Abstract
A GGGGCC (G4C2) hexanucleotide repeat expansion in C9ORF72 causes amyotrophic lateral sclerosis and frontotemporal dementia (C9ALS/FTD), while a CGG trinucleotide repeat expansion in FMR1 leads to the neurodegenerative disorder Fragile X-associated tremor/ataxia syndrome (FXTAS). These GC-rich repeats form RNA secondary structures that support repeat-associated non-AUG (RAN) translation of toxic proteins that contribute to disease pathogenesis. Here we assessed whether these same repeats might trigger stalling and interfere with translational elongation. We find that depletion of ribosome-associated quality control (RQC) factors NEMF, LTN1 and ANKZF1 markedly boost RAN translation product accumulation from both G4C2 and CGG repeats while overexpression of these factors reduces RAN production in both reporter assays and C9ALS/FTD patient iPSC-derived neurons. We also detected partially made products from both G4C2 and CGG repeats whose abundance increased with RQC factor depletion. Repeat RNA sequence, rather than amino acid content, is central to the impact of RQC factor depletion on RAN translation—suggesting a role for RNA secondary structure in these processes. Together, these findings suggest that ribosomal stalling and RQC pathway activation during RAN translation inhibits the generation of toxic RAN products. We propose augmenting RQC activity as a therapeutic strategy in GC-rich repeat expansion disorders.
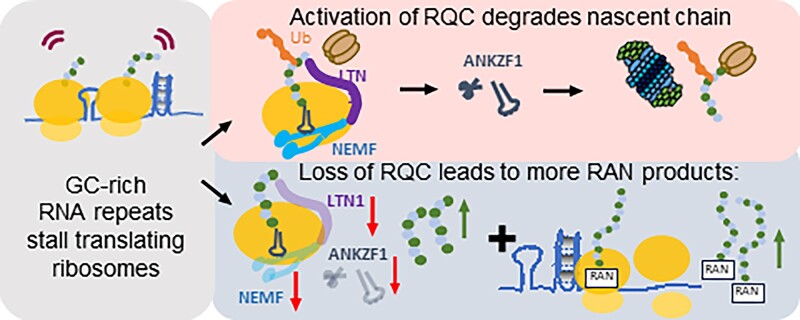
Graphical Abstract
Graphical Abstract.
Introduction
Short Tandem Repeat (STR) expansions cause more than 50 human neurodegenerative, neurodevelopmental, and neuromuscular disorders (1–6). Several STR expansions trigger a process known as repeat-associated non-AUG (RAN) translation, which is a non-canonical initiation process whereby proteins are generated from repeats without an AUG start codon (7–14). RAN translation-generated proteins accumulate within neurons and in patient tissues and elicit toxicity in both cellular and animal disease models (9,15–20). As at least 10 different neurodegenerative disease-associated STRs support RAN translation, there is significant interest in identifying selective inhibitors and regulators of this non-canonical translation process (6,13,20–30). However, to date, the precise mechanisms that regulate RAN translation remain largely unknown.
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are inexorably progressive and fatal neurodegenerative disorders. The most common genetic cause of FTD and ALS is an expanded GGGGCC (G4C2) hexanucleotide repeat within the first intron of C9ORF72 (C9ALS/FTD) (31–33). Fragile X-associated tremor/ataxia syndrome (FXTAS) is similarly a progressive adult-onset neurodegenerative disorder without effective treatments. FXTAS results from a transcribed CGG repeat expansion in the 5′ UTR of FMR1 that affects about 1/5000 people worldwide (34–37). The repeats that cause both C9ALS/FTD and FXTAS are highly GC-rich and are thought to form strong secondary structures as RNA, including RNA hairpins and G quadruplexes (38–43). Moreover, both repeat sequences support RAN translation (14,17,20,25,44–47). Without an AUG start codon, RAN translation can occur in all possible reading frames from both sense and antisense transcripts (46,48). For example, G4C2 repeats in C9ORF72 generate GA (glycine-alanine; +0 frame), GP (glycine-proline; +1 frame), and GR (glycine arginine; +2 frame) dipeptide repeat proteins (DPRs) while CGG repeats in FMR1 generate polyG (glycine; +1 frame), polyA (alanine; +2 frame), and polyR (arginine; +0 frame) homopolymeric peptides from the sense transcripts (46,49). However, the efficiency of RAN translation differs significantly across reading frames for each repeat, such that GA DPRs are produced more readily than GP or GR from G4C2 repeats, and polyG and polyA are more efficiently generated than polyR peptides from CGG repeats (17,20,44,45,49). While initial studies suggested that these differences might be due solely to near-cognate initiation sites in the sequences surrounding the repeats, more recent work suggests that these differences in translational efficiency persist even when each product is generated from an AUG codon in an ideal Kozak context (44,49–52). This suggests that differences in translational elongation efficiency may be critically important to RAN translation efficiency and as such could represent a therapeutic target.
Ribosomes stall during translational elongation in response to many cues, including RNA damage, mRNA secondary structures, the charge of the nascent polypeptide chain, and cellular stress (53–58). Stalled ribosomes trigger a series of mRNA and protein surveillance pathways known collectively as ribosome-associated protein quality control (RQC) (59–65). These quality control pathways ensure the fidelity of protein synthesis and degrade incompletely generated polypeptides and the mRNAs from which they are derived. Stalled ribosomes are sensed by RING-domain E3 ubiquitin ligase, zinc finger protein 598 (ZNF598, yeast Hel2), and receptor for activated C kinase 1 (RACK1, yeast Asc1) to dissociate the 80S ribosome (66–68). Alternatively, stalling at the 3′ end of mRNA is detected by the mRNA surveillance and ribosome rescue factor Pelota (PELO, yeast Dom34) and HBS1-like translational GTPase (HBS1L, yeast Hbs1) (69–71). The PELO:HBS1L complex subsequently recruits ATP binding cassette subfamily E member 1 (ABCE1, yeast Rli1) to disassemble the ribosome (72,73). Separation of 80S subunits releases 40S and 60S subunits. The 40S subunits contain the truncated mRNA, which is degraded by 5′–3′ exoribonuclease (XRN1) and the exosome complex (74). 60S subunits entrapped with tRNA-bound nascent polypeptide leads are engaged by the ribosomal quality control (RQC) complex, which targets partially made nascent polypeptides for proteasomal degradation (62,64,75–79). Assembly of the RQC complex is initiated by the binding of nuclear export mediated factor (NEMF, yeast Rqc2), which recruits and stabilizes binding by the E3 ligase listerin (LTN1, yeast Ltn1) (79–83). NEMF also synthesizes carboxy-terminal alanine and threonine tails (CAT-tails) on partially made polypeptide chains (84–87). This pushes amino acids on the nascent chain out from the ribosomal exit tunnel and triggers their ubiquitination by LTN1 (78,88–91). The ubiquitin chains signal for recruitment of the valosin-containing protein (VCP, yeast Cdc48) (92,93). The nascent polypeptide chain is then released by ankyrin repeat and zinc finger peptidyl tRNA hydrolase 1 (ANKZF1, yeast Vms1) and VCP targets the ubiquitylated polypeptide chain for proteasomal degradation (94–96).
Previous studies demonstrated that ribosomes stall during the synthesis of C9 ALS/FTD-associated GR and PR dipeptide proteins (97–99). These stalling events were not seen on similarly sized but uncharged DPR repeats such as GA and were therefore ascribed to positively charged arginine residues within these nascent polypeptide chains. Further studies suggest that the GR protein may also interact with protein surveillance pathways through other means than its directed translation (98,100,101). However, these studies were not done in the context of the GC-rich RNA repeats or in the absence of AUG start codons as occurs in RAN translation. As such, whether the dynamics of translation and/or the potential for repeat RNA structures to elicit ribosomal stalling and RQC pathway engagement remains largely unexplored.
In this study, we performed a targeted RAN translation modifier screen at two different GC-rich short tandem repeats with factors involved in the mRNA and protein surveillance pathways. We find that NEMF, LTN1, and ANKZF1 from the RQC pathway all act as significant modifiers of RAN translation from GC-rich repeat RNAs. This is true for the generation of both GA and polyG polypeptides, even though both products contain no runs of charged amino acids. Consistent with translational stalling in these reading frames, we detected partially made N-terminal peptides containing GA from G4C2 repeats and polyG from CGG repeats, and depletion of these RQC factors enhance the accumulation of these truncated repeat peptides. We see a similar effect that the RQC factor impacts on the accumulation of RAN products in patient-derived neurons and find that altering NEMF directly impacts repeat-associated toxicity in a Drosophila model system. Taken together, our results suggest that RQC factor engagement during translation through GC-rich short tandem repeats impacts both RAN translation efficiency and product generation through a repeat RNA structure-dependent mechanism. Moreover, as modulating RQC factor abundance modulates RAN product abundance, targeted augmentation of RQC activity is an intriguing target for further therapeutic evaluation.
Materials and methods
Plasmids
Plasmids and cloning primers used in this study and their sources are listed in Table 1. Primers and synthesized fragments used for cloning are listed in Table 2. ATG-V5-GA70-NLuc-3xFLAG was generated by the insertion of an ATG-V5 fragment flanked with NheI/EcoRV upstream of Intron-(GGGGCC)70-NLuc-3xFLAG (GA frame). ATG-V5-+1(CGG)100-NLuc-3xFLAG was generated by the insertion of an ATG-V5 sequence flanked by EcoRI and NarI upstream of +1(CGG)100-NLuc-3xFLAG (polyG frame). ATG-V5-(GGN)100 sequence was synthesized by GeneWiz with flanking restriction enzymes of EcoRI (5′) and XhoI (3′). The synthesized sequence was inserted into the ATG-V5-+1(CGG)100-NLuc-3xFLAG plasmid with EcoRI/XhoI to replace the ATG-V5-+1(CGG)100 sequence. pBI-dsRED/hNEMF R86S was a gift from Roger Sher. WT hNEMF was generated by mutating the serine at site 86 to an arginine using Q5 site-directed mutagenesis (NEB, E0554S). pBI-dsRED empty vector was cloned using annealing primers flanked by NheI and EcoRV. pCMV3TAG8li-hRNF160-3FLAG was a gift from Martin Dorf (Addgene plasmid #159138). pCMV6-hANKZF1-Myc-FLAG was purchased from Origene, RC201054. All other reporter sequences have been previously published (44,49).
Table 1.
Plasmids information
| Plasmid | Source | Cat# |
|---|---|---|
| pcDNA3.1-ATG-NLuc-3xFLAG | Kearse et al., 2016 | N/A |
| pcDNA3.1-Intron-(GGGGCC)3-NLuc-3xFLAG(+0 polyGA frame) | Green et al., 2017 | N/A |
| pcDNA3.1-Intron-(GGGGCC)35-NLuc-3xFLAG(+0 polyGA frame) | Green et al., 2017 | N/A |
| pcDNA3.1-Intron-(GGGGCC)70-NLuc-3xFLAG(+0 polyGA frame) | Green et al., 2017 | N/A |
| pcDNA3.1-Intron-(GGGGCC)70-NLuc-3xFLAG(+1 polyGP frame) | Green et al., 2017 | N/A |
| pcDNA3.1-Intron-(GGGGCC)70-NLuc-3xFLAG(+2 polyGR frame) | Green et al., 2017 | N/A |
| pcDNA3.1-(CGG)100-NLuc-3xFLAG (+0 polyR frame) | Kearse et al., 2016 | N/A |
| pcDNA3.1-(CGG)100-NLuc-3xFLAG (+1 polyG frame) | Kearse et al., 2016 | N/A |
| pcDNA3.1-(CGG)100-NLuc-3xFLAG (+2 polyA frame) | Kearse et al., 2016 | N/A |
| pcDNA3.1-ATG-V5-NLuc-3xFLAG | Kearse et al., 2016 | N/A |
| pcDNA3.1-ATG-V5-(GGGGCC)70-NLuc-3xFLAG (polyGA frame) | This paper | N/A |
| pcDNA3.1-ATG-V5-(CGG)100-NLuc-3xFLAG (polyG frame) | This paper | N/A |
| pcDNA3.1-ATG-V5-(GGN)100-NLuc-3xFLAG (polyG peptides) | This paper | N/A |
| pBI-CMV2-dsRED | This paper | N/A |
| pBI-CMV4-hNEMF/CMV2-dsRED | This paper | N/A |
| pCMV-FLAG | Sigma | E7908 |
| pCMV-3TAG8li-hRNF160-3xFLAG (hLTN1) | Addgene | 159138 |
| pCMV6-hANKZF1-Myc-FLAG | Origene | RC201054 |
Table 2.
Primers and fragments for cloning
| Plasmid | Forward | Reverse |
|---|---|---|
| ATG-V5-GA69-NLuc-3xFLAG | 5′-ctagctagctagtagatgggtaagcctatccctaaccctctcctcggtctcgattctacggat-3′ | 5′-cgtagaatcgagaccgaggagagggttagggataggcttacccatctactagctag-3′ |
| ATG-V5-+1(CGG)100-NLuc-3xFLAG | 5′-aattcgtagtatgggtaagcctatccctaaccctctcctcggtctcgattctacgg-3′ | 5′-cgccgtagaatcgagaccgaggagagggttagggataggcttacccatactacg-3′ |
| ATG-V5-(GGN)100-NLuc-3xFLAG | 5′-aattcgtagtatgggtaagcctatccctaaccctctcctcggtctcgattctacggcgccgctgccagggggcgtgcggcagcgcggtggcggaggaggtgggggtggtggaggaggaggcggtggtggaggtggtgggggaggaggcggaggagggggtggtggtggcggcggtggaggaggaggcggtggtggtggaggaggcggaggaggagggggtggaggaggaggtggcggtggtggtggaggaggcggaggaggaggtggcggtggtggaggtggcggtggagggggcggtggtggtggaggtgggggaggtggtggaggaggtggtggcggtggaggtggaggcggtggaggaggaggtggcggaggcggtggtggaggaggaggttgggcc-3′ | |
| pBI-dsRED | 5′-taggcggccgcatcgata-3′ | 5′-ggtggctggatccctagc-3′ |
| pBI-dsRED/hNEMF | 5′-gaagagtcggagattagtcagtg-3′ | 5′-aaatgttttcggcacttc-3′ |
For all repeat containing plasmids, repeat size was determined by a combination of restriction digest, sanger sequencing, and nanopore based plasmid sequencing. For larger repeats (e.g. 100 CGG or 70 GGGGCC repeat units), there is some intrinsic instability in repeat size- typically no more than 1–2 repeats from the mean size- that is apparent when plasmid preps undergo nanopore sequencing.
