strains (Figure 3D). The fold-reduction values of the tested gRNAs correlated with their depletion values in the screen (R2 = 0.763 for KPPR1 and R2 = 0.836 for NTUH-K2044) (Supplementary Table S2), supporting the validity of the screening results. We did observe generally large variability in the measured fold-reductions as part of our validation experiments, which we attribute to general variability between independent replicates when performing conjugation. The fold differences were higher in the screen than when testing individual gRNAs (Supplementary Table S2), which could be due to how the infrequent occurrence of escape mutants impacts either experimental setup.
We next applied machine learning to extract rules for guide design (41) (Figure 3E). We examined 738 features related to the predicted secondary structure, the sequence, or the dataset used (Supplementary Table S3). We found that the algorithm trained on both strains was more predictive than the one trained on single strains; for instance, the Spearman correlation coefficient (R) of the algorithm trained with KPPR1 and tested with KPPR1 is 0.589, whereas when trained on both strains and tested with KPPR1 R is 0.605. (Supplementary Figure S3B). The single-strain and mixed models yielded the strongest features determining guide depletion as determined by SHAP values (Figure 3F, Supplementary Figure S3C,D). The mixed algorithm's strongest features were the lower free energy of the gRNA secondary structure and the disfavoring of a cytosine before the PAM sequence (Figure 3F, Supplementary Figure S3D). These results corroborated the importance of gRNA folding and suggested a DTTTV PAM preference (D = A/G/T, V = A/C/G) in Kp for AsCas12a, which is normally associated with a TTTV motif (49,50,65). Furthermore, the presence of a G in the −1 PAM position and a T as the first nucleotide of the guide was disfavored together with other sequence features.
The identity of the specific strain also appeared as a relevant feature, strengthening the idea that the genetic background can influence the outcome of genomic targeting. However, when comparing guide depletion for the same guides in strains KPPR1 and NTUH-K2044, we do observe a strong correlation (Pearson correlation coefficient: R = 0.705), with only 0.9% of the guides having a significant differential activity between the strains (Figure 3G, Supplementary Table S4). There were no obvious features shared by these guides. Similarly, when we were validating the library experiment, we observed a similar behavior across strains for all the gRNAs (Figure 3D). While the results would suggest that differences in guide activity between strains is rare, more testing is needed to draw broader conclusions about the frequency in which CRISPR antimicrobial activities are strain-specific.
After training the machine-learning algorithm with the two strain datasets, we explored its use for guide design. First, we input previously tested guides (Figures 1B, 2A, D) into the algorithm (Supplementary Table S5). Guides that were active across strains (lacZ, rpoE, ftsZ2, rdgC2) had a predicted logFC < −3, while guides with variable activity (ftsZ, rdgC) had a predicted logFC > −3. Therefore, the logFC < −3 value was set as a threshold to distinguish efficient guides from average or poor guides for our experimental setup. Next, we designed six guide sequences not within the library with either high or low activity according to the algorithm (Supplementary Table S5) and tested them in the KPPR1 and in SB5442 strains. Guide activity predictions were accurate for both strains, with guides having a predicted logFC < −3 yielding a 103–104-fold reduction in transformants and less efficient gRNAs yielding a <10-fold decrease (Figure 3H). In conclusion, we generated a predictive algorithm for guide design that yields efficient antimicrobial activity (>99.9% elimination) across Kp strains.
