Abstract
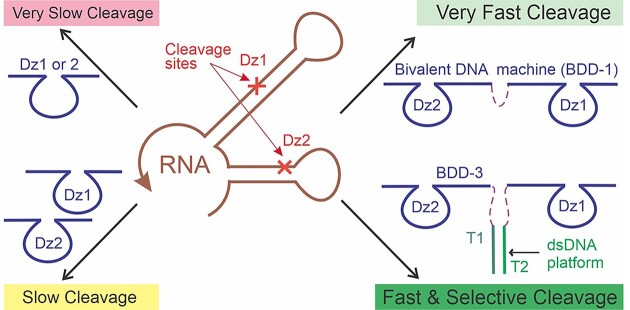
Multivalent recognition and binding of biological molecules is a natural phenomenon that increases the binding stability (avidity) without decreasing the recognition specificity. In this study, we took advantage of this phenomenon to increase the efficiency and maintain high specificity of RNA cleavage by DNAzymes (Dz). We designed a series of DNA constructs containing two Dz agents, named here bivalent Dz devices (BDD). One BDD increased the cleavage efficiency of a folded RNA fragment up to 17-fold in comparison with the Dz of a conventional design. Such an increase was achieved due to both the improved RNA binding and the increased probability of RNA cleavage by the two catalytic cores. By moderating the degree of Dz agent association in BDD, we achieved excellent selectivity in differentiating single-base mismatched RNA, while maintaining relatively high cleavage rates. Furthermore, a trivalent Dz demonstrated an even greater efficiency than the BDD in cleaving folded RNA. The data suggests that the cooperative action of several RNA-cleaving units can significantly improve the efficiency and maintain high specificity of RNA cleavage, which is important for the development of Dz-based gene knockdown agents.
Graphical Abstract
Graphical Abstract.
Introduction
Multivalent interactions between biological molecules continuously attract the attention of both fundamental sciences (1–3) and biotechnology (4,5). For example, multiple sequentially ordered zing finger motifs have been explored to bind genomic DNA with high affinity and specificity (6). Traditionally used in the form of single oligonucleotides, hybridization probes have been evolving into two- and multicomponent hybridization sensors, which enable both tight binding to DNA and RNA analytes and differentiating single nucleotide substitutions in their structures (7,8). The multivalent approach has not been adopted yet in the practice of oligonucleotide gene therapy (OGT) agents for suppressing specific mRNA. This approach, in general, can provide the advantages of both tight and selective mRNA-binding (7,8).
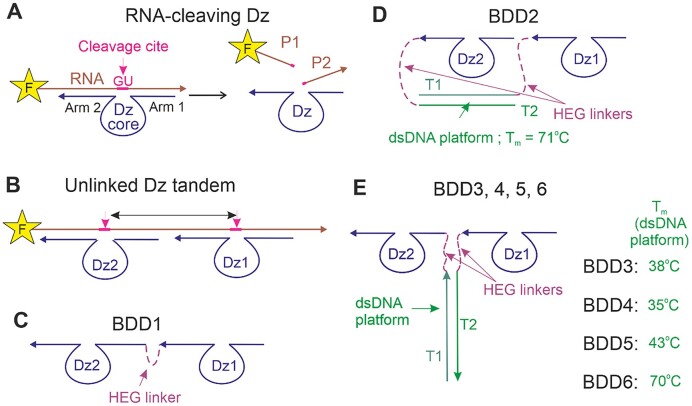
In this study, we explored the cleavage of folded RNA by RNA-cleaving DNAzymes (Dz) (9–12) organized in multicomponent cleaving agents of various designs (Figure 1). Traditional Dz agents bind RNA substrates by two RNA-binding arms followed by phosphodiester bond cleavage using nucleotides of the catalytic core (Figure 1A). A variety of Dz have been explored for gene knockdown in cell cultures and in vivo (11–13). Dz are known to be the most selective OGT agents: no hybridization-dependent off-target activity has been reported for Dz agents so far (14). Several Dz agents were tested in phase II of clinical trials (15–17).
Figure 1.
The design of RNA-cleaving monovalent and bivalent Dz agents. (A) Single RNA-cleaving Dz in complex with a 5′-fluorophore (F)-labelled RNA target. (B) A tandem of two Dz agents cleaving an RNA target at two closely located cleavage sites. (C) Bivalent Dz-based device BDD1 consisting of two Dz agents covalently linked to each other by a hexaethylene glycol (HEG) linker. (D) BDD2 with Dz1 and Dz2 attached to the opposite ends of a dsDNA platform made of T1 and T2 fragments. (E) BDD3, BDD4, BDD5 and BDD6 containing Dz1 and Dz2 linked to dsDNA platforms having different melting temperatures (Tm).
An important advantage of Dz over other OGT technologies such as RNAi and antisense oligonucleotides is their protein-independent mechanism of RNA cleavage. This is particularly attractive in the context of sophisticated DNA nanostructures for gene therapy (10,14,17–20) including those equipped with molecular computation functions for the analysis of cancer biomarkers (21). Such non-natural DNA structures often do not interact with protein enzymes but can be functionalized for RNA cleavage by protein-independent Dz.
Dz catalytic cycle includes RNA binding (k1), catalytic cleavage (k2), and product release (k3, Scheme 1). Poor target accessibility (low k1) is the major challenge that limits Dz therapeutic efficiency in vivo (14,21). It was shown earlier that natural RNA targets are poorly accessible by hybridization agents due to their secondary and tertiary structures (19,22). Among all OGT agents, Dz could be affected by the poor target accessibility the most since they use two relatively short RNA-binding arms, which provide less affinity (and higher selectivity) than longer, continuous sequences of ASO and siRNAs (14). The poor RNA target accessibility is a possible reason why Dz technology has not delivered a practically significant gene knockdown tool in cell culture or in vivo (11–13). To overcome this issue, two conventional solutions have been extensively explored. The first approach uses chemical modifications including 2′-OMe, 2′-F, 2′-2′-fluoro-arabino nucleic acid (FANA) and locked nucleic acids (LNA), which increase the thermodynamic stability of Dz/RNA hybrids (12,23,24). However, high affinity can slow down the product release stage of the catalytic cycle (14,25,26). Moreover, high-affinity arms may increase the probability of non-specific binding to an RNA sequence of a transcriptome size. Another ordinary solution of the RNA accessibility problem takes advantage of the computational analysis of the RNA-target regions to identify the least structured fragments (27). However, structures of long RNA and their protein-binding sites are poorly predictable by the currently available software (27). It would have been attractive to target RNA fragments independently on their folding energy. For example, recognition of specific sites carrying point mutations is needed for differentiating true targets from wild-type RNA including cancer-causing mutations in KRAS (28), BRCA1 and BRCA2 (29) genes, among others (30). There is no guarantee that these sites are situated in the most relaxed mRNA fragments.
Scheme 1.
The catalytic cycle of RNA-cleaving DNAzymes (Dz).
Here, we propose to achieve both tight and selective RNA binding using a nature-inspired multivalent approach. It was hypothesized that the cooperative binding of two or more Dz units in closely located positions in RNA would facilitate the RNA binding stage of the catalytic cycle due to the cooperative hybridization of the four (or more) RNA-binding arms. Such a tandem system may have a greater chance of RNA cleavage, which should linearly increase within the increase of the number of the targeted neighboring sites with single mRNA chain. Furthermore, a covalent or a noncovalent linkage between the two Dz should further increase binding cooperativity due to the entropy factor: the formation of a complex from the two (in the case of linked Dzs) vs three (for unlinked tandem of two Dzs) particles. We also expected that using multiple short RNA binding arms of the Dz tandem will allow an efficient product release (high k3), which will favor multiple catalytic turnovers.
The tighter the binding, the lower the selectivity of nucleic acid recognition, according to the affinity/specificity dilemma (31). To maintain the high selectivity of RNA recognition by bivalent Dz, we proposed to modulate the degree of Dz subunit association (and, therefore, Dz affinity to the targeted RNA) to determine the best balance of affinity/specificity during RNA target recognition.
In this work, we targeted two neighboring cleavage sites in a folded RNA by two Dz agents (Dz1 and Dz2) of a conventional design (Figure 1B). Furthermore, we joined Dz1 and Dz2 via a flexible linker to form bivalent Dz devices’ (BDD1 in Figure 1C) or by a variety of double-stranded (ds) DNA platforms (BDD 2–6 in Figure 1D and E, referred also to as ‘nanostructures’). It was found that both the Dz1 + Dz2 tandem and BDD can increase RNA cleavage efficiency in comparison with the traditional ‘monovalent’ Dz. We also demonstrated that BDD nanostructures maintain high selectivity of RNA recognition, which can be modulated by the change in melting temperature (Tm) of the dsDNA platform (Figure 1E).
