Abstract
Cerebral amyloid angiopathy (CAA) is associated with the accumulation of fibrillar Aβ peptides upon and within the cerebral vasculature, which leads to loss of vascular integrity and contributes to disease progression in Alzheimer’s disease (AD). We investigate the structure of human-derived Aβ40 fibrils obtained from patients diagnosed with sporadic or familial Dutch-type (E22Q) CAA. Using cryo-EM, two primary structures are identified containing elements that have not been observed in in vitro Aβ40 fibril structures. One population has an ordered N-terminal fold comprised of two β-strands stabilized by electrostatic interactions involving D1, E22, D23 and K28. This charged cluster is disrupted in the second population, which exhibits a disordered N-terminus and is favored in fibrils derived from the familial Dutch-type CAA patient. These results illustrate differences between human-derived CAA and AD fibrils, and how familial CAA mutations can guide fibril formation.
Keywords: Cerebral amyloid angiopathy, Alzheimer’s disease, Amyloid-β peptide, Vascular fibrils, Cryo-EM, Solid-state NMR
1. Introduction
Cerebral vascular pathologies including cerebral amyloid angiopathy (CAA) are a major contributor to the progression of neurodegenerative diseases, occurring in 80 % of Alzheimer’s Disease (AD) patients (Spina et al., 2021; Toledo et al., 2013). Despite its prevalence, CAA remains largely untreatable and is often overlooked (Sweeney et al., 2019). The hallmark clinical presentations of CAA include vascular cognitive impairment and dementia, recurring intracerebral hemorrhage and stroke (Reijmer et al., 2016). Vascular deposition of the amyloid-β (Aβ) peptide upon and within the cerebral vasculature is the pathological trigger of CAA. Although often considered concomitantly as both share Aβ deposition as a hallmark, AD and CAA are distinct diseases that can occur independently (Greenberg et al., 2020).
Aβ peptides originate from non-specific proteolytic cleavage of C99, the β-C-terminal fragment of the amyloid precursor protein (APP) (Wolfe et al., 1999). The cleavage event results in peptides of multiple lengths, with the major product being Aβ40 (Rummer and Heneka, 2014; Seino et al., 2021), the principal isoform in vascular deposits (Suzuki et al., 1994). The earliest familial form of CAA identified was linked to a Dutch family (Van Duinen et al., 1987) and originates from a glutamate to glutamine substitution at position 22 of the Aβ peptide (Van Broeckhoven et al., 1990). The resulting pathology, familial CAA Dutch (fCAA-Dutch), is an autosomal dominant form of vascular amyloidosis that is accompanied by accelerated cognitive impairment caused by extensive vascular amyloid deposition (Natté et al., 2001). fCAA-Dutch patients also spontaneously develop intracerebral hemorrhaging with the first stroke occurring on average at 25 years old (Van Duinen et al., 1987). Pathologically, the disorder is characterized by CAA type-2 Aβ deposition in cortical and leptomeningeal small arteries and arterioles with the absence of parenchymal cored amyloid plaques or neurofibrillary tangles that are key features of AD (Natté et al., 2001; Van Duinen et al., 1987).
The Aβ peptide is highly polymorphic, adopting a wide range of fibril conformations when incubated in solution (Tycko, 2014). However, it is unclear whether the structural variability observed in vitro reflects the presence of polymorphism in vivo (Cendrowska et al., 2020). The extent of structural variability of Aβ within a single patient remains debated. Some studies suggest one or two predominant structures exist per individual (Condello et al., 2018; Lu et al., 2013; Qiang et al., 2017), while others find variability within a patient (Rasmussen et al., 2017), or within amyloid plaques (Liu et al., 2016). Morphological differences between AD-specific parenchymal plaques and CAA-specific vascular deposits have been identified (Han et al., 2011; Rutgers et al., 2011; Schrag et al., 2011), suggesting a structural origin to the difference between the two pathologies. However, a robust link between structure and disease in the case of CAA has not been established, and the possibility of a structural origin to the variations in presentation, both between CAA subtypes and compared to AD, remains to be elucidated.