CRISPR-Cas12a packaged into phages with modified tails fibers function as antimicrobials against Kp
Following the development and validation of guide design against Kp, we next turned to delivery. We selected bacteriophages as a delivery method for AsCas12a and a designed gRNA, as they have been widely adopted for the delivery of antimicrobial phagemids and because they can naturally infect specific bacteria (66). In particular, we used T7 particles with modified tail fibers following an established technique to broaden the host range (30). Briefly, T7 phage lacking tail fiber genes was used to infect an E. coli strain carrying a plasmid encoding the alternative tail fibers, an antibiotic resistance gene and a packaging signal to produce transducing particles (Supplementary Figure S4A). The newly produced particles transduced the plasmid into a bacterial host and conferred antibiotic resistance, but only when the tail was compatible with the host receptors. We tested a collection of 26 T7-like bacteriophage tails (67) and evaluated their plasmid transduction efficiency to Kp strains (Supplementary Figure S4B, C). All tested tail fiber proteins supported plasmid transduction into an E. coli K-12 strain, whereas 11 supported transduction into the pathogenic E. coli strain MG5034. Among all the tail fibers though, only the previously characterized tail phiSG-JL2 (30) successfully delivered a plasmid to K. pneumoniae (Supplementary Figure S4C). In this case, measurable delivery was achieved for four of the six tested K. pneumoniae strains.
We then generated an all-in-one phagemid encoding both the AsCas12a nuclease and gRNA along with the phage packaging signal for phage-based delivery (Supplementary Figure S4D). We initially performed a transformation-based targeting assay to test its antimicrobial activity (Supplementary Figure S4E). The tested guides against ftsZ2, lacZ and lolD were selected either for their high activity in the library screen or because they were efficient in previous experiments (Figures 1B, 2A, D). The guides were selected to not match the E. coli genome to allow the production of the plasmid in this bacterium. While the lacZ-targeting gRNA did not yield a measurable reduction in transformation, the ftsZ and lolD gRNAs led respectively to a ∼103–104 and ∼102-fold drop in transformants and thus were used to test delivery (Supplementary Figure S4E). The lack of reduced colony counts when targeting lacZ could be linked to changes in the gRNA expression in the new plasmid setup.
We produced T7phiSG-JL2 phage particles carrying the tested CRISPR plasmids by transducing the genome of phages lacking tail-fiber genes in an E. coli strain expressing the tail fiber phiSG-JL2 and the validated CRISPR phagemid (Supplementary Figure S4D). We infected the three K. pneumoniae strains exhibiting high (>106) transduction efficiency (Supplementary Figure S4C) with these phages, and we observed a >102-fold reduction in transduction efficiency when selecting for transductants (Supplementary Figure S4F). One exception was the lolD-targeting gRNA in the SB5442 strain that exhibited highly variable transduction, which we attribute to intermediate targeting activity in this strain at the threshold of cell killing. In contrast, no reduction was observed for the pathogenic E. coli strain, which lacks the target site. Thus, AsCas12a and a gRNA can be delivered to different K. pneumoniae strains and actuate its antimicrobial function without undesired targeting of other bacteria.
Discussion
Through this study, we demonstrated that the effectiveness of CRISPR antimicrobials depends on numerous factors, including the selected Cas nuclease, the designed gRNA, and the specific target strain. One unique aspect of the study was comparing different CRISPR nucleases. While prior studies evaluated individual nucleases, we compared a diverse set using similar expression constructs. These comparisons resulted in DNA-targeting nucleases outperforming RNA-targeting nucleases. The underperformance of the RNA-targeting nuclease Cas13a may reflect the need for further optimization of the constructs (e.g. promoter strength, target site selection), as previous studies have reported potent antimicrobial activity with this nuclease type in other bacteria (21). Another possible explanation is that the target transcripts did not exceed a minimal expression threshold to enact cell dormancy (56), which could limit possible targets for Cas13-based antimicrobials. The lack of colony reduction when targeting non-essential genes with Cas12a2 was surprising given its ability to enact collateral cleavage of dsDNA that shuts down the cell regardless of the original target location (54,55). Instead, Cas12a2 may be acting through gene silencing, as targeting essential genes consistently resulted in colony reduction. More work will be needed to uncover the exact basis of antimicrobial activity for this nuclease. Finally, we note that the prevalent type III CRISPR-Cas systems that also recognize RNA targets could also be explored as potent antimicrobials (68). While these systems require a larger set of proteins, their accessory proteins activated through cyclic oligoadenylate molecules catalytically produced by triggered effector complexes could effectively eliminate this target expression threshold. Therefore, more exploration remains to be done to determine the range of CRISPR nucleases that can be applied as programmable antimicrobials, even if DNA-targeting nucleases already offer an effective starting point.