Materials and methods
Chemicals
Oligonucleotides were purchased from DNA-sintez (Moscow, Russian Federation) and Integrated DNA Technologies (IDT, Coralville, IA, USA). All the DNA and RNA sequences are listed in Supplementary Table S1. Other chemicals in the list are the following: tris(hydroxymethyl)aminomethane (TRIS), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), urea, tetramethylethylenediamine (TEMED)—Molekula (UK); magnesium chloride (MgCl2), ammonium persulfate (APS)—Carl Roth, (Germany); acrylamide 2k—Panreac Applichem (Germany); deionized formamide—Sigma-Aldrich (USA), N,N-methylene-(bis-acrylamide), ethylenediamine tetraacetic acid (EDTA), ethidium bromide (EtBr)—VMR (USA); sodium chloride (NaCl)—Vekton (Russia); boric acid—Techsnab (Russia); GelRed—biotium (USA); TryDye ultra-low range (ULR) ladder 10–700—New England Biolabs (USA); RNase H—TransGen Biotech (China). Milli-Q water was used to prepare all the buffers, and nuclease-free water was used to prepare the solutions of oligonucleotides.
Design of oligonucleotides
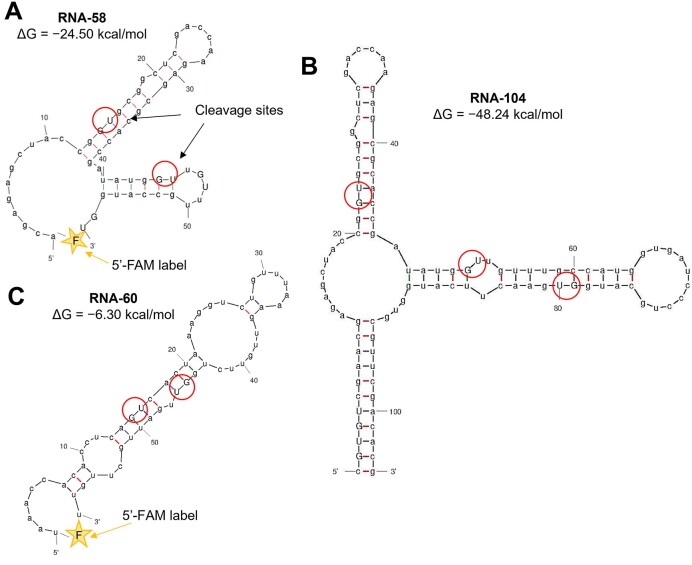
Thermodynamic parameters and predicted 2D-structure of the RNA fragments were estimated using the RNAFold application in the UNAFold web server (32). Dz and BDD agents were designed in accordance with their complementarity to the selected RNA fragments (Figure 2). Melting temperature (Tm) was calculated using the two-state melting hybridization application in the UNAFold web server (32). The design of Dz agents was carried out in a way to minimize the overlap between RNA-binding domains. While secondary structures did exist in certain regions of the BDDs, they consisted of short stems separated from each other within the secondary BDD structure, and presumably had little or no effect on BDD–RNA hybridization.
Figure 2.
Secondary structures of the three model RNA substrates used in this study. (A) Fragment of the strA gene RNA-58. (B) RNA-104 derived from RNA-58 by adding extra nucleotides to both 5′ and 3′ ends to increase the stability of its secondary structure. (C) RNA-60 is a fragment of the EIF3C gene with a relatively high folding energy. Red circles indicate Dz cleavage sites. RNA-58 and RNA-60 were labelled with a fluorescein (F) moiety at the 5′ ends.
Buffers for RNA-cleaving reaction
To perform the experiments on RNA cleavage by Dz, we used near-physiological Mg2+-containing Buffer 1: 15 mM NaCl, 150 mM KCl, 50 mM HEPES (pH 7.4), 2 mM MgCl2. Mg2+-free Buffer 2 contained 15 mM NaCl, 150 mM KCl, and 50 mM HEPES (pH 7.4). To terminate RNA cleavage reactions, 10 μl of Buffer 3 (8 M urea, 15% 2 × TBE was added) or Buffer 4 (95% deionized formamide, 25 mM EDTA) was added to 10 μl of the reaction mixture.
Assembling and analysis of BDD constructions
The assembly was done by mixing all components in Buffer 1. The mixture was heated at 95°C in a water bath followed by gradual cooling to room temperature overnight. The assembled BDD associations were mixed with the 4 × loading dye (Evrogen, Russia) and analyzed using native 12% polyacrylamide gel (AA:BA (29:1), TBE) for 90 min at 80 V. The gels were stained with 0.5 ug/mL EtBr dye followed by visualization using a gel documentation system GelDoc (BioRad, USA).
RNA cleavage under multiple-turnover conditions
To assess the efficiency of DNAzymes association, catalytic cleavage, and dissociation from the RNA substrate, we conducted experiments under multiple-turnover conditions ([Dz] < [RNA]). RNA substrates were incubated with Dz in the reaction Buffer 1 under 37°C for several time points: 1, 5 and 24 h. The cleavage at reaction was terminated by adding an equal volume (10 μl) of Buffer 3.
The samples were denatured at 95°C for 5 min followed by their incubation on ice for 5 min. Products of RNA cleavage were analysed using 20% denaturing PAGE (AA:BA (29:1), 7 M urea, 1× TBE) for 180 min at 80 V. The cleavage product bands in the gel were visualized based on the intrinsic fluorescence of the 5′-FAM labelled RNA substrates, or after GelRed dye staining.
RNA cleavage under single-turnover conditions
Dz agents (2 or 5 μM) and RNA substrates (1 μM) were annealed in the Mg2+-free Buffer 2 by heating at 95°C for 5 min, cooling on ice for 5 min followed by incubation at 37°C for 5 min. The cleavage reaction was initiated by the addition of 2 mM MgCl2. After 5- or 15-min incubation at 37°C, the reaction was stopped by the addition of an equal volume (10 μM) of loading Buffer 4 followed by 20% denaturing PAGE analysis, as described above.
RNA cleavage mediated with RNase H
Dz agents (0.1 μM) and the RNA substrates (1 μM) were incubated in Buffer 1 supplemented with 1 U of recombinant RNase H enzyme at 37°C for 20 min. To terminate the reaction, the formamide-containing loading Buffer 4 was added, and the tubes with mixed samples were preheated at 95 °C for 5 min to deactivate RNase H followed by cooling on ice. After stopping the incubation, RNA cleavage products were analyzed by 20% denaturing PAGE analysis.
Analysis of cleavage agent efficiency
The cleavage agent (CA) efficiency was characterized by the turnover number (TON) calculated as follows:
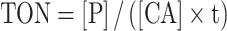 |
where [CA] is the total CA concentration and [P] is the concentration of the cleavage product(s), t is the reaction time (h). For monovalent Dz, [P] was calculated as the product band intensity divided by the sum of all bands (product plus the initial RNA substrate) multiplied by the starting concentration of RNA substrate [RNA]o. For bivalent and trivalent CA, [P] was calculated as a sum of the densities of the product bands (P1 + P2) divided by the sum of all bands (including RNA substrate) and multiplied by the starting concentration of RNA substrate [RNA]o. In the latter case, the calculation did not account for the cleavage of RNA at multiple sites in the assumption that any damage (single or double) is sufficient for RNA inactivation.
Under single-turnover conditions, the cleavage efficiency was evaluated by the pseudo-first-order reaction constant (kobs)
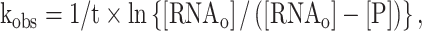 |
where [RNAo] is the starting concentration of the RNA substrate. In the analysis of time-dependence kinetics, the efficacies of cleavage agents were assessed with the initial rate (vo), which was calculated as follows:
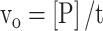 |
To evaluate the specificity of CA, enzymes the selectivity factor F(s) was calculated as follows:
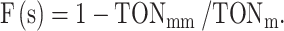 |
F(s) approaches 1 or 0 for the greatest or lowest selectivity, respectively. In this formula, TONmm and TONm correspond to the turnover numbers for the mismatched and matched CA, respectively.
Results and discussion
Selection of RNA targets
As model substrates, we used three synthetic RNA fragments with different folding energies: RNA-58, RNA-104 and RNA-60 (Figure 2). RNA-58 was a 58-nt fragment (nt – nucleotides) of streptomycin resistance strA gene cassette (33). This target has a low folding energy (−24.5 kcal/mol) and represents a challenging model for Dz-assisted RNA recognition and cleavage. RNA-104 was artificially derived from RNA-58 by adding extra nucleotides to further increase the secondary structure stability and introduce a third GU cleavage site located 25 nt from the nearest GU cleavage site. This separation of the cleavage sites might be important for the accommodation of the correctly folded Dz cores in proximity to each other. RNA-60 is a 60-nt fragment of the mRNA encoding the eukaryotic initiation translation factor 3 subunit С (EIF3C) (34). This sequence was used as a model substrate with a relatively high folding energy (−6.5 kcal/mol). RNA-58 and RNA-60 were 5′-fluorescein-labelled for convenient visualization in gel and quantification of the cleavage products.
