Abstract
The process of induced pluripotent stem cells (iPSCs) reprogramming involves several crucial events, including the mesenchymal-epithelial transition (MET), activation of pluripotent genes, metabolic reprogramming, and epigenetic rewiring. Although these events intricately interact and influence each other, the specific element that regulates the reprogramming network remains unclear. Dux, a factor known to promote totipotency during the transition from embryonic stem cells (ESC) to 2C-like ESC (2CLC), has not been extensively studied in the context of iPSC reprogramming. In this study, we demonstrate that the modification of H3K18la induced by Dux overexpression controls the metabolism-H3K18la-MET network, enhancing the efficiency of iPSC reprogramming through a metabolic switch and the recruitment of p300 via its C-terminal domain. Furthermore, our proteomic analysis of H3K18la immunoprecipitation experiment uncovers the specific recruitment of Brg1 during reprogramming, with both H3K18la and Brg1 being enriched on the promoters of genes associated with pluripotency and epithelial junction. In summary, our study has demonstrated the significant role of Dux-induced H3K18la in the early reprogramming process, highlighting its function as a potent trigger. Additionally, our research has revealed, for the first time, the binding of Brg1 to H3K18la, indicating its role as a reader of histone lactylation.
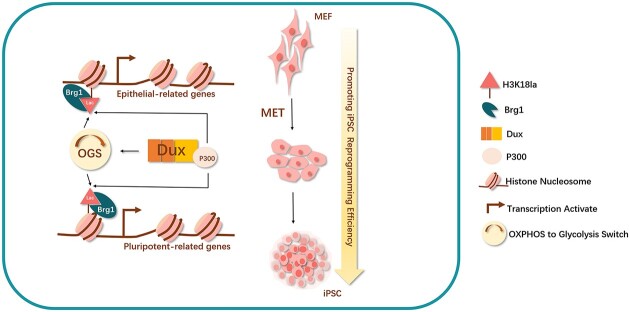
Graphical Abstract
Graphical Abstract.
Introduction
The role of Dux in embryonic development has garnered significant attention in recent years due to its ability to facilitate the conversion of ESCs into totipotent-like stem cells with enhanced developmental potential (1). In the context of mouse embryonic development, Dux mRNA exhibits high transcription levels specifically during the early 2-cell stage, coinciding with the initiation of the first wave of zygotic genome activation (ZGA) (2). While it has been demonstrated that Dux is not indispensable for ZGA, certain evidence suggests its crucial involvement in the transition of 2CLCs (3,4). 2CLCs represent a subset of ESCs that exhibit spontaneous expression of mouse 2-cell embryo-specific genes and repeats. In comparison to ESC, 2CLCs exhibit higher levels of H3K27ac, H3K4me3 and H3K4me1 modifications (5). Furthermore, 2CLCs have been shown to possess an expanded pluripotency compared to ESC, as they contribute not only to all three germ layers but also to the trophectoderm (TE) in chimera assays (6). Previous reports have indicated that the transition of 2CLCs is initiated by the downregulation of Myc and Dnmt1, leading to an increase in Dux expression. Furthermore, the exit from the 2CLCs state is initiated by the inhibition of Dux mRNA by Smg7/Upf1 (7,8). These findings emphasize the crucial role of Dux in the acquisition of totipotency.
Additionally, Dux makes significant contributions to the advancement of somatic cell nuclear transfer (SCNT) reprogramming. Dux demonstrates the ability to enhance the expression of ZGA genes in 2-cell SCNT embryos by improving the H3K9ac and H3K27ac modification, resulting in higher rates of pre- and post-implantation development (9–11). In a broader context, the N-terminus of the Dux protein consists of two highly conserved adjacent DNA binding homeodomains, while the C-terminus of Dux recruits CBP/p300, which facilitates the accumulation of histone H3 acetylation (10,12). The promotion of histone H3 acetylation has been previously established as a means to increase the efficiency of iPSC reprogramming (13). It has been demonstrated that Duxbl facilitates MET in muscle stem cells (14), which is a crucial step in the early stage of iPSC reprogramming (15). Furthermore, other characteristic genes known to promote 2CLC have also been discovered to enhance iPSC reprogramming through the promotion of H3 acylation or metabolic rewiring (16,17). Consequently, we propose the hypothesis that Dux may play a role in promoting iPSC reprogramming, particularly during the initial stages.
The establishment of iPSCs entails the reprogramming of differentiated somatic cells into a pluripotent state. Although Yamanaka factors possess the potential to reprogram all differentiated somatic cells, the successful completion of the reprogramming process necessitates the occurrence of multiple events in a sequential or simultaneous manner. Consequently, a significant proportion of cells fail to complete reprogramming and become trapped in the initial stages, leading to diminished reprogramming efficiency. The conventional reprogramming processes for iPSCs encompass various pivotal events operating at distinct regulatory levels. At the chromatin level, there is a rapid closure of many specific open sites related to fibroblasts during the early stage of reprogramming, while numerous sites associated with pluripotency become open in the later stage (18). At the epigenetic level, there is a decrease in the overall genomic methylation level, accompanied by a gradual increase in the acetylation of histone H3 and H4 (11). At the transcription level, there is a significant decrease in somatic-related genes during the first transcription wave, followed by a predominant increase in pluripotency-related genes during the second transcription wave (18–20). At the metabolic level, there is a transition in the primary energy supply pathway from oxidative phosphorylation (OXPHOS) commonly utilized by somatic cells to aerobic glycolysis, which is favoured by stem cells. This transition is referred to as the OXPHOS to glycolysis switch (OGS) (21). At the morphological level, cells undergo MET process, wherein they transform from fibroblast-like cells to epithelial-like cells. This transformation is accompanied by significant gene expression changes, particularly in cell adhesion molecules such as Cdh1, also known as E-cadherin (15). Hence, we posited the existence of a fundamental factor during the initial phase of reprogramming that propels the progression of stem cell-associated processes towards the attainment of pluripotency.
Epigenetic regulation encompasses various crucial mechanisms, among which histone post-translational modification (PTM) holds significance. Specifically, histone acetylation modification, extensively investigated, is presumed to facilitate transcription by counteracting the positive charge on the lysine side chain or enlisting specific reader proteins (22). In addition to t Dux, there is a consensus in the academic community that metabolites have the ability to influence the extent of histone post-translational modification (23). One such metabolite is lactate, which is a major product of glycolysis. Lactate-CoA, in particular, can serve as a substrate for histone modifications, thereby modulating the accessibility of DNA molecules and subsequently regulating gene expression (24). In a study conducted in 2019, Zhang et al. effectively employed high-performance liquid chromatography-mass spectrometry (LC/MS) tandem technology to successfully detect histone lysine lactylation. By utilizing the hypoxia and macrophage polarization model, they verified that lactate served as a substrate for the production of histone lactylation. Additionally, they made the groundbreaking discovery that the modification of lysine 18 on histone H3 (H3K18la) by lactate directly regulated gene expression at the chromosomal level and acted as an active transcriptional modification. Following this, scientists conducted a comparison of the enrichment of H3K18la, H3K27ac, H3K4me3, and H3K27me3 in cells derived from different germ layers, focusing on various transcription elements throughout the entire genome. The study revealed that H3K18la plays a crucial role in regulating the distribution of the transcriptional component, particularly at the enhancers of tissue-specific genes (25).
Previous investigations on the functionality of histone lactylation primarily concentrated on tissues and cells experiencing metabolite alterations within specific microenvironments, such as those associated with metabolic shifts in tumor cells, immune cells, and stem cells during specific biological activities (26). Stem cells, in particular, exhibit distinct metabolic traits compared to somatic cells, as they persistently rely on glycolysis as their primary energy source even under aerobic conditions. The phenomenon commonly known as the ‘Warburg Effect’ involves the pivotal role of lactate dehydrogenase (Ldha) as a significant regulator (27). Recent research has indicated that lactylation affects glycolysis-related genes, including Ldha, thereby enhancing their expression and facilitating the generation of additional substrates for lactylation. This process establishes a positive feedback loop, resulting in an increased lactylation modification (26). Notably, studies have demonstrated that promoting glycolysis during reprogramming enhances the efficiency of iPSC reprogramming (28). However, the role of histone lactylation in the process of iPSC reprogramming remains uncertain.
Numerous well-established histone modifying enzymes exist, including p300, which acts as a lactyltransferase responsible for histone lactylation (24). The reduction of histone lysine lactylation, specifically H3K18 and H4K5, is observed when classic HDAC1-3 and SIRT1-3 enzymes are present, indicating that these classic I HDACs function as effective ‘erasers’ of lysine lactylation in vitro (29). However, the identification of a ‘reader’ for lactylation remains unreported in the literature. In the context of histone acetylation and deacetylation, three primary domains have been implicated in the recognition process: the bromine domain (BD), the double PHD finger (DPF) domain, and the YEATS domain. The BD module, known for its conservation, binds to acetylated lysine residues on histones. Given the conservation of binding specificity among bromine domain homologs and the structural, functional, and evolutionary similarities between histone lactylation and histone acetylation, it is plausible to hypothesize that proteins containing the BD module may also serve as ‘readers’ for histone lactylation.
In the current investigation, it was observed that transient Dux overexpression (Dux OE) resulted in an enhancement of iPSC reprogramming efficiency through the upregulation of H3K18la level. The study established a connection between OGS, histone lactylation, and MET via H3K18la during the initial stages of reprogramming, highlighting the crucial role of H3K18la in successful reprogramming. Specifically, Dux facilitated an increase in H3K18la levels by promoting OGS and utilizing its C-terminal to recruit p300. Subsequently, Brg1 interacted with H3K18la, which was enriched on the promoters of MET-related genes, acting as a reader of lactylation to facilitate the reprogramming process.
Materials and methods
Cell culture
The animal feeding and experimental procedures were approved by the Ethics Committees of Harbin Medical University. The 4F2A-MEF cells, which expressed a Dox-inducible polycistronic 4F2A cassette, were derived from E13.5 embryos obtained from 4F2A mice purchased from Jackson Laboratory (USA). The Feeder-MEF cells were derived from E13.5 embryos obtained by crossing female C57BL/6 mice with male DBA/2 mice purchased from Vital River Laboratories (China). The MEF, HEK293T, and plate-E cells were cultured in DMEM/high glucose supplemented with FBS, penicillin-streptomycin, and L-glutamine. The mouse iPSC was maintained in Knockout DMEM supplemented with various components including KOSR and LIF.
Plasmids construction
Dux-related plasmids were constructed, amplified, purified, and quantified as previously described (10). The pMX-based retroviral vectors Oct3/4 (Addgene plasmid #13367), Sox2 (Addgene plasmid #13367), Klf4 (Addgene plasmid #13370), and c-Myc (Addgene plasmid #13375) were obtained from Addgene. The Lenti CRISPR V2 (Addgene Plasmid #52961), psPAX2 (Addgene Plasmid #12260), and pCMV-VSV-G (Addgene Plasmid #8454) were acquired from Addgene. The pHAGE2-TetOminiCMV-OSKM plasmid (Addgene plasmid #136541) was generously provided by Wu Guangming from Guangzhou Laboratory. Additionally, the pLV-CMV-mSmarca4 plasmid was purchased from Vector Builder.
Virus infection
Plat-E cells were transfected with Oct3/4, Sox2, Klf4 and c-Myc plasmids using the Lipofectamine® 3000 Transfection Kit. The supernatant containing the virus was collected and filtered at 48 h and 72 h post-infection, and subsequently used to infect target cells with polybrene for 48 h. HEK293T cells were transfected with pHAGE2-TetOminiCMV-OSKM plasmid along with packaging plasmids psPAX2 and pCMV-VSV-G. The sgBrg1 synthesized by Synbio Technologies following the Target Guide Sequence Cloning Protocol was cloned into the lentiviral vector lentiCRISPR v2.
In vitro transcription and mRNA transfection
The in vitro transcription of the reagent kits was conducted using the mMESSAGE-mMACHINE T7 Ultra Kit (ThermoFisher). The concentration of mRNA was determined using the Nanodrop 2000C (ThermoFisher) and the samples were stored at -80°C for future utilization. The Lipofectamine RNAiMax Reagent (Invitrogen) was employed according to the manufacturer's instructions for mRNA transfection, and the efficacy and dependability of transfection were assessed through quantitative real-time PCR.
iPSCs generation and AKP staining
Reprogramming efficiency was assessed through alkaline phosphatase (AKP) staining during the late stage of induction in this study. Three different methods were employed to induce iPSCs.
Firstly, the supernatants of retroviral-infected plate-E cells were used to infect MEF cells for 48 h. Subsequently, 4 × 104 cells were transferred into a 6-well plate with feeder cells and cultured until reprogramming Day 0. The resulting colonies were stained with AKP and counted on Day 10.