RNA T7 synthesis
pcDNA3.1(+) plasmids containing nano-luciferase and 3xFLAG (NLuc-3xFLAG) reporters were linearized after the 3′ FLAG tag with PspOMI. The efficiency of restriction enzyme digestion was confirmed with a DNA agarose gel. Linearized DNA plasmids were cleaned and concentrated with DNA Clean & Concentrator-25 (Zymo Research, D4033). RNAs were in vitro transcribed with HiScribe T7 ARCA mRNA Kit with tailing (NEB, E2060S) following the manufacturer's recommended protocol. mRNAs were then cleaned and concentrated with RNA Clean & Concentrator-25 Kit (Zymo Research, R1017) and run on a denaturing glyoxal RNA gel to verify mRNA size and integrity. To assure that truncated RNAs were not generated during in vitro transcription, mRNA was run on an agarose gel. The full-sized band (identified by SYBR Safe staining on a parallel-run lane of the same gel) was gel extracted and purified using electroelution per the manufacturer's protocol (BioRad, Model 422 Electro-eluter). mRNA was ethanol precipitated, then cleaned and concentrated with the RNA Clean & Concentrator-25 Kit (Zymo Research, R1017). Transcribed RNA sequences are shown in Table 3.
Table 3.
DNA plasmid sequence used to synthesize RNA transcript
| Reporter name | Sequence from T7 to PspOMI cut site |
|---|---|
| ATG-V5-NLuc-3xFLAG | GGAGACCCAAGCTGGCTAGCGTTTAAACTTAAGCTTGGCAATCCGGTACTGTTGGTAAAGCCACCATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGTCTTCACACTCGAAGATTTCGTTGGGGACTGGCGACAGACAGCCGGCTACAACCTGGACCAAGTCCTTGAACAGGGAGGTGTGTCCAGTTTGTTTCAGAATCTCGGGGTGTCCGTAACTCCGATCCAAAGGATTGTCCTGAGCGGTGAAAATGGGCTGAAGATCGACATCCATGTCATCATCCCGTATGAAGGTCTGAGCGGCGACCAAATGGGCCAGATCGAAAAAATTTTTAAGGTGGTGTACCCTGTGGATGATCATCACTTTAAGGTGATCCTGCACTATGGCACACTGGTAATCGACGGGGTTACGCCGAACATGATCGACTATTTCGGACGGCCGTATGAAGGCATCGCCGTGTTCGACGGCAAAAAGATCACTGTAACAGGGACCCTGTGGAACGGCAACAAAATTATCGACGAGCGCCTGATCAACCCCGACGGCTCCCTGCTGTTCCGAGTAACCATCAACGGAGTGACCGGCTGGCGGCTGTGCGAACGCATTCTGGCGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGTAAGGCCGCGACTCTAGAG |
| ATG-V5-GA70-NLuc-3xFLAG | GGGAGACCCAAGCTGGCTAGCTAGCTAGTAGATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGGATATCAAGATAGCGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCAGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGCTGGAAGCTTGGCAATCCGGTACTGTTGGTAAAGCCACCGGGGTCTTCACACTCGAAGATTTCGTTGGGGACTGGCGACAGACAGCCGGCTACAACCTGGACCAAGTCCTTGAACAGGGAGGTGTGTCCAGTTTGTTTCAGAATCTCGGGGTGTCCGTAACTCCGATCCAAAGGATTGTCCTGAGCGGTGAAAATGGGCTGAAGATCGACATCCATGTCATCATCCCGTATGAAGGTCTGAGCGGCGACCAAATGGGCCAGATCGAAAAAATTTTTAAGGTGGTGTACCCTGTGGATGATCATCACTTTAAGGTGATCCTGCACTATGGCACACTGGTAATCGACGGGGTTACGCCGAACATGATCGACTATTTCGGACGGCCGTATGAAGGCATCGCCGTGTTCGACGGCAAAAAGATCACTGTAACAGGGACCCTGTGGAACGGCAACAAAATTATCGACGAGCGCCTGATCAACCCCGACGGCTCCCTGCTGTTCCGAGTAACCATCAACGGAGTGACCGGCTGGCGGCTGTGCGAACGCATTCTGGCGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGTAAGGCCGCGACTCGAGAG |
| ATG-V5-+1(CGG)100-NLuc-3xFLAG | GGAGACCCAAGCTGGCTAGCGTTTAAACTTAAGCTTGGTACCGAGCTCGGATCCACTAGTCCAGTGTGGTGGAATTCGTAGTATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGCGCCGCTGCCAGGGGGCGTGCGGCAGCGCGGCGGCGGCGGCGGCGGCGGCGGAGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCTGGGCCTCGAGGATATCAAGATCTGGCCTCGGCGGCCAAGCTTGGCAATCCGGTACTGTTGGTAAAGCCACCGGGGTCTTCACACTCGAAGATTTCGTTGGGGACTGGCGACAGACAGCCGGCTACAACCTGGACCAAGTCCTTGAACAGGGAGGTGTGTCCAGTTTGTTTCAGAATCTCGGGGTGTCCGTAACTCCGATCCAAAGGATTGTCCTGAGCGGTGAAAATGGGCTGAAGATCGACATCCATGTCATCATCCCGTATGAAGGTCTGAGCGGCGACCAAATGGGCCAGATCGAAAAAATTTTTAAGGTGGTGTACCCTGTGGATGATCATCACTTTAAGGTGATCCTGCACTATGGCACACTGGTAATCGACGGGGTTACGCCGAACATGATCGACTATTTCGGACGGCCGTATGAAGGCATCGCCGTGTTCGACGGCAAAAAGATCACTGTAACAGGGACCCTGTGGAACGGCAACAAAATTATCGACGAGCGCCTGATCAACCCCGACGGCTCCCTGCTGTTCCGAGTAACCATCAACGGAGTGACCGGCTGGCGGCTGTGCGAACGCATTCTGGCGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGTAAGGCCGCGACTCTAGAG |
| ATG-V5-(GGN)100-NLuc-3xFLAG | GGAGACCCAAGCTGGCTAGCGTTTAAACTTAAGCTTGGTACCGAGCTCGGATCCACTAGTCCAGTGTGGTGGAATTCGTAGTATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGCGCCGCTGCCAGGGGGCGTGCGGCAGCGCGGTGGCGGAGGAGGTGGGGGTGGTGGAGGAGGAGGCGGTGGTGGAGGTGGTGGGGGAGGAGGCGGAGGAGGGGGTGGTGGTGGCGGCGGTGGAGGAGGAGGCGGTGGTGGTGGAGGAGGCGGAGGAGGAGGGGGTGGAGGAGGAGGTGGCGGTGGTGGTGGAGGAGGCGGAGGAGGAGGTGGCGGTGGTGGAGGTGGCGGTGGAGGGGGCGGTGGTGGTGGAGGTGGGGGAGGTGGTGGAGGAGGTGGTGGCGGTGGAGGTGGAGGCGGTGGAGGAGGAGGTGGCGGAGGCGGTGGTGGAGGAGGAGGTTGGGCCTCGAGGATATCAAGATCTGGCCTCGGCGGCCAAGCTTGGCAATCCGGTACTGTTGGTAAAGCCACCGGGGTCTTCACACTCGAAGATTTCGTTGGGGACTGGCGACAGACAGCCGGCTACAACCTGGACCAAGTCCTTGAACAGGGAGGTGTGTCCAGTTTGTTTCAGAATCTCGGGGTGTCCGTAACTCCGATCCAAAGGATTGTCCTGAGCGGTGAAAATGGGCTGAAGATCGACATCCATGTCATCATCCCGTATGAAGGTCTGAGCGGCGACCAAATGGGCCAGATCGAAAAAATTTTTAAGGTGGTGTACCCTGTGGATGATCATCACTTTAAGGTGATCCTGCACTATGGCACACTGGTAATCGACGGGGTTACGCCGAACATGATCGACTATTTCGGACGGCCGTATGAAGGCATCGCCGTGTTCGACGGCAAAAAGATCACTGTAACAGGGACCCTGTGGAACGGCAACAAAATTATCGACGAGCGCCTGATCAACCCCGACGGCTCCCTGCTGTTCCGAGTAACCATCAACGGAGTGACCGGCTGGCGGCTGTGCGAACGCATTCTGGCGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGTAAGGCCGCGACTCTAGAG |
Cell culture and transfections
HEK293 cells were maintained at 37°C, 5% CO2 in DMEM with high glucose (Gibco, 11965118) supplemented with 9% fetal bovine serum (50 ml FBS added to 500 ml DMEM; Bio-Techne, S11150). For siRNA transfection, HEK293 cells were seeded at 2 × 105 cells/ml with 100 μl in 96-well or 500 μl in 24-well plates. Cells were then transfected with siRNA at 1 nM/well in a mixture with Lipofectamine RNAiMax (Thermo Fisher, 13778075) following the manufacturer's recommended protocol on the same day while seeding the cells. siRNA used for this paper are listed in Table 4. For effector plasmid transfection, 24 hr after seeding the cells at 50–60% confluency, 100 and 500 ng of effectors were transfected in each well of 96-well and 24-well plate, respectively, with FuGeneHD (Promega, E2312), at a 3:1 ratio of FuGeneHD to DNA. Plasmid reporters were transfected into 70–80% confluent cells 48 h post-seeding. Each well of a 96-well and 24-well plate was transfected with 50 ng and 500 ng NL reporter DNA, respectively, with FuGeneHD as described above. Cells were harvested 24 h post-transfection for analysis. For mRNA transfection, T7 synthesized mRNA was transfected in cells at 70–80% confluency with TransIT-mRNA Transfection Kit (Mirus, MIR 2256) following the manufacturer's recommended protocol. Cells were harvested 24 hr post-transfection for analysis.
Table 4.
siRNA information
| siRNA target | Source | Sequence/ Cat#/ siRNA ID |
|---|---|---|
| NanoLuc #2 Custom (NLuc) | Horizon | F: 5′-gguuacgccgaacaugaucgacuuu-3′ R: 5′-Pagucgaucauguucggcguaaccuu-3′ |
| Negative Control No. 1 | Thermo silencer select | 4390843 |
| PELO | s28807 | |
| HBS1L | s21151 | |
| ABCE1 | s12088 | |
| XRN1 | s29016 | |
| RACK1 | s20340 | |
| ZNF598 | s40509 | |
| NEMF #1 | s17483 | |
| NEMF #2 | s17484 | |
| LTN1 #1 | s25003 | |
| LTN1 #2 | s25002 | |
| VCP | s14765 | |
| ANKZF1 #1 | s30259 | |
| ANKZF1 #2 | s30260 |
Luminescent assays
From a 96-well plate, cells were lysed with 60 μl Glo Lysis Buffer (Promega, E2661) for 5 min at room temperature with constant rocking. NanoGlo Substrate (Promega, N113B) was freshly diluted 1:50 in NanoGlo Buffer (Promega, N112A). For a nano-luciferase assay, 25 μl of the diluted NanoGlo substrate was added to 25 μl of cell lysate. To test cell viability, 25 μl of CellTiter-Glo (Promega, G7573) was added to another 25 μl of lysate. Both assays were mixed for 5 min in a covered opaque 96-well plate then luminescence was measured on a GloMax 96 Microplate Luminometer.
Immunoblotting
For HEK293, each well of cells from 24-well plates was lysed with 120 μl of RIPA buffer with protease inhibitor (Roche, 11836153001). All protein lysates were denatured with 12% β-mercaptoethanol in 6x SDS-loading dye, boiled at 95°C for 5 min. 20 μl lysate for each sample was loaded per well on 12% sodium dodecyl sulfate-polyacrylamide gels. To detect stalling products between 10–15 kDa, those samples were loaded on 15% sodium dodecyl sulfate-polyacrylamide gels. Gels were then transferred to PVDF membranes either overnight at 40 V at 4°C, or for 2.5 hr at 320 mAmps in the ice bucket at 4°C. Membranes were blocked with 5% non-fat dry milk for 30–60 min, and all antibodies were diluted in 5% non-fat dry milk. All primary antibody information and probing conditions are listed in Table 5. Washes were performed with 1x TBST 3 times, 5 min each. Horseradish peroxidase secondary antibodies were applied at 1:10 000, for 2 h at room temperature. Bands were then visualized on film. Mild stripping (1.5% glycine, 0.1% SDS, 1% Tween 20, pH to 2.2 with HCl) was performed with two 10 min incubations at room temperature. For immunoblot quantification, the background subtracted mean signal intensity of each band was measured using ImageJ. These values were then normalized to the mean of the non-targeting control-treated bands on the same blot to allow for blot-to-blot normalization across multiple experiments.
Table 5.
Antibody information
| Item | WB Conc. | Species | Company | Catalog number |
|---|---|---|---|---|
| FLAG M2 | 1:1000 | mouse | Sigma | F1804 |
| V5 | 1:2000 | mouse | Abcam | ab27671 |
| Tubulin | 1:1000 | mouse | DSHB | E7 |
| PELO | 1:1000 | rabbit | Abcam | ab154335 |
| HBS1L | 1:1000 | rabbit | Thermo Fisher | PIPA556241 |
| ABCE1 | 1:1000 | rabbit | Abcam | ab32270 |
| XRN1 | 1:1000 | rabbit | Thermo Fisher | PA5-41888 |
| RACK1 | 1:1000 | rabbit | Thermo Fisher | MA5-34809 |
| ZNF598 | 1:1000 | rabbit | Sigma | HPA041760 |
| NEMF | 1:1000 | rabbit | Proteintech | 11840–1-AP |
| LTN1 | 1:2000 | rabbit | Proteintech | 28452–1-AP |
| ANKZF1 | 1:2000 | rabbit | Proteintech | 20447–1-AP |
| VCP | 1:5000 | rabbit | Proteintech | 10736–1-AP |
Immunoprecipitation
Cells were lysed with NP40 lysis buffer (50 mM HEPES–KOH, 150 mM KCl, 0.5% NP40, 0.5 mM DTT, 2 mM EDTA, 1 mM PMSF, protease inhibitors) on ice and incubated at 4°C for 30 min. Lysates were cleared by centrifugation at 20 000 × g for 10 min at 4°C, and the supernatant was transferred into a new tube. Protein concentration for each sample was measured by BCA assay (Thermo Fisher, 23227). 1 mg of total protein was used for each immunoprecipitation. Each lysate was first incubated with 20 μl pre-washed FLAG M2magnetic beads (Sigma, M8823), rotating at 4°C for 2 h. Next, flow-through was collected and incubated with pre-washed 20 μl of V5-Trap magnetic beads (Proteintech, v5tma) rotating at 4°C for 2 h. 10% of input and 20% of the supernatant from each step was saved for immunoblot. Afterward, FLAG and V5 beads were washed with NP-40 lysis buffer until the absorbance of the wash supernatant at 280 nm is below 0.05. After the final wash, beads were resuspended with 2x SDS dye and boiled at 95°C for 5 min. The supernatant was collected for immunoblot.
Drosophila studies
Drosophila were crossed and maintained at 25°C on SY10 food supplemented with dry yeast. For eye phenotyping at a higher temperature, flies were crossed and maintained at 29°C. To measure G4C2 repeat RNA toxicity in flies, a previously characterized GMR-GAL4-driven UAS-(GGGGCC)28-EGFP reporter containing fly was used (102). NEMF knockdown flies were obtained from Bloomington Drosophila Stock Center (BDSC) with stock numbers BDSC 36955 and BDSC 25214. Rough eye phenotyping was performed as described earlier (103). In brief, 5–6 virgin female flies expressing the GMR-GAL4-driven UAS-(GGGGCC)28-EGFP transgene were crossed with male flies carrying a germline mutation (insertion/disruption) of the fly homolog of NEMF gene (Dmel\Clbn). The rough eye phenotype of flies in F1 progenies was determined at 1–2 days post-eclosion. Rough eye scores were given based on the following eye abnormalities: orientation of bristles, presence of supernumerary bristles, ommatidial fusion, and disarray, presence of necrosis, and shrinkage of the whole eye. Eye images were captured using a Leica M125 stereomicroscope with a Leica DFC425 digital camera. Eye images were scored and analyzed with ImageJ in a blinded manner.