Design of Dz cleaving agents (CA)
First, we designed Dz1 and Dz2 targeting the two cleavage sites separated by 29 nt within the RNA-58 sequence (Figure 2A). We used the catalytic core of Dz 10–23 because of its small size and high RNA cleavage activity (11,14,25). The 15-nt catalytic core was flanked with RNA-binding arms complementary to the substrate fragment at the R↓Y cleavage site with the greatest efficiency reported for G↓U sites (25). According to the common practice (11,12,25,26), RNA-binding arms were designed to have Tm lower than the reaction temperature (+37°C). Higher Tm would slow down the dissociation of the cleavage products (P1 and P2 in Figure 1A), thus slowing down the overall catalytic cycle shown in Scheme 1 (26). For Dz1, the lengths of both arms were 11 nt (11/11, Tm= 32°C for both arms), while for Dz2, the arms were 11 and 9 nt (11/9, Tm = 34 and 35°C, respectively). The sequences and characteristics of the studied Dz agents are listed in Supplementary Tables S2 and S3.
BDD1 consisted of two Dz1 and Dz2 linked to each other via a flexible hexaethylene glycol (HEG) linker. Covalently linked Dz1 and Dz2 should have an increased binding cooperativity to the RNA substrate in comparison with individual Dz1 and Dz2. The flexibility of HEG provides conformational freedom, thus allowing for the two catalytic cores to fold near each other in the BDD1/RNA complex. We also combined Dz1 and Dz2 via dsDNA platforms to form nanostructures shown in Figure 1D and E. Unlike BDD1, BDD2–6 can be integrated into more functionally complex DNA nanostructures capable of unwinding folded RNA and/or analyzing multiples cancer markers followed by target RNA cleavage (18–21). BDD2–6 a offered greater flexibility than BDD1 due to the presence of two rather than one HEG linkers. In BDD2, both Dz1 and Dz2 were linked to the common dsDNA platform via 3′-ends. The platform was stable (Tm = 71 °C) under the reaction condition. In BDD3–6, Dz1 and Dz2 were associated with the dsDNA platform via HEG-linkers at the neighboring 5′- and 3′-ends (Figure 1D and E). These four constructs differed from each other by the Tm values for the dsDNA platform, which was either higher (BDD5 and BDD6), roughly equal (BDD3), or lower (BDD4) than the reaction temperature. Such designs allowed us to gradually change the degree of Dz association and, therefore, study the impact of Dz cooperativity on both the efficiency and selectivity of the RNA cleavage.
The sequences, length, and the Tm values for all BDD-comprising oligonucleotides are listed in Supplementary Tables S2 and S3. The accuracy of BDD2, BDD5 and BDD6 assembly by annealing was verified using native PAGE (Supplementary Figure S1). Expectedly, BDD3 and BDD4 did not form a stable complex under the electrophoresis conditions (Supplementary Figure S1).
Dz1 + Dz2 tandem is more efficient in RNA cleavage than individual Dz1 or Dz2
Both individual Dz1 and Dz2 cleaved RNA-58 with kobs ≈ 0.1 h−1 (Figure 1B), which was lower than that reported for the cleavage of linear RNA substrates (kobs ∼10 h−1) (25,26). This correlates with the previous observations that stable RNA structures slow down the cleavage reaction (19,22) and illustrates that Dz agents of a conventional design are inefficient in cleaving folded RNA. This fact alone explains the lack of clinically significant Dz-based OGT agents (11–17).
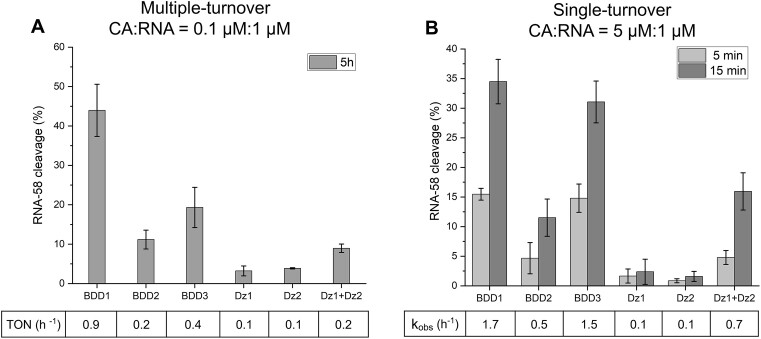
Under multiple-turnover conditions (Figure 3A), cleavage activity was characterized by the turnover number (TON), which is the number of RNA substrates cleaved by each agent in 1 h (see Materials and Methods for calculation details). The Dz1 + Dz2 tandem demonstrated 2-fold improvement in the cleavage efficiency. This reflects little or no cooperativity in the RNA-binding step and might be the result of independent (non-cooperative) RNA cleavage by the two Dz agents. We concluded that at low concentrations the Dz1 + Dz2 tandem is inefficient in RNA-binding.
Figure 3.
RNA-58 cleavage by Dz or BDD cleavage agent (CA). (A) Multiple-turnover conditions: RNA-58 (1 μM) was incubated with each of the cleavage agents (0.1 μM) in Buffer 1 at 37°C for 5 h followed by the analysis of the cleavage products by PAGE. Turnover number (TON) was calculated as described in Material and Methods. (B) Single-turnover conditions: RNA-58 (1 μM) was incubated with each cleaving agent (5 μM) at 37°C for either 5- or 15-min. Single-turnover data obtained for a Dz:RNA ratio of 2:1 is presented in Supplementary Figure S2. The data is the average values of three independent experiments. The parameters TON and kobs were as described in Materials and Methods and presented for 5 h and 15 min, respectively.
Under single-turnover conditions, the tandem of Dz1 and Dz2 improved RNA cleavage 7-fold in comparison with each individual Dz (Figure 3B, last group of bars). Therefore, high Dz1 and Dz2 concentrations enable cooperative binding and unwinding RNA-58 structure. This result suggested that an increase in (local) concentrations of Dz1 and Dz2 may facilitate cooperative RNA binding and inspired us to assemble Dz1 and Dz2 into BDD (Figure 1C–E).
BDD1 and BDD3 are more active than the tandem of unlinked Dz1 and Dz2
First, we studied different arrangements of Dz1 and Dz2 subunits by comparing the RNA cleavage efficiency of BDD1, 2, and 3. Under multiple-turnover conditions, all three BDD improved TON in comparison to both Dz1 and Dz2 acting separately or together (Figure 3A). BDD1 demonstrated a 9- and 4.5-fold increase in TON over individual Dz and Dz1 + Dz2 tandem, respectively. This high substrate turnover suggests that BDD1 was not inhibited by the RNA cleavage products (had high k3) due to the relatively short substrate binding arms.
Under single-turnover conditions, the greatest cleavage rates were achieved by BDD1 and BDD3 (kobs ∼1.7 and ∼1.5 h −1, respectively), which were 17- and 15-fold greater than that of individual Dz1 or Dz2. There was also a more than 2-fold improvement in BDD1 and BDD3 efficiencies in comparison with the Dz1 + Dz2 tandem. This result proves that joining two Dz units in a single nanostructure improves the cleavage efficiency of a folded RNA substrate likely due to facilitation of the binding stage (increasing k1).
Interestingly, BDD2 had a comparable or lower activity than the Dz1 + Dz2 tandem. We posit that the low cleavage profile of BDD2 is due to the lack of efficient cooperation between Dz1 and Dz2 because of their significant separation in the BDD2 nanostructure (Figure 1D). Shortening the dsDNA platform in BDD2 could improve the performance. However, we did not pursue this optimization route. Instead, we focused on the characterization of the more successful BDD1 and BDD3 constructs.
RNase H activity of BDD1 and BDD3
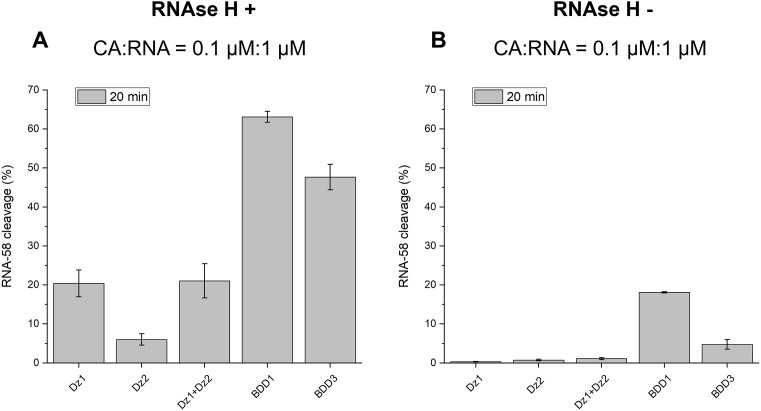
Considering BDD1 and BDD3 as more efficient bivalent agents in comparison with the individual Dzs, we evaluated the effect of multivalent Dz on RNA cleaving activity in the presence of RNase H. We hypothesized that BDDs having high affinity to RNA will improve RNase H dependent RNA degradation.