Secondly, the supernatants of lentiviral-infected HEK293T cells were used to infect MEF cells for 48 h. Following this, 4 × 104 cells were transferred into a 6-well plate with feeder cells the day before use, and treated with 0.1% doxycycline.
Thirdly, on the day prior to usage, 4 × 104 4F2A MEF cells were seeded onto a 6-well plate alongside feeder cells. Subsequently, the cells were subjected to a treatment of 0.1% doxycycline, resulting in their definitive reprogramming on Day 0. At Day 10, the colonies were stained using AP and subsequently quantified.
Glycolysis-stress tests
Glycolysis-stress tests were conducted using an XFe8 Analyzer system (Seahorse Bioscience) in accordance with the manufacturer's instructions. Prior to measurement, cells were seeded onto gelatin-coated 8-well plates the day before. The medium was then changed to XF Base Medium supplemented with 1 mM of l-glutamine and 10mM of glucose, and equilibrated for 1 h at 37°C without CO2 supply. ECAR was measured by sequentially adding 10 mM of glucose, 2μM of oligomycin, and 100 mM of 2-deoxyglucose to the medium.
Intracellular lactate concentration
The intracellular lactate concentrations were quantified utilizing a lactate assay kit (Sigma-Aldrich, cat. #MAK064) according to the manufacturer's guidelines. To summarize, cellular samples were homogenized in Lactate Assay Buffer and combined with a Master Reaction Mix containing Lactate Probes. The absorbance was assessed at 570 nm (A570), and the lactate concentration was determined by referencing a standard curve.
Immunoprecipitation (IP)
For each sample, cells were lysed using a Pierce™ Classic Magnetic IP/Co-IP Kit (Thermo Scientific #88804) according to the manufacturer's instructions. The resulting lysates were then incubated with a specific antibody overnight at 4ºC, followed by incubation with Pierce Protein A/G Magnetic Beads for 1 h at room temperature. The elution process was carried out using Lane Marker Sample Buffer, which was diluted 5-fold with purified water, for 30 min at room temperature. Protein loading buffer was subsequently added to each sample, and the samples were heated at 105ºC in a heating block for 10 min before being stored at –40ºC for future use.
Chromatin immunoprecipitation (ChIP) qPCR
ChIP assays were conducted using the SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) (Cell signaling technology #9003s) according to the manufacturer's instructions. Initially, the cells were cross-linked with 1% formaldehyde (Sigma) and subsequently quenched with 0.125 M of glycine. The lysed cells were then subjected to ChIP buffer and sonicated to obtain cross-linked chromatin. A portion of the chromatin samples was digested and the DNA concentration was determined. The diluted samples were incubated with specific antibodies, followed by the addition of protein G beads and further incubation. The chromatin was subsequently eluted and de-crosslinked. DNA was purified using a Purification Kit, followed by RT-qPCR analysis on the immunoprecipitated DNA using specific primers listed in Supplementary Table 1.
Immunofluorescence staining and images analysis
The cells were fixed in 4% paraformaldehyde at 4°C, then blocked with 2% BSA in PBST. Subsequently, the cells were incubated with primary antibodies overnight at 4°C and second antibodies for 1 h. After washing, the cells were mounted with ProLong™ Gold Antifade Mountant with DAPI and imaged using Nikon TE2000-U and Nikon ECLIPSE Ti-U microscopes. Image analysis was conducted using ImageJ.
Western blots and analysis
Protein samples were subjected to separation using 12.5% SDS-polyacrylamide electrophoresis gels, followed by transfer onto a polyvinyl difluoride membrane (Millipore). The membranes were subsequently blocked in a solution of 5% skim milk diluted in PBST (0.1% Tween 20 in PBS) for a duration of 1 h at room temperature. Following this, the membranes were incubated with specific primary antibodies at a dilution recommended in the accompanying instructions, overnight at a temperature of 4°C. The blots were then washed and incubated with the corresponding secondary antibody for a duration of 1 h at room temperature. All images were captured using the ChemiDoc™ Imaging System (BIO-RAD) and subsequently analyzed using ImageJ.
Total RNA extraction and RT-PCR
The extraction of total cellular RNA was conducted in accordance with a TRIzol-Based protocol, and subsequent reverse transcription was carried out using the TransScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) kit (Transgen #AT341-03) following the guidelines provided by the manufacturer. Real-time PCR was performed using the TransStart® Top Green qPCR SuperMix (Transgen #AQ131-01) as the manufacturer's instructions, and the analysis was conducted on the CFX96 Real-time System (Bio-Rad). The primers are listed in Supplementary Table 1.
RNA-seq
RNA extraction was conducted using a TRIzol-based protocol. Subsequently, libraries were prepared following the guidelines provided by the Illumina TruSeq RNA Sample Preparation kit. Sequencing was carried out on a MiSeq instrument at Shanghai Majorbio Bio-Pharm Technology. The obtained data were subjected to analysis using Trimmatic software (v0.39), hisat2 software (v2.1.0), and feature Counts software (v1.6.0). The repeat sequence was annotated utilizing the RepeatMask GTF file available on the UCSC website. The Counts matrix obtained is utilized as the input data file for gene differential expression analysis through the utilization of the R package DESeq2. Transcripts meeting the criteria of | log2FC | ≥ 1.5 and FDR < 0.05 are identified and labeled as differential transcripts. The data used in this study are at Supplementary Table 2.
CUT&Tag
To perform CUT & Tag analysis, we utilized the Hyperactive InSitu Universal CUT&Tag Assay Kit for Illumina Pro (Vazyme Biotech #TD904-02). A total of 10 000 cells were collected as samples for the experimental group, with two replicates conducted. Subsequently, the prepared libraries underwent PCR amplification after extraction using phenol-chloroform and ethanol precipitation. The PCR cycling conditions consisted of an initial step at 72°C for 5 min, followed by denaturation at 98°C for 30 s. This was succeeded by 14 cycles of denaturation at 98°C for 10 s and annealing at 60°C for 30 s. The amplification process was completed by performing a final extension step at a temperature of 72°C for a duration of 1 min, followed by holding the samples at a temperature of 4°C. For the purpose of post-PCR cleanup, a volume of DNA clean beads equal to 1.5 times the volume of the DNA was utilized. In order to guarantee a thorough and precise sequencing analysis, all libraries that were prepared underwent sequencing on the Illumina Nova Seq 6000 platform, while strictly adhering to the manufacturer's guidelines.
ChIP-seq and CUT&Tag analysis
The ChIP-seq and CUT&Tag data utilized in this study were obtained from publicly available databases, specifically the ENA (https://www.ebi.ac.uk/ena/browser/) or NCBI (https://www.ncbi.nlm.nih.gov/sra/) Download. The acquired data underwent analysis using Trimmatic software (v0.39), Bowtie2 software (v2.3.4.1) and Macs2 software (v2.1.1). The R package of ChIPseeker was employed for the analysis and processing of the ‘.mappedPeak’ file. Additionally, the bedGraphToBigWig tool was utilized to convert ‘.bdg’ files to ‘.bw’ files for compatibility with UCSC. Finally, the sorted bam file or bw file was visualized using IGV software (v2.8.2). The data used in this study are at Supplementary Table 2.
4D-FastDIA quantitative proteomics
The 4D-FastDIA quantitative proteomics analysis experiments were conducted at PTM BIO. The data are available in Pride database (PXD043547). In brief, the project commenced by subjecting raw LC–MS datasets to a database search using DIA-NN 1.8, yielding matrices with Normalized protein intensity. The intensities (I) were then centralized and converted to relative quantitative values (R) using the specified formula (i for samples, j for proteins). Rij = Iij/Mean (Ij).
Sample preparation involved enzymolysis, adding DTT at 56°C for 30 min, followed by IAM treatment in the dark for 15 min. TEAB-diluted urea and trypsin (1:50 ratio) facilitated overnight digestion, followed by a 1:100 trypsin addition for a 4-h hydrolysis. The resulting trypsin peptide, dissolved in solvent A, was injected into a self-made column. Peptide separation on a nanoElute UHPLC system employed a gradient from 6% to 80% solvent B over 70 min, with subsequent analysis on a timsTOF Pro mass spectrometer. Operating in PASEF mode, the system focused on precursor ions within a defined m/z range.
Molecular docking
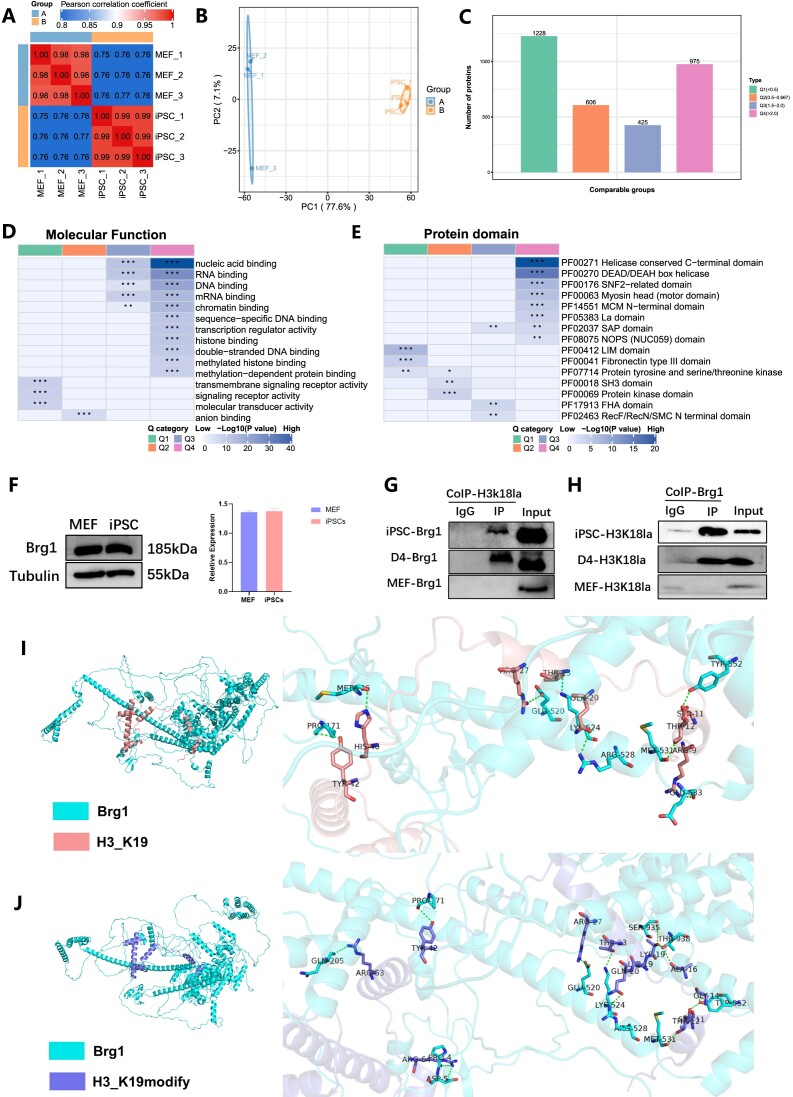
Molecular docking encompasses the utilization of computational software to simulate the interplay between molecules. In the present investigation, the AlphaFold software was employed to simulate the protein sequences of Brg1 and H3. Subsequently, the HDCOK program was utilized for protein–protein and protein–DNA/RNA docking, whereby the structure with the most favorable score was chosen for subsequent analysis. The Docking core employs iterative functions ITScorePP or ITScorePR to compute scores, with more negative values indicating heightened binding and interaction. Additionally, a confidence score is defined to denote the probability of molecule binding. The Brg1 protein and unmodified H3 protein exhibited an optimal docking score of –257.78, accompanied by a confidence score of 0.896. Conversely, the Brg1 protein and K19-modified H3 protein displayed an optimal docking score of –310.50, with a confidence score of 0.961. These findings substantiate the reliability of the complex models and indicate a substantial likelihood of binding.
Statistics
The WB, Co-IP, AKP staining and IF experiments were repeated at least three times. The qPCR and ChIP-qPCR experiments were repeated at least three times with three replicates every time. The CUT&Tag experiment was conducted on two occasions, while RNA-seq and IP-MS experiments were conducted three times. The resulting data were analyzed using a bioinformatic approach. Additional data were subjected to Student's t-test and presented as mean ± standard deviation. Statistical significance was determined based on P values, with *P< 0.05 considered statistically significant, **P< 0.01 considered highly significant, and ***P< 0.001 considered extremely significant. All statistical analyses were carried out using GraphPad Prism 8.0.2.