One surprising observation was that CRISPR antimicrobial activity can vary between similar strains despite using the same Cas12a nuclease and expression constructs as well as targeting the same genomic site. Specifically, we identified and characterized two gRNAs targeting either rdgC or ftsZ that exhibited distinct antimicrobial activity between strains. In both cases, we found that resolving predicted secondary structures that would interfere with gRNA function restored antimicrobial activity. Such an impact on gRNA folding has been observed previously for different CRISPR-Cas systems (56,59,60,62,69–71). However, the connection between gRNA folding and strain-specific antimicrobial activity is likely dependent on other factors. Specifically, we posit that the different strains exhibit distinct sensitivities to CRISPR antimicrobials due to differences in DNA repair and cell survival, where the activity of poorly folding gRNAs would be sufficiently attenuated to allow survival in some strains but not in others. While strain-specific antimicrobial activity was rare in our genome-wide screen in two distinct Kp strains, this possibility should be considered when developing CRISPR antimicrobials intended to eradicate diverse strains of the same genus and species—at least until more data are available.
As part of the work, we performed a genome-wide gRNA screen to elucidate factors influencing antimicrobial activity across strains. These efforts parallel a prior unpublished screen with Cas9 that similarly sought to elucidate factors for gRNA design based on antimicrobial activity (72). By performing a similar screen but using AsCas12a and across strains, we found that the vast majority of guides showed similar depletion efficiencies between the two analyzed strains. The factors that most contributed to guide functionality were gRNA folding, in line with gRNA folding leading to poor performance for the rdgC- and ftsZ-targeting gRNAs, although other factors such as PAM-flanking nucleotides were also important. The resulting prediction algorithm based on machine learning can be applied to more readily design new gRNAs as antimicrobials. Future work could evaluate how well these predictions extend to other bacterial pathogens and whether new screens and subsequent machine learning is necessary when deviating beyond Kp. Such work should also take into account escaper cells enriching otherwise cytotoxic guide sequences as part of the screen, even if the effect is likely small given their low frequency when testing individual gRNAs.
Finally, our work explored the delivery of Cas12a-based antimicrobials to Kp using T7 phage particles packaging engineered phagemids. Phagemid delivery has represented the most common mode to introduce CRISPR antimicrobials into bacterial cells (17,22,73,74), although other delivery approaches have been explored including plasmid conjugation and infection with a temperate or lytic phage genetically manipulated to express the CRISPR antimicrobial (20,25,26,75). Benefits of phagemid delivery include the ease of cloning new constructs, the ability to express tail fiber proteins in trans, and the lack of other phage components that could otherwise negatively impact non-target cells receiving the CRISPR antimicrobial. We specifically leveraged tail fiber protein expression in trans by expressing a large set of tail fibers compatible with T7 phages but with the potential to infect Kp strains. This effort revealed a single tail fiber protein that could infect several Kp strains, laying the groundwork to incorporate more diverse or engineered tail fibers (76). With further efforts to enhance delivery and incorporate insights into Cas nuclease selection and gRNA design, CRISPR antimicrobials could become more prominent tools in the ongoing battle against antibiotic-resistant infections.
Supplementary Material
Acknowledgements
We thank Oleg Dmytrenko for providing SuCas12a2 and MpCas12a2 plasmids used to generate the plasmids tested in this study and Chunyu Liao for sharing several plasmids used in this work. The SpCas9 plasmid was a gift from Luciano Maraffini (Addgene plasmid # 42876; http://n2t.net/addgene:42876; RRID:Addgene_42876).
Author contributions: Conceptualization: E.V., S.M., M.G., U.Q., S.B., D.B., C.L.B.; Methodology: E.V., S.M., M.G., Y.Y., L.B., D.B., C.L.B.; Software: Y.Y., L.B.; Investigation: E.V., S.M., M.G., J.Y., D.C., B.B.; Experiments analysis: E.V., S.M., M.G., Y.Y., L.B., D.B.; Resources: S.B., M.L.; Writing - Original Draft: E.V., C.L.B.; Writing - Review and Editing: all authors; Images preparation: E.V., Y.Y., C.L.B.; Supervision: L.B., U.Q., D.B., C.L.B; Project administration: C.L.B.; Funding acquisition: S.B., L.B., U.Q., D.B., C.L.B.
