Abstract
Herpesvirus ateles is an agent indigenous to spider monkeys (Ateles spp.) and causes fulminant lymphomas in various New World primates. Structural and genetic relatedness led to the classification of this virus as a member of the genus Rhadinovirus. It is most closely related to Herpesvirus saimiri. The 108,409-bp light DNA segment of the herpesvirus ateles strain 73 genome has two genes for U-RNA-like transcripts and 73 open reading frames, of which at least 6 show significant homologies to cellular genes (encoding complement control proteins, apoptosis-regulatory proteins, D-type cyclins, interleukin-8 receptors, and enzymes involved in nucleotide metabolism). The left terminal region of the light DNA segment bears the putative rhadinovirus oncogene tio.
Herpesvirus ateles strain 810 was isolated from a primary kidney cell culture of a mature male spider monkey (Ateles geoffroyii) imported from Guatemala (47). This isolate, taxonomically classified as Ateline herpesvirus 2 (AtHV-2) (49), was found to be oncogenic in marmosets (Saguinus oedipus) and owl monkeys (Aotus trivirgatus), which develop malignant lymphomas with leukemia (25, 37, 46). More AtHVs (strains 73, 87, 93, and 94) were isolated from lymphocytes of Columbian spider monkeys (Ateles paniscus) by cocultivation with permissive cell cultures (18). Strain 73 was classified as AtHV-3 (49). Cotton-topped and white-lipped marmosets, owl monkeys (24), and distinct rabbit strains (M. D. Daniel, R. D. Hunt, N. W. King, and J. K. Ingalls, Abstr. 3rd Int. Symp. Oncogenesis and Herpesviruses, p. 213, 1977), infected with AtHV-3 or with AtHV strain 93 or 94 developed lymphomas from which continuous cell lines can be established (18). AtHV-3 was also shown to transform T lymphocytes from S. oedipus and S. fuscicollis in vitro (16, 17, 32).
Members of the genus Rhadinovirus have been classified by their genetic organization. Complete genomic sequences are known for Alcelaphine herpesvirus 1 (15), Murine herpesvirus 68 (60), Human herpesvirus 8 (50, 54), Rhesus rhadinovirus (56), and Herpesvirus saimiri A11 (Saimiriine herpesvirus 2 [SaHV-2]) (5), the type species of the genus. Equine herpesvirus 2 is closely genetically related to the rhadinoviruses; however, its overall genome structure corresponds to that of betaherpesviruses (58). All of these viruses are clearly distinct from Epstein-Barr virus, which defines the type species of the genus Lymphocryptovirus within the subfamily Gammaherpesvirinae.
Since AtHV-2 and -3 and SaHV-2 have been proven unique in their ability to transform monkey T cells to a phenotype of permanent growth in vitro and in vivo and represent the only available model system for studying viral T-cell lymphoma induction, I explored the genetic content of AtHV-3. In this report, the genetic relationship of AtHV-3 to the family of herpesviruses is established by the presentation of the primary structure of its genome.
AtHV-3 was obtained from a frozen virus stock (18) and was propagated on owl monkey kidney cells (10) as described elsewhere (19). Viral DNA was digested with a restriction endonuclease (SacI, EcoRI, PvuII, or HindIII) and subcloned into pBluescript (Stratagene, LaJolla, Calif.) by standard procedures. Authentic clones were confirmed as such by hybridization with AtHV-3 DNA, partial sequencing, and comparison with the SaHV-2 prototype sequence (5). A set of 61 distinct overlapping clones covering the whole light DNA segment (L-DNA) as well as several individual heavy-DNA (H-DNA) repeat clones were obtained and sequenced by using a combination of the shotgun and primer walking strategies. The final AtHV-3 L-DNA sequence was generated from a total of 728,899 bases, resulting in a redundancy of 6.63 per base pair. Computer analyses were performed as described previously (5, 15). Potential open reading frames (ORFs) were defined by applying the following criteria: (i) a minimum of 60 amino acids in the derived polypeptide, (ii) a codon preference like those of unambiguously identified viral genes, (iii) the presence of a typical translational start signal, (iv) potential promoter and transcriptional terminator elements, or (v) sequence homologies to known reading frames of viral or cellular origin.
Earlier studies of the genome structure of AtHV revealed that its genomic DNA is composed of a unique DNA segment of low G+C content (L-DNA), comprising 74% of the genome, flanked by multiple copies of a tandemly repeated element of high G+C content (H-DNA) (19). This resembles the genome structure of SaHV-2 (9), the prototype of the genus Rhadinovirus (5). The unique L-DNA region of AtHV-3 was determined to have a total of 108,409 bp (36.6% G+C), while the prototypic H-DNA repeat unit was found to contain 1,582 bp (77.1% G+C).