Results
Dux increases iPSCs reprogramming efficiency by metabolism rewiring
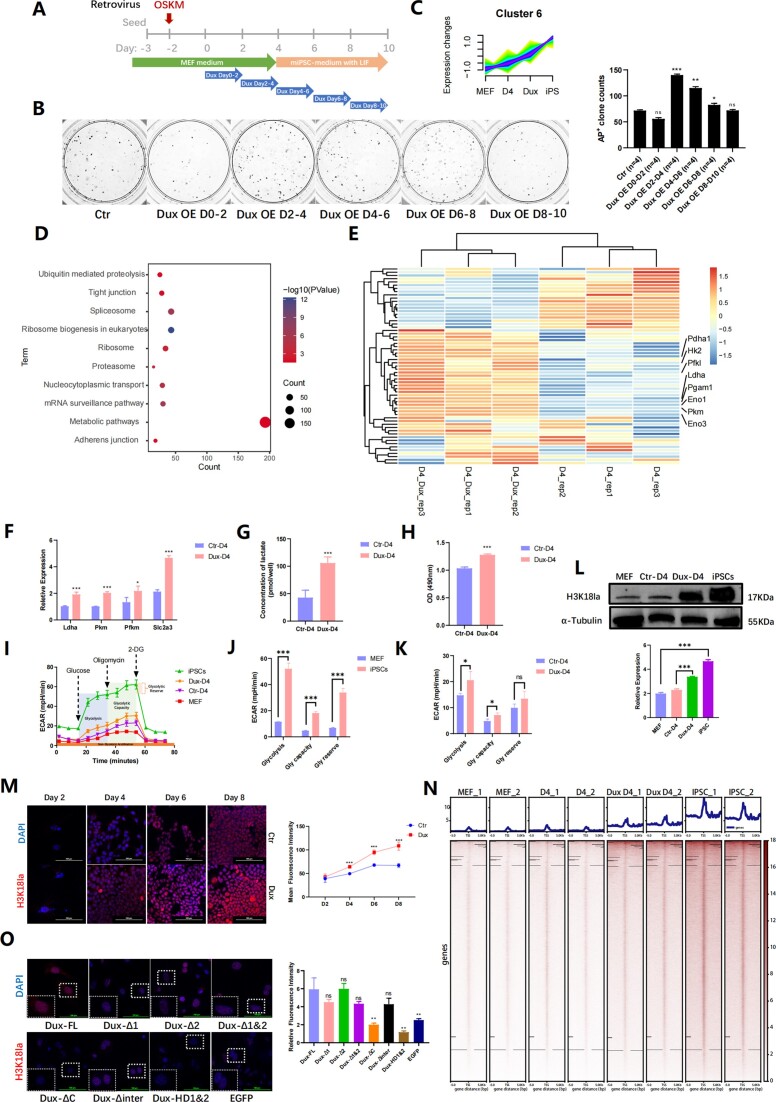
Multiple studies have demonstrated the ability of Dux to reduce DNA methylation across the entire genome (30) and recruit p300 to enhance acetylation (10). However, the potential of Dux to effectively promote iPSCs reprogramming remains uncertain due to the induction of apoptosis with prolonged expression of Dux (7). The precise dosage and timing of Dux expression have been shown to be crucial for regulating the developmental process (31). In this study, we used a retrovirus strategy to create iPSCs and a lentiviral system to overexpress Dux at specific times during reprogramming. We found that Dux OE on Days 2–4 significantly increased reprogramming efficiency (Figure 1A, B), with iPSC colonies showing normal pluripotency marker expression. Additionally, we conducted immunofluorescence and HE staining on embryonic bodies and Nude mice teratoma assay, which revealed the presence of three-layer markers both in vivo and in vitro (Supplementary Figure 1A–C). Our experiments showed that Dux OE during early reprogramming stages positively affects iPSCs generation.
Figure 1.
Dux promoted iPSCs reprogramming efficiency by metabolic remodeling. (A) Schematic diagram of classic Yamanaka factors reprogramming method with Dux OE on different time points. (B) AKP staining was conducted to visualize iPSCs clones with Dux OE at different time points. A statistical chart was generated to display the number of clones in each dish. Bars represent the mean and SD of n = 4 independent experiments. (C) RNA-seq analyses showed gene expression in Cluster 6 by Mfuzz clustering. MEF, D4, and iPSC each represented distinct stages of reprogramming, while Dux represented Dux OE during reprogramming on Days 2–4, and sampled on D4. (D) KEGG analysis showed genes enrichment in Cluster 6. (E) RNA-seq analyses showed glycolysis-related gene expression with Dux OE. (F) qPCR showed gene expression on glycolysis-related genes with Dux OE. Bars represent the mean and SD of n = 3 independent experiments. (G) Lactic probe assay indicated the concentration level of lactate with Dux OE. Bars represent the mean and SD of n = 3 independent experiments. (H) LDH assay showed at optical density at 490nm with Dux OE. Bars represent the mean and SD of n = 3 independent experiments. (I) The ECAR difference between MEF, D4, Dux D4 and iPSC were diagramed. ECAR presented parameters of the glycolytic pathway: glycolysis, glycolytic capacity, glycolytic reserve. (J) The statistical chart represented the ECAR difference between MEF and iPSC. Bars represent the mean and SD of n = 3 independent experiments. (K) The statistical chart showed the ECAR difference between Dux D4, and the control group. Bars represent the mean and SD of n = 3 independent experiments. (L) WB showed H3K18la levels in MEF, iPSCs, and cells during iPSCs reprogramming with Dux OE. The histogram analysis showed the relative statistical values of H3K18la for each group. Bars represent the mean and SD of n = 3 independent experiments. (M) Immunofluorescence staining showed the level of H3K18la modification during reprogramming. The line graph displayed the mean fluorescence intensity at every time point. Bars represent the mean and SD of n = 3 independent experiments. (N) CUT&Tag analysis showed genomic enrichment for H3K18la on MEF, D4, Dux D4 and iPSCs. (O) Immunofluorescence staining indicated H3K18la modification with Dux mutants. The histogram analysis displayed mean fluorescence intensity in each group. Bars represent the mean and SD of n = 3 independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001. t-test.
Subsequently, we examined the gene expression patterns in the non-reprogrammed state (MEFs), early reprogrammed state (Day 4), and fully reprogrammed state (iPSCs). Stable reprogramming progress was achieved using tetO-4F2A MEF. Reprogramming was initiated with doxycycline induction for 48h, followed by Dux OE for another 48h. The time window of reprogramming Day 2–Day 4 was studied for its impact on iPSCs reprogramming using RNA-seq. We performed a pseudo-time analysis by Fuzzy c-means algorithm (Mfuzz), a temporal cluster method that enables the calculation of the probability of each gene in every cluster (32). We supposed Dux OE on reprogramming Day 2–Day 4 (Dux D4) as an intermediate state between Day 4 and iPSC, and we got 10 distinct clusters (Supplementary Figure 1D). Cluster6, which including 4632 genes, exhibited a consistent increase in expression throughout the reprogramming process (Figure 1C). Genes in cluster6 revealed that a majority of them were found to be grouped within metabolic pathways according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Figure 1D). This finding implies that as reprogramming progresses, the expression of these genes increases gradually, facilitated by Dux and linked to the metabolism pathway in iPSCs development. We analyzed public data and used KEGG to find that genes enriched in Dux in ESC are also enriched in MAPK and HIF-1 signaling pathways, whereas the p38 MAPK pathway could enhance glycolysis (33), and the HIF-1/LDH pathway could promote lactate production (34), suggesting a link between Dux and glycolysis (Supplementary Figure 1E). Heatmap showed the expression level of glycolysis-related genes in Dux D4 and D4 groups (gene set was downloaded from REACTOME [REACTOME_GLYCOLYSIS, R-MMU-70171]), Log2(FPKM + 1) matrix was used as input, and scaled by rows. We found that glycolysis-related genes were mainly upregulated with Dux OE (Figure 1E). The correlation between Dux and metabolic processes suggested that Dux might promote reprogramming by regulating glycolysis metabolism pathway.
Notably, quantitative polymerase chain reaction (qPCR) analysis demonstrated that Dux significantly augmented the transcriptional activity of glycolytic genes in the early phases of reprogramming (Figure 1F). Moreover, Dux OE led to an augmentation in lactate secretion (Figure 1G). The activity of lactate dehydrogenase was enhanced with Dux OE, as assessed through a colorimetric assay (Figure 1H). The extracellular acidification rate (ECAR) was measured using the Seahorse assay, whereby the glycolytic capacity was determined by calculating the disparity between post-Oligomycin ECAR and baseline ECAR. The outcomes revealed a significant disparity in the levels of Glycolysis, Glycolytic capacity and Glycolytic reserve between MEF and iPSCs. The administration of Dux OE resulted in a notable elevation in the overall levels of ECAR, particularly in terms of Glycolysis level and Glycolytic capacity, surpassing those observed in the control group (Figure 1I–K). These collective observations indicated that Dux played a significant role in promoting the reprogramming of iPSCs by facilitating OGS.
During iPSCs reprogramming, OGS induces changes in metabolic products, leading to the accumulation of glycolysis-derived products, which in turn triggers the accumulation of a newly discovered type of histone modification—histone lactylation (24). Dux, known to promote OGS, was also found to facilitate the promotion of H3K18la during reprogramming (Figure 1L, Supplementary Figure 1F). Furthermore, we observed a gradual accumulation of H3K18la as reprogramming progressed, with significantly higher fluorescence intensity detected after Dux OE at each time point (Figure 1M). Subsequently, we performed Cleavage Under Targets and Tagmentation (CUT&Tag) detection on MEF, D4, Dux D4 and iPSCs, our data indicated a gradual accumulation of H3K18la enrichment on genes throughout the programming process, with Dux promoting the enrichment of H3K18la on the genome (Figure 1N).
p300 functions as a lactyltransferase, specifically targeting H3K18 for both acetylation and lactylation (24). Additionally, the C-terminal domain of Dux plays a crucial role in recruiting p300, thereby increasing the levels of genomic H3K27ac and H3K9ac (10). In order to investigate the specific contribution of Dux domains in promoting H3K18la, a series of Dux mutant plasmids were generated, each lacking different domain (Supplementary Figure 1G). These mutant plasmids were then utilized to produce mRNA through an in vitro transcription process, and subsequently transfected into cells. Our study revealed that Dux mutants lacking both N-terminal homology domains (DuxΔ1&2) exhibited a loss of nuclear localization (Supplementary Figure 1H). Conversely, the absence of the C-terminal domain (Dux ΔC and Dux HD1&2) prevented Dux mutant mRNA from elevating the level of H3K18la (Figure 1O). Following this, we conducted an analysis of the CUT&Tag data pertaining to mutations in the Dux domain in ESC available in publicly accessible databases. Our findings revealed a notable decrease in the binding capacity of Dux to genes associated with glycolysis and pluripotency in instances where the last 14 amino acids in the C-terminus of Dux were mutated (Supplementary Figure 2A). These results indicate that H3K18la accumulates during iPSCs reprogramming, and Dux facilitates the increase in H3K18la levels through its C-terminal domain.