Contributor Information
Elena Vialetto, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), 97080 Würzburg, Germany.
Solange Miele, Institut Pasteur, Université Paris Cité, Synthetic Biology, Paris, France.
Moran G Goren, Faculty of Medicine, Tel Aviv University, 69978 Tel Aviv, Israel.
Jiaqi Yu, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), 97080 Würzburg, Germany.
Yanying Yu, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), 97080 Würzburg, Germany.
Daphne Collias, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), 97080 Würzburg, Germany.
Beatriz Beamud, Institut Pasteur, Université Paris Cité, Synthetic Biology, Paris, France.
Lisa Osbelt, Helmholtz Centre for Infection Research (HZI), 38124 Braunschweig, Germany; German Center for Infection Research (DZIF), partner site Hannover-Braunschweig, 38124 Braunschweig, Germany.
Marta Lourenço, Institut Pasteur, Université Paris Cité, Biodiversity and Epidemiology of Bacterial Pathogens, Paris, France.
Till Strowig, Helmholtz Centre for Infection Research (HZI), 38124 Braunschweig, Germany; German Center for Infection Research (DZIF), partner site Hannover-Braunschweig, 38124 Braunschweig, Germany.
Sylvain Brisse, Institut Pasteur, Université Paris Cité, Biodiversity and Epidemiology of Bacterial Pathogens, Paris, France.
Lars Barquist, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), 97080 Würzburg, Germany; Department of Biology, University of Toronto Mississauga, Mississauga, Ontario L5L 1C6, Canada; University of Würzburg, Medical Faculty, 97080 Würzburg, Germany.
Udi Qimron, Faculty of Medicine, Tel Aviv University, 69978 Tel Aviv, Israel.
David Bikard, Institut Pasteur, Université Paris Cité, Synthetic Biology, Paris, France.
Chase L Beisel, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz Centre for Infection Research (HZI), 97080 Würzburg, Germany; University of Würzburg, Medical Faculty, 97080 Würzburg, Germany.
Data availability
Information on the used plasmids is listed in Supplementary Table S6. Plasmids will be provided by the lead contact upon reasonable request. The library sequencing data have been deposited at GEO under accession # GSE237136. All original code has been deposited at Mendeley and are publicly available through the following DOI: 10.17632/6sy9szgwtj.1.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Joint Programming Initiative on Antimicrobial Resistance [01KI1824 to C.L.B. and T.S., ANR-18-JAM2-0004-04 to S.B. and D.B., 15370 to U.Q.]; Donation from the Pfizer Foundation (to D.B. and S.B.); Bavarian State Ministry for Science and the Arts through the research network bayresq.net (to L.B.); U.Q. has received funding from the European Research Council [818878]. Funding for open access charge: Internal funding.
Conflicts of interest statement
C.L.B. is a co-founder and member of the Scientific Advisory Board for Locus Biosciences as well as a member of the Scientific Advisory Board for Benson Hill. D.B. is a co-founder and Scientific Advisory Board member for Eligo Bioscience. U.Q. is the CTO and member of the Scientific Advisory Board for Trobix-Bio. The other authors have no conflicts of interest to declare.