Under multiple-turnover conditions with RNase H (20 min), BDD1 cleaved ∼65% of RNA, which is more than 3-fold greater than individual Dz1 or Dz1 + Dz2 tandem (Figure 4). Dz2 demonstrated less efficiency in comparison to Dz1. We explain the difference in Dz1 and Dz2 performances by the difference in RNA binding arms of the two agents: Dz1 has a longer RNA-binding arm (11 + 11 nt, Dz1 in Supplementary Table S1) in comparison to Dz2 (11 + 9 nt, Dz2 in Supplementary Table S1). It was shown earlier that the differences in the lengths of the RNA-binding domains can impact RNase H-mediated RNA cleavage (35).
Figure 4.
BDD1 and BDD3 demonstrate greater RNase H-dependent RNA cleaving than Dz1, Dz2 or Dz1 + Dz2. RNA-58 (1 μM) was incubated with each of the cleavage agents (0.1 μM) in Buffer 1 at 37 °C for 20 min with or without RNase H. The cleavage products were separated from RNA substrate by denaturing PAGE (Supplementary Figure S3) followed by quantification as detailed in Materials and Methods. (A) RNA-58 cleavage in the presence of RNase H (RNase H+). (B) RNA-58 cleavage by the Dz cleaving agents in the absence of RNase H (RNase H–). The data is the average values of three independent experiments.
T m of the dsDNA platform affects RNA cleavage
The primary reason for further studying BDD1 and BDD3 was their high RNA cleaving activity and, therefore, the greatest potential as RNA cleaving agents among all Dz constructs studied here. In addition, BDD1 had the most straightforward connection of the two Dz units. Despite somewhat lower cleavage efficiency, BDD3 design allows rational control of the association of Dz1 and Dz2 units by changing the melting temperature of the DNA scaffold, thus offering a means to study the effect of cooperativity on both the efficiency and selectivity of the bivalent Dz constructions. Moreover, the BDD3 design allows integration of the multicomponent agent into complex DNA nanostructures, which could be the next step in the development of smart DNA devices for gene knockdown (10,18–21).
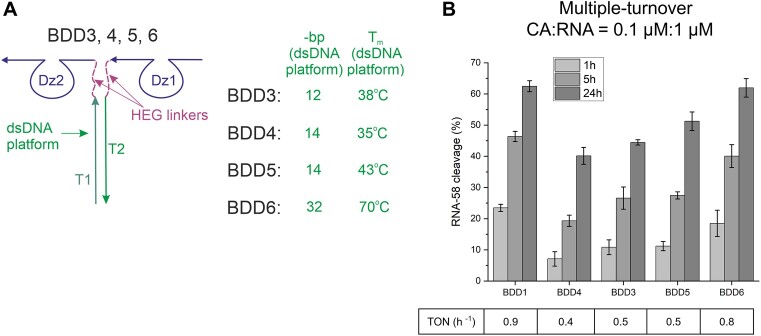
Under multiple-turnover conditions, BDD1 outperformed BDD3 (Figure 3), which can be explained by the low stability of the dsDNA platform (Tm ∼ 38 °C) in BDD3 at the reaction conditions (37 °C). This Tm indicates that less than half of the Dz1 and Dz2 molecules are associated in BDD3 under the experimental conditions. Increasing the melting temperature of the dsDNA platform of BDD3 should increase the cooperativity of Dz1 and Dz2 in RNA binding and thus bring its performance closer to that of BDD1. To verify this hypothesis, we designed a series of BDD4–6 with different Tm of the dsDNA platform (Figure 5A).
Figure 5.
RNA cleavage activity depends on the stability of the dsDNA platform in BDDs. (A) Design of BDD constructs with different lengths of the dsDNA platform with noted number of base pairs (bp) and melting temperature (Tm). (B) RNA-58 cleavage by BDD1,3–6 under multiple-turnover conditions. TON was calculated after 5 h of incubation as described in the Materials and Methods. The data is the average values of three independent experiments.
As expected, increasing the Tm of the dsDNA platform caused a consecutive increase in BDD activity, with the most stable dsDNA platform of BDD6 having TON comparable with that of BDD1 under multiple-turnover conditions (Figure 5 and Supplementary Figure S2). This proves our hypothesis that the limiting stage in the cleavage of a folded RNA is RNA-binding, and this problem can be addressed by increasing binding cooperativity by stabilizing the dsDNA platform.
BDD1 has a greater RNA-cleaving activity than the optimized individual Dz
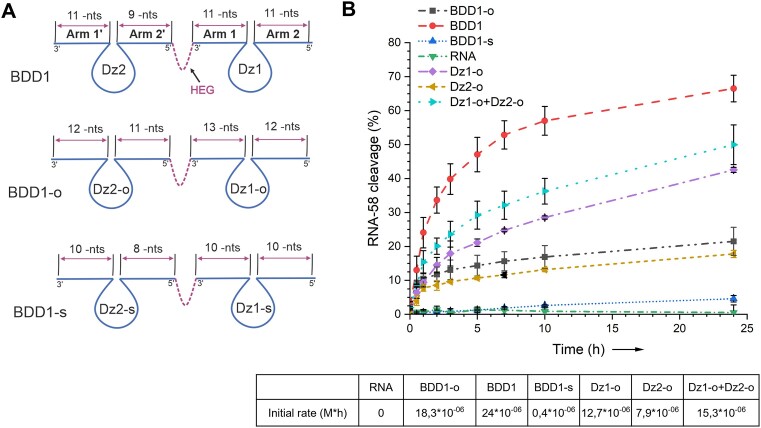
It was shown earlier that the increase in the affinity of RNA-binding arms in Dz agents increases the cleavage efficiency of folded RNA substrates (11–14). However, none of the prior studies with one recent exception (36) have optimized the structures of Dz agents for the cleavage of folded RNA. We reasoned here that a fair comparison of the cleavage efficiency and selectivity of the bivalent agents should be done with optimal rather than suboptimal Dz of the traditional design. Therefore, we performed optimization of Dz1 and Dz2 RNA-binding arms under multiple-turnover conditions (Supplementary Figure S4).
The most active Dz1_13/12 and Dz2_12/11 had TONs of 0.35 h−1 and 0.13 h−1, respectively, which was a 3.5- and 1.3 times higher than suboptimal Dz1 and Dz2, respectively. These two Dz were named ‘Dz1-o’ and ‘Dz2-o’, where ‘o’ stands for ‘optimized’. In tandem, Dz1-o and Dz2-o cleaved RNA-58 with a TON of 0.4 h−1. This demonstrates little or no cooperativity in RNA-58 binding (Supplementary Figures S4 and S5), which is similar to that of the suboptimal Dz1 and Dz2 under multiple-turnover conditions (Figure 3A). Time dependence of RNA-58 cleavage demonstrated comparable initial reaction rates for BDD1 than for Dz1-o and Dz2-o. However, after 24 h, BDD1 outperformed Dz1-o and Dz2-o acting individually or in tandem (Figure 6B). This data reflects product inhibition of long-armed Dz1-o and Dz2-o. Therefore, BDD1 is likely to suffer less from product inhibition than the optimized Dz. This confirms our hypothesis that the BDD design can help find a balance between tight substrate binding and efficient product release (Scheme 1).
Figure 6.
Cleavage of RNA-58 by different Dz agents under multiple-turnover conditions. (A) Designs of BDD and Dz agents with initial (BDD1), optimized (BDD1-o) and shortened (BDD1-s) variants. (B) Time-dependence of RNA-58 cleavage. RNA-58 (1 μM) was incubated with Dz agents (0.1 μM) in Buffer 1 at 37°C for 0–24 h (9 time points). The cleavage products were separated from RNA-58 by PAGE (Supplementary Figure S4) followed by their quantification as described in Materials and Methods. The initial cleavage rates are in the table below the graph. The data is the average values of three independent experiments.
Attempt to optimize BDD1
Next, we varied the length of RNA-binding arms to further improve the RNA-cleaving efficiency of BDD1 under multiple-turnover conditions (Figure 6A). BDD1-o was designed by linking Dz1-o and Dz2-o via a polyethylene glycol linker (HEG); BDD1-s contained all four arms shortened by 1 nt (Figure 6A). BDD1-o was deemed to increase RNA-58 binding affinity, while BDD1-s was supposed to improve product release, as compared to BDD1. Still, BDD1 had the greatest initial cleavage rate (Figure 6B). The time dependence curve for BDD1-o reached a plateau at an earlier time point than for BDD1, thus indicating significant product inhibition. BDD1-s demonstrated reduced activity, likely due to the lower affinity to RNA-58 substrate. This data indicates that Dz agents of traditional design (with Tm of RNA binding arms ≤ 37 °C) might be good candidates for building tandems of Dz in BDD-like constructions, which have the balance of tight RNA-binding and efficient product release.