H3K18la modification affects iPSC reprogramming efficiency
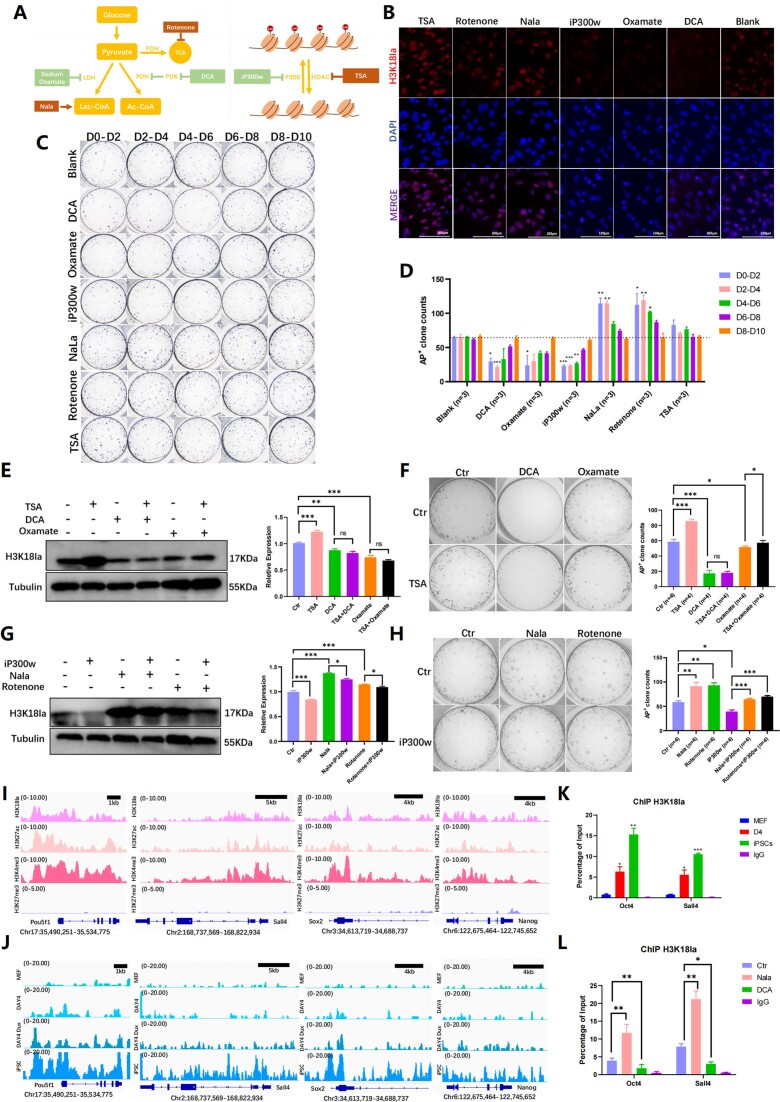
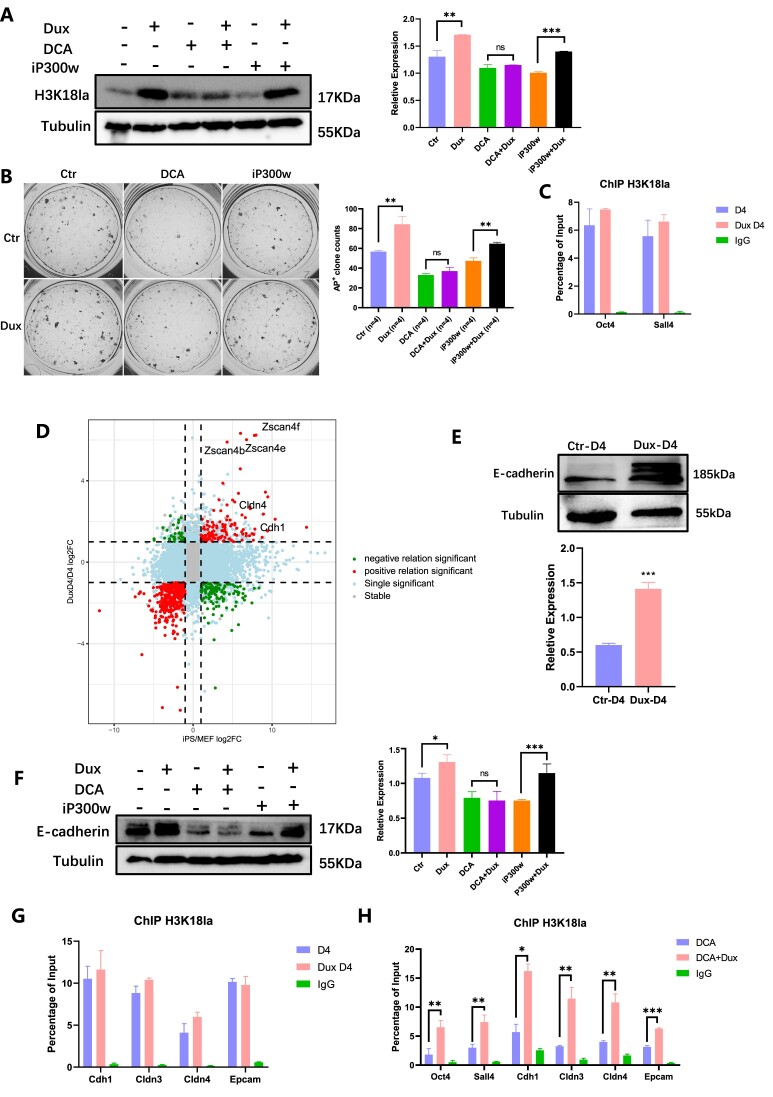
The findings suggest that Dux may regulate H3K18la levels, potentially enhancing reprogramming. Various lactylation regulators were used to study H3K18la's role in iPSCs reprogramming including sodium dichloroacetate (DCA), sodium oxamate, rotenone, sodium lactate (NaLa), iP300w and TSA (Supplementary Table 3). DCA and sodium oxamate inhibit glycolysis by affecting pyruvate dehydrogenase and lactate dehydrogenase, while Rotenone and Nala promote glycolysis. These compounds can impact reprogramming and pluripotency by regulating glycolysis efficiency (21,35,36). Importantly, it should be noted that DCA, sodium oxamate, and Rotenone have been shown to regulate genomic pan lactylation but not pan acylation (24). The enzymes p300 and HDAC1-3 were previously identified as ‘writers’ and ‘erasers’ of lactylation, respectively. To modulate the H3K18la levels, we utilized the p300 inhibitor iP300w and the HDAC inhibitor TSA (Figure 2A).
Figure 2.
The dynamic change of H3K18la during reprogramming was regulated by modifying the histone modification enzymes and glycose substrates. (A) The diagram showed H3K18la regulators by regulating the main glycolysis metabolism process and lactylation modification enzyme. (B) Immunofluorescence showed H3K18la levels with different regulators in the early reprogramming. (C) AKP staining indicated clones monitored by different regulators at various time points. (D) The histogram showed AP+ clone numbers with H3K18la regulators on different time points during reprogramming. Bars represent the mean and SD of n = 3 independent experiments. (E) WB showed the level of H3K18la treated with TSA combined with DCA or Oxamate on reprogramming Day 2–Day 4. The histogram analysis showed the relative statistical values of H3K18la for each group. Bars represent the mean and SD of n = 4 independent experiments. (F) AKP staining showed clones treated with TSA combined with DCA or Oxamate. The histogram presented the AP + clone numbers. Bars represent the mean and SD of n = 4 independent experiments. (G) WB showed the level of H3K18la treated with iP300w combined with Nala or Rotenone on reprogramming Day 2–Day 4. The histogram analysis showed the relative statistical values of H3K18la for each group. Bars represent the mean and SD of n = 4 independent experiments. (H) AKP staining showed clones treated with iP300w combined with Nala or Rotenone. The histogram presented the AP + clone numbers. Bars represent the mean and SD of n = 4 independent experiments. (I) Public bioinformatic data analysis showed the enrichment of H3K18la, H3K27ac, H3K4me3 and H3K27me3 on pluripotent-related genes. (J) CUT&Tag analysis showed the enrichment of H3K18la on pluripotent-related genes in MEF, D4, Dux D4 and iPSC. (K) ChIP-qPCR of H3K18la enrichment on pluripotent-related genes during reprogramming. Bars represent the mean and SD of n = 3 independent experiments. (L) ChIP-qPCR of H3K18la enrichment on pluripotent-related genes in reprogramming Day 4 with H3K18la regulated. Bars represent the mean and SD of n = 3 independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001. t-test.
To investigate the impact of H3K18la, inhibitors were utilized during the initial reprogrammed stage to assess the efficiency of Dux. The subsequent immunofluorescence and Western blotting analyses demonstrated that DCA, sodium oxamate, and iP300w led to a reduction in H3K18la levels, whereas Nala, Rotenone, and TSA resulted in an increase in H3K18la levels (Figure 2B, Supplementary Figure 2B). Furthermore, the impact of DCA, sodium oxamate, Nala, and rotenone on H3K18ac was found to be negligible, as indicated by the absence of any noticeable effect on its level. Conversely, iP300w was observed to decrease the level of H3K18ac, while TSA was found to enhance its level (Supplementary Figure 2C). Additionally, DCA and sodium oxamate were found to decrease the secretion of lactate by cells, whereas Nala and rotenone were found to increase lactate secretion. Notably, the administration of iP300w or TSA did not yield any significant alteration in lactate secretion (Supplementary Figure 2D).
Henceforth, we examine a groupset of small molecule drugs including DCA, Sodium oxamate, Rotenone, and Nala that can regulate H3K18la by affecting OGS. Additionally, another groupset of small molecule drugs including, iP300w and TSA, can regulate H3K18la modification by modulating histone lactyltransferase P300 and histone delactyltransferase HDAC (modification enzyme). Consequently, we designate these drugs as the regulators of H3K18la. We subjected cells to treatment with said regulators at various time intervals and quantified the quantity of AP+ clones on Day 10. Our findings demonstrated a notable alteration in the number of AP+ clones following administration of these regulators during the initial stages of reprogramming (Days 0–2 and Days 2–4). The downregulation of H3K18la resulted in a decrease in the number of AP+ clones, while the upregulation of H3K18la using Nala and Rotenone significantly increased the number of AP+ clones in the early stages of reprogramming. Although TSA also increased the number of AP+ clones, this effect was not statistically significant (Figure 2C, D). These findings suggested a crucial role for H3K18la in iPSC reprogramming, particularly in the initial stages of the process.
The above results demonstrated the regulation of H3K18la by affecting through the modulation of OGS and histone modification enzyme, ultimately impacting iPSCs reprogramming efficiency. Our approach involved the utilization of a drug combination strategy to manipulate H3K18la via histone modification enzymes based on OGS regulation. Subsequently, we evaluated the effects of early-stage H3K18la regulation on the efficiency of reprogramming. To investigate this matter, we devised two contrasting sets of experiments. The initial group encompassed negative regulators of glycolysis, including DCA and sodium oxamate, with the inclusion of TSA to enhance H3K18la modification levels. Conversely, the second group comprised positive regulators of glycolysis, including Nala and Rotenone, with the use of iP300w to reduce H3K18la modification levels. The 4F2A MEF cells were subjected to treatment with these regulators from reprogramming Day 2 to Day 4. The first set of experiments indicated that when treated with DCA or Sodium oxamate to reduce cellular H3K18la levels, TSA was unable to restore the levels of H3K18la modification (Figure 2E). In the second set of experiment, Nala and Rotenone were able to reverse the decrease in H3K18la caused by iP300w (Figure 2G). Both experiments emphasize the importance of OGS in H3K18la modification. Consistent with expectations, the number of AP+ clones exhibited a similar pattern to the level of H3K18la. The reprogramming efficiency treated with TSA is not restored following the impact of DCA and Sodium oxamate, whereas Nala or Rotenone can mitigate the influence of iP300w on reprogramming efficiency. The observed changes in H3K18la levels during early reprogramming further emphasize the significant role of H3K18la in early reprogramming. Additionally, it is noteworthy that elevated glycolytic levels can regulate H3K18la modification, even in the absence of p300 inhibition. It is possible that other lactyltransferase compensated for the depletion of p300 and increased the H3K18la level (Figure 2F, H).
To confirm H3K18la's role in reprogramming, we compared our CUT&Tag data with publicly ChIP-seq data and found enrichment of H3K18la on glycolysis and pluripotency genes, which increased as reprogramming advanced (Supplementary Figure 2E). The utilization of the Integrated Genome Viewer (IGV) facilitated the identification of histone modification enrichment on genes. Our analysis revealed a significant enrichment of active histone modifications, specifically H3K18la, H3K27ac, and H3K4me3, on the promoters of pluripotency marker genes Pou5f1 (also referred to as Oct4), Sall4, Sox2 and Nanog in mouse ESC. Additionally, we observed a lack of enrichment of the repressive histone modification H3K27me3 at the loci of pluripotency-related genes (Figure 2I). To analysis these results, we conducted a CUT&Tag analysis of H3K18la enrichment. Our findings demonstrated a consistent pattern of H3K18la enrichment at the pluripotent genes’ promoters, in line with publicly available data. Notably, as the reprogramming process advanced, there was a gradual accumulation of H3K18la at the pluripotent gene promoter, with a significant enrichment observed in iPSCs (Figure 2J, Supplementary Figure 4G). Subsequently, an analysis was conducted on the ChIP-qPCR data pertaining to loci enrichment, revealing a notable disparity in H3K18la enrichment at the promoters of Oct4 and Sall4 between MEF and iPSCs. Specifically, on reprogramming Day 4, the level of H3K18la enrichment surpassed that of MEF, yet it did not attain the level observed in iPSCs (Figure 2K). This observation implies a gradual accumulation of H3K18la at promoters of genes associated with pluripotency throughout the reprogramming process. Lastly, Nala exhibited an upregulation in the enrichment of H3K18la on Oct4 and Sall4 promoters, whereas DCA downregulated it during early reprogramming (Figure 2L). In conclusion, our findings highlight the specific enrichment of H3K18la in genes associated with pluripotency through the actions of lactyltransferase and glycolysis substrates during the early stage of reprogramming.