References
- 1. Cook M.A., Wright G.D. The past, present, and future of antibiotics. Sci. Transl. Med. 2022; 14:eabo7793. [DOI] [PubMed] [Google Scholar]
- 2. Hutchings M.I., Truman A.W., Wilkinson B. Antibiotics: past, present and future. Curr. Opin. Microbiol. 2019; 51:72–80. [DOI] [PubMed] [Google Scholar]
- 3. Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization Antimicrobial Resistance: global report on surveillance. 2014;
- 5. De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020; 33:e00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018; 18:318–327. [DOI] [PubMed] [Google Scholar]
- 7. Paterson D.L., Bonomo R.A. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 2005; 18:657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uribe R.V., Rathmer C., Jahn L.J., Ellabaan M.M.H., Li S.S., Sommer M.O.A. Bacterial resistance to CRISPR-Cas antimicrobials. Sci. Rep. 2021; 11:17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donskey C.J., Chowdhry T.K., Hecker M.T., Hoyen C.K., Hanrahan J.A., Hujer A.M., Hutton-Thomas R.A., Whalen C.C., Bonomo R.A., Rice L.B. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N. Engl. J. Med. 2000; 343:1925–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Selle K., Fletcher J.R., Tuson H., Schmitt D.S., McMillan L., Vridhambal G.S., Rivera A.J., Montgomery S.A., Fortier L.-C., Barrangou R. et al. Targeting of Clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio. 2020; 11:e00019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jansen R., van Embden J.D.A., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002; 43:1565–1575. [DOI] [PubMed] [Google Scholar]
- 12. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 13. Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008; 322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009; 139:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P. et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bikard D., Euler C.W., Jiang W., Nussenzweig P.M., Goldberg G.W., Duportet X., Fischetti V.A., Marraffini L.A. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014; 32:1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bikard D., Hatoum-Aslan A., Mucida D., Marraffini L.A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012; 12:177–186. [DOI] [PubMed] [Google Scholar]
- 19. Gomaa A.A., Klumpe H.E., Luo M.L., Selle K., Barrangou R., Beisel C.L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014; 5:e00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park J.Y., Moon B.Y., Park J.W., Thornton J.A., Park Y.H., Seo K.S. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 2017; 7:44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiga K., Tan X.-E., Ibarra-Chávez R., Watanabe S., Aiba Y., Sato’o Y., Li F.-Y., Sasahara T., Cui B., Kawauchi M. et al. Development of CRSIPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020; 11:2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Citorik R.J., Mimee M., Lu T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014; 32:1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reuter A., Hilpert C., Dedieu-Berne A., Lematre S., Gueguen E., Launay G., Bigot S., Lesterlin C. Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity. Nucleic Acids Res. 2021; 49:3584–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang Y.K., Kwon K., Ryu J.S., Lee H.N., Park C., Chung H.J. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjug. Chem. 2017; 28:957–967. [DOI] [PubMed] [Google Scholar]
- 25. Hamilton T.A., Pellegrino G.M., Therrien J.A., Ham D.T., Bartlett P.C., Karas B.J., Gloor G.B., Edgell D.R. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat. Commun. 2019; 10:4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodrigues M., McBride S.W., Hullahalli K., Palmer K.L., Duerkop B.A. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob. Agents Chemother. 2019; 63:e01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruotsalainen P., Penttinen R., Mattila S., Jalasvuori M. Midbiotics: conjugative plasmids for genetic engineering of natural gut flora. Gut Microbes. 2019; 10:643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki Y., Onuma H., Sato R., Sato Y., Hashiba A., Maeki M., Tokeshi M., Kayesh M.E.H., Kohara M., Tsukiyama-Kohara K. et al. Lipid nanoparticles loaded with ribonucleoprotein–oligonucleotide complexes synthesized using a microfluidic device exhibit robust genome editing and hepatitis B virus inhibition. J. Controlled Release. 2021; 330:61–71. [DOI] [PubMed] [Google Scholar]