Recent studies have demonstrated structural differences between amyloidogenic proteins incubated in solution and those extracted from human patients (Schweighauser et al., 2020), including Aβ (Ghosh et al., 2021b; Kollmer et al., 2019; Wickramasinghe et al., 2021; Yang et al., 2022), suggesting that the cellular environment may influence fibril formation. The differences can arise in the shape or fold of the fibrils or in more subtle differences involving specific electrostatic or side chain packing interactions. For example, the in vitro structures of Aβ40 and Aβ42 reveal that electrostatic interactions of Lys28 may play a large role in fibril formation. In Aβ40, Lys28 interacts with Asp23, an interaction that allows the hydrophobic C-terminus to bend around and interact with the hydrophobic LVFF sequence. One interesting exception is the structure of the Osaka mutant (Schütz et al., 2015), which lacks E22. In this case, Lys28 interacts with Asp1 at the N-terminus of a neighboring protofilament within the fibril, yielding a very different fibril fold. In brain-derived Aβ40 fibrils, the D23-K28 interaction is generally preserved, but with differences in packing of the N-terminus or C-terminus within the fibril (see below). In Aβ42 with two extra C-terminal amino acids, Lys28 is oriented outward from the fibril center in in vitro fibrils and interacts with the C-terminal carboxyl group. This interaction induces a third β-strand in the overall fibril fold. In recent cryo-EM structures of brain-derived Aβ42 (Yang et al., 2022), the third β-strand is observed, Lys28 is oriented away from the fibril center, but the packing interactions within the fibril and between protofibrils are different than observed in in vitro structures.
Studies using electron cryo-microscopy (cryo-EM) (Ghosh et al., 2021a; Kollmer et al., 2019; Yang et al., 2022) and solid-state nuclear magnetic resonance (NMR) spectroscopy (Ghosh et al., 2021b; Lu et al., 2013) have investigated ex vivo Aβ fibril structures. Kollmer et al. (2019) (Kollmer et al., 2019) targeted vascular amyloid from cerebral meninges. They observed structural variability but were able to obtain a 4.4 A structure of the predominant fibril population. The resulting structure revealed an ordered N-terminus and intermolecular hydrophobic packing, differing from structures of in vitro Aβ40 fibrils. The structure had some elements in common with fibrils seeded from parenchymal brain tissue using Aβ40 by Ghosh and colleagues (Ghosh et al., 2021a), although the latter exhibited a disordered N-terminal segment. It is not clear whether these structures are specific to the individuals studied or characteristic of their respective disease states.
In this work, we compare the structures of Aβ40 fibrils seeded from vascular amyloid deposits isolated from sporadic CAA (sCAA) and fCAA-Dutch patients. We have recently shown that laser capture microdissection can be used to selectively isolate vascular amyloid deposits from brain slices (Irizarry et al., 2021). We use cryo-EM to determine fibril structures and identify structural elements unique to brain vascular-derived fibrils and use solid-state NMR spectroscopy to confirm the presence of these structural features. We find two structures prominently populate the patient-derived samples. Both structures exhibit a similar fold of the fibril core, characterized by an electrostatic cluster composed of residues D1-E22-D23-K28. Importantly, the highly ordered N-terminal segment composed of two β-strands in one population is unique to ex vivo fibrils. A structured N-terminus is generally not observed in fibrils formed in vitro. However, the N-terminus of C99 is folded into a two-stranded β-hairpin prior to cleavage by γ-secretase. We show here that K28 stabilizes the structured N-terminus in both the γ-secretase substrate and brain-derived fibrils, suggesting a role of the electrostatic cluster in APP proteolytic processing and in fibril formation in the human brain. The comparison of the fibril structures from sCAA and fCAA-Dutch patients highlights the importance of the D1-E22-D23-K28 electrostatic cluster in fibril formation and in disease progression in CAA and AD.