H3K18la promotes mesenchymal-epithelial transition
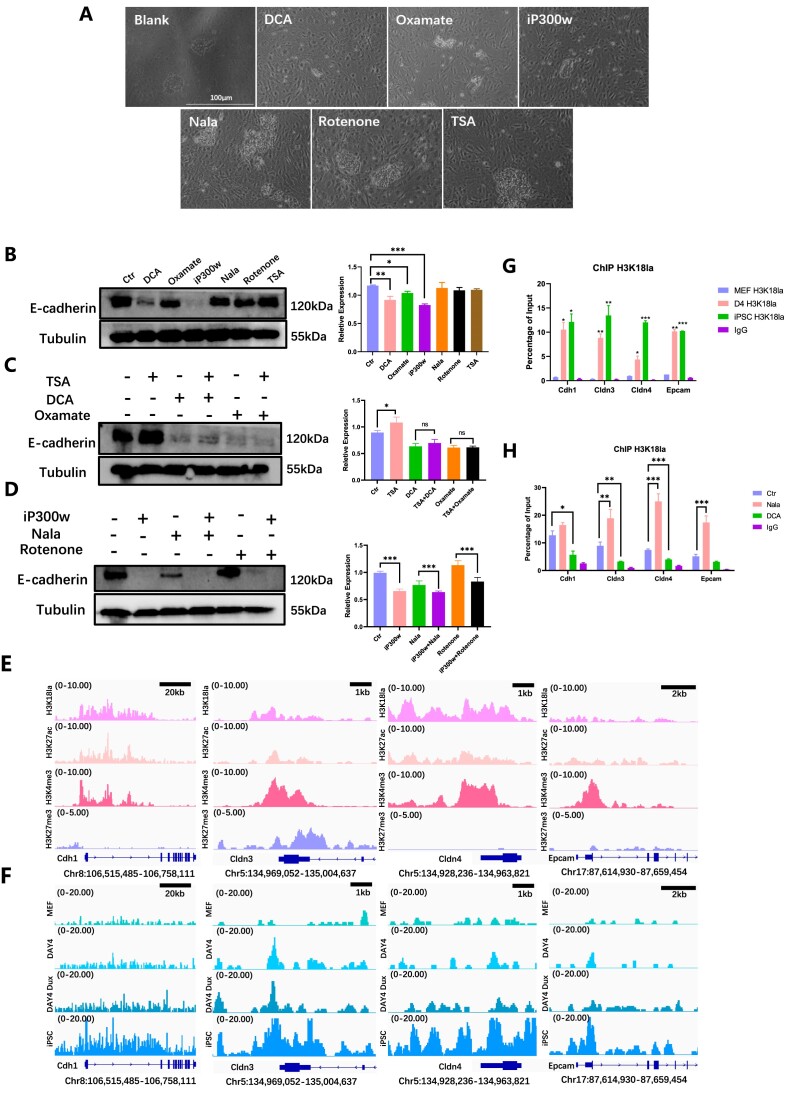
MET played a pivotal role in the initial reprogramming of iPSCs. Considering the observed impacts of H3K18la regulators during the early phase of reprogramming, it raised the question of whether their influence on early reprogramming occurred through the regulation of MET. Activation of cadherin 1 (Cdh1 or E-cadherin) is a well-established indicator of MET. Consequently, this section aimed to assess whether H3K18la facilitated early reprogramming by promoting MET. The pHAGE2-TetOminiCMV-OSKM plasmid, which was transfected with lentivirus, was prepared for the purpose of this study. In this reprogramming system, the observation of cell morphology was facilitated. Throughout the reprogramming process, the fibroblast-like shape cells gradually transformed into a paving stone-like appearance (Supplementary Figure 3A). Additionally, E-cadherin protein exhibited a significant increase (Supplementary Figure 3B).
Through the application of H3K18la regulators to cells from reprogramming Day 0–4, we observed a variation in the quantity of cells undergoing MET (Figure 3A). Additionally, the downregulation of E-cadherin was observed when treated with DCA, sodium oxamate, and iP300w, as illustrated in Figure 3B. The modulation of E-cadherin appeared to align with the levels of H3K18la when utilized in conjunction with H3K18la regulators (Figure 3C, D).
Figure 3.
H3K18la promotes mesenchymal-epithelial transition. (A) The cellular morphology was observed under light microscopy following treatment with H3K18la regulators during the early stages of reprogramming. (B) WB showed E-cadherin level treated with H3K18la regulators in early reprogramming. The statistical graph showed E-cadherin relative expression. Bars represent the mean and SD of n = 4 independent experiments. (C) WB showed E-cadherin level when treated with TSA combined with DCA or sodium oxamate in the early reprogramming. The histogram analysis indicated the relative statistical values of E-cadherin for each group. Bars represent the mean and SD of n = 4 independent experiments. (D) WB showed E-cadherin level when treated with iP300w combined with Nala or Rotenone in the early reprogramming. The histogram analysis showed the relative statistical values of E-cadherin for each group. Bars represent the mean and SD of n = 4 independent experiments. (E) Public bioinformatic analysis showed the enrichment of H3K18la, H3K27ac, H3K4me3, and H3K27me3 epithelial-related genes. (F) CUT&Tag analysis showed the enrichment of H3K18la on epithelial-related genes in MEF, D4, Dux D4 and iPSC. (G) ChIP-qPCR showed H3K18la enrichment on epithelial-related genes during reprogramming. Bars represent the mean and SD of n = 3 independent experiments. (H) ChIP-qPCR showed H3K18la enrichment on epithelial-related genes in reprogramming Day 4 with H3K18la regulators. Bars represent the mean and SD of n = 3 independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001. t-test.
Subsequent investigations confirmed the presence of H3K18la enrichment at gene loci associated with epithelial junction. Based on the findings of bioinformatics analysis, it was observed that epithelial-related gene locations in ESCs exhibited significant enrichments of H3K18la, H3K27ac and H3K4me3, while no significant enrichment of H3K27me3 was observed (Figure 3E). We compared CUT&Tag data for H3K18la during reprogramming and found gradual accumulation at epithelial-related genes. In iPSC, H3K18la was significantly enriched at the epithelial genes’ promoters (Figure 3F, Supplementary Figure 4H). Mesenchymal-related gene loci did not show significant enrichments of H3K18la, H3K27ac and H3K4me3. Enrichment was observed at the Snail gene locus, but the distinction between activated and repressed modifications was unclear. Bivalent histone modification was observed at the Twist1 gene locus, indicating transcriptional suppression (Supplementary Figure 3C). Our data showed that, there was no significant difference in the enrichment of H3K18la at these sites during the reprogramming process (Supplementary Figure 3D). Subsequent ChIP-qPCR assays demonstrated notable disparities in H3K18la enrichment on genes associated with epithelial junction. H3K18la levels reached a magnitude similar to that observed in iPSCs. Remarkably, the accumulation of H3K18la progressively intensified at the promoters of epithelial-related genes during reprogramming process (Figure 3G). The results of this study also demonstrated that the administration of H3K18la regulators led to a notable increase in the presence of H3K18la at the promoters of genes associated with epithelial junction. Specifically, Nala was observed to enhance the enrichment of H3K18la in epithelial-related genes, whereas DCA resulted in a decrease in H3K18la enrichment at these specific sites (Figure 3H). Comparing the outcomes suggests that H3K18la modification boosts reprogramming efficiency by promoting cellular MET. This research highlights the importance of a network involving OGS, H3K18la and MET, with H3K18la being crucial in the early stages of reprogramming.
Dux facilitates H3K18la dynamic changes during reprogramming by promoting H3K18la enrichment on epithelial-related genes
These aforementioned outcomes established the regulatory role of H3K18la in epithelial-related and pluripotent-related genes during the early stages of reprogramming. To investigate whether Dux modulated H3K18la through OGS or the recruitment of p300, additional experiments were conducted. Dux increased the level of H3K18la modification during reprogramming D2-D4, but H3K18la level could not promoted by Dux with the existence of DCA. However, when iP300w consumed p300, Dux rescued H3K18la levels (Figure 4A). AP+ clones displayed a similar pattern in the level of H3K18la when treated with H3K18la regulators and Dux (Figure 4B). We also tested the effect of Dux on H3K18la enrichment, and ChIP-qPCR demonstrated that in the early stages of reprogramming, Dux OE led to a slight enhancement in the enrichment of H3K18la on pluripotent genes, although this increase was not statistically significant (Figure 4C). The findings suggested that Dux facilitated H3K18la modification mostly by upregulating p300 rather than promoting OGS.
Figure 4.
Dux facilitates H3K18la dynamic changes during reprogramming. (A) WB showed the level of H3K18la in combination with Dux OE and DCA or iP300w. The histogram analysis displayed the relative statistical values of H3K18la for each group. Bars represent the mean and SD of n = 4 independent experiments. (B) AKP staining showed clones treated with Dux OE, DCA, and iP300w. The histogram analysis of the AP + clones in each group. Bars represent the mean and SD of n = 4 independent experiments. (C) ChIP-qPCR of H3K18la enrichment on pluripotent-related genes on Dux D4. Bars represent the mean and SD of n = 3 independent experiments. (D) Nine-quadrant scatter plot showed the relationship between differential expression genes (DEGs) in Dux D4 compared to D4 (Log2FoldChange > 1 & Padj < 0.05), and the DEGs in iPSC compared to MEF. (E) WB showed the level of E-cadherin with Dux OE. The histogram analysis indicated the relative statistical values of E-cadherin for each group. Bars represent the mean and SD of n = 3 independent experiments. (F) WB showed the level of E-cadherin in combination with Dux OE and DCA or iP300w. The histogram analysis displayed the relative statistical values of E-cadherin for each group. Bars represent the mean and SD of n = 4 independent experiments. (G) ChIP-qPCR showed H3K18la enrichment on epithelial-related genes with Dux OE. Bars represent the mean and SD of n = 3 independent experiments. (H) ChIP-qPCR showed H3K18la enrichment on epithelial-related genes and pluripotent-related genes with H3K18la regulated. Bars represent the mean and SD of n = 3 independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001. t-test.
Nine-quadrant scatter plot showed the relationship between differential expression genes (DEGs) in Dux D4 compared to D4 (Log2FoldChange > 1 & Padj < 0.05), and the DEGs in iPSC compared to MEF. Dux downstream genes Zscan4 cluster and epithelial-related genes Cdh1 and Cldn4 were found in the upper right quadrant, which represented the both up-regulated genes in the two groups (Figure 4D). Consequently, we speculated that Dux contributed to MET through H3K18la. Dux OE resulted in an elevation of E-cadherin (Figure 4E) and an upregulation of genes associated with epithelial junction, while simultaneously downregulating genes associated with mesenchymal characteristics (Supplementary Figure 4A, B). Additionally the regulation of E-cadherin exhibited a similar pattern to that of H3K18la in response to various regulators. DCA and iP300w decreased the level of E-cadherin, and Dux was able to rescue the effect of iP300w on E-cadherin, while not having the same rescuing effect on DCA (Figure 4F). Furthermore, ChIP-qPCR analysis revealed a slight increase in the enrichment of H3K18la with Dux OE, although this increase did not reach statistical significance (Figure 4G). The administration of DCA resulted in a reduction in the enrichment of H3K18la on genes linked to pluripotency and epithelial junction, whereas Dux OE successfully restored the levels of H3K18la (Figure 4H). Additionally, the enrichment of H3K27ac increased with reprogramming, and Dux significantly enhanced the enrichment of H3K27ac on these genes (Supplementary Figure 4C–F). Interestingly, the enrichment of H3K18la on epithelial-related genes on early reprogramming showed a pronounced changes compared to H3K27ac (Figure 3G, Supplementary Figure 4E). This suggests that H3K18la plays a unique role in regulating pluripotent and epithelial-related genes, particularly in the early stage of reprogramming.
H3K18la has distinct binding proteins in MEF and iPSC
Our study has identified an OGS-H3K18la-MET network that is involved in a series of reprogramming events, which can be modulated by manipulating the H3K18la modification. To explore the proteins that interact with H3K18la, we employed co-immunoprecipitation (Co-IP) experiments in conjunction with 4D-FastDIA LC–MS/MS analysis. Our focus was specifically on investigating the proteins that bind to H3K18la in iPSCs and MEFs. The Pearson correlation coefficient test was utilized to evaluate the correlations, demonstrating the experiment samples′ commendable reproducibility (Figure 5A). Furthermore, principal component analysis revealed significant distinctions between the proteins associated with H3K18la in iPSC and MEF (Figure 5B). Based on variations in protein level, we categorized proteins with distinct concentrations in MEF and iPSC into four groups (Q1–Q4). Subsequently, we conducted a screening of differential proteins, identifying 975 proteins (Q4) with intensity in iPSC more than double that in MEF, at a significance level of P< 0.05 (Figure 5C). Our analysis of molecular function indicated that the Q4 proteins primarily participate in RNA/DNA binding, chromatin binding, and histone binding (Figure 5D, Supplementary Figure 5A). H3K18la exhibited a tendency to interact specifically with proteins containing the Helicase conserved C-terminal domain, DEAD/DEAH box helicase, and SNF2-related domain in iPSCs (Figure 5E, Supplementary Figure 5B).
Figure 5.