Sequence analysis of the standard H-DNA repeat unit of AtHV-3 uncovered numerous internal direct and inverted repeat structures, among them an internal 154-bp direct repeat sequence whose presence has been suggested by endonuclease digestion data (19). The H-DNA of SaHV-2 and AtHV-2 and -3 had been found not to be homologous by heteroduplex analysis (19); this was confirmed here by comparison of the AtHV-3 and SaHV-2 H-DNA sequences. The only common motifs found are related to pac-1 and pac-2 sequences corresponding to genome cleavage recognition motifs conserved among herpesviruses (11) and to the cleavage-packaging site which defines the genomic termini and the junctions between H- and L-DNA sequences. These junctions, which were defined by the first nucleotide that diverges from the standard H-DNA repeat unit, can be localized to a single nucleotide of H-DNA (position 1336H) at both ends of the L-DNA. Like in SaHV-2, the far-left terminal region of L-DNA consists of H-DNA (2, 4), which in the case of AtHV-3 is not rearranged but continuous.
Analysis of the genomic sequence of the L-DNA of AtHV-3 revealed 73 ORFs that potentially code for at least 73 proteins (Fig. 1; Table 1). Forty-eight reading frames were found to be conserved among most herpesviruses. A minimum of 14 deduced amino acid sequences are specific for the subfamily Gammaherpesvirinae (ORFs 3, 10, 11, 23, 27, 28, 45, 48 to 52, 58, and 75); 6 were found in rhadinoviruses only (ORFs 1, 4, 14, 71, 72, and 73), some of which appear to be restricted to specific virus species. Molecular piracy of cellular genes appears to be a common feature of rhadinovirus genomes and is most apparent in the genomes of SaHV-2 (5) and the recently isolated human Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8 [HHV-8]) (50, 54). Analysis of the entire 108.4 kbp of the AtHV-3 L-DNA also revealed a number of cellular homologs which might contribute to viral pathogenicity. These include a virus-encoded interleukin-8 receptor (IL-8R) (1), a D-type cyclin (29, 57), FLICE-inhibitory protein (FLIP) (59), thymidylate synthetase (TS) (23, 53), ie14/vsag (34, 63), complement-regulatory proteins (2, 20), and two U-RNA-like transcripts (3, 39, 61) (Fig. 1; Table 1). Notably, AtHV-3 does not code for a homolog of dihydrofolate reductase, IL-17, or CD59, which are encoded by SaHV-2. Homologs for IL-6, macrophage-inhibitory protein 1-α and -β chemokines, and viral interferon-regulatory factors, all of which are encoded by HHV-8, were not identified. However, apparently all identified ORFs of AtHV-3 are conserved in SaHV-2 (Table 1), with amino acid sequence identities ranging from 30.4 to 92.5% (average, 75.1%). A gapped alignment of their complete L-DNAs revealed an average DNA sequence conservation of 76.4%.
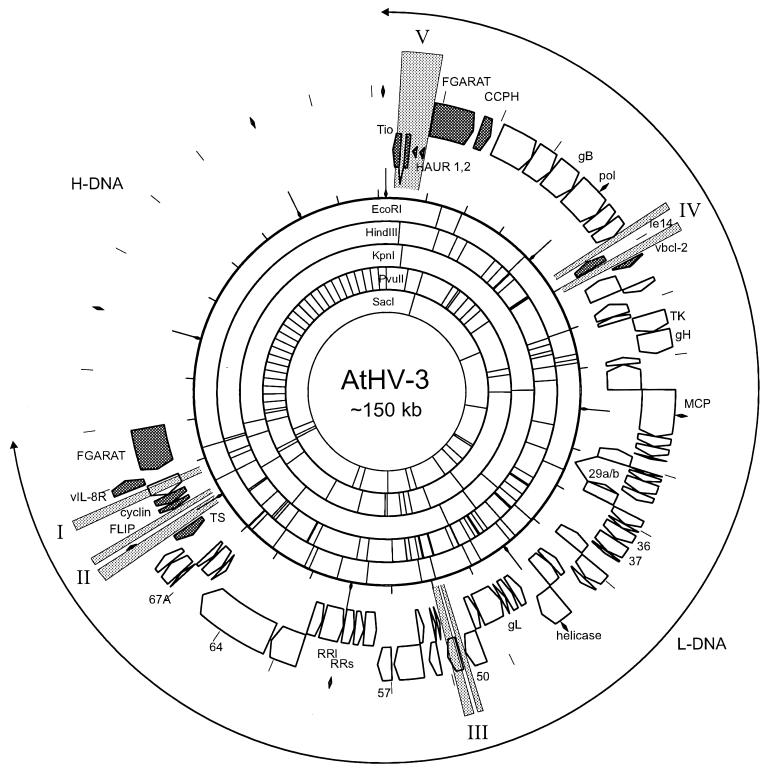
FIG. 1.