For GR-GFP quantification, virgin females carrying GMR-GAL4; UAS-LDS-(G4C2)44-GFP (140) were crossed to males harboring RNAi constructs from two libraries. A) TRIP RNAi lines: w; clbn-RNAi (derived from BL62402) and its control w;; mCherry-RNAi (derived from BL35785). B) KK RNAi lines: clbn (VDRC 103351), ltn1 (VDRC 104233) and the control vkg-RNAi (VDRC 106812). We used vkg RNAi as a control for KK library, as knockdown of vkg doesn’t affect toxicity of G4C2 repeats. Eyes of female progeny were imaged through 8–10 z-stacks under UV light using a Leica M125 stereomicroscope with a Leica DFC425 digital camera. GFP levels were quantified from the maximum projection images in a blinded manner with ImageJ.
Maintenance of iPSCs and differentiation into forebrain-like neurons (iNs)
C9 patient-derived induced pluripotent stem cells (iPSCs; CS52iALS-C9n6) and isogenic controls (CS52iALS-C9n6.ISOC3) were obtained from the Cedars-Sinai iPSC Core. iPSCs were maintained in TeSR-E8 media (Stemcell Technologies, 05990) on vitronectin-coated plates, and passaged every 4–5 days using EDTA as described in Weskamp et al. (104). A doxycycline-inducible cassette for induced expression of Ngn1/2 was integrated into the CLYBL safe harbor locus of each line, as per Weskamp et al, enabling directed differentiation into forebrain-like glutamatergic iNeurons (iNs) (104–107). Neural progenitor cells (NPCs) were frozen on day 2 of differentiation and stored in liquid N2 until needed. One day before plating NPCs (1 × 106 cells/well in a 6-well plate), each well was coated overnight at 37°C with 1 ml of 100 μg/ml poly-L-ornithine hydrobromide (PLO, Sigma, P3655) prepared in filter sterilized 0.1 M borate buffer (Fisher Chemical, A73-500) at pH 8.4. PLO was removed by washing with sterile water 3× and air-dried for at least 1 h. The remainder of the iNs differentiation procedure was as described in Weskamp et al. (104).
Lentivirus transduction
Lentiviral shRNA plasmids against NEMF, LTN1 and ANKZF1 were purchased from Horizon Discovery. Lentiviral overexpression of NEMF and ANKZF1 was purchased from VectorBuilder (Table 6). Lentiviral overexpression of LTN1 was cloned with NEBuilder Gibson Assembly. In brief, the RPL22 sequence from the plasmid pLV-Ef1a-RPL22-3XHA-P2A-EGFP-T2A-Puro (gift from Hemali Phatnani, Addgene plasmid # 170317) was removed by restriction enzymes digestion with EcoRI-HF and AgeI-HF. The human LTN1 was amplified from pCMV-3TAG8li-hRNF160-3xFLAG with primers (forward: 5′- tttgccgccagaacacaggaccggttaatctgcgctgccaccatgggcggz-3′, Reverse 5′-aattcgtggcgccagatccgggctcgacatcgatgaaaaacg-3′) designed using NEBuilder Assembly Tool v2.7.1. pLV-Ef1a-hLTN1-3XHA-P2A-EGFP-T2A-Puro was generated by Gibson cloning with the hLTN1 fragment generated as described above. Lentiviruses were packed at the University of Michigan Vector Core with HIV lentivirus and then 10x concentrated in 10 ml of DMEM. A GFP vector control was purchased from the University of Michigan Vector Core. Transduction efficiencies as measured by GFP fluorescence were tested in HEK293 cells with 10 μg/ml polybrene following the lentiviral transduction protocol provided by the Vector Core. Knockdown and overexpression of the gene were confirmed with immunoblot. C9 and its isogenic control iNeurons were transduced with lentivirus on Day 6 post differentiation. Cell media were replaced with fresh B27 media on Day 8, and cells were harvested on Day 14.
Table 6.
Lentiviral constructs
| Plasmid | Source | Cat# |
|---|---|---|
| Lenti-EV-GFP-VSVG | UM Vector Core | UMICHVC-YT-4612 |
| SMARTvector Lentiviral Human NEMF hEF1a-TurboGFP shRNA |
Horizon | V3SH11240-224809840 |
| SMARTvector Lentiviral Human LTN1 hEF1a-TurboGFP shRNA |
Horizon | V3SH11240-226558312 |
| SMARTvector Lentiviral Human ANKZF1 hEF1a-TurboGFP shRNA |
Horizon | V3SH11240-224939332 |
| pLV-EGFP-EF1A-hNEMF [NM_004713.6] | VectorBuilder | VB221216-1307gkc |
| pLV-EF1A-hLTN1-3XHA-P2A-EGFP-T2A-Puro | This paper | N/A |
| pLV-EGFP-EF1A-hANKZF1 [NM_001042410.2] |
VectorBuilder | VB900124-1663tuj |
Gp Msd
From a 6-well plate, cells were washed with 1x PBS on ice and then harvested by scraping with 200 μl of Co-IP buffer (50 mM Tris–HCl, 300 mM NaCl, 5 mM EDTA, 0.1% triton-X 100, 2% SDS, protease inhibitors, phosphoSTOP) on ice. Lysates were passed through a 28.5 G syringe 10 times, spun at 16 000 × g for 20 min at 15°C, and then the supernatant was collected. Levels of polyGP proteins in cell lysates were measured using the Meso Scale Discovery (MSD) electrochemiluminescence detection technology as previously described (108). Briefly, a purified mouse monoclonal polyGP antibody was used as both the capture and detection antibody (Target ALS Foundation, TALS 828.179). For capture, the antibody was biotinylated and used to coat a 96-well MSD Small Spot Streptavidin plate (Meso Scale Discovery, L45SA-2), whereas the detection antibody was tagged with an electro-chemiluminescent label (MSD GOLD SULFO-TAG). An equal amount of each lysate was diluted in TBS and tested in duplicate in a blinded fashion. For each well, the intensity of emitted light, which is reflective of GP abundance and presented as arbitrary units, was acquired upon electrochemical stimulation of the plate.
Quantitative real-time reverse transcription PCR (qRT-PCR)
RNA from iN lysates was isolated and collected using Quick-RNA MiniPrep Kit (Zymo Research, R1054). 2 μg of RNA per sample was treated with 2 U of TURBO DNase (Thermo Fisher Scientific, AM2238) for 30 min at 37°C twice to remove contaminating genomic and plasmid DNA, and then recovered using the RNA Clean & Concentrator-5 Kit (Zymo Research, R1015). cDNA from each sample was generated from 250 ng of RNA from the previous step with a mixture of oligo(dT) and random hexamer primers (iScript cDNA Synthesis Kit, Bio-Rad, 1708891). Finally, cDNA abundance was measured using iQ SYBR Green Supermix (Bio-Rad, 1708882) from an iQ5 qPCR system (Bio-Rad), and the appropriate primers at 100 nM. Primer information is listed in Table 7. cDNA abundance was quantified using a modified ΔΔCt method recommended by the manufacturer.
Table 7.
Primer sets for qRT-PCR
| Primer name | Sequence | Source |
|---|---|---|
| C9 Intron-F | 5′-ctccccactacttgctctcacagta-3′ | Rodriguez et al., 2021 |
| C9 Intron-R | 5′-tagcgcgcgactcctgagttcca-3′ | Rodriguez et al., 2021 |
| NEMF-F | 5′-aacttgcggtagtgaccctc-3′ | This paper |
| NEMF-R | 5′-attcagctccgcgagtacg-3′ | This paper |
| LTN1-F | 5′-tgtccaagccaaacctcttga-3′ | This paper |
| LTN1-R | 5′-ggacatgccttggttaga-3′ | This paper |
| ANKZF1-F | 5′-gcagaaatccggcaatcgac-3′ | This paper |
| ANKZF1-R | 5′-gcccttagaagacgcaccaa-3′ | This paper |
| GAPDH-F | 5′-aaggtgaaggtcggagtcaa-3′ | Tseng et al., 2021 |
| GAPDH-R | 5′-ggaagatggtgatgggattt-3′ | Tseng et al., 2021 |
Polysome profiling
HEK293 cells at 85–95% confluency in a 15 cm culture dish were treated with 100 μg/ml cycloheximide (CHX) for 5 min at 37°C. The culture dish was placed on ice, washed with 5 ml ice-cold PBS containing 100 μg/ml CHX, harvested by scraping with another 5 ml cold PBS + CHX, and centrifuged at 1200 × g at 4°C for 5 min. PBS was aspirated and the pellet was resuspended in polysome-profiling lysis buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 15 mM MgCl2, 8% (vol/vol) glycerol, 20 U/ml SUPERase, 80 U/ml murine RNase inhibitor, 0.1 mg/ml heparin, 100 μg/ml CHX, 1 mM DTT,1x EDTA-free protease inhibitor cocktail, 20 U/ml Turbo DNase, 1% Triton X-100). Lysates were vortexed for 30 s, passed through a 21 G needle 10 times, and incubated on ice for 5 min. Cellular debris was pelleted at 14 000 × g at 4°C for 10 min, and the supernatant was transferred to a pre-cooled tube. Total lysate RNA was estimated by NanoDrop. Lysates were flash-frozen in liquid nitrogen and stored at –80°C until fractionation.
Sucrose gradients were prepared by sequentially freezing 2.7 ml of 50, 36.7, 23.3 and 10% sucrose (wt/vol) in 13.2 ml thin wall polypropylene tubes (Beckman Coulter, 331372). Sucrose-gradient buffer consisted of 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 15 mM MgCl2, 10 U/ml SUPERase, 20 U/ml murine RNase inhibitor, 100 μg/ml CHX and 1 mM DTT. Before use, gradients were thawed and linearized overnight at 4°C. For fractionation, approximately 50 μg total RNA was applied to the top of the sucrose gradient. Gradients were spun at 35 000 rpm at 4°C for 3 h using a Beckman Coulter Optima L-90 K ultracentrifuge and SW 41 Ti swinging-bucket rotor. Gradients were fractionated with Brandel's Gradient Fractionation System, measuring absorbance at 254 nm. The detector was baselined with 60% sucrose chase solution and its sensitivity was set to 0.05. For fractionation, 60% sucrose was pumped at a rate of 1.5 ml/min. Brandel's PeakChart software was used to collect the profile data.
Reporter RNA stability assay
Reporter mRNA stability was assessed as previously described (49). Briefly, we measured reporter mRNA levels serially by qRT-PCR after treatment with actinomycin D at 5 μg/ml for 0, 2, 4 or 6 h starting 24 h post reporter plasmid transfection. We extracted total RNA using the Quick-RNA MiniPrep kit (Zymo Research, R1054). 10 μg of total RNA was treated twice with 1 μl Turbo DNase (2 U/μl; Ambion, AM2238) at 37 ºC for 30 min and was subsequently purified using RNA Clean and Concentrator-25 (Zymo Research, R1017). cDNA was generated as described above qRT-PCR method section. qRT-PCR was performed on a Taqman system (QuantStudio™ 3 System) using custom-designed TaqMan gene expression assays for nano-luciferase (NL TaqMan primer set/probe: AP7DWMP, actin primer/probe set: Hs99999903_m1).
Reporter protein stability assay
Reporter protein stability was assessed as previously described (44). Briefly, 24 h post reporter plasmid transfection, performed as above, HEK293 cells were treated with 10 μg/ml puromycin for 0, 4, 6 and 12 h. After each time point, cells were lysed in 60 μl Glo Lysis Buffer for 5 min at room temperature and stored at −20 °C. After all time points were collected, nano-luciferase activities were measured simultaneously.
To assess the effect of ubiquitin proteasome system activity on generation of full-length and truncated translation products, HEK293 cells were reverse transfected with the indicated siRNAs. After 48 h, these cells were transfected with ATG-V5-+1(CGG)100-NLuc-3xFLAG plasmids. twenty-four hours later, cells were treated with 10 μM MG132 for 0, 2 or 4 h. After each timepoint, cells were lysed in RIPA buffer and loaded on a 15% SDS-PAGE gel and quantified as described above.
Data analysis
Prediction of RNA structure and calculation of the minimum free energy was computed by The Vienna RNA Website, RNAfold 2.5.1. (109). Statistical analyses were performed with GraphPad Prism9.5.1. All luciferase activity was calculated by normalizing the nano-luciferase signal with cell titer. For comparison of NLuc reporter luciferase activity assays, GP MSD response, and fly eye phenotype quantification, we used two-tailed unpaired Student's t-test with Welch's correction for multiple comparisons to confirm the statistical difference between control and multiple experimental groups within each sample. To assess group effects, we used a two-way ANOVA with Dunnett's multiple comparison tests to compare the differences between groups within different samples. Fly eye necrosis and width measurements were done with reviewer genotype- blinded analysis by at least two independent investigators to avoid subjective bias. Fly eye width measurement was performed with ImageJ (www.imagej.nih.gov/ij/). Experiments were performed with a minimum of three independent biological samples (n > 3) with technical replication of results from each sample. Fly experiments were done using multiple crosses with a minimum of 10 flies analyzed per group per cross. Further statistical analysis details are included in figure legends including the numbers of analyzed samples, statistical tests, and P values.
Results
RQC pathway factor depletion enhances RAN translation from GC-rich repeat sequences
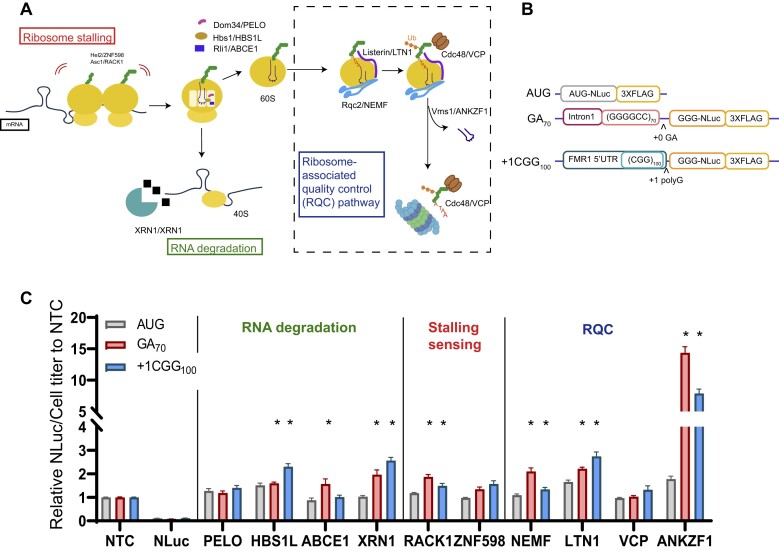
Slowing or stalling of translational elongation can result in ribosomal collisions and engagement of ribosomal quality control pathways (Figure 1A). We hypothesized that GC-rich repetitive elements in mRNAs would be prone to triggering such events. We therefore performed a targeted modifier screen at two disease-associated repetitive elements (G4C2 hexanucleotide or CGG trinucleotide repeats) for factors involved in mRNA and protein surveillance pathways to evaluate their role in regulating RAN translation. Specifically, we used commercially generated siRNAs (Table 4) to lower the expression of RNA degradation pathway factors PELO, HBS1L, ABCE1, and XRN1; ribosome stalling sensing factors RACK1 and ZNF598, or RQC pathway factors NEMF, LTN1, VCP, and ANKZF1. Validation of gene knockdown for each factor was confirmed by immunoblotting (Supplementary Figure S1A–J).