BDD-1 remains active at lower concentrations
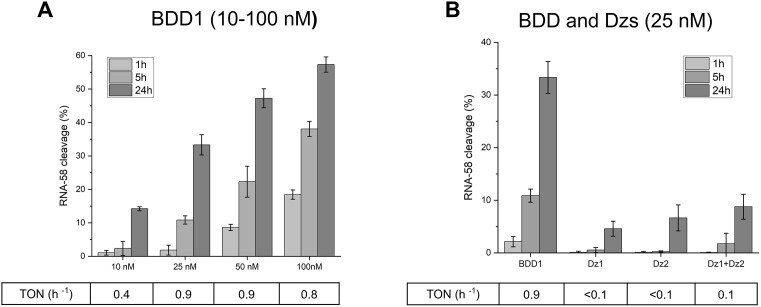
When OGT agents act in the intracellular environment, their concentrations are poorly controlled because of inefficient delivery, poor exosomal escape, and nuclease degradation (15,16). Therefore, OGT agents active at low concentrations are preferable. Here, we investigated the efficiency of RNA-58 cleavage by reduced concentrations of BDD1. The TON for BDD1-catalyzed RNA-58 cleavage remained nearly constant as the concentration was reduced from 100 to 25 nM and dropped 2-fold only at 10 nM BDD1 (Figure 7A). At the same time, Dz1 and Dz2 acting individually or in a tandem exhibited a reduced TON of 0.1 h−1 or lower when their concentrations were reduced to 50 nM (Supplementary Figure S6). Figure 7B compares the performances of BDD1, Dz1 and Dz2 at 25 nM. BDD1 significantly outperformed Dz1 + Dz2 tandem after 5 h of reaction (0.9 h−1 versus 0.1 h−1). This experiment suggests that at low concentrations of cleaving agents in cells, BDD1 design may offer an even more pronounced advantage than in the model system explored in this study.
Figure 7.
BDD1 remains active at low concentrations. (A) The efficiency of RNA-58 cleavage by BDD1 at different concentrations (10, 25, 50, or 100 nM) of the cleaving agent. (B) The efficiency of RNA-58 cleavage in the presence of 25 nM BDD1, Dz1, Dz2, Dz1 + Dz2. TON was calculated after 5 h of incubation. The data is the averaged values of three independent experiments.
BDD selectivity can be modulated by changing the Tm of the dsDNA platform
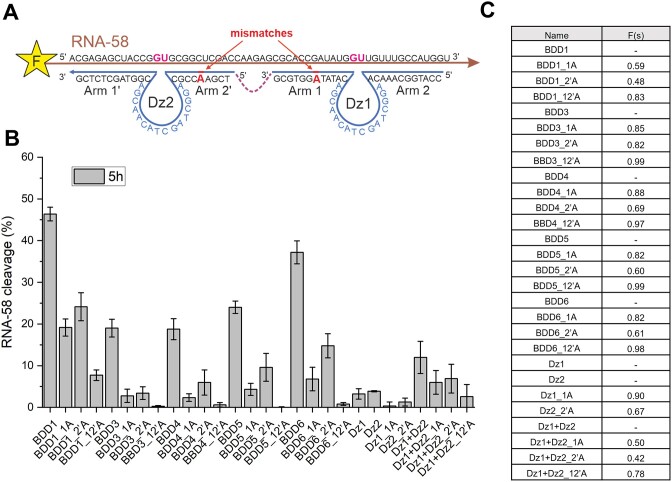
To assess the selectivity of BDD constructs, we incorporated mismatches at the same position of the RNA-binding arms in Dz1, Dz2, BDD1, 3 and 4′ as shown in Figure 8A. Fully matched BDD constructs were compared with their mismatched counterparts having single-nucleotide substitutions in Arm 1 or Arm 2′ or both (Figure 8). The selectivity of each Dz agent was quantified by the selectivity factor F(s), where F(s) approaching 1 reflects the greatest selectivity (Figure 8С). BDD1 demonstrated only moderate selectivity: only the presence of both mismatches resulted in a high F(s) of 0.83 (Figure 8B and C, Supplementary Figure S7). This is in line with the affinity/specificity dilemma: the high affinity of BDD1 to RNA-58 results in its low selectivity (14,31). BDD3 and BDD4 demonstrated the highest F(s) of all BDD showing no significant RNA cleavage in the presence of single mismatches (Figure 8C, Supplementary Figure S7), which correlates with the lowest Tm of their dsDNA scaffolds (38°C and 35°C, respectively). BDD3 and BDD4 can be used as highly selective alternatives of BDD1 if high specificity of gene silencing needs to be achieved.
Figure 8.
Selectivity of RNA cleavage by different Dz agents. (A) RNA-58 was cleaved by Dz agents having one nucleotide mismatch in one (1A or 2′A) or two (12′A) arms. (B) Relative efficiency of RNA-58 cleavage under multiple-turnover conditions (5 h, 1 μM RNA, 0.1 μM Dz agents). (C) Table with selectivity factors F(s) for the RNA-cleaving BDDs and Dz with or without mismatches. F(s) was calculated as (1 – TONmm / TONm), where TONmm and TONm and the turnover numbers for mismatched and matched cleaving agents, respectively. The data is the averaged values of three independent experiments.
Both RNA binding and increased chance of RNA cleavage contribute to BDD1 cleavage efficiency
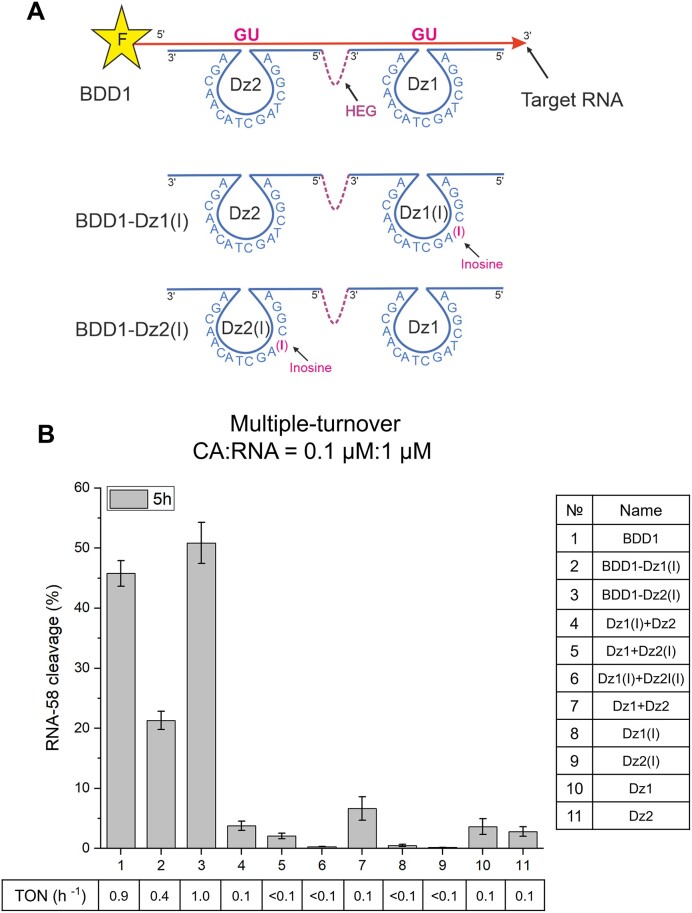
According to our hypothesis, two factors contribute to the efficiency of BDD: (i) facilitated binding of a folded RNA and (ii) increased (by a factor of 2) probability of RNA cleavage due to the action of the two catalytic cores. In this part of the study, we assessed the relative contributions of the two factors to the BDD1 cleavage activity by designing BDD1-Dz1(I) and BDD1-Dz2(I) with a catalytically inactive Dz1 and Dz2 fragments, respectively. Thymidine in position 4 (T4) of the catalytic core was substituted with inosine (Figure 9A), as described earlier by Zaborowska et al. (37). Dz1(I) and Dz2(I) were completely inactive in RNA cleavage (Figure 9B, bars 9 and 10, Supplementary Figure S8). No significant cleavage improvement was observed when inactivated Dzs were added to their corresponding active partner in tandem (compare bar 5 with 12 and bar 6 with 11 in Figure 9B). On the contrary, both BDD1 versions containing one inactive Dz maintained visible RNA-cleaving activity: BDD1 with deactivated Dz1 showed a <2-fold decrease, while BDD1 with inactivated Dz2 demonstrated comparable to BDD1 activity (Figure 9B, Compare bars 12 and 3 to bar 1). We also conducted additional experiments with BDD containing catalytic cores fully substituted with non-nucleotide HEG linkers (Supplementary Figure S9). The experiments revealed that RNA cleavage in BDD with HEG substitutions was 2 times lower than that of BDD versions with inosine substitutions. This difference can be explained by the contribution of the nucleotides of the catalytic core to RNA binding by Dz, a phenomenon reported recently (23,38). At the same time, the relative cleavage activity of HEG-core BDDs and Dz remained similar to that of inosine BDDs and Dz (Figure 9B, 3 and 4; Supplementary Figure S9). No changes were observed with tandem or single Dz. We, therefore, concluded that both factors: (i) the increased RNA binding affinity due to cooperative binding of the target by multiple RNA binding arms and (ii) the greater chance of RNA cleavage by the two catalytic cores contribute to the improved efficiency of BDD1 in cleaving RNA-58.