Circular representation of the AtHV-3 genome. H-DNA sequences were added to L-DNA to give a total genome size of approximately 150 kbp as estimated by earlier studies. ORFs are shown as directed boxes around circular restriction maps, and cellular homologs are shaded dark gray. Areas significantly different from the SaHV-2 genome are depicted as light-gray-shaded areas I to V. The scale is 5 kbp/unit, with a diamond sign located every 20 kbp. ORF numbers are given for orientation and reference to Table 1. Abbreviations: HAUR, herpesvirus ateles U-like RNA; CCPH, complement control protein homolog; gB, glycoprotein B; pol, DNA polymerase; ie14/vsag, viral superantigen; TK, thymidine kinase; gH, glycoprotein H; MCP, major capsid protein; gL, glycoprotein L; RR, ribonucleotide reductase (l, large subunit; s, small subunit); cyclin, viral D-type cyclin; vIL-8R, viral IL-8 receptor; FGARAT, formylglycineamide ribotide amidotransferase.
TABLE 1.
Description of AtHV-3 ORFs and comparison to SaHV-2 and HHV-8
| AtHV-3 ORF | Stranda | Position of:
|
No. of amino acids encoded | Calculated molecular mass of product (kDa) | % Identity of product with:
|
Product description/homologyb | ||
|---|---|---|---|---|---|---|---|---|
| Start/exon 1 | Exon 2/stop | SaHV-2 | KSHV (HHV-8) | |||||
| 1 | c | 90–730 | 1339–1507 | 269 | 29.2 | 32.8/40.0 | —c | Tio; homologous to StpC and Tip |
| 3 | 3224 | 6961 | 1,245 | 138.1 | 70.0 | 25.4 | FGARAT | |
| 4a | 7470 | 8552 | 360 | 40.2 | 73.1 | 28.3 | mCCPH | |
| 4b | 7470–8331 | 8525–8571 | 302 | 33.7 | 72.2 | 30.6 | sCCPH | |
| 6 | 9205 | 12591 | 1,128 | 127.4 | 86.7 | 54.9 | Major single-stranded-DNA binding protein | |
| 7 | 12598 | 14640 | 680 | 78.5 | 82.3 | 45.2 | Transport protein | |
| 8 | 14627 | 17050 | 807 | 91.1 | 85.9 | 55.7 | Glycoprotein B | |
| 9 | 17123 | 20152 | 1,009 | 114.4 | 87.5 | 60.8 | DNA polymerase | |
| 10 | 20192 | 21412 | 406 | 45.4 | 82.0 | 24.0 | Homologous to EBV Raji LF1/LF2 in B95-8 deletion, EHV-2 ORF10 | |
| 11 | 21415 | 22632 | 405 | 45.8 | 75.6 | 28.4 | Homologous to EBV Raji LF2 in B95-8 deletion, EHV-2 ORF11 | |
| 14 | c | 23207 | 24028 | 273 | 30.6 | 47.0 | — | ie14/vsag |
| 16 | 25075 | 25599 | 174 | 19.5 | 65.0 | 19.0 | bcl2 family | |
| 17 | c | 25629 | 27128 | 499 | 55.9 | 77.6 | 41.7 | Protease, minor capsid scaffold protein |
| 18 | 27121 | 27891 | 256 | 29.7 | 82.8 | 48.0 | ||
| 19 | c | 27885 | 29519 | 544 | 61.2 | 79.7 | 46.2 | Virion protein |
| 20 | c | 29149 | 30060 | 303 | 34.5 | 67.3 | 41.8 | Fusion protein |
| 21 | 30059 | 31642 | 527 | 59.7 | 76.4 | 28.1 | Thymidine kinase | |
| 22 | 31639 | 33789 | 716 | 83.2 | 87.6 | 34.3 | Glycoprotein H | |
| 23 | c | 33782 | 34549 | 255 | 29.2 | 83.7 | 33.5 | |
| 24 | c | 34559 | 36751 | 730 | 82.5 | 82.1 | 47.4 | |
| 25 | 36757 | 40872 | 1,371 | 153.8 | 89.4 | 65.0 | Major capsid protein | |
| 26 | 40888 | 41802 | 304 | 34.5 | 89.8 | 55.6 | Capsid protein VP23 | |
| 27 | 41809 | 42627 | 272 | 31.7 | 72.1 | 31.5 | ||
| 28 | 42692 | 42916 | 74 | 7.9 | 56.3 | 36.1 | ||
| 29a | c | 42929 | 43984 | 351 | 39.0 | 90.6 | 52.3 | DNA-packaging protein, terminase |
| 30 | 44089 | 44316 | 75 | 8.3 | 81.3 | 33.3 | ||
| 31 | 44283 | 44909 | 208 | 24.4 | 87.5 | 44.3 | ||
| 32 | 44855 | 46180 | 441 | 51.2 | 71.4 | 29.8 | ||
| 33 | 46173 | 47165 | 330 | 36.8 | 79.7 | 36.2 | ||