Figure 1.
NEMF, LTN1, and ANKZF1 act as genetic modifiers of RAN translation from both G4C2 and CGG repeats. (A) Stalled or collided ribosomes are sensed by ZNF598 and RACK1 to separate 80S ribosome to 60S and 40S. The 40S subunit with truncated mRNA is then released by PELO in concert with HBS1L and ABCE1 and subsequently degraded by XRN1. NEMF CAT-tails partially generated peptides within the 60S subunit, allowing for degradation by the proteasome through a process that involves ubiquitination by LTN1 and VCP. ANKZF1 removes the tRNA, allowing for ribosomal recycling. Schematic adapted with permission from (63). (B) Schematic of transiently expressed nano-luciferase (NLuc) reporters used to assess RAN translation product abundance. Either the first intron of C9orf72 including 70 GGGGCC (G4C2) repeats or the 5′UTR of FMR1 containing 100 CGG repeats were placed 5′ to a modified NLuc with the AUG initiator codon mutated to GGG. (C) Results from a targeted screen of key RQC pathway factors. The relative expression of NLuc was normalized to cell titer. These normalized values were then expressed as a fold change compared to a non-targeting siRNA control (NTC). The effect of transient transfection of each siRNA or luciferase reporter on cell titer is shown in Supplemental Figure S1 (Supplementary Figure S1). Data represent mean with error bars ± SEM of 6 biological replicates from at least 2 independent experiments. *P< 0.05; one-way ANOVA with Dunnett's multiple comparison test compared to NTC.
To assess the impact of these factors on RAN translation, we used two well-characterized transiently transfected RAN translation-specific nano-luciferase reporters for G4C2 repeats and CGG repeats, respectively (Figure 1B) (44,49). For each, the repeat is surrounded by its native sequence (for G4C2, this is the intronic sequence from human C9orf72 that surrounds the repeat; for CGG, this is the 5′UTR of human FMR1). The repeat and surrounding sequence is embedded 5′ to a modified nano-luciferase that has its initiation codon changed to GGG. This mutation and removal of internal AUG codons allow the luciferase to selectively report on RAN translational initiation within or just 5′ to the repeat (44,49). An AUG-initiated nano-luciferase serves as a positive control for canonical translation initiation and elongation. As depletion of some RQC factors suppressed global protein translation (Supplementary Figure S2A–E) and lowered expression of a co-transfected firefly luciferase, we utilized cell titer as a normalization control (Supplementary Figure S1K). This global effect on translation was not driven by activation of the integrated stress response (Supplementary Figure S2F).
For factors involved in ribosome-associated RNA degradation, depletion of XRN1 and HSB1L increased RAN translation at both G4C2 and CGG repeats (Figure 1C). However, the largest effects were observed for factors involved in the RQC pathway that classically engages the ribosomes after stall detection and ribosome splitting. Depletion of LTN1, NEMF and ANKZF1 in particular increased the accumulation of RAN translation products from both G4C2 and CGG repeats, with increases between 2 and 5-fold compared to a non-targeting siRNA control (Figure 1C). We therefore focused our attention on these RQC pathway factors and their effect on RAN translation.
Reduction of NEMF, LTN1 or ANKZF1 enhances RAN translation in a repeat-length dependent manner
Previous studies found that depletion of NEMF and LTN1 either increase (98) or had no significant effect (100) on the production of GR and PR DPRs from non-GC-rich sequences. This effect was attributed to the positively charged arginine residues in PR and GR DPR tracks, which can interact with the ribosome exit tunnel to elicit ribosomal stalling (97,99,110). However, RNA secondary structures can also elicit ribosomal stalling (111–113). As GC-rich repeats form stable secondary structures and the RNA helicases that resolve these structures are implicated in RAN translation (114–117), we hypothesized that ribosome stalling would occur during RAN translation from GC-rich repeats, leading to RQC pathway activation. To assess this, we first validated the effects of NEMF, LTN1, and ANKZF1 on RAN translation with a second set of siRNAs. Knockdown of NEMF, LTN1, or ANKZF1 with this second set of siRNAs elicited increases in GA DPR RAN production from 70 G4C2 repeats and FMRpolyG (glycine) production from 100 CGG repeats (Figure 2, Supplementary Figure S3). To assure that these effects were not driven by effects of RQC pathway manipulation on mRNA stability, we measured GA70 and + 1CGG reporter RNA levels over time after treatment of cells with actinomycin D to halt new transcription. As expected, there was a slow decline in the level of both reporters over 6 hours. However, this change in reporter mRNA levels was not impacted by knockdown of either NEMF or ANKZF1 (Supplementary Figure S4A, D). To assess whether the effects of RQC pathway alterations might be impacting the stability of the RAN translation reporter proteins, we performed a parallel analysis of the levels of each reporter over 12 hours after treatment with puromycin to block new reporter synthesis. Knockdown of either NEMF or ANKZF1 had no impact on the signal from either reporter at 12 hours post-treatment (Supplementary Figure S4B, C, E, F). These data suggest that reduction of these RQC factors is enhancing RAN translation through direct effects on translation.
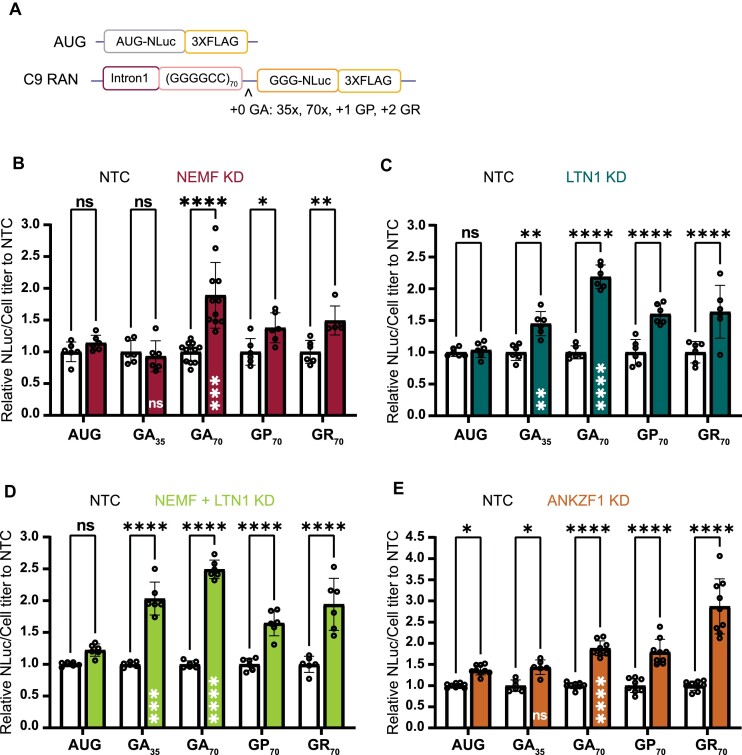
Figure 2.
Depletion of NEMF, LTN1, and ANKZF1 enhances RAN translation in a repeat length-dependent manner and across all reading frames. (A) Schematic of C9orf72 RAN G4C2 repeat length and reading frame reporters. Single nucleotide insertions to shift the reading frame and repeat contractions allowed for measurement of products from all potential reading frames and across 2 repeat sizes. (B–E) Luciferase assays after NEMF, LTN1, both NEMF and LTN1 or ANKZF1 depletion. All graphs show mean with error bars ± SD. Each N is shown as an open circle (n = 6–9/group across at least two independent experiments). Asterisks above each bar are comparisons of expression between NTC and gene(s) knockdown. ns = not significant; *P< 0.05; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001, as determined with two-way ANOVA with Sidak's multiple comparison test. Asterisks placed inside each bar are comparisons between AUG-driven no-repeat control and different repeat lengths of the GA frame of cells treated with gene(s) knockdown. ns = not significant; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001, represent unpaired t-test.
We next assessed whether this impact by RQC factors was dependent on the length of the G4C2 repeat. At 35 G4C2 repeats, we did not see any effect of NEMF knockdown on GA frame RAN translation, suggesting repeat-length dependence (Figure 2B, Supplementary Figure S3B). Similarly, the depletion of LTN1 significantly increased RAN translation from both GA35 and GA70 repeat reporters, but the effect on the larger repeat was greater (Figure 2C, Supplementary Figure S3B). As NEMF helps recruit LTN1 to assemble the RQC complex, we asked if there is a synergetic effect of NEMF and LTN1 knockdown on RAN translation. When we performed the knockdown of both NEMF and LTN1 together, we observed a mild additive effect, with more RAN translation production from GA35 and GA70 reporters compared to NEMF alone (Figure 2D, Supplementary Figure S3A). Similarly, the depletion of ANKZF1 had a modest effect on translation from an AUG reporter with no repeat and from our GA35 reporter, but a larger effect on product generation from the GA70 reporter (Figure 2E, Supplementary Figure S3B).
If RNA secondary structure contributes to ribosomal stalling, then the effect of knockdown of RQC factors should influence RAN translation production from all 3 potential reading frames. To address this, we utilized (G4C2)70 RAN translation reporters, where the NLuc tag was in the GP (+1) or GR (+2) reading frames (Figure 2A). Knockdown of NEMF increased RAN from (G4C2)70 sequences in all 3 reading frames (Figure 2B). Knockdown of LTN1 increased RAN product accumulation from (G4C2)70 sequences in GA, GP, and GR frames (Figure 2C). Knockdown of ANKZF1 significantly increased RAN production from (G4C2)70 sequences in all three reading frames, with greater effects on the GR frame (Figure 2E). ANKZF1 knockdown also enhanced production in all three potential CGG repeat reading frames (Supplementary Figure S3E). Taken together, these data suggest that depleting RQC factors increase RAN translation in a repeat length-dependent but repeat reading-frame independent fashion.
Detection of short/truncated translation products from G4C2 and CGG transcripts
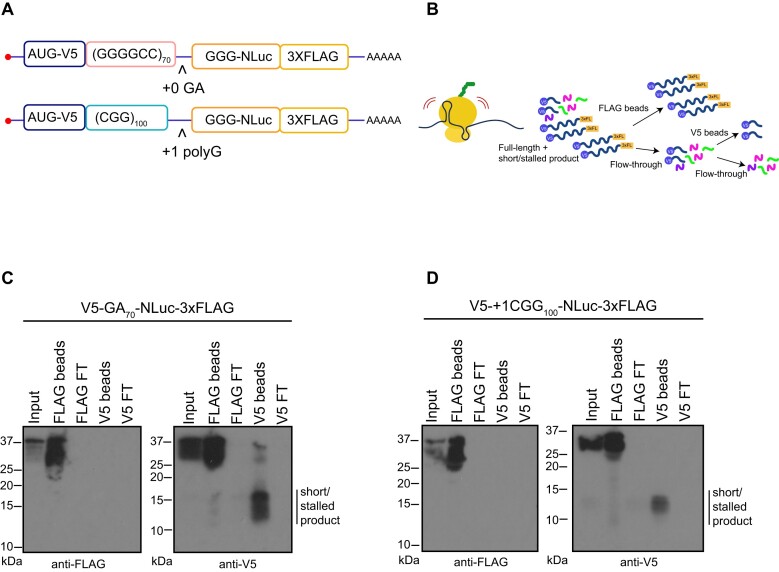
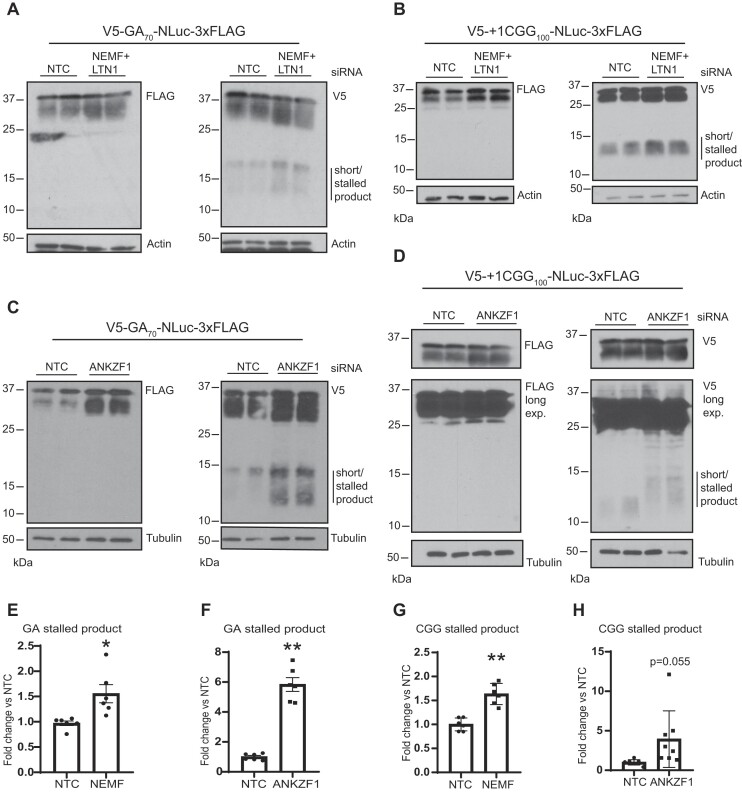
RQC pathway activation typically triggers the degradation of partially made translation products through a proteosome-dependent process. Prior studies utilized a dual fluorescent tagging system with GFP and RFP bracketing a ribosome-stall inducing sequence coupled with flow cytometry to measure stalling during translation (91). In this set up ‘stalling’ events are measured by detecting the signal of one (stalled/truncated) or both reporters (full-length). However, truncated products would not be detectable by either carboxyl-terminal nano-luciferase (NLuc) signal or FLAG tag western blots. We therefore generated a new set of reporter constructs with an AUG-initiated V5 tag at the amino-terminus of 70 G4C2 repeats in the GA frame or 100 CGG in the polyG frame. For each construct, we retained an in-frame NLuc and 3xFLAG at the carboxyl-terminus (Figure 3A). To avoid the generation of potentially aberrant RNA products from plasmids that might complicate our data interpretation, we transfected in vitro synthesized mRNAs with either G4C2 repeats or CGG repeats into HEK293 cells. We then performed a dual IP experiment where we first used FLAG magnetic beads to pull down the full-length products and then performed a second IP with V5 on the flowthrough to enrich for partially made products that only have the amino terminus of the protein (Figure 3B). We detected partially made products, approximately 10–15 kDa in size, only with the V5 antibody for both GA (Figure 3C) and polyG (Figure 3D). Importantly, constructs lacking the GC-rich repeats did not generate any partially made products (Supplementary Figure S5A-C).