H3K18la interacts with chromatin remodeling complex Brg1 in early reprogramming. (A) Pearson's correlation coefficient was analyzed to evaluate the repeatability of individual samples. (B) Principal component analysis indicated the difference between MEF and iPSC. (C) The protein was divided into four groups according to the differential expression (Q1–Q4), and Q4 represented that the protein expression in iPSC was twice that in MEF, and P< 0.05. (D)The molecular function of proteins interacted with H3K18la. (E) Protein domains interacted with H3K18la. (F) WB showed Brg1 protein level in MEF and iPSC. The histogram analysis displayed the relative statistical values of Brg1 in MEF and iPSC. Bars represent the mean and SD of n = 3 independent experiments. (G) Co-IP showed H3K18la interaction with Brg1. (H) Co-IP showed Brg1 interaction with H3K18la. (I) Interaction mode of Brg1 protein and unmodified H3 protein. (J) Interaction mode of Brg1 protein and K19 lactated modified H3 protein *P< 0.05, **P< 0.01, ***P< 0.001. t-test.
It is worth noting that the switch/sucrose non-fermentable (SWI/SNF) complex, also known as the Brg1/Brm-associated factor (BAF), is considered essential for chromatin remodelling (37). The complex includes an ATPase subunit, a conserved core, and a variant subunit. Several BAF complex family members were found among Q4 proteins, including Smarca4 (also known as Brg1), Smarcb1, Smarcc1 (also known as Baf155), Smarce, and Arid1a/b. Furthermore, it has been established that ESC harboring a distinct variant of the BAF complex, referred to as esBAF, exhibit the presence of Smarca4 but not Smarca2 (also known as Brm) (38). Our findings, however, did not detect Brm among the Q4 proteins. The esBAF complex plays a crucial role as a fundamental constituent of the core pluripotent transcriptional machinery. Its specific constituents are capable of interacting with pluripotent factors such as Oct4 (39), Nanog (40) and Sox2 (41), thereby facilitating the maintenance of ESCs′ pluripotency and iPSCs reprogramming. Brg1, as the enzymatic subunit of BAF complexes, played a crucial role in the function of the BAF complex. Previous studies have demonstrated that chromatin remodeling factors, such as Brg1, Baf155 and INO80, which interact with Oct4, promote reprogramming at the chromatin level (42). Furthermore, the Brg1-containing bromodomain interacts with chromatin through its binding to acetylated histones or transcription factors (43). Considering the potential role of H3K18la as a binding site for Brg1, we hypothesized that Brg1 functions as a reader of lactylation, thereby enhancing the accessibility and transcriptional activity of nucleosomes.
To validate this hypothesis, we investigated the differential protein levels of Brg1 between MEF and iPSC, and evaluated the binding of H3K18la in both cell types. However, our findings indicated that there was no significant distinction in the levels of Brg1 protein between somatic cells and stem cells (Figure 5F). Co-IP assays revealed a strong association between H3K18la and Brg1 in iPSCs, not in MEFs (Figure 5G and H). The interaction occurred early in reprogramming, indicating a difference in Brg1-H3K18la interaction between somatic and pluripotent cells.
The protein sequences of Brg1 and H3 were computationally generated using the AlphaFold software, and their binding capabilities were compared both before and after the introduction of the lactic acid group (notably, the model construction incorporated methionine at the first translation start site, resulting in lysine being positioned at position 19). The findings revealed that the unmodified H3 protein established nine distinct groups of hydrogen bond interactions with the N-terminal region of the Brg1 protein, encompassing Thr11, Met106, Pro171, Glu520, Lys524, Arg528, Met531, Glu533 and Tyr552. However, the binding of K19 carrying modified H3 protein to Brg1 protein resulted in the formation of 13 distinct groups of hydrogen bond interactions at the same binding position, involving Pro5, Asp6, Pro171, Gln205, Glu520, Lys524, Arg528, Met531, Tyr552, Ser935 and Thr938. The presence of a higher number of hydrogen bond interactions suggested a stronger binding affinity between Brg1 and K19-modified H3 protein (Figure 5I, J). Given the known involvement of Brg1 in acetylation and the similarities between acetylation and lactylation, it was plausible to hypothesize that Brg1 might act as a ‘reader’ for lactylation. The findings of our study demonstrated that Brg1 exhibited a stronger affinity for the H3K18la-modified histone variant compared to unmodified H3, implying its potential role as a histone lactylation reader. Furthermore, our results suggest that Brg1’s interaction with H3K18la is likely involved in chromatin remodeling complexes, thereby potentially facilitating the accessibility of chromatin regions associated with genes related to epithelial function.
Brg1 works as a reader of H3K18la in the early reprogramming
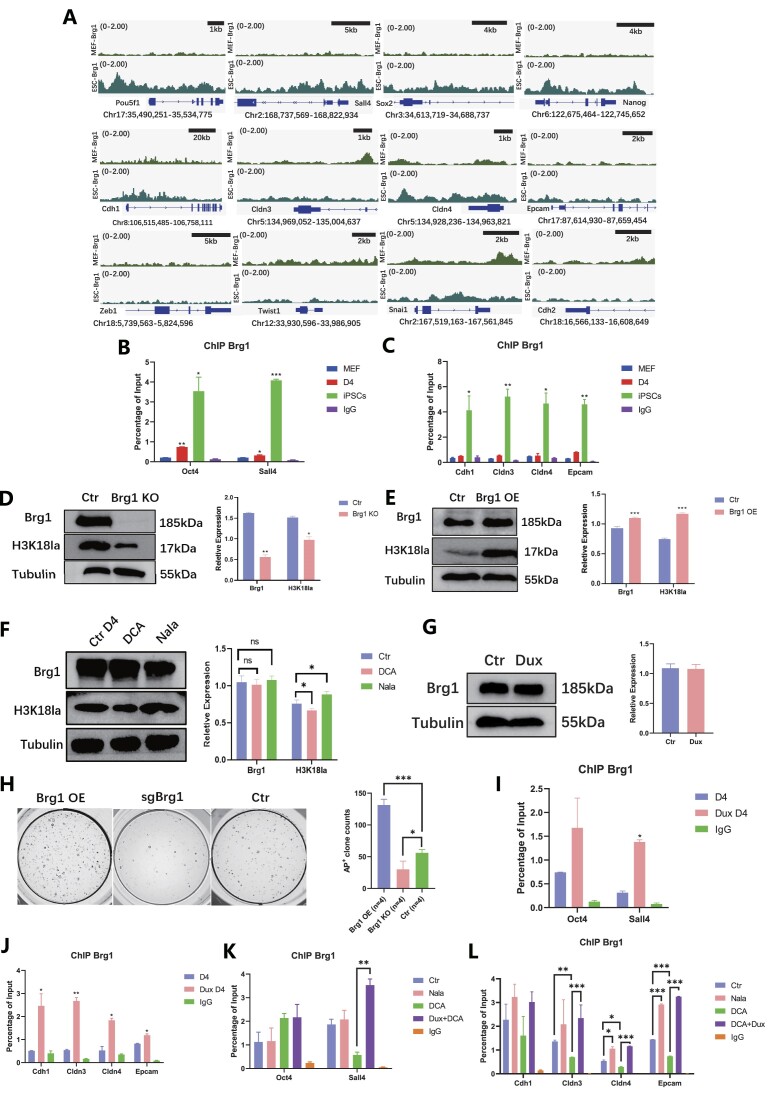
Based on the published data, a distinct disparity in the enrichment of Brg1 at pluripotent genes and epithelial genes was evident between MEF and ESC. Conversely, no significant variation in Brg1 enrichment was observed in mesenchymal-related genes between MEF and ESC (Figure 6A). Although there was some enrichment of Brg1 on the Snail gene locus, the differentiation between MEF and ESCs in terms of Brg1 enrichment was not statistically significant (Figure 6A). Consequently, ChIP-qPCR analysis was conducted on loci that exhibited enrichment according to the bioinformatics data. Brg1 was found to be associated with the process of reprogramming at the pluripotent genes and epithelial genes, including Oct4, Sall4, Cdh1, Cldn3, Cldn4 and Epcam (Figure 6B, C).
Figure 6.
Brg1 works as a reader of H3K18la in the early reprogramming. (A) Public bioinformatic data analysis displayed the enrichment of Brg1 in MEF and ESCs on the epithelial-related genes and pluripotent-related genes. (B) ChIP-qPCR showed Brg1 enrichment on pluripotent-related genes during reprogramming. Bars represent the mean and SD of n = 3 independent experiments. (C) ChIP-qPCR showed Brg1 enrichment on epithelial-related genes during reprogramming. Bars represent the mean and SD of n = 3 independent experiments. (D) WB showed Brg1 and H3K18la level with Brg1 KO. The histogram analysis showed the relative statistical values of Brg1 and H3K18la for each group. Bars represent the mean and SD of n = 3 independent experiments. (E) WB showed Brg1 and H3K18la level with Brg1 OE. The histogram analysis showed the relative statistical values of Brg1 and H3K18la for each group. Bars represent the mean and SD of n = 3 independent experiments. (F) WB showed Brg1 level regulated by DCA and Nala. The histogram analysis showed the relative statistical values of Brg1 and H3K18la in each group. Bars represent the mean and SD of n = 3 independent experiments. (G) WB showed Brg1 level regulated by Dux OE. The histogram analysis displayed the relative statistical values of Brg1 and H3K18la for each group. Bars represent the mean and SD of n = 3 independent experiments. (H) AKP stain showed clones with Brg1OE/KO. The histogram analysis showed the number of AP + clones for each group. Bars represent the mean and SD of n = 4 independent experiments. (I) ChIP-qPCR showed Brg1 enrichment on pluripotent-related genes. Bars represent the mean and SD of n = 3 independent experiments. (J) ChIP-qPCR showed Brg1 enrichment on epithelial-related genes. Bars represent the mean and SD of n = 3 independent experiments. (K) ChIP-qPCR showed H3K18la enrichment on pluripotent-related genes with H3K18la regulated. Bars represent the mean and SD of n = 3 independent experiments. (L) ChIP-qPCR showed H3K18la enrichment on epithelial-related genes with H3K18la regulated. Bars represent the mean and SD of n = 3 independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001. t-test.
Subsequently, the relationship between H3K18la and Brg1 was investigated through the examination of Brg1 OE or knockout (KO). It was observed that during the early stages of reprogramming, Brg1 OE led to an increase in H3K18la levels, whereas Brg1 knockout resulted in a decrease in H3K18la levels (Figure 6D, E). However, no significant changes in Brg1 were observed in relation to the H3K18la regulators or Dux OE (Figure 6F, G). The present study aimed to examine the impact of regulating Brg1 during the initial stages of reprogramming on the efficacy of iPSC reprogramming. The findings revealed that Brg1 OE resulted in the anticipated enhancement of iPSC reprogramming efficiency. Conversely, knockout of Brg1 led to a reduction in the formation of AP+ clones during iPSCs reprogramming (Figure 6H).
Furthermore, the outcomes indicated that while Dux OE did not alter the level of Brg1, it did promote the accumulation of Brg1 at genes associated with pluripotency and epithelial junction (Figure 6I, J). In a similar vein, the impact of H3K18la on the protein level of Brg1 was found to be negligible. However, it was observed that H3K18la played a crucial role in modulating the enrichment of Brg1 (Figure 6J, K). Although no significant differences were detected in the enrichment on pluripotent-related genes, the accumulation of Brg1 at epithelial-related genes was found to be regulated by H3K18la and could be restored through Dux OE (Figure 6L). These findings provide insight into the dual functionality of Brg1, serving as both a regulator and a reader of H3K18la.
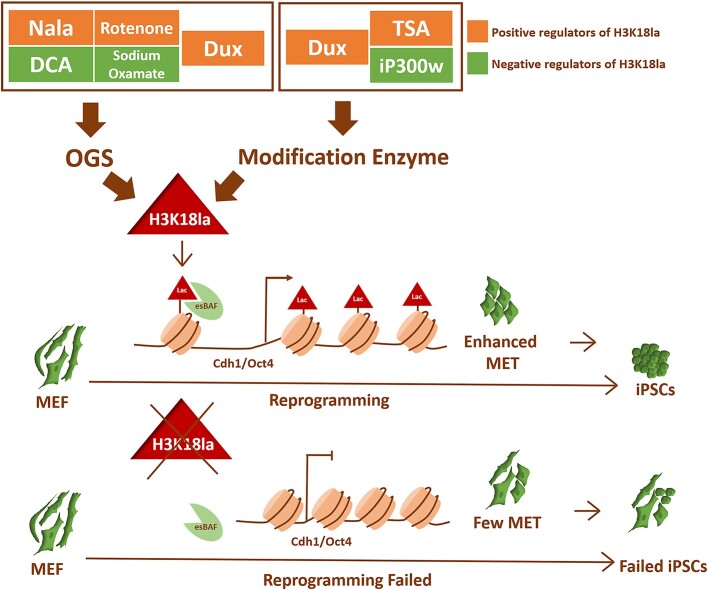
The co-localization of H3K18la and Brg1 was observed at pluripotent-related and epithelial-related genes, leading to the facilitation of cellular MET. The modulation of H3K18la during the initial phases of reprogramming ultimately influenced the efficacy of iPSCs reprogramming (Figure 7).