| 29b | c | 47107 | 48063 | 318 | 36.4 | 81.8 | 61.8 | |
| 29 | c | 42929–44071 | 47155–48063 | 683 | 77.0 | 87.3 | 58.9 | DNA-packaging protein, terminase |
| 34 | 48062 | 49012 | 316 | 36.2 | 87.0 | 44.3 | ||
| 35 | 48999 | 49454 | 151 | 17.4 | 76.0 | 29.9 | ||
| 36 | 49339 | 50631 | 430 | 48.4 | 81.4 | 30.2 | Phosphotransferase | |
| 37 | 50631 | 52082 | 483 | 55.9 | 87.4 | 50.9 | Alkaline exonuclease | |
| 38 | 52037 | 52237 | 66 | 7.7 | 69.7 | 40.0 | Homologous to myristylated tegument protein in EHV-2 | |
| 39 | c | 52263 | 53360 | 365 | 42.2 | 84.1 | 49.3 | Integral membrane protein |
| 40 | 53445 | 54773 | 442 | 50.7 | 68.6 | 26.9 | Helicase-primase complex | |
| 41 | 54926 | 55408 | 160 | 18.5 | 74.4 | 27.5 | Helicase-primase complex | |
| 42 | c | 55400 | 56197 | 265 | 29.7 | 80.0 | 37.7 | |
| 43 | c | 56163 | 57863 | 566 | 64.6 | 89.2 | 58.8 | Minor capsid protein, virion protein |
| 44 | 57829 | 60174 | 781 | 88.0 | 89.5 | 61.1 | Helicase-primase complex, helicase | |
| 45 | c | 60214 | 60996 | 260 | 29.1 | 61.0 | 29.9 | |
| 46 | c | 61004 | 61762 | 252 | 29.1 | 82.9 | 59.1 | Uracil DNA glycosidase |
| 47 | c | 61740 | 62162 | 140 | 15.9 | 67.2 | 32.1 | Glycoprotein L |
| 48 | c | 62159 | 64537 | 792 | 92.6 | 50.7 | 21.6 | Contains a large acidic repeat |
| 49 | c | 64768 | 65682 | 304 | 35.8 | 73.9 | 20.2 | |
| 50 | 65682 | 67250 | 522 | 58.2 | 68.7 | 21.7 | Transcriptional control; homologous to EBV Rta | |
| 51 | 67818 | 68867 | 349 | 37.0 | 40.5 | 23.9 | Virus-specific glycoprotein | |
| 52 | c | 68906 | 69253 | 115 | 13.2 | 66.7 | 24.3 | |
| 53 | c | 69295 | 69564 | 89 | 10.0 | 61.8 | 25.8 | |
| 54 | 69636 | 70499 | 287 | 32.2 | 74.9 | 35.0 | dUTPase | |
| 55 | c | 70537 | 71139 | 200 | 22.5 | 83.5 | 46.4 | |
| 56 | 71118 | 73625 | 835 | 96.0 | 81.3 | 44.3 | Helicase-primase complex, primase | |
| 57 | 73782 | 75053 | 423 | 47.5 | 76.2 | 27.6 | Transcriptional control; homologous to EBV Mta | |
| 58 | c | 75434 | 76495 | 353 | 40.7 | 83.3 | 29.3 | |
| 59 | c | 76504 | 77604 | 366 | 40.1 | 77.0 | 30.8 | Processivity factor of DNA polymerase |
| 60 | c | 77719 | 78636 | 305 | 35.3 | 92.5 | 63.0 | Ribonucleotide reductase small subunit |
| 61 | c | 78642 | 80945 | 767 | 87.0 | 90.0 | 51.9 | Ribonucleotide reductase large subunit |
| 62 | c | 80945 | 81937 | 330 | 37.5 | 86.1 | 40.1 | Probable capsid assembly, DNA maturation protein |
| 63 | 81944 | 84643 | 899 | 103.4 | 75.9 | 30.7 | Tegument protein | |
| 64 | 84643 | 92058 | 2,471 | 280.0 | 72.0 | 30.5 | Large tegument protein | |
| 65 | c | 92062 | 92463 | 133 | 14.4 | 69.7 | 29.3 | Capsid protein |
| 66 | c | 92426 | 93760 | 444 | 50.9 | 79.5 | 34.6 | |
| 67 | c | 93679 | 94380 | 233 | 26.5 | 84.1 | 50.6 | Tegument protein |
| 67A | c | 94377 | 94634 | 85 | 9.7 | 84.0 | 41.6 | |
| 68 | 94627 | 95937 | 436 | 49.3 | 77.8 | 47.0 | Probable major envelope protein | |
| 69 | 95939 | 96724 | 261 | 29.8 | 88.1 | 49.0 | ||
| 70 | c | 96897 | 97769 | 290 | 32.9 | 84.8 | 65.5 | TS |
| 71 | c | 100187 | 100528 | 113 | 13.4 | 42.5 | 28.3 | FLIP |
| 72 | c | 100529 | 101317 | 262 | 29.6 | 74.8 | 29.6 | D-type cyclin |
| 73 | c | 101349 | 102692 | 447 | 46.5 | 35.0 | 15.0 | Glycine rich, repetitive structure |
| 74 | 103148 | 104113 | 321 | 36.6 | 70.6 | 31.5 | Viral IL-8 receptor | |
| 75 | c | 104198 | 108097 | 1,299 | 143.1 | 73.6 | 36.0 | FGARAT |
c, complementary strand.