Figure 3.
Detection of partially made products translated from GC-rich transcripts. (A, B) Dual-tagged AUG initiated constructs were generated to allow for capture of N-terminal fragments generated during translation through the repeats. These were generated to detect AUG-V5-(G4C2)70-NLuc-3xFLAG in GA frame and AUG-V5-+1(CGG)100-NLuc-3xFLAG in polyG frame. A dual FLAG and V5 IP workflow was used to enrich for incomplete products generated from GC-rich transcripts. 3xFL: 3xFLAG. Green, pink, and purple elements represent other proteins from cell lysate that were cleared from the flow-through. (C, D) V5 antibody IP reveals truncated/stalled products (line next to the blot) generated from G4C2 (panel C) or CGG repeats (panel D) that do not contain the carboxyl-terminal FLAG tag. FT: Flow-through. Representative images from at least two independent experiments.
To determine if these smaller products could result from truncated mRNA species generated during in vitro transcription of the reporters (42), we gel-purified the full-length mRNA post-transcription (Supplementary Figure S6A). These purified mRNAs still generated the smaller product (Supplementary Figure S6B). After activation of the RQC pathway, partially generated products are targeted for degradation by the Ubiquitin Proteosome system (54–56). To determine if these smaller products might represent incomplete degradation of proteins generated by ribosomal stalling, we inhibited the ubiquitin proteasome system by treating cells with MG132 and measured small product abundance. Smaller N-terminal products accumulated after MG132 treatment (Supplementary Figure S6C, D). These data suggest that these smaller proteins are stalled translation products that are cleared in a proteasome dependent fashion.
To assess whether NEMF or ANKZF1 affect the generation or accumulation of these truncated repeat products, we repeated the studies as above after depletion of NEMF + LTN1 or ANKZF1. From immunoblot analysis, we detected an increase in both the abundance of the full-length products and the truncated products after knockdown of NEMF or after knockdown of ANKZF1 from both (G4C2)70 and (CGG)100 repeats (Figure 4). We did not observe any change in the expression or generation of products from a no-repeat control (Supplementary Figure S5B, C). These observations suggest that the accumulation of these short/stalled products is impacted by RQC factor abundance.
Figure 4.
Depletion of NEMF, LTN1, and ANKZF1 enhances generation of truncated GA and polyG products from GC-rich transcripts. (A–D) Immunoblots of HEK293 cells transfected with in vitro synthesized G4C2 (GA frame) and CGG repeat RNA reporters (polyG frame) in the presence and absence of NEMF + LTN1, and ANKZF1. Blots represent two biological replicates. Lines next to the blots indicate short/stalled product products. (E–H) Quantification of V5 positive truncated products generated by GA or + 1CGG translation reporters after depletion of NEMF/LTN1 (E and G) or ANKZF1 (F and H). Data represents fold change from non-template control stalled product abundance. Error bars ± SEM of n = 6–8 from at least three independent experiments, *P≤ 0.05; ***P≤ 0.005. t-test with Welch's correction.
Repeat RNA sequence determines the impact of RQC factor depletion
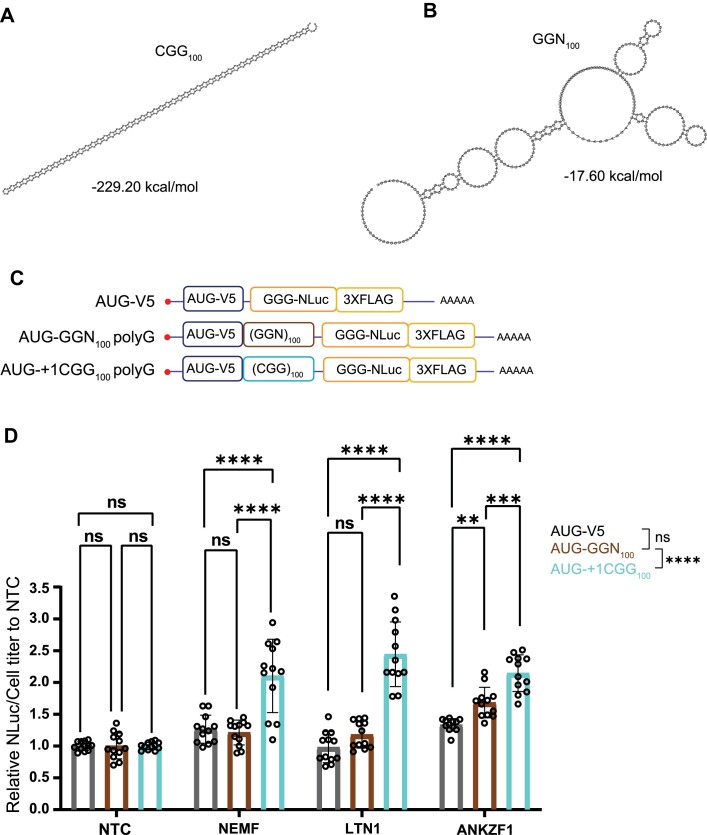
Prior studies are conflicted as to whether GA DPRs generated from non-repetitive mRNA sequences induce ribosomal stalls (98,100). Our data suggests that the repeat RNA sequence and structure may be sufficient to elicit ribosomal stalling and RQC activation (Figures 3 and 4). To assess this more formally, we took advantage of the redundant nature of RNA codons to generate a reporter that would make a polyG product but lack the CGG repeat RNA structure (Figure 5A, B). Glycine is encoded by GGN, where N is any nucleotide. Therefore, we generated constructs with identical AUG initiation codons upstream of either a GGC repeat or a GGN repeat that should not form a strong hairpin structure (Figures 5A–C). Both constructs generate the same polyG-containing protein that we can measure using NLuc (Figure 5C). Compared to a non-targeting siRNA control, depletion of NEMF, LTN1, or ANKZF1 significantly enhanced the production of the polyG protein from the AUG-V5-(CGG)100 reporter (Figure 5D). As RAN translation from CGG repeats lacking near-cognate initiation codons is much less efficient than AUG initiation (49,52), this result suggests that the modulation of these factors is acting primarily at the stage of translational elongation rather than RAN translational initiation. In contrast, depletion of either NEMF or LTN1 had no impact on polyG protein generation from the AUG-V5-(GGN)100 construct or an AUG-V5-NLuc no-repeat control (Figure 5D). Depletion of ANKZF1 did modestly enhance polyG production from the AUG-V5-(GGN)100 construct, but the effect was significantly weaker than that seen for polyG protein production from the AUG-V5-(CGG)100 reporter (Figure 5D). Together, these results suggest that the repeat sequence plays a key role in eliciting ribosomal stalling and the interplay of RQC factors with CGG repeat translation.
Figure 5.
Enhancement of polyG production with NEMF, LTN1 and ANKZF1 depletion requires the CGG repeat RNA structure. (A, B) Prediction of the optimal RNA secondary structure and calculation of the minimum free energy in CGG100 and GGN100 repeats. Only the repeat region from each repeat was used to predict the RNA secondary structure and calculate the minimum free energy. The results were computed by RNAfold 2.5.1. (C)Schematics of AUG-V5-NLuc-3xFLAG, AUG-V5-+1(CGG)100-NLuc-3xFLAG, and AUG-V5-(GGN)100-NLuc-3xFLAG transcripts. (D) Knockdown of NEMF, LTN1, and ANKZF1 in HEK293 with no-repeat control, polyG from GGN repeats, and CGG repeats RNA transfection. Data represent means with error bars ± SD of n = 12, ns = not significant; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001. The statistic result placed in the legend is group comparisons by two-way ANOVA with Sidak's multiple comparisons tests.
Depletion of NEMF aggravates toxicity in Drosophila model of C9ALS/FTD
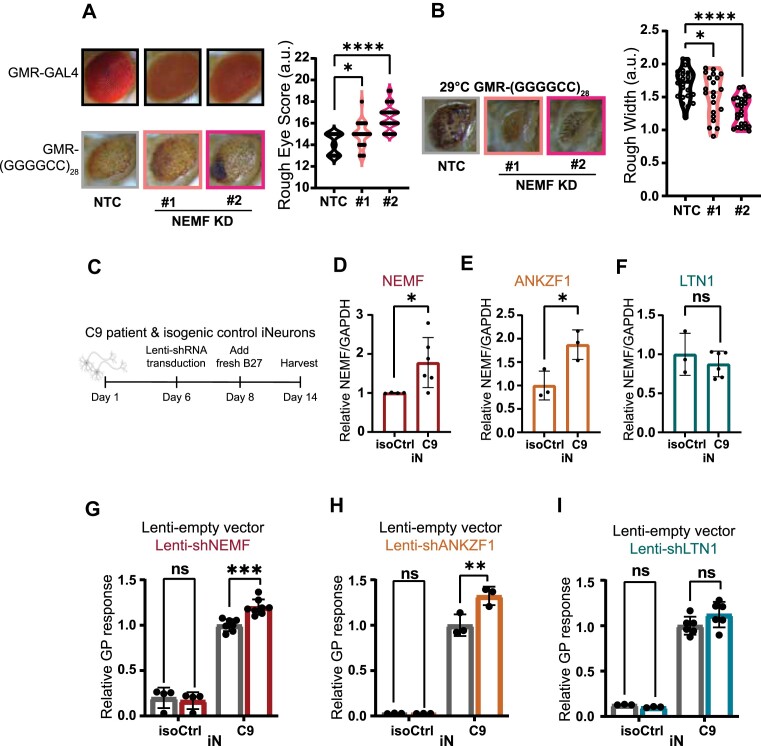
A prior study demonstrated worsening of rough eye phenotypes with depletion of NEMF in Drosophila expressing dipeptide GR or PR from constructs lacking a G4C2 repeat, which again suggested a direct role for serial arginine translation in RQC pathway activation (98,101). To assess the impact of modulating RQC factor expression on G4C2 repeat-associated phenotypes in vivo, we utilized an established Drosophila model of C9ALS/FTD that supports RAN translation from G4C2 repeats and elicits significant toxicity resulting in a rough eye phenotype when expressed using a GMR-GAL4 driver (102). Genetic knockdown of NEMF led to a more severe rough eye phenotype compared to the control cross in multiple NEMF depletion lines and siRNA lines (Figure 6A, Supplementary Figure S7). A similar effect enhancement was observed with knockdown of LTN1 (Supplementary Figure S7B, C). Rearing the G4C2 repeat-expressing flies at a higher temperature (29°C) led to more severe eye degeneration as marked by physical constriction of eye width (103,118). Depletion of NEMF mitigated this decrease in eye width (Figure 6B). In a second Drosophila model where G4C2 repeat production can be monitored by GFP expression, genetic knockdown of NEMF or LTN1 led to enhanced accumulation of GR-GFP translation products (Supplementary Figure S7A, B, D). Together these data suggest that depletion of NEMF or LTN1 enhances G4C2 repeat elicited toxicity in Drosophila via enhanced RAN translation.
Figure 6.
Depletion of NEMF enhances repeat-associated toxicity in a fly model of C9 ALS/FTD and DPR accumulation in human neurons. (A) Representative images of Drosophila eyes expressing (G4C2)28 repeats under the GMR-GAL4 driver in the presence or absence of NEMF at 25°C [BDSC36955 #1 and BDSC25214 #2]. Rough eye phenotypes quantified using an established nominal scoring system are shown as violin plots on the right. Individual flies are represented by single data points. n = 30–33/genotype. (B) (G4C2)28 repeats expressed with a GMR-GAL4 driver at 29°C show decreased eye width that is enhanced by the depletion of NEMF, as quantified on the right. Graphs represent the mean with error bars ± SD, n = 21–24. For A and B, *P< 0.05; ****P≤ 0.0001 by one-way ANOVA with Dunnett's multiple comparison test. (C) Schematic workflow for studies with C9ALS patient-derived iNeurons (iN). (D–F) RNA Expression of NEMF, LTN1, and ANKZF1 transcripts from C9ALS and isogenic control iN lysates. Data represent means with error bars ± SD. n = 3–6/gene, ns = not significant; *P< 0.05 by Student's t-test. (G–I) Quantification of GP by MSD assay from C9ALS and isogenic control iNs treated with lenti-empty vector or lenti-shRNA of NEMF, LTN1 or ANKZF1. Data represent mean ± SD; n = 3–6, ns = not significant; **P≤ 0.01; ***P≤ 0.001 by two-way ANOVA with Sidak's multiple comparison test.
Depletion of NEMF and ANKZF1 enhances RAN translation in C9ORF72 human neurons
We next assessed whether modulating RQC factors also influence RAN translation product accumulation from the endogenous repeat locus in C9ORF72 ALS patient-derived neurons. To this end, we utilized a well-characterized pair of C9 patient iPSC-derived iNeurons (iN) with an accompanied isogenic control line (119). These iN lines contain a doxycycline inducible Ngn1/2 cassette that allows for rapid neuronal differentiation (106). After two weeks of differentiation, we harvested cell lysates and performed an MSD assay to measure GP DPR product abundance (Figure 6C). In parallel, we performed a qRT-PCR analysis of RNA abundance for different RQC factors. We observed a significant increase of NEMF and ANKZF1 transcript expression in C9ALS iNeurons compared to isogenic controls differentiated in parallel, suggesting that there could be a compensatory upregulation of RQC machinery elicited by the repeat expansion (Figure 6D, E). There was no change in LTN1 transcript abundance between these C9ALS iNeurons and their isogenic control (Figure 6F).
As expected, we were able to reliably measure GP DPR abundance in C9ALS iNeurons but not in isogenic control neurons across multiple differentiations (Figure 6G-I). To assess whether altering RQC factor abundance could impact endogenous RAN product abundance, we utilized lentiviral delivery of shRNAs against each factor. In line with our reporter assays, we observed a significant increase in GP DPR abundance following the depletion of NEMF or ANKZF1 in C9 iN compared to iNeurons treated with a lentiviral control (Figure 6G, H). However, the depletion of LTN1 did not significantly impact GP DPR abundance (Figure 6I). The RQC pathway typically pairs with the mRNA surveillance pathway after the separation of 80S ribosomes into 40S and 60S subunits to degrade the template mRNA. Therefore, we assessed whether endogenous C9 RNA abundance might be impacted by the depletion of NEMF, LTN1, and ANKZF1. Using primers that specifically target the first intron of C9RNA that contains the repeat, we observed an increase of C9 transcript abundance with depletion of NEMF or LTN1 (Supplementary Figure S8A-B). However, we observed no change in C9 transcript abundance with depletion of ANKZF1 compared to a lentiviral control (Supplementary Figure S8C). Taking our data together with previous studies, it remains unclear whether the activation of RQC degrades endogenous repeat containing C9 RNA (98).