Figure 7.
Models for the functional outcomes of H3K18la modification in the early reprogramming. Dux facilitated iPSCs reprogramming by enhancing the level of H3K18la modification, a process regulated by OGS and histone modification enzymes. Brg1, acting as a reader in the regulatory network, recognized lactylated histone H3. The combination of H3K18la and Brg1 accumulated on the promoters of genes related to pluripotency and epithelial function, thereby promoting the process of cellular MET. The regulation of H3K18la during the early stages of reprogramming ultimately impacted the efficiency of iPSCs reprogramming.
Discussion
The efficiency of reprogramming iPSCs has not yet reached a satisfactory level. Previous research has primarily concentrated on crucial events occurring during the initial stages of reprogramming, aiming to enhance iPSC efficiency by controlling key processes. These include the transient activation of mitoflashes to initiate Nanog activation (44), the modulation of histone lysine demethylation through the temporary opening of mitochondrial permeability transition pores (45), and the regulation of essential enzymes in the TCA cycle to facilitate histone H3 acetylation, ultimately leading to the relaxation of pluripotent gene chromatin (46). Furthermore, it is worth mentioning that the induction process of chemical-induced pluripotent stem cells (ciPSC) differs fundamentally from the conventional Yamanaka factor-induced reprogramming (47). Notably, while one study has highlighted the role of Dux in promoting ciPSC reprogramming, it is important to acknowledge that this process involves the emergence of an intermediate state resembling extra-embryonic endodermal cells, commonly referred to as the XEN-like phase. This particular stage of development distinguishes ciPSC from traditional reprogramming methods and provides notable benefits by enhancing their effectiveness. The presence of Dux has the potential to enhance ciPSC reprogramming efficiency by promoting the formation of this crucial state (48). KLF-4 has been shown to be the main driver for a metabolic switch in iPSCs and other reprogramming systems (49,50), and KLF-4 has also believed to have a positive role in regulating MET (15). Therefore, a comparative analysis of the genes regulated by KLF-4 and Dux during reprogramming was conducted, revealing distinct regulatory patterns for each transcription factor (Data not shown). Our RNA-seq analysis further demonstrated a significant upregulation of Klf4 expression upon Dux overexpression during reprogramming. Additionally, CUT&tag data from ESC indicated enrichment of Dux in the distal region of Klf4, suggesting a potential regulatory relationship between the two factors (51) (data not shown). Our research sheds light on the role of Dux in augmenting the efficiency of iPSCs reprogramming induced by the classic Yamanaka factors. The temporary Dux OE was observed to improve the efficiency of conventional iPSCs reprogramming by upregulating H3K18la modification during the initial stage of reprogramming.
The findings of our study elucidate the significant role played by H3K18la during the initial phase of reprogramming. It is worth noting that prior research has only briefly touched upon the association between histone lactylation and iPSC reprogramming. Specifically, Glis1 was found to induce metabolic remodeling in the early stages of reprogramming, while the late stages were characterized by multi-level epigenetic modifications, including the accumulation of H3K18la and H3K27ac in pluripotency genes such as Oct4 and Sall4 (52). However, the complete elucidation of the specific mechanism through which H3K18la enhances reprogramming efficiency remains insufficient. Our investigation has contributed to the advancement of our comprehension regarding the functional importance of H3K18la in the initial phases of iPSC reprogramming, thereby highlighting its indispensable role in this procedure. In summary, our research has unveiled a complex OGS-H3K18la-MET cascade network that is activated by H3K18la during the initiation of iPSC reprogramming. This study investigates the regulatory mechanisms of glycolysis metabolism involving regulators and Dux OE, as well as the impact of H3K18la modification on reprogramming efficiency. Furthermore, our results reveal the interaction between the bromine-containing protein Brg1 and H3K18la, leading to its accumulation on MET-related gene promoters and functioning as a lactylation ‘reader’ through its bromine domain binding lactyl-lysine residues.
The focus of histone modification lies in the examination of the ‘Writer,’ ‘Reader,’ and ‘Eraser’ components involved in this process. Notably, there exist three prominent families of histone acetyltransferases: p300/CREB binding protein (p300/CBP) (53), MYST (Moz, Ybf2, Sas2 and Tip60) (54), and GNAT (GCN5 related N-acetyltransferases) (55). Recent investigations into various complexes formed by p300 with crotonyl coenzyme A, propionyl coenzyme A, and butyryl coenzyme A have provided insights into the structural foundation of p300-mediated histone lysine acylation (56). Hence, the function of p300/CBP has been expanded beyond acetyltransferase to encompass more prevalent acyltransferase activities. A recent investigation has identified additional histone lactylation ‘Writers’ apart from p300/CBP—MOF and GCN5 (57). In line with this study, our own research demonstrated that even when the p300 inhibitor iP300w was administered to decrease the modifying enzyme of H3K18la and increase La-CoA, there was still a significant enhancement in the level of H3K18la. This suggests the existence of other lactylation Writers, aside from p300, that regulate the level of H3K18la.
The protein that possesses the ability to identify and bind to specific epigenetic modifications is commonly referred to as ‘readers’. Within the context of histone acetylation and deacetylation reading, three primary domains are involved: bromine domain (BD) (43), double PHD finger (DPF) (58) and YEATS (59). Recent research has demonstrated that mutations in acetylated readers can result in aberrant cell fate (60). Furthermore, investigations are currently underway to explore novel acylation-modified readers, such as DPF and YEATS, which serve as specific benzoylation readers (61). In recent years, the study of the BAF complex has progressed gradually. The structure of the BAF complex was precisely described in a 2020 Nature article using cryo-electron microscopy. It was demonstrated that the bromodomain of SMARCA2/4 has the ability to bind directly to histone nucleosomes (37). In our experiments, we utilized IP combined with mass spectrometry to identify most of the components of the BAF complex in iPSC. Concurrently, we explored the increasing binding ability of Brg1 to H3K18la as the reprogramming progresses. Further research could investigate whether there is a recruitment relationship between Brg1 and H3K18la on target genes, and whether the binding of Brg1 and H3K18la is domain-specific.
However, the lactylation ‘reader’ remains an area of limited research. Our research, in conjunction with previous studies, has identified numerous parallels between histone lactylation and acetylation, such as shared mechanisms of action, common origins, and the enzymes responsible for modifying histones (25,62,63). In fact, acetylation and lactylation exhibit distinct regulatory mechanisms, as evidenced in our manuscript. In our study, a series of protocols were used to alter lactylation without affecting acetylation. Specifically, small molecules capable of modulating lactylation through OGS do not exert the same influence on acetylation (24) (Supplementary Figure 2B, C). Furthermore, the temporal dynamics of acetylation and lactylation differ in the context of iPSC reprogramming. Our findings indicate that H3K27ac levels are elevated slightly on Oct4/Cdh1 and other genes in the initial stages of reprogramming (Supplementary Figure 4E/F). However, H3K18la showed a significantly increase on Oct4/Cdh1 (Figures 3G, 2K). Consequently, we posit that H3K18la and H3K27ac play distinct roles in enhancing the expression of pluripotent genes and MET-related genes during iPSC reprogramming. Nevertheless, it is imperative to validate these observations through comprehensive assessment of reprogramming progress at a genomic scale and the overall extent of multi-modification site changes.
The metabolic disturbance not only affects epigenetic changes through metabolic rewiring, but it also involves the transfer of various metabolic enzymes from the cytoplasm or mitochondria to the nucleus. These enzymes actively participate in modifying histones and DNA through processes such as methylation, acetylation, succinylation, crotonylation, phosphorylation, and glycosylation. Consequently, these modifications alter the chromatin structure and regulate the transcription of genes. For instance, the activation of growth factor receptors such as Egfr, Fgfr and Pdgfr can induce the translocation of pyruvate kinase M2 (Pkm2) into the nucleus. This translocated Pkm2 then binds and activates β-catenin (23) and phosphorylated histone H3 (64), which subsequently promotes the expression of genes related to glycolysis. This activation also enhances glucose uptake and lactic acid production, known as the Warburg effect (65,66). Additionally, during reprogramming, the intermediates of the TCA cycle, namely Pyruvate Dehydrogenase E1 Subunit Alpha 1 (Pdha1), pyruvate carboxylase (Pcb), aconitase 2 (Aco2), citrate synthase (Cs) and isocitrate dehydrogenase 3 (Idh3), gradually translocate to the nucleus. The nuclear translocation of Pdha1 resulted in an elevation of acetyl coenzyme A levels within the nucleus, thereby facilitating histone H3 acetylation and inducing chromatin remodeling of pluripotent genes. Additionally, nuclear-located Pdha1 played a crucial role in promoting the transition from the Prime-Naïve state to a more differentiated state, as well as activating genes associated with the 2-cell stage, ultimately enabling cells to acquire totipotency (46). These findings collectively suggest that the nuclear translocation of glycolytic metabolic enzymes operates as a positive feedback mechanism in the regulation of glycolytic processes. However, the presence of nuclear translocation in Ldha remains uncertain due to the regulation of cell metabolism during reprogramming. It is hypothesized that the translocation of Ldha into the nucleus may induce the presence of histone lactylation substrate La-CoA in the nucleus. Further investigation can be conducted to explore this aspect of the findings.
Supplementary Material
Notes
Present address: Xinglin Hu, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Xingwei Huang, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Yue Yang, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Yuchen Sun, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Yanhua Zhao, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Zhijing Zhang, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Dan Qiu, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Yanshaung Wu, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Present address: Guangming Wu, Guangzhou Laboratory, No. 9 XingDaoHuanBei Road, Guangzhou International Bio Island, Guangzhou 510005 Guangdong Province, P.R. China.
Present address: Lei Lei, Department of Histology and Embryology, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, P.R. China.
Contributor Information
Xinglin Hu, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Xingwei Huang, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China; Guangzhou Laboratory, No. 9 XingDaoHuanBei Road, Guangzhou International Bio Island, Guangzhou 510005 Guangdong Province, China; Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510320, China.
Yue Yang, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Yuchen Sun, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Yanhua Zhao, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Zhijing Zhang, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Dan Qiu, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Yanshuang Wu, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Guangming Wu, Guangzhou Laboratory, No. 9 XingDaoHuanBei Road, Guangzhou International Bio Island, Guangzhou 510005 Guangdong Province, China; Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510320, China.
Lei Lei, Department of Histology and Embryology, Basic Medical Science College, Harbin Medical University, 194 Xuefu Rd, Nangang District, Harbin, Heilongjiang Province 150081, China.
Data availability
IP/MS that supports the findings of this study are available in Pride database (PXD043547), RNA-sequencing and CUT&Tag data are available through GEO (GSE251642). Data from public databases are available in this table.
| Data type | Data source | Introduction |
|---|---|---|
| CUT&Tag | GSE195856 (25) | H3K18la enrichment in mESCs |
| ChIP-seq | GSE87279 (67) | Dux enrichment in C2C12 cell lines |
| CUT&Tag | GSE195856 (25) | H3K27ac enrichment in mESCs |
| CUT&Tag | GSE195856 (25) | H3K4me3 enrichment in mESCs |
| CUT&Tag | GSE195856 (25) | H3K27me3 enrichment in mESCs |
| ChIP-seq | GSE111264 (68) | Brg1 enrichment in mESCs |
| ChIP-seq | GSE83295 (38) | Brg1 enrichment in MEF cells |
| CUT&Tag | GSM7019131 (51) | Dux C3 + 14 rep 1 mCherry in mESCs |
| CUT&Tag | GSM7019132 (51) | Dux C3 + 14 rep 2 mCherry in mESCs |
| CUT&Tag | GSM7019133 (51) | Dux C3delt14 rep 1 mCherry in mESCs |
| CUT&Tag | GSM7019134 (51) | Dux C3delt14 rep 2 mCherry in mESCs |
| CUT&Tag | GSM7019127 (51) | Dux FL rep 1 mCherry in mESCs |
| CUT&Tag | GSM7019128 (51) | Dux FL rep 2 mCherry in mESCs |
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Key projects of Heilongjiang Natural Science Foundation [ZD2021C005 to L.L.]. Funding for open access charge: Key projects of Heilongjiang Natural Science Foundation [ZD2021C005].
Conflict of interest statement. None declared.