Taken in part from descriptions of other sequenced herpesvirus ORF products. Abbreviations: FGARAT, formylglycineamide ribotide amidotransferase; mCCPH and sCCPH, membrane-bound and soluble complement control protein homologs; EBV, Epstein-Barr virus; EHV-2, equine herpesvirus 2.
—, not present.
At least five genomic loci of AtHV-3 were found to be quite distinct from those of SaHV-2 (Fig. 1). Region I corresponds to a repetitive DNA structure within ORF73 which is present in most rhadinoviruses (5, 15, 54). The function of the ORF73 protein is not known; however, a protein encoded at an analogous position in the HHV-8 genome has been demonstrated to be a nuclear antigen expressed during latency (52). Region II is characterized by variability within the 5′ noncoding region of the thymidylate synthetase (TS) gene, which has been tentatively mapped as the origin of lytic replication in SaHV-2 (55), and by the presence of an additional 168-bp repeat sequence, composed of 15.3 copies of an 11-bp unit, downstream of ORF71. Independent of this repeat insertion, ORF71 of AtHV-3 is likely to be nonfunctional due to multiple frame shifts at the 3′ end of this ORF, which encodes a FLIP in other rhadinoviruses (45, 59). This appears to be a specific feature of AtHVs, since similar results were obtained for AtHV-2 after PCR amplification and sequencing of the corresponding genomic region. Region III is composed of two insertions, of 347 and 240 bp, into the AtHV-3 genome, which affect the 3′ noncoding region of ORF50 and most of ORF51, respectively. Although no obvious sequence homology was detected, AtHV-3 ORF51 is a positional homolog of HHV-8 K8.1, murine herpesvirus 68 M7, bovine herpesvirus 4 BORFD1, alcelaphine herpesvirus 1 A8, and SaHV-2 ORF51. All of these ORFs code for a typical type I transmembrane glycoprotein with a high content of Ser and Thr residues and multiple N-linked glycosylation sites (NxT/S). Region IV of AtHV-3 has four deletions relative to the SaHV-2 genome: one of 177, affecting ORF12; one of 900 bp, in the gene encoding a viral IL-17 (5, 33); and two, of 632 and 84 bp, in the gene coding for the CD59 homologue in SaHV-2 (6).
Region V corresponds to a highly variable region of SaHV-2 (44) which has been shown to mediate the oncogenic and transforming phenotype (12–14, 31, 35, 42). The high degree of variability among different strains of SaHV-2 has led to their subdivision into three subgroups, A, B, and C (43). Group A, represented by the SaHV-2 prototype strain A11, encodes a single protein, termed StpA (for saimiri transformation-associated protein of group A), which has been demonstrated to be transforming in cell culture and in transgenic mice (30, 36). StpA has been shown to become phosphorylated by cellular Src and to bind to the Src SH2 domain by a phosphotyrosine-dependent mechanism (38). It has also been suggested that StpA interacts with the T-cell-specific Src family kinases Lck and Fyn (38). Group C viruses C484 and C488 encode two proteins, StpC and Tip, within this variable genomic region (7, 21). On the one hand, StpC has been shown to be transforming in cell culture (30) and to cause epithelial tumors in transgenic mice (48). It interacts directly with cellular Ras and competes with cellular Raf for binding to Ras, thereby affecting the signal transduction pathway of Ras (22, 26). On the other hand, Tip, a tyrosine kinase-interacting protein of SaHV-2 group C viruses, interacts with T-cell-specific kinases of the Src family, predominantly Lck (8, 41). Although its influence on Lck activity has been controversial (28, 40, 51, 62), Tip has been shown to associate with Lck by binding to the SH3 domain of Lck via its SH3 binding motif and, additionally, to the kinase domain of Lck via its CSKH motif (27, 28; U. Friedrich, unpublished data). In transformed monkey T cells, a spliced mRNA is transcribed within this variable genomic region of AtHV-3, which encodes the two-in-one protein Tio, a protein that exhibits homologies with both StpC and Tip (4). Homologous sequences were identified in AtHV-2, indicating a very close relationship between AtHV-2 and AtHV-3. AtHV-3-encoded Tio has been demonstrated to combine functions of Tip and StpA/B: Tio interacts with cellular Src family kinases by binding to their SH3 domains via an SH3 binding motif related to Tip and by binding to the SH2 domains of Lck, Src, and Fyn in a phosphotyrosine-dependent manner, like StpA (4). Sequence homology to StpC also suggests an additional function related to StpC; however, this has not yet been supported by experimental evidence. Tio appears to be the single rhadinovirus oncoprotein encoded in the entire AtHV-3 genome which is a multifunctional protein involved in signal transduction.