Enhancing the expression of RQC factors suppresses RAN product accumulation
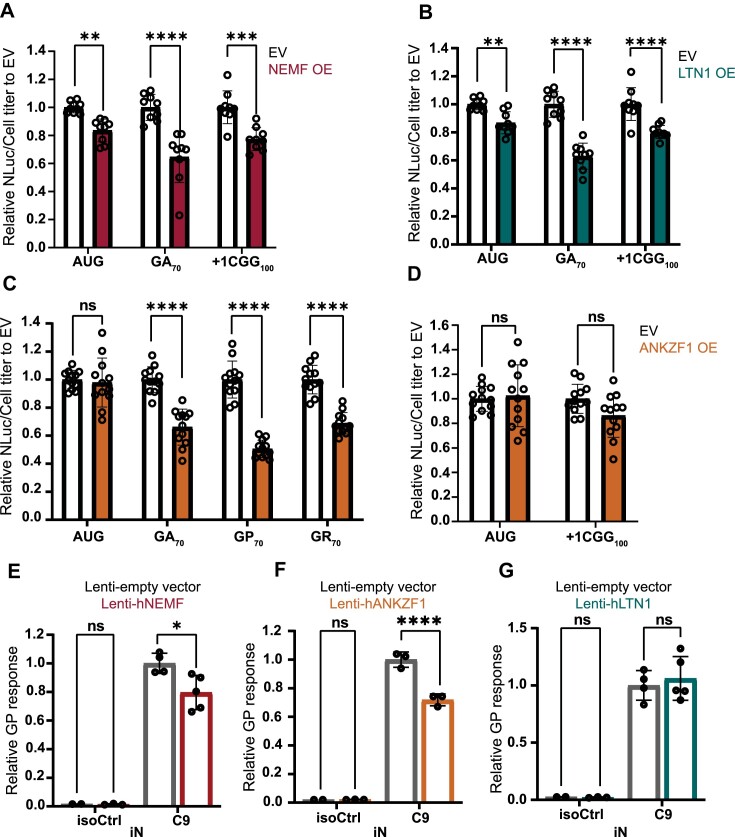
As depletion of NEMF, LTN1, and ANKZF1 increase GA and polyG product accumulation from GC-rich repeats, we wondered if boosting NEMF, LTN1, or ANKZF1 expression in cells might suppress RAN translation or lower RAN product accumulation. To assess this, we overexpressed each of the RQC factors (NEMF, LTN1, and ANKZF1) in conjunction with the AUG-driven no repeat control, G4C2 GA reporter, or the CGG polyG reporter. Overexpression of these factors did not significantly impact global translation based on polysome profiling (Supplementary Figure S9). Overexpression of NEMF and LTN1 decreased both GA and polyG RAN product accumulations (Figure 7A-B). Overexpression of ANKZF1 decreased the accumulation of RAN products from all three (GA, GP and GR) reading frames from G4C2 repeats without impacting an AUG-driven no-repeat control (Figure 7C). In contrast, overexpression of ANKZF1 did not significantly decrease polyG RAN product abundance (Figure 7D). Next, we transduced C9 patient iPSC-derived iNeurons (iN) with lentivirus expressing hNEMF, hLTN1 or hANKZF1 and subsequently measured the GP DPR abundance (Figure 6C). Overexpression of either NEMF or ANKZF1 in C9 iN significantly decreased GP abundance compared to a lentiviral control (Figure 7E, F). However, there was no significant increase in GP abundance seen with hLTN1 expression (Figure 7G). Overexpression of NEMF, LTN1, or ANKZF1 did not affect C9 intronic transcript abundance (Supplementary Figure S8D-F). Taken together, these data suggest that modulation of NEMF and ANKZF1 directly impact RAN product accumulation from both reporters and endogenous loci in patient-derived neurons.
Figure 7.
Overexpression of NEMF, LTN1 and ANKZF1 decreases RAN translation from G4C2 and CGG repeats. (A, B) Relative expression of AUG-driven no repeats, (G4C2)70 repeats in the GA frame, and (CGG)100 repeats in the polyG frame when overexpressing empty vector (EV) versus hNEMF or hLTN1. (C) Relative expression of AUG-driven no-repeat control and (G4C2)70 repeats in the GA, GP, and GR frames when overexpressing empty vector (EV) versus hANKZF1. (D) Relative expression of AUG-driven no-repeat control and (CGG)100 repeats in the polyG frame when overexpressing empty vector (EV) versus hANKZF1. Data represent means with error bars ± SD of n = 9–12, ns = not significant; **P≤ 0.01; ***P≤ 0.001; ****P≤ 0.0001 by two-way ANOVA with Sidak's multiple comparison test. (E–G) Relative GP response of C9 and isogenic control iN treated with lenti-empty vector or lenti-hNEMF, hLTN1, and hANKZF1. The level of GP was measured by MSD. Data represent means with error bars ± SD of n = 3–5, ns = not significant; *P< 0.05; ****P≤ 0.0001 by two-way ANOVA with Sidak's multiple comparison test.
Discussion
Transcribed GC-rich short tandem repeat expansions in C9ORF72 and FMR1 form strong RNA secondary structures as either RNA hairpins or G-quadruplexes (38,39,42,43,120). These structures are critical for repeat-associated non-AUG (RAN) translation (8,20,44,45,121–123). mRNA secondary structures can impede elongating ribosomes leading to stalling or collision, and activation of ribosome-associated quality-control pathways (56,63,65,77). To understand how these pathways interplay with RAN translation across GC-rich repeat RNAs, we performed a targeted screen on factors from the mRNA and protein surveillance pathways associated with ribosomal stalling. We identified NEMF, LTN1, and ANKZF1 from the RQC pathway as robust inhibitors of RAN translation on G4C2 mRNA repeats in the GA frame and CGG repeats in the polyG frame. Depletion of NEMF, LTN1, and ANKZF1 increased the abundance of RAN products while overexpression decreased detectable RAN products, with similar genetic compensation effects at endogenous loci in human C9 patient-derived neurons (iNs). Intriguingly, these same factors are upregulated in C9 iNs, suggesting that these pathways may be activated by the translation of expanded repeats. With an N-terminal tagging system, we observed that both G4C2 and CGG repeat sequences generate partially made truncated products, suggesting peptide release associated with RQC degradation pathways activated during repeat translation. Importantly, these effects are not mediated by the charge of the amino acids associated with the peptides (both GA and polyG are uncharged), but by the mRNA structures themselves, as we can abrogate this genetic interaction by changing the repeat sequence to diminish the predicted RNA hairpin structures formed by the CGG repeat element. These data suggest a role for RQC pathways in both RAN translation and AUG-initiated translation of GC-rich structured repeat RNAs, with implications for disease pathogenesis and therapeutic development in this currently untreatable class of disorders.
Several studies implicate RQC activation as associated with neuronal death and human neurodegenerative disease (88,124–128). NEMF mutations in mice are sufficient to trigger neurodegenerative phenotypes and variants of NEMF in humans are associated with neuromuscular disease (129,130). LTN1 mutations in mice also trigger movement disorder phenotypes and motor neuron degeneration (131). Factors from protein surveillance pathways directly associate with ALS/FTD-causing toxic arginine-rich dipeptide repeat (DPR) proteins, GR, and PR (98,100,101,110). AUG-initiated arginine-rich proteins from GR and PR encoding RNAs induce ribosomal stalling in an RNA-independent and DPR protein length-dependent manner in both mammalian cells and GR and PR DPR protein-expressing flies (97,132). In these cases, ZNF598, NEMF and LTN1 regulate the expression of GR protein in a fashion that does not require the G4C2 repeat RNA sequence or structure, and these effects are thought to be due to interactions of the charged arginine residue within the DPR protein. Importantly, in these studies, AUG-initiated translation of GA or PA sequences from a non-repetitive sequence was insufficient to elicit ribosomal stalling (99). In contrast, we observe evidence for ribosomal stalling impacting the expression of GA, GP and GR generated through RAN translation on G4C2 repeat transcripts that include the native upstream endogenous intronic sequence of C9ORF72. We also demonstrate that RAN translation of polyglycine from CGG repeats but not AUG-initiated translation of polyglycine from a codon optimized non-repetitive sequence is highly sensitive to RQC factor expression manipulation. As such, our findings suggest that RNA secondary structures formed by GC-rich STRs contribute to ribosomal stalling and activation of downstream surveillance mechanisms. As RAN translation can proceed in multiple reading frames on the same repeat simultaneously, our data cannot rule out a second contribution to ribosomal stalling by rare polyGR translation events on G4C2 or polyR CGG repeats as a stall trigger in other reading frames.
Prior studies utilized P2A-based polycistronic reporter systems encoding two fluorescent proteins to assess translation elongation stalling (91). In these assays, the efficiency of the translation is measured by the ratio of two fluorescent proteins by flow cytometry. However, on GC-rich repeats, the relative loss of expression from a C-terminal tagged fluorescent protein could also result from translational frameshifting, which occurs on both CGG and G4C2 repeats (51,133). In addition, fluorescent protein systems cannot exclude a contribution to C-terminal tag fluorescent protein expression from internal ribosome entry site (IRES) initiation within these GC-rich repeats as is proposed as a potential mechanism for RAN translation at both CGG and G4C2 repeats (45,122,123). To overcome these limitations, we used an N-terminal AUG-V5 tag upstream of G4C2 and CGG repeats to successfully detect short, truncated V5-tagged peptides, which are likely to be released from stalled ribosomes and targeted for proteasomal degradation. Consistent with this concept, these truncated products accumulate in response to UPS impairment. Future studies will be needed to see if such peptides are independently toxic and contribute to disease.
Among the modifiers studied, loss of ANKZF1 (yeast Vms1) had the greatest impact on RAN translation product accumulation from both CGG and G4C2 repeats. This factor was not previously assessed with PR or GR-elicited ribosome stalling, presumably because it was thought to act at a very late stage in the RQC pathway. Overexpression of ANKZF1 in mammalian cells selectively decreased expression of GA, GP and GR from G4C2 repeats and we see a similar of this effect in C9 patient iPSC-derived iNeurons. ANKZF1 is conserved from yeast to humans, where it is thought to act as a hydrolase and/or nuclease to release the nascent peptide chain from the peptidyl-tRNA on the 60S ribosome (95,96,134–137). In addition, Vms1 in yeast may have additional roles related to ribosomal stalling that are independent of the canonical RQC pathway, as it can cleave peptidyl-tRNA chains on the leading stalled ribosome independent of 40S and 60S subunit splitting (96). Consistent with this earlier role in stall resolution, Vms1 is found across the entire gradient in yeast polysome profiles, suggesting an association with polysomes as well as isolated and split 60S subunits (96). In the absence of ANKZF1, as with other RQC factors, 60S subunits cannot be recycled for further rounds of translation, leading to a decrease in global translation (Supplementary Figure S2).
In this work, we highlight NEMF, LTN1 and ANKZF1 as regulators of RAN translation product accumulation from G4C2 and CGG repeats. By detecting truncated products generated from GC-rich transcripts, we suggest that translational stalling occurs within the short tandem repeat region of these mRNAs. We propose that these stalled ribosomes are triggered at least in part by the mRNA repeat secondary structure and that they recruit RQC complexes to assist with ribosomal recycling and clearance of aberrant mRNA and proteins (63). How exactly this ribosomal recycling and clearance would work on repeats, however, is somewhat unclear. Classically, the RQC pathway degrades nascent peptide chains after ribosomal splitting and disassembly through a process that is CAT-tailing and NEMF-dependent (85,86). This CAT-tailing is thought to allow for lysine residues to exit the ribosome and then be (84,85) ubiquitinated in an LTN1-dependent fashion to allow for targeting of nascent peptide chain to proteasomal degradation (58,61,89,91,134). However, there is no lysines available for ubiquitination in either of the sequences that serve as RAN translation templates from C9orf72-associated GA, GP and GR reading frames from the sense transcript or the upstream endogenous 5′UTR, including the CGG repeat, of the FMR1 sequence. Without an available lysine, we hypothesize that even when RQC pathways are triggered during RAN translation, the truncated products generated by RQC processing will likely be resistant to ubiquitination and degradation. In contrast, the V5-tagged constructs used to identify these N-terminal fragments do contain lysine within that introduced tag. Consistent with this, the abundance of these truncted proteins increases with MG132 treatment, which impairs proteasomal degradation (Supplementary Figure S6). This data is consistent with prior work suggesting that UPS impairment enhances CGG repeat-elicited phenotypes (138). What role such truncated degradation-resistant products might play in repeat-associated toxicity is an important future research direction.
Carboxyl-terminal reporters are often used to measure the efficiency of RAN translation initiation in different settings (44,45,49). These approaches face a limitation in that they assume translational elongation is held constant across reporters. As RAN translation potentially happens in multiple reading frames of the same transcript with different rates for both initiation and elongation, interpretation of results from the use of such carboxyl terminal tags as their sole readouts will need to be re-evaluated. Here, we observed largely similar results from both reporters using such a C-terminal tag reporter system as well as from MSD-based assays that directly measure DPR product generation (and as such are not reliant on translation elongation extending completely through the repeat to the NLuc reporter).
The increase in generation of both partially made products from stalled ribosomes and complete protein products which contain the C-terminal reporter in the absence of key RQC factors (Figure 4) suggests that ribosomal stalling triggers multiple events that exist in an equilibrium with each other. When the surveillance pathway is activated, stalled or collided ribosomes will be removed from the repeat mRNA. If the repeat mRNA is not degraded during this process, then removal of stalled ribosomes would also allow trailing ribosomes to complete translation through the repeat at increased rates, resulting in greater C-terminal reporter signal. When RQC factors are depleted, this will result in a failure to clear the nascent peptides from the 60S subunit. This will result in accumulation of stall products and their detection on denaturing gels as observed here. However, reduction of these same RQC factors will also slow the rate at which such stall events are recognized and cleared. This delay should create a longer time window during which stalled ribosomes can resume elongation through the repeat without engagement of the RQC machinery, resulting in more initiating ribosomes generating complete products. This may also impact the rates of ribosomal frameshifts on the repeat (133,139).
While past studies (97–99,132) and our data are largely consistent with loss of RQC factors acting on translational elongation, it is important to note that our data are consistent with an alternative model where RQC factor depletion reduces global 60S ribosomal turnover and the resultant ribosomal stress then acts to enhance RAN translational initiation. There is a precedent for such an idea, as activation of the integrated stress response augments RAN translation in the face of global decreases in protein synthesis (44,45). While RQC factor depletion did not trigger ISR induction (Supplementary Figure S2F), it is intruiging that RAN translation is enhanced by multiple different processes that inhibit global protein translation- including low-dose treatment with the protein synthesis elongation inhibitor cycloheximide (141). To explain this, Kearse et al. proposed that ribosomal pre-initiation complexes queue on 5′ leaders behind stalled ribosomes to favor non-AUG initiation by allowing for greater dwell times over near-AUG cognate codons (141). Further work will be needed to assess if a similar process may be at play with RQC factor depletion and RAN translation.
In summary, we find that RQC pathways inhibit accumulation of RAN translation products generated from GC-rich transcripts. Depletion of NEMF, LTN1 and ANKZF1 increases the accumulation of proteins made via RAN translation from GC-rich transcripts while overexpression of NEMF, LTN1, and ANKZF1 decreases the production of these proteins. These data suggest that augmenting RQC activity could have therapeutic benefits in GC-rich nucleotide repeat expansion diseases and that translational elongation needs to be considered in studies of RAN translation and other initiation-dependent processes.