Figure 9.
Influence of mutation in the Dz catalytic core on RNA cleavage efficiency. (A) Localization I > T mutation in BDD1. (B) Relative efficiency of RNA-58 cleavage by different Dz agents under multiple turnover conditions and corresponding turnover numbers (TON). The data is the averaged values of three independent experiments.
From bivalent to trivalent Dz agents
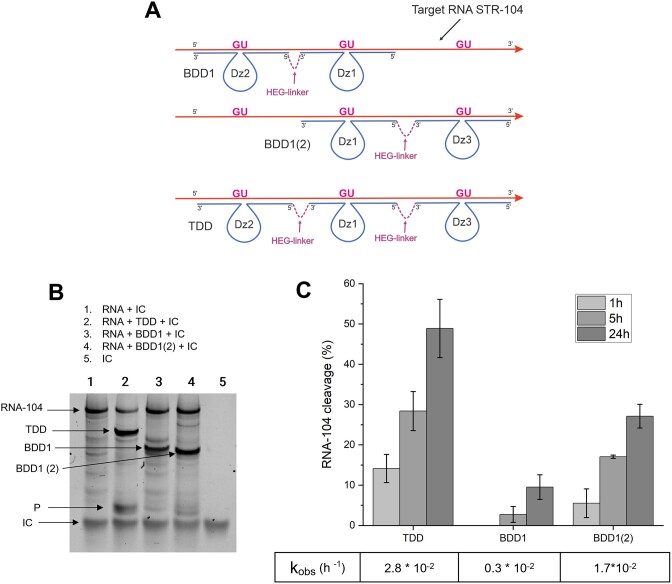
Next, we investigated if the success of BDD1 can be further extended to trivalent agents for cleaving an RNA with an even more stable secondary structure. We, therefore, designed RNA-104, having ΔG of −48 kcal/mol, Figure 2B. In addition to BDD1, we designed BDD1(2) and a trivalent Dz device (TDD), as illustrated in Figure 10A. Dz3 fragment of TDD was equipped with RNA-binding arms having Tm of 32 and 33°C (Supplementary Table S3). TDD demonstrated the highest catalytic activity among the three constructs, with kobs = 2.8 × 10−2 h−1, which was greater than the sum of the rate constants for BDD1 and BDD1(2). We concluded that the incorporation of additional Dz cores in a cooperative tandem provides a way to improve RNA cleavage efficiency by Dz cleaving agents. Reduction in the concentration of the cleaving agents significantly slowed down the RNA-104 cleavage reactions (Supplementary Figure S10). However, the limitations of this strategy require further investigation.
Figure 10.
TDD cleaves RNA-104 more efficiently than BDD1. (A) Designs for TDD, BDD1 and BDD1(2). BDD1(2) repeats the design of BDD1 and targets the RNA-104 region at the 3′-end. The experiments were performed with RNA:BDD ratio = 1:2 (RNA = 1 μM, BDD = 2 μM). (B) PAGE analysis of the RNA-104 cleavage products in the presence of different cleaving agents. The gel was stained with GelRed. The cleavage efficiency was quantified based on the reduction of the RNA-104 band intensity. IC is the band corresponding to the internal control oligonucleotide used for normalization for quantification of the RNA cleavage activity. Band P corresponds to the expected cleavage products (not used for quantification). (C) RNA cleavage efficiency in the presence of TDD, BDD1 or BDD1(2) with kobs values indicated in the table below the graph. The data is the averaged values of three independent experiments.
Bivalent Dz agents have additive efficiency in cleaving a ‘relaxed’ RNA-60 substrate
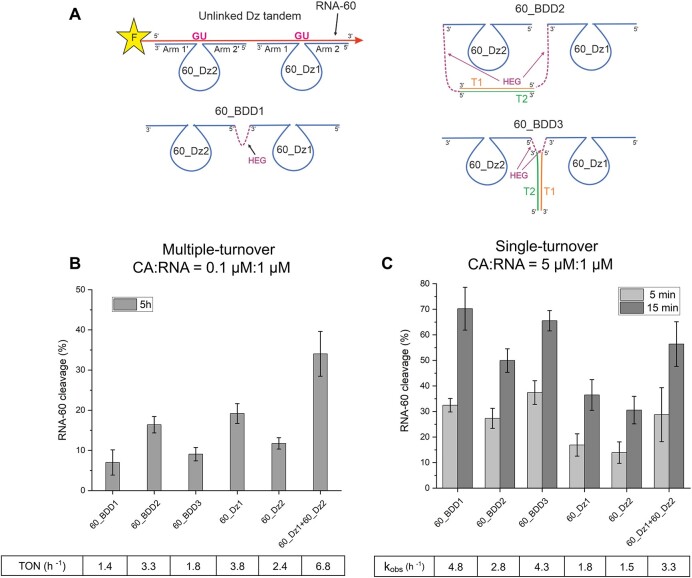
According to our results, bivalent Dz agents increased RNA cleavage rate by facilitating the binding stage of the catalytic cycle. In this scenario, the Dz tandem approach should demonstrate a less pronounced improvement in cleaving relaxed RNA substrates as compared with conventional Dz agents. We, therefore, targeted RNA-60 (folding ΔG of −6.30 kcal/mol, Figure 2C) of a sequence unrelated to RNA-58. Cleaving agents 60_Dz1 and 60_Dz2 were designed according to the conventional rules (11,12,36) and then were linked together to form 60_BDD1, 2 and 3 constructions.
For both multiple and single turnover experiments, the tandem of 60_Dz1 or 60_Dz2 cleaved RNA-60 with additive efficiency equal to the sum of the efficiencies of 60_Dz1 and 60_Dz2, which suggests the absence of cooperativity in RNA binding by the two agents (Figure 11B and C). Under multiple-turnover conditions, 60_BDD1, 2 and 3 did not improve the activity of individual 60_Dz1 or 60_Dz2 and performed less efficiently than the tandem of 60_Dz1 or 60_Dz2 (Figure 11B), which can be explained by the inhibition of BDD by the RNA cleavage products. Under single-turnover conditions, the kobs values obtained for BDD1 and BDD3 (4.8 h−1 and 4.3 h−1, respectively) were twice as high as those for individual Dz1 and Dz2 (1.8 h−1 and 1.5 h−1, Figure 11C), which reflects a tighter binding of the RNA substrate by the BDD constructs in comparison with the Dz of a traditional design. This data experimentally proves the efficiency of a conventional Dz design in cleaving relaxed RNA fragments. However, such a success demonstrated by the conventional Dz agents in these experiments may not be translated to the intracellular conditions with low mRNA concertation when the RNA binding step limits the reaction. Under such conditions BDD agents might still be preferable over Dz of traditional design.
Figure 11.
Bivalent agents only moderately improve cleavage of RNA-60 substrate. (A) Dz agents for cleaving RNA-60. (B) Percent of RNA-60 (1 μM) cleavage by Dz agent (0.1 μM) under multiple-turnover condition. (C) Percent of RNA-60 (1 μM) cleavage by Dz agents (5 μM) under single-turnover conditions. Single-turnover data obtained for a Dz:RNA ratio of 2:1 is presented in Supplementary Figure S11. Incubation time was 5 and 15 min, for dark grey and light grey bars, respectively. The values for kobs were calculated as described in the Materials and Methods. The data is the averaged values of three independent experiments.
Discussion
Bivalent and multivalent recognition may improve both the affinity and selectivity of biochemical recognition (4–8). This approach is well known in hybridization probes for nucleic acid analysis (7,8,39) but it has been underexplored in mRNA recognition by OGT agents. Bivalent ASO and siRNA have been suggested earlier with moderate success. For example, Kandimalla's group investigated the cooperative binding of two antisense oligonucleotides (ASO) to inhibit HIV-1 replication (40). Although cooperativity did not increase knockdown efficiency, it increased specificity. Recently, Khvorova and colleagues studied divalent small interfering RNA (di-siRNA). Di-siRNA targeting the HTT gene in tissues of the CNS demonstrated promising results with a low rate of off-target effects (41). Both technologies relied on protein-dependent mRNA binding, which is hard to predict since mRNA/OGT agent complexes would be stabilized by the protein (RNase H or RISC complex) binding in the intracellular environment. Therefore, the balance of selectivity/affinity is difficult to control as it will depend on intracellular enzyme concentrations. Dz agents can bind and cleave mRNA in a protein-independent manner and, therefore, open a route to the rational design of tunable fully artificial DNA nanoconstructs with an arbitrary architecture, as exemplified by recent studies (10,17–21).