Nucleotide sequence accession numbers.
The nucleotide sequences of the AtHV-3 unique L-DNA region, AtHV-3 H-DNA repeat unit, AtHV-2 ORF71, and AtHV-2 tio gene were submitted to the GenBank database and assigned accession no. AF083424, AF126541, AF133729, and AF135064, respectively.
Acknowledgments
I thank B. Fleckenstein and S. M. Lang for critical readings of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 466.
Footnotes
To the memory of my father.
REFERENCES
- 1.Ahuja S K, Murphy P M. Molecular piracy of mammalian interleukin-8 receptor type B by herpesvirus saimiri. J Biol Chem. 1993;268:20691–20694. [PubMed] [Google Scholar]
- 2.Albrecht J-C, Fleckenstein B. New member of the multigene family of complement control proteins in herpesvirus saimiri. J Virol. 1992;66:3937–3940. doi: 10.1128/jvi.66.6.3937-3940.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht J-C, Fleckenstein B. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 1992;20:1810. doi: 10.1093/nar/20.7.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht J-C, Friedrich U, Kardinal C, Koehn J, Fleckenstein B, Feller S M, Biesinger B. Herpesvirus ateles gene product Tio interacts with nonreceptor protein tyrosine kinases. J Virol. 1999;73:4631–4639. doi: 10.1128/jvi.73.6.4631-4639.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albrecht J-C, Nicholas J, Cameron K R, Newman C, Fleckenstein B, Honess R W. Herpesvirus saimiri has a gene specifying a homologue of the cellular membrane glycoprotein CD59. Virology. 1992;190:527–530. doi: 10.1016/0042-6822(92)91247-r. [DOI] [PubMed] [Google Scholar]
- 7.Biesinger B, Trimble J J, Desrosiers R C, Fleckenstein B. The divergence between two oncogenic herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology. 1990;176:505–514. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 8.Biesinger B, Tsygankov A Y, Fickenscher H, Emmrich F, Fleckenstein B, Bolen J B, Broker B M. The product of the herpesvirus saimiri open reading frame 1 (tip) interacts with T cell-specific kinase p56lck in transformed cells. J Biol Chem. 1995;270:4729–4734. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 9.Bornkamm G W, Delius H, Fleckenstein B, Werner F-J, Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976;19:154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel M D, Melendez L V, Hunt R D, King N W, Anver M, Fraser C E, Barahona H, Baggs R B. Herpesvirus saimiri. VII. Induction of malignant lymphoma in New Zealand White rabbits. J Natl Cancer Inst. 1974;53:1803–1807. [PubMed] [Google Scholar]
- 11.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers R C, Bakker A, Kamine J, Falk L A, Hunt R D, King N W. A region of the herpesvirus saimiri genome required for oncogenicity. Science. 1985;228:184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers R C, Silva D P, Waldron L M, Letvin N L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986;57:701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelaphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falk L, Johnson D, Deinhardt F. Transformation of marmoset lymphocytes in vitro with herpesvirus ateles. Int J Cancer. 1978;21:652–657. doi: 10.1002/ijc.2910210517. [DOI] [PubMed] [Google Scholar]
- 17.Falk L, Wright J, Wolfe L, Deinhardt F. Herpesvirus ateles: transformation in vitro of marmoset splenic lymphocytes. Int J Cancer. 1974;14:244–251. doi: 10.1002/ijc.2910140213. [DOI] [PubMed] [Google Scholar]
- 18.Falk L A, Nigida S M, Deinhardt F, Wolfe L G, Cooper R W, Hernandez-Camacho J I. Herpesvirus ateles: properties of an oncogenic herpesvirus isolated from circulating lymphocytes of spider monkeys (Ateles sp.) Int J Cancer. 1974;14:473–482. doi: 10.1002/ijc.2910140407. [DOI] [PubMed] [Google Scholar]