Supplementary Material
Acknowledgements
We thank current and former members of the Todd lab and the Barmada lab for critical discussions, and technical advice in the pursuit of this study. We thank the University of Michigan Vector Core for packaging lentivirus for this study. We thank Dr. Claudio Joazeiro for sharing cell lines and materials for the initial studies and Dr James Bardwell for his generous support. We thank the University of Michigan Mass Spectrometry-Based Proteomics Resource Facility, especially Dr Venkatesha Basrur, for the assistance and use of equipment.
Author contributions: Y-J.T., I.M., R.S. and P.K.T. conceived the project. Y-J.T., I.M. and P.K.T. designed the experiments with input from A.K. Y-J.T. performed, optimized, and visualized experiments with help from A.K. S.O. and R.K.; K.J.-W. and L.P. performed the GP MSD assay; E.M.H.T. and S.J.B. assisted with patient-derived iPSC iNeurons. I.M. and E.Y. performed Drosophila experiments and assisted with the design and analysis of these studies. X.D. performed polysome profiling experiments. N.B.G. performed initial studies. Y.-J.T. wrote the initial draft of the manuscript with input from I.M. and P.K.T. P.K.T. and A.K. re-wrote portions of the manuscript in response to reviewer requests with input from Y.-J.T. All authors reviewed and edited the manuscript.
Contributor Information
Yi-Ju Tseng, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; Cellular and Molecular Biology Graduate Program, University of Michigan, Ann Arbor, MI 48109, USA.
Amy Krans, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; Ann Arbor Veterans Administration Healthcare, Ann Arbor, MI 48109, USA.
Indranil Malik, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; Department of Biotechnology, Indian Institute of Technology Hyderabad, Kandi, Sangareddy, 502284 Telangana, India.
Xiexiong Deng, Department of Molecular, Cellular and Developmental Biology, Howard Hughes Medical Institute, University of Michigan, Ann Arbor, MI 48109, USA.
Evrim Yildirim, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Sinem Ovunc, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Elizabeth M H Tank, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Karen Jansen-West, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Ross Kaufhold, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; Medical Scientist Training Program, University of Michigan, Ann Arbor, MI 48109, USA.
Nicolas B Gomez, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; Medical Scientist Training Program, University of Michigan, Ann Arbor, MI 48109, USA.
Roger Sher, Department of Neurobiology and Behavior & Center for Nervous System Disorders, Stony Brook University, Stony Brook, NY 11794, USA.
Leonard Petrucelli, Department of Neuroscience, Mayo Clinic, Jacksonville, FL 32224, USA.
Sami J Barmada, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Peter K Todd, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; Ann Arbor Veterans Administration Healthcare, Ann Arbor, MI 48109, USA.
Data availability
All data points are included in the main figures or supplementary data. Raw data used for figure generation are available upon request.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
NIH [P50HD104463, R01NS099280, R01NS086810 to P.K.T., R01 NS097542, R01 NS113943, R56 NS128110 to S.J.B., RF1AG062171, R35NS097273, P01NS084974, AG062077 to L.P.]; P.K.T. and A.K. were also supported by VA BLRD [BX004842]; Y-J.T. was supported by the Cellular and Molecular Biology Graduate Program, University of Michigan; I.M. was supported by an Alzheimer's Association Research Fellowship.
Conflict of interest statement. P.K.T. served as a consultant to Denali Therapeutics and Reata Therapeutics. He holds a shared patent on antisense oligonucleotide technology with Ionis Pharmaceuticals. He has lisenced technology to Denali Therapeutics and to Millipore. None of the studies here are directly related to these potential conflicts. None of these parties had any role in the research, funding, interpretation or writing of this manuscript.
References
- 1. Nelson D.L., Orr H.T., Warren S.T. The unstable repeats-three evolving faces of neurological disease. Neuron. 2013; 77:825–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koob M.D., Moseley M.L., Schut L.J., Benzow K.A., Bird T.D., Day J.W., Ranum L.P.W. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat. Genet. 1999; 21:379–384. [DOI] [PubMed] [Google Scholar]
- 3. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., Fitzhugh W. et al. Initial sequencing and analysis of the human genome: international Human Genome Sequencing Consortium. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 4. Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P.W. Myotonic dystrophy type 2 caused by a CCTG expansion in intron I of ZNF9. Science. 2001; 293:864–867. [DOI] [PubMed] [Google Scholar]
- 5. Verkerk A.J.M.H., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P.A., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F. et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991; 65:905–914. [DOI] [PubMed] [Google Scholar]
- 6. Malik I., Kelley C.P., Wang E.T., Todd P.K. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 2021; 22:589–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Green K.M., Linsalata A.E., Todd P.K. RAN translation—What makes it run?. Brain Res. 2016; 1647:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A.C. et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. 2011; 108:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cleary J.D., Ranum L.P.W. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013; 22:R45–R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zu T., Pattamatta A., Ranum L.P.W. Repeat-associated non-ATG translation in neurological diseases. Cold Spring Harb. Perspect. Biol. 2018; 10:a033019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cleary J.D., Ranum L.P.W. Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr. Opin. Genet. Dev. 2014; 26:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao F.B., Richter J.D. Microsatellite expansion diseases: repeat toxicity found in translation. Neuron. 2017; 93:249–251. [DOI] [PubMed] [Google Scholar]
- 13. Bañez-Coronel M., Ayhan F., Tarabochia A.D., Zu T., Perez B.A., Tusi S.K., Pletnikova O., Borchelt D.R., Ross C.A., Margolis R.L. et al. RAN translation in Huntington Disease. Neuron. 2015; 88:667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mori K., Arzberger T., Grässer F.A., Gijselinck I., May S., Rentzsch K., Weng S.-M., Schludi M.H., van der Zee J., Cruts M. et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013; 126:881–893. [DOI] [PubMed] [Google Scholar]
- 15. Freibaum B.D., Lu Y., Lopez-Gonzalez R., Kim N.C., Almeida S., Lee K.-H., Badders N., Valentine M., Miller B.L., Wong P.C. et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015; 525:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xinmei W., Wenzhi T., Thomas W., Karthik K., Shamamandri M.S., Yingxiao S., Shaoyu L., Shneider N.A., Monaghan J., Pandey U.B. et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72- ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014; 84:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sellier C., Buijsen R.A.M., He F., Natla S., Jung L., Tropel P., Gaucherot A., Jacobs H., Meziane H., Vincent A. et al. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron. 2017; 93:331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buijsen R.A.M., Visser J.A., Kramer P., Severijnen E.A.W.F.M., Gearing M., Charlet-Berguerand N., Sherman S.L., Berman R.F., Willemsen R., Hukema R.K. Presence of inclusions positive for polyglycine containing protein, FMRpolyG, indicates that repeat-associated non-AUG translation plays a role in fragile X-associated primary ovarian insufficiency. Hum. Reprod. 2016; 31:158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buijsen R.A.M., Sellier C., Severijnen L.A.W.F.M., Oulad-Abdelghani M., Verhagen R.F.M., Berman R.F., Charlet-Berguerand N., Willemsen R., Hukema R.K. FMRpolyG-positive inclusions in CNS and non-CNS organs of a fragile X premutation carrier with fragile X-associated tremor/ataxia syndrome. Acta Neuropathol. Commun. 2014; 2:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todd P.K., Oh S.Y., Krans A., He F., Sellier C., Frazer M., Renoux A.J., Chen K.c., Scaglione K.M., Basrur V. et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013; 78:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mori K., Weng S.-M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013; 339:1335–1338. [DOI] [PubMed] [Google Scholar]
- 22. Ishiguro T., Sato N., Ueyama M., Fujikake N., Sellier C., Kanegami A., Tokuda E., Zamiri B., Gall-Duncan T., Mirceta M. et al. Regulatory role of RNA chaperone TDP-43 for RNA misfolding and repeat-associated translation in SCA31. Neuron. 2017; 94:108–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soragni E., Petrosyan L., Rinkoski T.A., Wieben E.D., Baratz K.H., Fautsch M.P., Gottesfeld J.M. Repeat-associated non-ATG (RAN) translation in fuchs’ endothelial corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2018; 59:1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zu T., Cleary J.D., Liu Y., Bañez-Coronel M., Bubenik J.L., Ayhan F., Ashizawa T., Xia G., Clark H.B., Yachnis A.T. et al. RAN translation regulated by muscleblind proteins in myotonic dystrophy type 2. Neuron. 2017; 95:1292–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ash P.E.A., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.-L., DeJesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., Rademakers R. et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013; 77:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong H.E., Lim J., Linsalata A., Kang Y., Malik I., Allen E.G., Cao Y., Shubeck L., Johnston R., Huang Y. et al. Identification of PSMB5 as a genetic modifier of fragile X–associated tremor/ataxia syndrome. Proc. Natl. Acad. Sci. U.S.A. 2022; 119:e2118124119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodman L.D., Prudencio M., Kramer N.J., Martinez-Ramirez L.F., Srinivasan A.R., Lan M., Parisi M.J., Zhu Y., Chew J., Cook C.N. et al. Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat. Neurosci. 2019; 22:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jovičić A., Mertens J., Boeynaems S., Bogaert E., Chai N., Yamada S.B., Paul J.W., Sun S., Herdy J.R., Bieri G. et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015; 18:1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamada S.B., Gendron T.F., Niccoli T., Genuth N.R., Grosely R., Shi Y., Glaria I., Kramer N.J., Nakayama L., Fang S. et al. RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat. Neurosci. 2019; 22:1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujino Y., Mori K., Nagai Y. Repeat-associated non-AUG translation in neuromuscular diseases: mechanisms and therapeutic insights. J. Bioch. 2023; 173:273–281. [DOI] [PubMed] [Google Scholar]
- 31. DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011; 72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Zee J., Gijselinck I., Dillen L., Van Langenhove T., Theuns J., Engelborghs S., Philtjens S., Vandenbulcke M., Sleegers K., Sieben A. et al. A Pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum. Mutat. 2013; 34:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majounie E., Renton A.E., Mok K., Dopper E.G.P., Waite A., Rollinson S., Chiò A., Restagno G., Nicolaou N., Simon-Sanchez J. et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012; 11:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001; 57:127–130. [DOI] [PubMed] [Google Scholar]
- 35. Tassone F., Iong K.P., Tong T.H., Lo J., Gane L.W., Berry-Kravis E., Nguyen D., Mu L.Y., Laffin J., Bailey D.B. et al. FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Med. 2012; 4:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W. et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004; 291:460–469. [DOI] [PubMed] [Google Scholar]
- 37. Hagerman R., Hagerman P. Fragile X-associated tremor/ataxia syndrome: pathophysiology and management. Curr. Opin. Neurol. 2021; 34:541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Z., Ursu A., Childs-disney J.L., Rice J.E., Petrucelli L., Disney M.D., Wang Z., Ursu A., Childs-disney J.L., Guertler R. et al. The hairpin form of r (G 4 C 2) exp in c9ALS /FTD is repeat-associated non-ATG translated and a target for bioactive small molecules article The hairpin form of r (G 4 C 2) exp in c9ALS / FTD is repeat-associated non-ATG translated and a target for B. Cell Chem Biol. 2019; 26:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Asamitsu S., Yabuki Y., Ikenoshita S., Kawakubo K., Kawasaki M., Usuki S., Nakayama Y., Adachi K., Kugoh H., Ishii K. et al. CGG repeat RNA G-quadruplexes interact with FMRpolyG to cause neuronal dysfunction in fragile X-related tremor/ataxia syndrome. Sci. Adv. 2021; 7:eabd9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife. 2016; 5:e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zamiri B., Reddy K., Macgregor R.B., Pearson C.E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNAbinding proteins. J. Biol. Chem. 2014; 289:4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A.J., Shaw P.G., Kim M.-S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M.C., Parkinson G., Isaacs A.M. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012; 2:1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Green K.M., Glineburg M.R., Kearse M.G., Flores B.N., Linsalata A.E., Fedak S.J., Goldstrohm A.C., Barmada S.J., Todd P.K. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun. 2017; 8:2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cheng W., Wang S., Mestre A.A., Fu C., Makarem A., Xian F., Hayes L.R., Lopez-Gonzalez R., Drenner K., Jiang J. et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun. 2018; 9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zu T., Liu Y., Banez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gendron T.F., Bieniek K.F., Zhang Y.-J., Jansen-West K., Ash P.E.A., Caulfield T., Daughrity L., Dunmore J.H., Castanedes-Casey M., Chew J. et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013; 126:829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krans A., Kearse M.G., Todd P.K. Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome. Ann. Neurol. 2016; 80:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kearse M.G., Green K.M., Krans A., Rodriguez C.M., Linsalata A.E., Goldstrohm A.C., Todd P.K. CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell. 2016; 62:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwab S.R., Shugart J.A., Horng T., Malarkannan S., Shastri N. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004; 2:e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tabet R., Schaeffer L., Freyermuth F., Jambeau M., Workman M., Lee C.Z., Lin C.C., Jiang J., Jansen-West K., Abou-Hamdan H. et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat. Commun. 2018; 9:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y., Glineburg M.R., Basrur V., Conlon K., Wright S.E., Krans A., Hall D.A., Todd P.K. Mechanistic convergence across initiation sites for RAN translation in fragile X associated tremor ataxia syndrome. Hum. Mol. Genet. 2022; 31:2317–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wolin S.L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988; 7:3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buskirk A.R., Green R. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. Roy. Soc. B: Biol, Sci. 2017; 372:20160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brandman O., Hegde R.S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016; 23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Inada T. Quality controls induced by aberrant translation. Nucleic Acids Res. 2020; 48:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ikeuchi K., Izawa T., Inada T. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front. Genet. 2019; 10:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matsuo Y., Uchihashi T., Inada T. Decoding of the ubiquitin code for clearance of colliding ribosomes by the RQT complex. Nat. Commun. 2023; 14:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Filbeck S., Cerullo F., Pfeffer S., Joazeiro C.A.P. Ribosome-associated quality-control mechanisms from bacteria to humans. Mol. Cell. 2022; 82:1451–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shoemaker C.J., Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K. et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017; 8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shao S., Brown A., Santhanam B., Hegde R.S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell. 2015; 57:433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Joazeiro C.A.P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 2019; 20:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lyumkis D., Dos Passosb D.O., Tahara E.B., Webb K., Bennett E.J., Vinterbo S., Potter C.S., Carragher B., Joazeiro C.A.P. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl. Acad. Sci. USA. 2014; 111:15981–15986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ikeuchi K., Tesina P., Matsuo Y., Sugiyama T., Cheng J., Saeki Y., Tanaka K., Becker T., Beckmann R., Inada T. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019; 38:e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., Bennett E.J. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell. 2017; 65:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Garzia A., Jafarnejad S.M., Meyer C., Chapat C., Gogakos T., Morozov P., Amiri M., Shapiro M., Molina H., Tuschl T. et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 2017; 8:16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sitron C.S., Park J.H., Brandman O. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA. 2017; 23:798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saito S., Hosoda N., Hoshino S.I. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 2013; 288:17832–17843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guydosh N.R., Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014; 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shoemaker C.J., Eyler D.E., Green R. Dom34:hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science (1979). 2010; 330:369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pisareva V.P., Skabkin M.A., Hellen C.U.T., Pestova T.V., Pisarev A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011; 30:1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pisarev A.V., Skabkin M.A., Pisareva V.P., Skabkina O.V., Rakotondrafara A.M., Hentze M.W., Hellen C.U.T., Pestova T.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010; 37:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Orban T.I., Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the ski complex, and the exosome. RNA. 2005; 11:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Defenouillère Q., Fromont-Racine M. The ribosome-bound quality control complex: from aberrant peptide clearance to proteostasis maintenance. Curr. Genet. 2017; 63:997–1005. [DOI] [PubMed] [Google Scholar]