- 29. Ando H., Lemire S., Pires D.P., Lu T.K. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015; 1:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yosef I., Goren M.G., Globus R., Molshanski-Mor S., Qimron U. Extending the host range of bacteriophage particles for DNA transduction. Mol. Cell. 2017; 66:721–728. [DOI] [PubMed] [Google Scholar]
- 31. Yehl K., Lemire S., Yang A.C., Ando H., Mimee M., Torres M.D.T., de la Fuente-Nunez C., Lu T.K. Engineering phage host-range and suppressing bacterial resistance through phage tail Fiber mutagenesis. Cell. 2019; 179:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nassif X., Sansonetti P.J. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 1986; 54:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huynh B.-T., Passet V., Rakotondrasoa A., Diallo T., Kerleguer A., Hennart M., Lauzanne A.D., Herindrainy P., Seck A., Bercion R. et al. Klebsiella pneumoniae carriage in low-income countries: antimicrobial resistance, genomic diversity and risk factors. Gut Microbes. 2020; 11:1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu K.-M., Li L.-H., Yan J.-J., Tsao N., Liao T.-L., Tsai H.-C., Fung C.-P., Chen H.-J., Liu Y.-M., Wang J.-T. et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 2009; 191:4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferrières L., Hémery G., Nham T., Guérout A.-M., Mazel D., Beloin C., Ghigo J.-M. Silent mischief: bacteriophage mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol. 2010; 192:6418–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gomes A.É.I., Stuchi L.P., Siqueira N.M.G., Henrique J.B., Vicentini R., Ribeiro M.L., Darrieux M., Ferraz L.F.C. Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using reverse transcription quantitative real-time PCR. Sci. Rep. 2018; 8:9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lam M.M.C., Wick R.R., Watts S.C., Cerdeira L.T., Wyres K.L., Holt K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021; 12:4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu Y., Gawlitt S., de Andrade E Sousa L.B., Merdivan E., Piraud M., Beisel C.L., Barquist L Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration. Genome Biol. 2024; 25:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feurer M., Klein A., Eggensperger K., Springenberg J.T., Blum M., Hutter F. Auto-sklearn: efficient and robust automated machine learning. Automated Machine Learning: The Springer Series on Challenges in Machine Learning. 2019; Cham: Springer; 113–134. [Google Scholar]
- 43. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorenz R., Hofacker I.L., Bernhart S.H. Folding RNA/DNA hybrid duplexes. Bioinformatics. 2012; 28:2530–2531. [DOI] [PubMed] [Google Scholar]
- 45. Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020; 2:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Modrich P., Richardson C.C. Bacteriophage T7 deoxyribonucleic acid replication in vitro. Bacteriophage T7 DNA polymerase: an enzyme composed of phage- and host-specific subunits. J. Biol. Chem. 1975; 250:5515–5522. [PubMed] [Google Scholar]
- 47. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishimasu H., Ann Ran F., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016; 165:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marino N.D., Pinilla-Redondo R., Bondy-Denomy J. CRISPR-Cas12a targeting of ssDNA plays no detectable role in immunity. Nucleic Acids Res. 2022; 50:6414–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meeske A.J., Nakandakari-Higa S., Marraffini L.A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature. 2019; 570:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016; 353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dmytrenko O., Neumann G.C., Hallmark T., Keiser D.J., Crowley V.M., Vialetto E., Mougiakos I., Wandera K.G., Domgaard H., Weber J. et al. Cas12a2 elicits abortive infection through RNA-triggered destruction of dsDNA. Nature. 2023; 613:588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bravo J.P.K., Hallmark T., Naegle B., Beisel C.L., Jackson R.N., Taylor D.W. RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2. Nature. 2023; 613:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vialetto E., Yu Y., Collins S.P., Wandera K.G., Barquist L., Beisel C.L. A target expression threshold dictates invader defense and prevents autoimmunity by CRISPR-Cas13. Cell Host Microbe. 2022; 30:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cui L., Bikard D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 2016; 44:4243–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Collias D., Vialetto E., Yu J., Co K., Almási É.d.H., Rüttiger A.