- 19.Fleckenstein B, Bornkamm G W, Mulder C, Werner F J, Daniel M D, Falk L A, Delius H. Herpesvirus ateles DNA and its homology with Herpesvirus saimiri nucleic acid. J Virol. 1978;25:361–373. doi: 10.1128/jvi.25.1.361-373.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fodor W L, Rollins S A, Bianco-Caron S, Rother R P, Guilmette E R, Burton W V, Albrecht J-C, Fleckenstein B, Squinto S P. The complement control protein homolog of herpesvirus saimiri regulates serum complement by inhibiting C3 convertase activity. J Virol. 1995;69:3889–3892. doi: 10.1128/jvi.69.6.3889-3892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geck P, Whitaker S A, Medveczky M M, Medveczky P G. Expression of collagenlike sequences by a tumor virus, herpesvirus saimiri. J Virol. 1990;64:3509–3515. doi: 10.1128/jvi.64.7.3509-3515.1990. . (Erratum, 65:7084, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J, Williams K, Duboise S M, Alexander L, Veazey R, Jung J U. Substitution of ras for the herpesvirus saimiri STP oncogene in lymphocyte transformation. J Virol. 1998;72:3698–3704. doi: 10.1128/jvi.72.5.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honess R W, Bodemer W, Cameron K R, Niller H H, Fleckenstein B, Randall R E. The A+T-rich genome of herpesvirus saimiri contains a highly conserved gene for thymidylate synthase. Proc Natl Acad Sci USA. 1986;83:3604–3608. doi: 10.1073/pnas.83.11.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt R D, Barahona H, Daniel M D. Herpesvirus ateles malignant lymphoma in owl monkeys: a new susceptible primate. In: Yohn D S, Bentvelzen P, editors. Advances in comparative leukemia research. Amsterdam, The Netherlands: Elsevier/North-Holland; 1978. pp. 198–200. [Google Scholar]
- 25.Hunt R D, Melendez L V, Garcia F G, Trum B F. Pathologic features of herpesvirus ateles lymphoma in cotton-topped marmosets (Saguinus oedipus) J Natl Cancer Inst. 1972;49:1631–1639. doi: 10.1093/jnci/49.6.1631. [DOI] [PubMed] [Google Scholar]
- 26.Jung J U, Desrosiers R C. Association of the viral oncoprotein STP-C488 with cellular ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung J U, Lang S M, Friedrich U, Jun T, Roberts T M, Desrosiers R C, Biesinger B. Identification of Lck-binding elements in tip of herpesvirus saimiri. J Biol Chem. 1995;270:20660–20667. doi: 10.1074/jbc.270.35.20660. [DOI] [PubMed] [Google Scholar]
- 28.Jung J U, Lang S M, Jun T, Roberts T M, Veillette A, Desrosiers R C. Downregulation of Lck-mediated signal transduction by tip of herpesvirus saimiri. J Virol. 1995;69:7814–7822. doi: 10.1128/jvi.69.12.7814-7822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung J U, Stäger M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamine J, Bakker A, Desrosiers R C. Mapping of RNA transcribed from a region of the Herpesvirus saimiri genome required for oncogenicity. J Virol. 1984;52:532–540. doi: 10.1128/jvi.52.2.532-540.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiyotaki M, Solomon K R, Letvin N L. Herpesvirus ateles immortalizes in vitro a CD3+ CD4+ CD8+ marmoset lymphocyte with NK function. J Immunol. 1988;140:730–736. [PubMed] [Google Scholar]
- 33.Knappe A, Hiller C, Niphuis H, Fossiez F, Thurau M, Wittmann S, Kuhn E-M, Lebecque S, Banchereau J, Rosenwirth B, Fleckenstein B, Heeney J, Fickenscher H. The interleukin-17 gene of herpesvirus saimiri. J Virol. 1998;72:5797–5801. doi: 10.1128/jvi.72.7.5797-5801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knappe A, Hiller C, Thurau M, Wittmann S, Hofmann H, Fleckenstein B, Fickenscher H. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J Virol. 1997;71:9124–9133. doi: 10.1128/jvi.71.12.9124-9133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koomey J M, Mulder C, Burghoff R L, Fleckenstein B, Desrosiers R C. Deletion of DNA sequence in a nononcogenic variant of Herpesvirus saimiri. J Virol. 1984;50:662–665. doi: 10.1128/jvi.50.2.662-665.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kretschmer C, Murphy C, Biesinger B, Beckers J, Fickenscher H, Kirchner T, Fleckenstein B, Ruther U. A herpes saimiri oncogene causing peripheral T-cell lymphoma in transgenic mice. Oncogene. 1996;12:1609–1616. [PubMed] [Google Scholar]
- 37.Laufs R, Melendez L V. Oncogenicity of herpesvirus ateles in monkeys. J Natl Cancer Inst. 1973;51:599–608. [PubMed] [Google Scholar]