2. Materials and methods
2.1. Synthetic Aβ40
Wild-type, mutant, and isotope-edited Aβ peptides were synthesized using tBOC-chemistry ERI-Amyloid (Waterbury, CT) and purified by high-performance liquid chromatography using linear water-acetonitrile gradients containing 0.1 % (v/v) trifluoroacetic acid. The mass of the purified peptide was measured using matrix-assisted laser desorption or electrospray ionization mass spectrometry and was consistent with the calculated mass for the peptide. On the basis of analytical reverse-phase high-performance liquid chromatography and mass spectrometry, the purity of the peptides was 95–99 %.
2.2. Isolation and templated growth of cerebral vascular amyloid deposits from human brain
Tissues used in these studies were from a 71-year-old male sCAA patient (University of California Irvine Alzheimer’s Disease Research Center) and a 76-year-old fCAA-Dutch patient (Leiden University Medical Center, Leiden, NL). The samples used here were prepared as described in Irizarry et al. (2021). Briefly, for each case, individual amyloid-containing cerebral vessels were identified, excised, and captured using a LMD6 laser capture microdissection microscope LMD6 (Leica Microsystems). The dissected vascular amyloid deposits were collected into sodium phosphate buffer, pH 7.2. Three rounds of templated growth were then performed using synthetic wild-type Aβ (1–40) at 15 % seed content in sodium phosphate buffer.
2.3. Fourier-transformed infrared spectroscopy
FTIR measurements were made with a Bruker Vertex 70v spectrometer with a room temperature detector and attenuated total reflectance (ATR) accessory. Samples were layered on a 2 mm germanium ATR plate (Pike Technologies) by drying 100 μL of peptide sample on the Ge surface with a stream of air. The spectral resolution was 4 cm−1.
The C55 peptides were co-solubilized in DMPC, DMPG, and octyl-β-glucoside in hexafluoroisopropanol. The molar ratio of peptide:lipid was 1:60 and the molar ratio of DMPC:DMPG was 10:3. The solution was incubated overnight at 37 °C, after which the solvent was removed under a stream of argon gas. The dried mixture was rehydrated in HEPES buffer (10 mM HEPES, 50 mM NaCl, pH 7.0) gently mixed at 37 °C for 6 h. The octyl-β-glucoside (2 % w/v) was removed by dialysis using Spectra-Por dialysis tubing with a 3500 MW cutoff.
2.4. Nuclear magnetic resonance spectroscopy
Room temperature solid-state magic angle spinning (MAS) NMR experiments were performed at a 13C frequency of 125 MHz on a Bruker AVANCE spectrometer using 4 mm MAS probes. The MAS spinning rate was set to either 9 or 12 kHz to prevent rotational sidebands from covering target cross-peaks. Ramped amplitude cross polarization was used with a contact time of 2 ms. The 13C field strength was 54.4 kHz and ramped 1H field was centered at approximately 50 kHz. Two-pulse phase-modulated decoupling was used during the evolution and acquisition periods with a radiofrequency field strength of 82.7 kHz. Inter-nuclear 13C-13C distance constraints were obtained from 2D dipolar assisted rotational resonance (DARR) NMR experiments using a mixing time of 600 ms. Each data set contained 64 t1 increments and 1024 complex t2 points with spectral widths of 27.7 kHz in both dimensions. 512 scans were averaged per t1 increment. All 13C solid-state MAS NMR spectra were externally referenced to the 13C resonance of neat TMS at 0 ppm at room temperature. Using TMS as the external reference, we calibrated the carbonyl resonance of solid glycine at 176.46 ppm. The chemical shift difference between 13C of DSS in D2O relative to neat TMS is 2.01 ppm.
Samples were prepared via templated growth as described in Irizarry et al. (2021) using 13C-labeled peptides. The sample used as the template was the one used for cryo-EM acquisition, except for the NMR experiments using the ring–13C F19, 2-13C G33, U-13C L34, 5-13C M35 labeled peptide for the CAA patient (shown in Fig. 4), which used generation 5 fibrils as the template.