-S., Achmedov T., Strowig T., Beisel C.L. Systematically attenuating DNA targeting enables CRISPR-driven editing in bacteria. Nat. Commun. 2023; 14:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Creutzburg S.C.A., Wu W.Y., Mohanraju P., Swartjes T., Alkan F., Gorodkin J., Staals R.H.J., van der Oost J. Good guide, bad guide: spacer sequence-dependent cleavage efficiency of Cas12a. Nucleic Acids Res. 2020; 48:3228–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liao C., Ttofali F., Slotkowski R.A., Denny S.R., Cecil T.D., Leenay R.T., Keung A.J., Beisel C.L. Modular one-pot assembly of CRISPR arrays enables library generation and reveals factors influencing crRNA biogenesis. Nat. Commun. 2019; 10:2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moreb E.A., Hoover B., Yaseen A., Valyasevi N., Roecker Z., Menacho-Melgar R., Lynch M.D. Managing the SOS response for enhanced CRISPR-Cas-based recombineering in E. coli through transient inhibition of host RecA activity. ACS Synth. Biol. 2017; 6:2209–2218. [DOI] [PubMed] [Google Scholar]
- 62. Wessels H.-H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020; 38:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Konstantakos V., Nentidis A., Krithara A., Paliouras G. CRISPR–Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning. Nucleic Acids Res. 2022; 50:3616–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jacobsen T., Ttofali F., Liao C., Manchalu S., Gray B.N., Beisel C.L. Characterization of Cas12a nucleases reveals diverse PAM profiles between closely-related orthologs. Nucleic Acids Res. 2020; 48:5624–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lam K.N., Spanogiannopoulos P., Soto-Perez P., Alexander M., Nalley M.J., Bisanz J.E., Nayak R.R., Weakley A.M., Yu F.B., Turnbaugh P.J. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep. 2021; 37:109930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lourenço M., Osbelt L., Passet V., Gravey F., Megrian D., Strowig T., Rodrigues C., Brisse S. Phages against noncapsulated Klebsiella pneumoniae: broader host range, slower resistance. Microbiol. Spectr. 2023; 11:e0481222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. van Beljouw S.P.B., Sanders J., Rodríguez-Molina A., Brouns S.J.J. RNA-targeting CRISPR–Cas systems. Nat. Rev. Microbiol. 2022; 21:21–34. [DOI] [PubMed] [Google Scholar]
- 69. Thyme S.B., Akhmetova L., Montague T.G., Valen E., Schier A.F. Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat. Commun. 2016; 7:11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhu H., Liang C. CRISPR-DT: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Bioinformatics. 2019; 35:2783–2789. [DOI] [PubMed] [Google Scholar]
- 71. Wong N., Liu W., Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015; 16:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gutierrez B., Ng J.W., Cui L., Becavin C., Bikard D. Genome-wide CRISPR-Cas9 screen in E. coli identifies design rules for efficient targeting. 2018; bioRxiv doi:30 April 2018, preprint: not peer reviewed 10.1101/308148. [DOI]
- 73. Kiga K., Tan X.-E., Ibarra-Chávez R., Watanabe S., Aiba Y., Sato’o Y., Li F.-Y., Sasahara T., Cui B., Kawauchi M. et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020; 11:2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gencay Y.E., Jasinskytė D., Robert C., Semsey S., Martínez V., Petersen A.Ø., Brunner K., de Santiago Torio A., Salazar A., Turcu I.C. et al. Engineered phage with antibacterial CRISPR-Cas selectively reduce E. coli burden in mice. Nat. Biotechnol. 2023; 42:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Selle K., Fletcher J.R., Tuson H., Schmitt D.S., McMillan L., Vridhambal G.S., Rivera A.J., Montgomery S.A., Fortier L.-C., Barrangou R. et al. In Vivo targeting of clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio. 2020; 11:e00019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Apjok G., Számel M., Christodoulou C., Seregi V., Vásárhelyi B.M., Stirling T., Eszenyi B., Sári T., Vidovics F., Nagrand E. et al. Characterization of antibiotic resistomes by reprogrammed bacteriophage-enabled functional metagenomics in clinical strains. Nat. Microbiol. 2023; 8:410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Information on the used plasmids is listed in Supplementary Table S6. Plasmids will be provided by the lead contact upon reasonable request. The library sequencing data have been deposited at GEO under accession # GSE237136. All original code has been deposited at Mendeley and are publicly available through the following DOI: 10.17632/6sy9szgwtj.1.