- 38.Lee H, Trimble J J, Yoon D-W, Regier D, Desrosiers R C, Jung J U. Genetic variation of herpesvirus saimiri subgroup A transforming protein and its association with cellular src. J Virol. 1997;71:3817–3825. doi: 10.1128/jvi.71.5.3817-3825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S I, Murthy S C, Trimble J J, Desrosiers R C, Steitz J A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 40.Lund T, Medveczky M M, Medveczky P G. Herpesvirus saimiri Tip-484 membrane protein markedly increases p56lck activity in T cells. J Virol. 1997;71:378–382. doi: 10.1128/jvi.71.1.378-382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lund T, Medveczky M M, Neame P J, Medveczky P G. A herpesvirus saimiri membrane protein required for interleukin-2 independence forms a stable complex with p56lck. J Virol. 1996;70:600–606. doi: 10.1128/jvi.70.1.600-606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medveczky M M, Geck P, Sullivan J L, Serbousek D, Djeu J Y, Medveczky P G. IL-2 independent growth and cytotoxicity of herpesvirus saimiri-infected human CD8 cells and involvement of two open reading frame sequences of the virus. Virology. 1993;196:402–412. doi: 10.1006/viro.1993.1495. [DOI] [PubMed] [Google Scholar]
- 43.Medveczky M M, Szomolanyi E, Hesselton R, DeGrand D, Geck P, Medveczky P G. Herpesvirus saimiri strains from three DNA subgroups have different oncogenic potentials in New Zealand White rabbits. J Virol. 1989;63:3601–3611. doi: 10.1128/jvi.63.9.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medveczky P, Szomolanyi E, Desrosiers R C, Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984;52:938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meinl E, Fickenscher H, Thome M, Tschopp J, Fleckenstein B. Anti-apoptotic strategies of lymphotropic viruses. Immunol Today. 1998;19:474–479. doi: 10.1016/s0167-5699(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 46.Melendez L V, Hunt R D, Garcia F G, Daniel M D, Fraser C E, Barahona H H, King N W. Herpesvirus ateles, the second lymphoma virus of monkeys. Bibl Haematol. 1973;39:410–415. doi: 10.1159/000427870. [DOI] [PubMed] [Google Scholar]
- 47.Melendez L V, Hunt R D, King N W, Barahona H H, Daniel M D, Fraser C E, Garcia F G. Herpesvirus ateles, a new lymphoma virus of monkeys. Nat New Biol. 1972;235:182–184. doi: 10.1038/newbio235182b0. [DOI] [PubMed] [Google Scholar]
- 48.Murphy C, Kretschmer C, Biesinger B, Beckers J, Jung J, Desrosiers R C, Muller-Hermelink H K, Fleckenstein B W, Ruther U. Epithelial tumours induced by a herpesvirus oncogene in transgenic mice. Oncogene. 1994;9:221–226. [PubMed] [Google Scholar]
- 49.Murphy F A, Fauquet C M, Mayo M A, Jarvis A W, Ghabrial S A, Summers M D, Martelli G P, Bishop D H L. The classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. [Google Scholar]
- 50.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noraz N, Saha K, Ottones F, Smith S, Taylor N. Constitutive activation of TCR signaling molecules in IL-2-independent herpesvirus saimiri-transformed T cells. J Immunol. 1998;160:2042–2045. [PubMed] [Google Scholar]
- 52.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richter J, Puchtler I, Fleckenstein B. Thymidylate synthase gene of herpesvirus ateles. J Virol. 1988;62:3530–3535. doi: 10.1128/jvi.62.9.3530-3535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schofield A. Investigations of the origins of replication of herpesvirus saimiri. Ph.D. thesis. London, United Kingdom: The British Library, Open University; 1994. [Google Scholar]
- 56.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 58.Telford E A, Watson M S, Aird H C, Perry J, Davison A J. The DNA sequence of equine herpesvirus 2. J Mol Biol. 1995;249:520–528. doi: 10.1006/jmbi.1995.0314. [DOI] [PubMed] [Google Scholar]
- 59.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 60.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wassarman D A, Lee S I, Steitz J A. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 1989;17:1258. doi: 10.1093/nar/17.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiese N, Tsygankov A Y, Klauenberg U, Bolen J B, Fleischer B, Broker B M. Selective activation of T cell kinase p56lck by herpesvirus saimiri protein tip. J Biol Chem. 1996;271:847–852. doi: 10.1074/jbc.271.2.847. [DOI] [PubMed] [Google Scholar]
- 63.Yao Z, Maraskovsky E, Spriggs M K, Cohen J I, Armitage R J, Alderson M R. Herpesvirus saimiri open reading frame 14, a protein encoded by T lymphotropic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J Immunol. 1996;156:3260–3266. [PubMed] [Google Scholar]