- 76. Matsuo Y., Tesina P., Nakajima S., Mizuno M., Endo A., Buschauer R., Cheng J., Shounai O., Ikeuchi K., Saeki Y. et al. RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1. Nat. Struct. Mol. Biol. 2020; 27:323–332. [DOI] [PubMed] [Google Scholar]
- 77. Hashimoto S., Sugiyama T., Yamazaki R., Nobuta R., Inada T. Identification of a novel trigger complex that facilitates ribosome-associated quality control in mammalian cells. Sci. Rep. 2020; 10:3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012; 151:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yonashiro R., Tahara E.B., Bengtson M.H., Khokhrina M., Lorenz H., Chen K.C., Kigoshi-Tansho Y., Savas J.N., Yates J.R., Kay S.A. et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife. 2016; 5:e11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mizuno M., Ebine S., Shounai O., Nakajima S., Tomomatsu S., Ikeuchi K., Matsuo Y., Inada T. The nascent polypeptide in the 60S subunit determines the Rqc2-dependency of ribosomal quality control. Nucleic Acids Res. 2021; 49:2102–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shen P.S., Park J., Qin Y., Li X., Parsawar K., Larson M.H., Cox J., Cheng Y., Lambowitz A.M., Weissman J.S. et al. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015; 347:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bengtson M.H., Joazeiro C.A.P. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010; 467:470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Doamekpor S.K., Lee J.W., Hepowit N.L., Wu C., Charenton C., Leonard M., Bengtson M.H., Rajashankar K.R., Sachs M.S., Lima C.D. et al. Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits. Proc. Natl. Acad. Sci. USA. 2016; 113:E4151–E4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kostova K.K., Hickey K.L., Osuna B.A., Hussmann J.A., Frost A., Weinberg D.E., Weissman J.S. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science. 2017; 357:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sitron C.S., Brandman O. CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat. Struct. Mol. Biol. 2019; 26:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lytvynenko I., Paternoga H., Thrun A., Balke A., Müller T.A., Chiang C.H., Nagler K., Tsaprailis G., Anders S., Bischofs I. et al. Alanine tails signal proteolysis in bacterial ribosome-associated quality control. Cell. 2019; 178:76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thrun A., Garzia A., Kigoshi-Tansho Y., Patil P.R., Umbaugh C.S., Dallinger T., Liu J., Kreger S., Patrizi A., Cox G.A. et al. Convergence of mammalian RQC and C-end rule proteolytic pathways via alanine tailing. Mol. Cell. 2021; 81:2112–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Udagawa T., Seki M., Okuyama T., Adachi S., Natsume T., Noguchi T., Matsuzawa A., Inada T. Failure to degrade CAT-tailed proteins disrupts neuronal morphogenesis and cell survival. Cell Rep. 2021; 34:108599. [DOI] [PubMed] [Google Scholar]
- 89. Shao S., Von der Malsburg K., Hegde R.S. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 2013; 50:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shao S., Hegde R.S. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell. 2014; 55:880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Juszkiewicz S., Hegde R.S. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell. 2017; 65:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Defenouillère Q., Yao Y., Mouaikel J., Namane A., Galopier A., Decourty L., Doyen A., Malabat C., Saveanu C., Jacquier A. et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. USA. 2013; 110:5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Verma R., Oania R.S., Kolawa N.J., Deshaies R.J. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife. 2013; 2013:e00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Su T., Izawa T., Thoms M., Yamashita Y., Cheng J., Berninghausen O., Hartl F.U., Inada T., Neupert W., Beckmann R. Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis. Nature. 2019; 570:538–542. [DOI] [PubMed] [Google Scholar]
- 95. Zurita Rendón O., Fredrickson E.K., Howard C.J., Van Vranken J., Fogarty S., Tolley N.D., Kalia R., Osuna B.A., Shen P.S., Hill C.P. et al. Vms1p is a release factor for the ribosome-associated quality control complex. Nat. Commun. 2018; 9:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Verma R., Reichermeier K.M., Burroughs A.M., Oania R.S., Reitsma J.M., Aravind L., Deshaies R.J. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature. 2018; 557:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kriachkov V., Ormsby A.R., Kusnadi E.P., McWilliam H.E.G., Mintern J.D., Amarasinghe S.L., Ritchie M.E., Furic L., Hatters D.M. Arginine-rich C9ORF72 ALS proteins stall ribosomes in a manner distinct from a canonical ribosome-associated quality control substrate. J. Biol. Chem. 2023; 299:102774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Park J., Lee J., Kim J.H., Lee J., Park H., Lim C. ZNF598 co-translationally titrates poly(GR) protein implicated in the pathogenesis of C9ORF72-associated ALS/FTD. Nucleic Acids Res. 2021; 49:11294–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Radwan M., Ang C.S., Ormsby A.R., Cox D., Daly J.C., Reid G.E., Hatters D.M. Arginine in C9ORF72 dipolypeptides mediates promiscuous proteome binding and multiple modes of toxicity. Mol. Cell. Proteomics. 2020; 19:640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Viera Ortiz A.P., Cajka G., Olatunji O.A., Mikytuck B., Shalem O., Lee E.B. Impaired ribosome-associated quality control of C9orf72 arginine-rich dipeptide-repeat proteins. Brain. 2022; 146:2897–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li S., Wu Z., Tantray I., Li Y., Chen S., Dong J., Glynn S., Vogel H., Snyder M., Lu B. Quality-control mechanisms targeting translationally stalled and C-terminally extended poly(GR) associated with ALS/FTD. Proc. Natl. Acad. Sci. USA. 2020; 117:25104–25115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. He F., Flores B.N., Krans A., Frazer M., Natla S., Niraula S., Adefioye O., Barmada S.J., Todd P.K. The carboxyl termini of RAN translated GGGGCC nucleotide repeat expansions modulate toxicity in models of ALS/FTD. Acta Neuropathol Commun. 2020; 8:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Malik I., Tseng Y., Wright S.E., Zheng K., Ramaiyer P., Green K.M., Todd P.K. SRSF protein kinase 1 modulates RAN translation and suppresses CGG repeat toxicity. EMBO Mol. Med. 2021; 13:e14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Weskamp K., Tank E.M., Miguez R., McBride J.P., Gómez N.B., White M., Lin Z., Gonzalez C.M., Serio A., Sreedharan J. et al. Shortened TDP43 isoforms upregulated by neuronal hyperactivity drive TDP43 pathology in ALS. J. Clin. Invest. 2020; 130:1139–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cerbini T., Funahashi R., Luo Y., Liu C., Park K., Rao M., Malik N., Zou J. Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines. PLoS One. 2015; 10:e0116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Busskamp V., Lewis N.E., Guye P., Ng A.H., Shipman S.L., Byrne S.M., Sanjana N.E., Murn J., Li Y., Li S. et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 2014; 10:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Habibey R., Striebel J., Schmieder F., Czarske J., Busskamp V. Long-term morphological and functional dynamics of human stem cell-derived neuronal networks on high-density micro-electrode arrays. Front. Neurosci. 2022; 16:951964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Andrade N.S., Ramic M., Esanov R., Liu W., Rybin M.J., Gaidosh G., Abdallah A., Del’olio S., Huff T.C., Chee N.T. et al. Dipeptide repeat proteins inhibit homology-directed DNA double strand break repair in C9ORF72 ALS/FTD. Mol Neurodegener. 2020; 15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Loveland A.B., Svidritskiy E., Susorov D., Lee S., Park A., Zvornicanin S., Demo G., Gao F.B., Korostelev A.A. Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM. Nat. Commun. 2022; 13:2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wieland M., Hartig J.S. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007; 14:757–763. [DOI] [PubMed] [Google Scholar]
- 112. Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001; 20:4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Cheng W., Wang S., Zhang Z., Morgens D.W., Hayes L.R., Lee S., Portz B., Xie Y., Nguyen B.V., Haney M.S. et al. CRISPR-Cas9 screens identify the RNA helicase DDX3X as a repressor of C9ORF72 (GGGGCC)n repeat-associated non-AUG translation. Neuron. 2019; 104:885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Linsalata A.E., He F., Malik A.M., Glineburg M.R., Green K.M., Natla S., Flores B.N., Krans A., Archbold H.C., Fedak S.J. et al. DDX 3X and specific initiation factors modulate FMR 1 repeat-associated non-AUG-initiated translation. EMBO Rep. 2019; 20:e47498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Tseng Y.J., Sandwith S.N., Green K.M., Chambers A.E., Krans A., Raimer H.M., Sharlow M.E., Reisinger M.A., Richardson A.E., Routh E.D. et al. The RNA helicase DHX36–G4R1 modulates C9orf72 GGGGCC hexanucleotide repeat–associated translation. J. Biol. Chem. 2021; 297:100914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liu H., Lu Y.-N., Paul T., Periz G., Banco M.T., Ferré-D’Amaré A.R., Rothstein J.D., Hayes L.R., Myong S., Wang J. A helicase unwinds hexanucleotide repeat RNA G-quadruplexes and facilitates repeat-associated non-AUG translation. J. Am. Chem. Soc. 2021; 143:7368–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Green K.M., Miller S.L., Malik I., Todd P.K. Non-canonical initiation factors modulate repeat-associated non-AUG translation. Hum. Mol. Genet. 2022; 31:2521–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sareen D., O’Rourke J.G., Meera P., Muhammad A.K.M.G., Grant S., Simpkinson M., Bell S., Carmona S., Ornelas L., Sahabian A. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 2013; 5:208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Fry M., Loeb L.A. The fragile X syndrome d(CGG)(n) nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. USA. 1994; 91:4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kearse M.G., Todd P.K. Repeat-associated non-AUG translation and its impact in neurodegenerative disease. Neurotherapeutics. 2014; 11:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sonobe Y., Ghadge G., Masaki K., Sendoel A., Fuchs E., Roos R.P. Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiol. Dis. 2018; 116:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Chiang P.W., Carpenter L.E., Hagerman P.J. The 5′-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J. Biol. Chem. 2001; 276:37916–37921. [DOI] [PubMed] [Google Scholar]
- 124. Wu Z., Wang Y., Lim J., Liu B., Li Y., Vartak R., Stankiewicz T., Montgomery S., Lu B. Ubiquitination of ABCE1 by NOT4 in response to mitochondrial damage links Co-translational quality control to PINK1-directed mitophagy. Cell Metab. 2018; 28:130–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Li Y., Geng J., Rimal S., Wang H., Liu X., Lu B., Li S. The mTORC2/AKT/VCP axis is associated with quality control of the stalled translation of poly(GR) dipeptide repeats in C9-ALS/FTD. J. Biol. Chem. 2023; 299:102995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lu B. Translation stalling and ribosome collision leading to proteostasis failure: implications for neurodegenerative diseases. Neural Regen Res. 2023; 18:111–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rimal S., Li Y., Vartak R., Geng J., Tantray I., Li S., Huh S., Vogel H., Glabe C., Grinberg L.T. et al. Inefficient quality control of ribosome stalling during APP synthesis generates CAT-tailed species that precipitate hallmarks of Alzheimer's disease. Acta Neuropathol Commun. 2021; 9:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wu Z., Tantray I., Lim J., Chen S., Li Y., Davis Z., Sitron C., Dong J., Gispert S., Auburger G. et al. MISTERMINATE mechanistically links mitochondrial dysfunction with proteostasis failure. Mol. Cell. 2019; 75:835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Martin P.B., Kigoshi-Tansho Y., Sher R.B., Ravenscroft G., Stauffer J.E., Kumar R., Yonashiro R., Müller T., Griffith C., Allen W. et al. NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease. Nat. Commun. 2020; 11:4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ahmed A., Wang M., Bergant G., Maroofian R., Zhao R., Alfadhel M., Nashabat M., AlRifai M.T., Eyaid W., Alswaid A. et al. Biallelic loss-of-function variants in NEMF cause central nervous system impairment and axonal polyneuropathy. Hum. Genet. 2021; 140:579–592. [DOI] [PubMed] [Google Scholar]
- 131. Chu J., Hong N.A., Masuda C.A., Jenkins B.V., Nelms K.A., Goodnow C.C., Glynne R.J., Wu H., Masliah E., Joazeiro C.A.P. et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. USA. 2009; 106:2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mizielinska S., Gronke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O.C., Pietrzyk J. et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014; 345:1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wright S.E., Rodriguez C.M., Monroe J., Xing J., Krans A., Flores B.N., Barsur V., Ivanova M.I., Koutmou K.S., Barmada S.J. et al. CGG repeats trigger translational frameshifts that generate aggregation-prone chimeric proteins. Nucleic Acids Res. 2022; 50:8674–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kuroha K., Zinoviev A., Hellen C.U.T., Pestova T.V. Release of ubiquitinated and non-ubiquitinated nascent chains from stalled mammalian ribosomal complexes by ANKZF1 and Ptrh1. Mol. Cell. 2018; 72:286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Inada T. tRNA recycling on stalled ribosomes. Nat. Struct. Mol. Biol. 2019; 26:340–342. [DOI] [PubMed] [Google Scholar]
- 136. Yip M.C.J., Keszei A.F.A., Feng Q., Chu V., McKenna M.J., Shao S. Mechanism for recycling tRNAs on stalled ribosomes. Nat. Struct. Mol. Biol. 2019; 26:343–349. [DOI] [PubMed] [Google Scholar]
- 137. Izawa T., Park S.H., Zhao L., Hartl F.U., Neupert W. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell. 2017; 171:890–903. [DOI] [PubMed] [Google Scholar]
- 138. Oh S.-Y., He F., Krans A., Frazer M., Taylor J.P., Paulson H.L., Todd P.K. RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Hum. Mol. Genet. 2015; 24:4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Latallo M.J., Wang S., Dong D., Nelson B., Livingston N.M., Wu R., Zhao N., Stasevich T.J., Bassik M.C., Sun S. et al. Single-molecule imaging reveals distinct elongation and frameshifting dynamics between frames of expanded RNA repeats in C9ORF72-ALS/FTD. Nat. Commun. 2023; 14:5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Goodman L.D., Prudencio M., Srinivasan A.R., Rifai O.M., Lee V.M.-Y., Petrucelli L., Bonini N.M. EIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. Acta Neuropathol. Commun. 2019; 7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kearse M.G., Goldman D.H., Choi J., Nwaezeapu C., Liang D., Green K.M., Goldstrohm A.C., Todd P.K., Green R., Wilusz J.E. Ribosome queuing enables non-AUG translation to be resistant to multiple protein synthesis inhibitors. Genes Dev. 2019; 33:871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data points are included in the main figures or supplementary data. Raw data used for figure generation are available upon request.