Earlier, a hairpin ribozyme-based 'twin ribozyme’ with two joint catalytic cores linked by a nucleotide bridge was reported to maintain its activity in cleaving a specific RNA substrate (42). In another study, a hammerhead ribozyme-based construction (Maxizyme) that targeted two distinct sites of two different mRNAs was proposed (43). However, no systematic optimization of the cleavage of folded RNA targets was done in these works. These studies were not continued probably because ribozymes were considered to be less promising gene therapy agents than Dz due to their lower biochemical stability and higher cost (10,11). Unwalla and colleagues used two Dz agents to target two remote cite of a human immunodeficiency virus (HIV) RNA. However, the double Dz system did not show improvement in the RNA-cleaving activity in comparison with a single Dz (44). Chen et al. used a similar approach for targeting the β-lactamase gene in ampicillin-resistant bacteria and achieved almost 2-fold improvement in gene inhibition (45). Recently, Chaput and colleagues used biologically stable XNAzymes X10-23, which were designed to target two distant sites of GFP mRNA (12). Two non-tandem XNAzymes targeting internal and 3′-UTR sites of GFP mRNA caused successful suppression of GFP fluorescence in HEK293 cells, thus demonstrating higher effectiveness compared to individual single XNAzyme (12). In all these studies, double Dz systems did not pursue cooperativity in RNA binding since the targeted sites were separated from each other by dozens of nucleotides. To date, neither multivalency nor polyvalency has been explored to achieve cooperative RNA binding by Dz agents.
Here, we used two Dz agents targeting two cleavage sites separated by 30 nt in the primary structure of RNA. Under single-turnover conditions, the Dz1 + Dz2 tandem demonstrated a 7-fold improvement in RNA cleavage activity over Dz1 and Dz2 acting separately. Considering that the catalytic activity did not depend on the product release stage (k3), we concluded that the majority of the 7-fold increase was due to the cooperative binding of the RNA target by the two Dz. Indeed, when we applied the same approach for cleavage of a ‘relaxed’ RNA-60 (Figure 10), we observed an additive effect, where the sum of the cleavage rates of Dz1 and Dz2 working separately was equal to that of Dz1 + Dz2 tandem. We, therefore, concluded that the use of tandems of Dz agents targeting neighboring RNA sites is a promising strategy for improving the cleavage of folded RNA substrates including natural mRNA.
Furthermore, Dz1 and Dz2 were covalently linked to form BDD1 or combined into DNA nanostructures (BDD2–6, Figure 1). Under single-turnover conditions, BDD1 and BDD3 demonstrated about 17-fold improvement in kobs in comparison with individual Dz1 or Dz2 and about 2.4-fold improvement over the Dz1 + Dz2 tandem. Under multiple-turnover conditions, BDD1 outperformed individual Dz1 and Dz2, as well as the Dz1 + Dz2 tandem 9 and 4.5 times, respectively. Therefore, assembling a tandem of Dz in a single structure can further improve mRNA cleavage efficiency due to the increase in RNA binding cooperativity.
Expectedly, the selectivity of BDD1 was reduced, as demonstrated by the moderate differentiation factors of 0.48 and 0.59, while Dz1 or Dz2 alone had a greater selectivity (F(s) of 0.90 and 0.67, Figure 8). This phenomenon is known as the affinity/specificity dilemma: the higher the hybridization probe affinity to a target, the lower the specificity (31). We further demonstrated that the affinity and specificity of BDD can be adjusted by changing the dsDNA platform Tm (Figures 1E, 5 and 8). For example, BDD3 was expected to have less than 50% of Dz1 and Dz2 associated in the nanostructure under the reaction conditions. This expectedly reduced its affinity to the RNA substrate, which was reflected by a 2.3-fold decrease in the turnover number in comparison with the best-performing BDD1. At the same time, the selectivity of BDD1 was comparable with that of individual Dz1 and Dz2 (Figure 8). Since the BDD3 was still 7 times more active than individual Dz1 or Dz2, we concluded that BDD3 design is optimal in terms of the affinity/selectivity balance.
Further, we demonstrated that the idea of the bivalent Dz can be extended to trivalent Dz agents if there is a need to cleave RNA with even greater structural stability. TDD agents outperformed BDD up to 3-fold. Overall, we concluded that the multivalent design could offer improvement in RNA cleavage in comparison with a single Dz depending on the thermodynamic stability of the RNA structure - the greater the stability of RNA, the greater improvement by multivalent Dz can be expected. Therefore, multivalent Dz is a promising tool for cleavage of folded biological RNA.
This study was possible due to using synthetic RNA oligonucleotide model substrates. Such RNA substrates have the sequences of natural mRNA targets, but poorly reflect their secondary structures. However, we believe that experiments with such folded RNA substrates closer reflect the kinetics of biological RNA cleavage than the experiments with short (15–20 nt) linear RNA substrates as they provide an energy barrier for Dz agents to bind RNA before cleavage. This binding step might be a limiting stage for suppression of natural mRNA in vivo. We, therefore, strongly advocate for using long model RNA substrates in optimization of Dz agents, as was expressed by us earlier (36) and supported recently by Zhang et al. (46).
In conclusion, multivalent Dz constructions increase cleavage rates of RNA folded in complex structures both under single and multiple turnover conditions due to facilitation of the RNA binding stage without suffering from product inhibition. The specificity of multivalent Dz agents can be modulated by controlling the melting temperature of a DNA platform connecting them. Extremely stable RNA fragments can be targeted by tri- and multivalent Dz agents. Overall, this study introduces multivalent Dz agents as an efficient and specific alternative to the traditional Dz agents. The concept can be extended towards ribozymes and antisense agents for mRNA suppression in living cells. Future studies should include the protection of multivalent Dz constructions from nuclease degradation by chemical modifications and assessing their efficiency in mRNA suppression in cell culture.
Supplementary Material
Acknowledgements
D.M.K. is grateful to Dr. Yulia V. Gerasimova for the discussion and careful reading of the manuscript.
Contributor Information
Mikhail V Dubovichenko, Laboratory of Frontier Nucleic Acid Technologies in Gene Therapy of Cancer, SCAMT Institute, ITMO University, Saint-Petersburg, 191002, Russia.
Michael Batsa, Laboratory of Frontier Nucleic Acid Technologies in Gene Therapy of Cancer, SCAMT Institute, ITMO University, Saint-Petersburg, 191002, Russia.
Gleb A Bobkov, Laboratory of Frontier Nucleic Acid Technologies in Gene Therapy of Cancer, SCAMT Institute, ITMO University, Saint-Petersburg, 191002, Russia.
Gleb S Vlasov, Laboratory of Frontier Nucleic Acid Technologies in Gene Therapy of Cancer, SCAMT Institute, ITMO University, Saint-Petersburg, 191002, Russia.
Ahmed A El-Deeb, Laboratory of Frontier Nucleic Acid Technologies in Gene Therapy of Cancer, SCAMT Institute, ITMO University, Saint-Petersburg, 191002, Russia.
Dmitry M Kolpashchikov, Laboratory of Frontier Nucleic Acid Technologies in Gene Therapy of Cancer, SCAMT Institute, ITMO University, Saint-Petersburg, 191002, Russia; Chemistry Department, University of Central Florida, Orlando, FL 32816, USA; Burnett School of Biomedical Sciences, University of Central Florida, Orlando, FL 32816, USA; National Center for Forensic Science, University of Central Florida, Orlando, FL, 32816, USA.
Data availability
Secondary structures of RNA and melting temperature (Tm) of oligonucleotides were estimated by using UNAFold Web Server (http://www.unafold.org/) with RNAfold (http://www.unafold.org/mfold/applications/rna-folding-form.php) and Two State Melting Hybridization applications (http://www.unafold.org/Dinamelt/applications/two-state-melting-hybridization.php).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
The project was supported by project of ITMO University's priority 2030 Development strategy №922017. Funding for open access charge: personal contribution.
Conflict of interest statement. The authors declare that they have no conflict of interest.