References
- 1. Hendrickson P.G., Dorais J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L. et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017; 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falco G., Lee S.L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007; 307:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eckersley-Maslin M., Alda-Catalinas C., Blotenburg M., Kreibich E., Krueger C., Reik W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019; 33:194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017; 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu Y., Yu J., Gu J., Xue C., Zhang L., Chen J., Shen L. Relaxed 3D genome conformation facilitates the pluripotent to totipotent-like state transition in embryonic stem cells. Nucleic Acids Res. 2021; 49:12167–12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012; 487:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu X., Djekidel M.N., Zhang Y. A transcriptional roadmap for 2C-like-to-pluripotent state transition. Sci. Adv. 2020; 6:eaay5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu X., Wu X., Djekidel M.N., Zhang Y. Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat. Cell Biol. 2019; 21:835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang G., Zhang L., Liu W., Qiao Z., Shen S., Zhu Q., Gao R., Wang M., Wang M., Li C. et al. Dux-mediated corrections of aberrant H3K9ac during 2-cell genome activation optimize efficiency of somatic cell nuclear transfer. Cell Stem Cell. 2021; 28:150–163. [DOI] [PubMed] [Google Scholar]
- 10. Huang X., Hu X., Jiang Q., Cao Q., Wu Y., Lei L. Functional study of distinct domains of Dux in improving mouse SCNT embryonic developmentdagger. Biol. Reprod. 2021; 105:1089–1103. [DOI] [PubMed] [Google Scholar]
- 11. Yang L., Song L., Liu X., Bai L., Li G. KDM6A and KDM6B play contrasting roles in nuclear transfer embryos revealed by MERVL reporter system. EMBO Rep. 2018; 19:e46240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi S.H., Gearhart M.D., Cui Z., Bosnakovski D., Kim M., Schennum N., Kyba M. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 2016; 44:5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A.E., Melton D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008; 26:795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Preussner J., Zhong J., Looso M., Braun T., Kim J. Connect-four: genomic analyses of regenerating stem cells identifies zygotic Dux factors as tumor initiators. Mol. Cell Oncol. 2019; 6:1565469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010; 7:51–63. [DOI] [PubMed] [Google Scholar]
- 16. Hernandez C., Wang Z., Ramazanov B., Tang Y., Mehta S., Dambrot C., Lee Y.W., Tessema K., Kumar I., Astudillo M. et al. Dppa2/4 facilitate epigenetic remodeling during reprogramming to pluripotency. Cell Stem Cell. 2018; 23:396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng Z.L., Zhang M.L., Lin H.P., Gao C., Song J.B., Zheng Z., Li L., Zhang Y., Shen X., Zhang H. et al. The Zscan4-Tet2 transcription nexus regulates metabolic rewiring and enhances proteostasis to promote reprogramming. Cell Rep. 2020; 32:107877. [DOI] [PubMed] [Google Scholar]
- 18. Li D., Liu J., Yang X., Zhou C., Guo J., Wu C., Qin Y., Guo L., He J., Yu S. et al. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017; 21:819–833. [DOI] [PubMed] [Google Scholar]
- 19. Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 20. Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012; 151:1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Folmes C.D., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011; 14:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011; 480:118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M. et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019; 574:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Galle E., Wong C.W., Ghosh A., Desgeorges T., Melrose K., Hinte L.C., Castellano-Castillo D., Engl M., de Sousa J.A., Ruiz-Ojeda F.J. et al. H3K18 lactylation marks tissue-specific active enhancers. Genome Biol. 2022; 23:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan R.Y., He L., Zhang J., Liu X., Liao Y., Gao J., Liao Y., Yan Y., Li Q., Zhou X. et al. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer's disease. Cell Metab. 2022; 34:634–648. [DOI] [PubMed] [Google Scholar]
- 27. Xu K., Yin N., Peng M., Stamatiades E.G., Shyu A., Li P., Zhang X., Do M.H., Wang Z., Capistrano K.J. et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science. 2021; 371:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010; 7:651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreno-Yruela C., Zhang D., Wei W., Baek M., Liu W., Gao J., Dankova D., Nielsen A.L., Bolding J.E., Yang L. et al. Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci. Adv. 2022; 8:eabi6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang Z., Yu J., Cui W., Johnson B.K., Kim K., Pfeifer G.P. The chromosomal protein SMCHD1 regulates DNA methylation and the 2c-like state of embryonic stem cells by antagonizing TET proteins. Sci. Adv. 2021; 7:eabb9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo M., Zhang Y., Zhou J., Bi Y., Xu J., Xu C., Kou X., Zhao Y., Li Y., Tu Z. et al. Precise temporal regulation of Dux is important for embryo development. Cell Res. 2019; 29:956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar L., M E.F Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007; 2:5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mager C.E., Mormol J.M., Shelton E.D., Murphy P.R., Bowman B.A., Barley T.J., Wang X., Linn S.C., Liu K., Nelin L.D. et al. p38 MAPK and MKP-1 control the glycolytic program via the bifunctional glycolysis regulator PFKFB3 during sepsis. J. Biol. Chem. 2023; 299:103043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang Z., Su W., Wei X., Qu S., Zhao D., Zhou J., Wang Y., Guan Q., Qin C., Xiang J. et al. HIF-1alpha drives resistance to ferroptosis in solid tumors by promoting lactate production and activating SLC1A1. Cell Rep. 2023; 42:112945. [DOI] [PubMed] [Google Scholar]
- 35. Cao Y., Guo W.T., Tian S., He X., Wang X.W., Liu X., Gu K.L., Ma X., Huang D., Hu L. et al. miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J. 2015; 34:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015; 21:392–402. [DOI] [PubMed] [Google Scholar]
- 37. He S., Wu Z., Tian Y., Yu Z., Yu J., Wang X., Li J., Liu B., Xu Y. Structure of nucleosome-bound human BAF complex. Science. 2020; 367:875–881. [DOI] [PubMed] [Google Scholar]
- 38. Vierbuchen T., Ling E., Cowley C.J., Couch C.H., Wang X., Harmin D.A., Roberts C.W.M., Greenberg M.E. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell. 2017; 68:1067–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. King H.W., Klose R.J. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife. 2017; 6:e22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu W., Stein P., Cheng X., Yang W., Shao N.Y., Morrisey E.E., Schultz R.M., You J. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 2014; 21:1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singhal N., Graumann J., Wu G., Arauzo-Bravo M.J., Han D.W., Greber B., Gentile L., Mann M., Scholer H.R. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010; 141:943–955. [DOI] [PubMed] [Google Scholar]
- 42. Chen K., Long Q., Xing G., Wang T., Wu Y., Li L., Qi J., Zhou Y., Ma B., Scholer H.R. et al. Heterochromatin loosening by the Oct4 linker region facilitates Klf4 binding and iPSC reprogramming. EMBO J. 2020; 39:e99165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999; 399:491–496. [DOI] [PubMed] [Google Scholar]
- 44. Ying Z., Chen K., Zheng L., Wu Y., Li L., Wang R., Long Q., Yang L., Guo J., Yao D. et al. Transient activation of mitoflashes modulates nanog at the early phase of somatic cell reprogramming. Cell Metab. 2016; 23:220–226. [DOI] [PubMed] [Google Scholar]
- 45. Ying Z., Xiang G., Zheng L., Tang H., Duan L., Lin X., Zhao Q., Chen K., Wu Y., Xing G. et al. Short-term mitochondrial permeability transition pore opening modulates histone lysine methylation at the early phase of somatic cell reprogramming. Cell Metab. 2019; 29:502. [DOI] [PubMed] [Google Scholar]
- 46. Li W., Long Q., Wu H., Zhou Y., Duan L., Yuan H., Ding Y., Huang Y., Wu Y., Huang J. et al. Nuclear localization of mitochondrial TCA cycle enzymes modulates pluripotency via histone acetylation. Nat. Commun. 2022; 13:7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao T., Fu Y., Zhu J., Liu Y., Zhang Q., Yi Z., Chen S., Jiao Z., Xu X., Xu J. et al. Single-Cell RNA-Seq reveals dynamic early embryonic-like programs during chemical reprogramming. Cell Stem Cell. 2018; 23:31–45. [DOI] [PubMed] [Google Scholar]
- 48. Yang L., Liu X., Song L., Di A., Su G., Bai C., Wei Z., Li G. Transient Dux expression facilitates nuclear transfer and induced pluripotent stem cell reprogramming. EMBO Rep. 2020; 21:e50054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nishimura K., Aizawa S., Nugroho F.L., Shiomitsu E., Tran Y.T.H., Bui P.L., Borisova E., Sakuragi Y., Takada H., Kurisaki A. et al. A role for KLF4 in promoting the metabolic shift via TCL1 during induced pluripotent stem cell generation. Stem Cell Rep. 2017; 8:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weng M., Hu H., Graus M.S., Tan D.S., Gao Y., Ren S., Ho D.H.H., Langer J., Holzner M., Huang Y. et al. An engineered Sox17 induces somatic to neural stem cell fate transitions independently from pluripotency reprogramming. Sci. Adv. 2023; 9:eadh2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith C.M., Grow E.J., Shadle S.C., Cairns B.R. Multiple repeat regions within mouse DUX recruit chromatin regulators to facilitate an embryonic gene expression program. 2023; bioRxiv doi:08 June 2023, preprint: not peer reviewed 10.1101/2023.03.29.534786. [DOI]
- 52. Li L., Chen K., Wang T., Wu Y., Xing G., Chen M., Hao Z., Zhang C., Zhang J., Ma B. et al. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat. Metab. 2020; 2:882–892. [DOI] [PubMed] [Google Scholar]
- 53. Liu X., Wang L., Zhao K., Thompson P.R., Hwang Y., Marmorstein R., Cole P.A. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008; 451:846–850. [DOI] [PubMed] [Google Scholar]
- 54. Yuan H., Rossetto D., Mellert H., Dang W., Srinivasan M., Johnson J., Hodawadekar S., Ding E.C., Speicher K., Abshiru N. et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012; 31:58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brownell J.E., Zhou J., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996; 84:843–851. [DOI] [PubMed] [Google Scholar]
- 56. Maksimoska J., Segura-Pena D., Cole P.A., Marmorstein R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry. 2014; 53:3415–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang N., Wang W., Wang X., Mang G., Chen J., Yan X., Tong Z., Yang Q., Wang M., Chen L. et al. Histone lactylation boosts reparative gene activation post-myocardial infarction. Circ. Res. 2022; 131:893–908. [DOI] [PubMed] [Google Scholar]
- 58. Zeng L., Zhang Q., Li S., Plotnikov A.N., Walsh M.J., Zhou M.M. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010; 466:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao D., Guan H., Zhao S., Mi W., Wen H., Li Y., Zhao Y., Allis C.D., Shi X., Li H. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016; 26:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wan L., Chong S., Xuan F., Liang A., Cui X., Gates L., Carroll T.S., Li Y., Feng L., Chen G. et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature. 2020; 577:121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ren X., Zhou Y., Xue Z., Hao N., Li Y., Guo X., Wang D., Shi X., Li H. Histone benzoylation serves as an epigenetic mark for DPF and YEATS family proteins. Nucleic Acids Res. 2021; 49:114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pasini D., Malatesta M., Jung H.R., Walfridsson J., Willer A., Olsson L., Skotte J., Wutz A., Porse B., Jensen O.N. et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010; 38:4958–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W.K., Lu Z PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012; 150:685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., Lyssiotis C.A., Aldape K., Cantley L.C., Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012; 14:1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang W., Xia Y., Cao Y., Zheng Y., Bu W., Zhang L., You M.J., Koh M.Y., Cote G., Aldape K. et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell. 2012; 48:771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017; 49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gatchalian J., Malik S., Ho J., Lee D.S., Kelso T.W.R., Shokhirev M.N., Dixon J.R., Hargreaves D.C. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 2018; 9:5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
IP/MS that supports the findings of this study are available in Pride database (PXD043547), RNA-sequencing and CUT&Tag data are available through GEO (GSE251642). Data from public databases are available in this table.
| Data type | Data source | Introduction |
|---|---|---|
| CUT&Tag | GSE195856 (25) | H3K18la enrichment in mESCs |
| ChIP-seq | GSE87279 (67) | Dux enrichment in C2C12 cell lines |
| CUT&Tag | GSE195856 (25) | H3K27ac enrichment in mESCs |
| CUT&Tag | GSE195856 (25) | H3K4me3 enrichment in mESCs |
| CUT&Tag | GSE195856 (25) | H3K27me3 enrichment in mESCs |
| ChIP-seq | GSE111264 (68) | Brg1 enrichment in mESCs |
| ChIP-seq | GSE83295 (38) | Brg1 enrichment in MEF cells |
| CUT&Tag | GSM7019131 (51) | Dux C3 + 14 rep 1 mCherry in mESCs |
| CUT&Tag | GSM7019132 (51) | Dux C3 + 14 rep 2 mCherry in mESCs |
| CUT&Tag | GSM7019133 (51) | Dux C3delt14 rep 1 mCherry in mESCs |
| CUT&Tag | GSM7019134 (51) | Dux C3delt14 rep 2 mCherry in mESCs |
| CUT&Tag | GSM7019127 (51) | Dux FL rep 1 mCherry in mESCs |
| CUT&Tag | GSM7019128 (51) | Dux FL rep 2 mCherry in mESCs |