References
- 1. Xia X., Zhang G., Ciamarra M.P., Jiao Y., Ni R. The role of receptor uniformity in multivalent binding. JACS Au. 2023; 3:1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubacheva G.V., Curk T., Richter R.P. Determinants of superselectivity practical concepts for application in biology and medicine. Acc. Chem. Res. 2023; 56:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 4. Chittasupho C. Multivalent ligand: design principle for targeted therapeutic delivery approach. Ther. Deliv. 2012; 3:1171–1187. [DOI] [PubMed] [Google Scholar]
- 5. Böhmer V.I., Szymanski W., Feringa B.L., Elsinga P.H. Multivalent probes in molecular imaging: reality or future?. Trends Mol. Med. 2021; 27:379–393. [DOI] [PubMed] [Google Scholar]
- 6. Durai S., Mani M., Kandavelou K., Wu J., Porteus M.H., Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005; 33:5978–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolpashchikov D.M. Binary probes for nucleic acid analysis. Chem. Rev. 2010; 110:4709–4723. [DOI] [PubMed] [Google Scholar]
- 8. Kolpashchikov D.M. Evolution of hybridization probes to DNA machines and robots. Acc. Chem. Res. 2019; 52:1949–1956. [DOI] [PubMed] [Google Scholar]
- 9. Breaker R.R., Joyce G.F. A DNA DNAzyme that cleaves RNA. Chem. Biol. 1994; 1:223–229. [DOI] [PubMed] [Google Scholar]
- 10. Su J., Sun C., Du J., Xing X., Wang F., Dong H. RNA-cleaving DNAzyme-based amplification strategies for biosensing and therapy. Adv. Healthc. Mater. 2023; 12:e2300367. [DOI] [PubMed] [Google Scholar]
- 11. Fokina A.A., Stetsenko D.A., François J.C. DNA DNAzymes as potential therapeutics: towards clinical application of 10-23 DNAzymes. Expert Opin. Biol. Ther. 2015; 15:689–711. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y., Nguyen K., Spitale R.C., Chaput J.C. A biologically stable DNAzyme that efficiently silences gene expression in cells. Nat. Chem. 2021; 13:319–326. [DOI] [PubMed] [Google Scholar]
- 13. Thomas I.B.K., Gaminda K.A.P., Jayasinghe C.D., Abeysinghe D.T., Senthilnithy R. DNAzymes, novel therapeutic agents in cancer therapy: a review of concepts to applications. J. Nucleic Acids. 2021; 2021:9365081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nedorezova D.D., Dubovichenko M.V., Belyaeva E.P., Grigorieva E.D., Peresadina A.V., Kolpashchikov D.M. Specificity of oligonucleotide gene therapy (OGT) agents. Theranostics. 2022; 12:7132–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larcher L.M., Pitout I.L., Keegan N.P., Veedu R.N., Fletcher S. DNAzymes: Expanding the Potential of Nucleic Acid Therapeutics. Nucleic Acid Ther. 2023; 33:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao L., Zhao Y., Yang M., Luan G., Du T., Deng S., Jia X. A promising nucleic acid therapy drug: DNAzymes and its delivery system. Front. Mol. Biosci. 2023; 10:1270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan J., Ran M., Shen X., Zhang H. Therapeutic DNAzymes: from structure design to clinical applications. Adv. Mater. 2023; 35:e2300374. [DOI] [PubMed] [Google Scholar]
- 18. Spelkov A.A., Goncharova E.A., Savin A.M., Kolpashchikov D.M. Bifunctional RNA-targeting deoxyribozyme nanodevice as a potential theranostic agent. Chem. - A Eur. J. 2020; 26:3489–3493. [DOI] [PubMed] [Google Scholar]
- 19. Nedorezova D.D., Fakhardo A.F., Nemirich D.V., Bryushkova E.A., Kolpashchikov D.M. Towards DNA nanomachines for cancer treatment: achieving selective and efficient cleavage of folded RNA. Angew. Chem. Int. Ed. 2019; 58:4654–4658. [DOI] [PubMed] [Google Scholar]
- 20. Molden T.A., Niccum C.T., Kolpashchikov D.M. Cut and paste for cancer treatment: a DNA nanodevice that cuts out an RNA marker sequence to activate a therapeutic function. Angew. Chem. Int. Ed. 2020; 59:21190–21194. [DOI] [PubMed] [Google Scholar]
- 21. Gomes de Oliveira A.G., Dubovichenko M.V., ElDeeb A.A., Wanjohi J., Zablotskaya S., Kolpashchikov D.M. RNA-cleaving DNA thresholder controlled by concentrations of miRNA cancer marker. ChemBioChem. 2021; 22:1750–1754. [DOI] [PubMed] [Google Scholar]
- 22. Donini S., Clerici M., Wengel J., Vester B., Peracchi A. The advantages of being locked. J. Biol. Chem. 2007; 282:35510–35518. [DOI] [PubMed] [Google Scholar]
- 23. Gupta A., Mishra A., Puri N. Peptide nucleic acids: advanced tools for biomedical applications. J. Biotechnol. 2017; 259:148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen K., Malik T.N., Chaput J.C. Chemical evolution of an autonomous DNAzyme with allele-specific gene silencing activity. Nat. Commun. 2023; 14:2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santoro S.W., Joyce G.F. A general purpose RNA-cleaving DNA DNAzyme. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santoro S.W., Joyce G.F. Mechanism and utility of an RNA-cleaving DNA DNAzyme. Biochemistry. 1998; 37:13330–13342. [DOI] [PubMed] [Google Scholar]
- 27. Fu L., Cao Y., Wu J., Peng Q., Nie Q., Xie X. UFold: fast and accurate RNA secondary structure. Nucleic Acids Res. 2022; 50:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hobbs G.A., Der C.J., Rossman K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016; 129:1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lima Z.S., Ebadi M.R., Amjad G., Younesi L. Application of imaging technologies in breast cancer detection: a review article open access maced. J. Med. Sci. 2019; 7:838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waarts M.R., Stonestrom A.J., Park Y, Levine R.L. Targeting mutations in cancer. J. Clin. Invest. 2022; 132:e154943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Demidov V.V., Frank-Kamenetskii M.D. Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem. Sci. 2004; 29:62–71. [DOI] [PubMed] [Google Scholar]
- 32. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sundin G.W., Bender C.L. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 1996; 5:133–143. [DOI] [PubMed] [Google Scholar]
- 34. Song N., Wang Y., Gu X.D., Chen Z.Y., Shi L.B. Effect of siRNA-mediated knockdown of eIF3c gene on survival of colon cancer cells. J. Zhejiang Univ. Sci. B. 2013; 14:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drozd V.S., Eldeeb A.A., Kolpashchikov D, Nedorezova D.D. Binary antisense oligonucleotide agent for cancer marker-dependent degradation of targeted RNA. Nucleic Acid Ther. 2022; 32:412–420. [DOI] [PubMed] [Google Scholar]
- 36. Nedorezova D.D., Dubovichenko M.V., Kalnin A.J., Nour M.A.Y., Eldeeb A.A., Ashmarova A.I., Kurbanov G.F., Kolpashchikov D.M Cleaving folded RNA with DNAzyme 10-23 agents. ChemBioChem. 2023; 25:e202300637. [DOI] [PubMed] [Google Scholar]
- 37. Zaborowska Z., Fürste J.P., Erdmann V.A., Kurreck J. Sequence requirements in the catalytic core of the ‘10-23’ DNA DNAzyme. J. Biol. Chem. 2002; 277:40617–40622. [DOI] [PubMed] [Google Scholar]
- 38. Borggräfe J., Gertzen C.G.W., Viegas A., Gohlke H., Etzkorn M. The architecture of the 10-23 DNAzyme and its implications for DNA-mediated catalysis. FEBS J. 2023; 290:2011–2021. [DOI] [PubMed] [Google Scholar]
- 39. Hussein Z., Nour M.A.Y., Kozlova A.V., Kolpashchikov D.M., Komissarov A.B., El-Deeb A.A. DNAzyme Nanomachine with Fluorogenic Substrate Delivery Function: Advancing Sensitivity in Nucleic Acid Detection. Anal Chem. 2023; 95:8667–18672. [DOI] [PubMed] [Google Scholar]
- 40. Kandimalla E.R., Manning A., Lathan C., Byrn R.A., Agrawal S. Design, biochemical, biophysical and biological properties of cooperative antisense oligonucleotides. Nucleic Acids Res. 1995; 23:3578–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alterman J.F., Godinho B.M.D.C., Hassler M.R., Ferguson C.M., Echeverria D., Sapp E., Haraszti R.A., Coles A.H., Conroy F., Miller R. et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 2019; 37:884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmidt C., Rüdiger W., Müller S RNA double cleavage by a hairpin-derived twin ribozyme. Nucleic Acids Res. 2000; 28:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iyo M., Kawasaki H., Taira K. Maxizyme technology. Methods Mol. Biol. 2004; 252:257–265. [DOI] [PubMed] [Google Scholar]
- 44. Unwalla H., Banerjea A.C. Novel mono- and di-DNA-DNAzymes targeted to cleave TAT or TAT-REV RNA inhibit HIV-1 gene expression. Antiviral Res. 2001; 277:127–139. [DOI] [PubMed] [Google Scholar]
- 45. Chen F., Wang R., Zeng Z., Zhang H., Zhang J. Inhibition of ampicillin-resistant bacteria by novel mono-DNAzymes and Di-DNAzyme targeted to B-lactamase mRNA. Oligonucleotides. 2004; 14:80–89. [DOI] [PubMed] [Google Scholar]
- 46. Yan J., Ran M., Zhang H. A scientific debate: the sword that cleaves chaos of DNAzyme catalysis research. Biomed. Technol. 2023; 4:21–23. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Secondary structures of RNA and melting temperature (Tm) of oligonucleotides were estimated by using UNAFold Web Server (http://www.unafold.org/) with RNAfold (http://www.unafold.org/mfold/applications/rna-folding-form.php) and Two State Melting Hybridization applications (http://www.unafold.org/Dinamelt/applications/two-state-melting-hybridization.php).