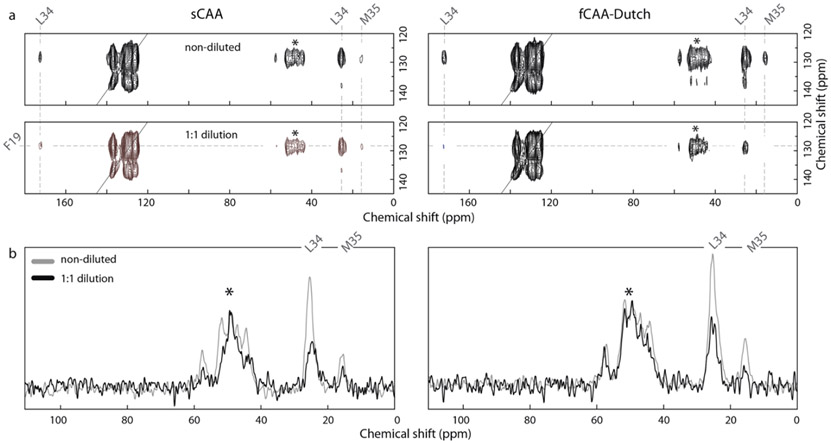
Fig. 4.
Probing brain-derived Aβ40 fibrils for population B with solid-state NMR. Two-dimensional 13C DARR NMR measurements of fibrils derived from sCAA and fCAA patients to probe the contact between F19 and L34. The population B fibril structure is distinct from most in vitro fibrils by the linear shape of the inner Aβ peptides. This structure results in close proximity of F19 and L34 at the intermolecular interface. (a) Representative 2D 13C DARR NMR spectra are shown for the sCAA and fCAA-Dutch patient samples after the third round of seeding (G3). (top) Region of the 2D NMR spectrum showing the diagonal resonance of ring-13C–Phe19 and crosspeaks to U-13C–Leu34 reflecting close Phe19-Leu34 packing in both sCAA and fCAA-Dutch fibrils. (bottom) 1:1 isotope dilution indicate a marked intensity decrease of the ring-13C–Phel9 – U-13C–Leu34 crosspeaks in both patient samples. (b) Row through the diagonal ring-13C–Phe19 resonance showing cross peaks to Leu34. In order to assess whether the contacts are intra- or inter-molecular a parallel experiment was undertaken with 13C-labeled Aβ peptide diluted 50% with unlabeled Aβ peptide. Both samples exhibit a reduction in cross peak intensity consistent with inter-molecular interactions. Asterisks indicate MAS rotational side-bands.
2.5. Thioflavin T fluorescence spectroscopy
Fluorescence measurements were taken using a Spectra Max iD3 spectrometer (Molecular Devices). A final concentration of 37.5 μM thioflavin-T was used with an excitation wavelength of 440 nm and an emission wavelength of 490 nm in a 96-well clear Greiner microplate. Measurements were taken every 10 min for 80 h with 2 s of low orbital shaking in-between reads. The OD setting was set to 1. Negative blank wells and thioflavin T controls were included. All conditions were replicated three times, and the results averaged with standard deviation used to assess error. The overall experiments were repeated twice to test reproducibility; we show only one iteration.
The fluorescence kinetic experiments used samples prepared via templated growth as described in Irizarry et al. (2021) but using a 10 % seed content to enhance potential differences in efficiency.
2.6. Negative stain transmission electron microscopy
Samples were diluted, deposited onto freshly glow-discharged carbon-coated copper 400 mesh grids (Electron Microscopy Sciences). Excess sample and buffer were removed by wicking with a Whatman 1 filter paper and immediately followed by negative staining with 2 % (w/v) freshly filtered uranyl formate solution and air dried. The samples were imaged on a FEI Tecnai 12 BioTwin 80 kV transmission electron microscope with micrographs captured with an Advanced Microscopy Techniques camera at nominal magnification of 250,000×, corresponding to 2 Å/pixel.
2.7. Cryo-electron microscopy
Following two-fold dilution in deionized water, 3.5 μL of sample were applied on a glow-discharged holey gold grid (UltrAuFoil Au R1.2/1.3, 300 mesh, Electron Microscopy Sciences), blotted for 3 sec, at 4 °C and 100 % relative humidity, followed by plunge freezing in liquid ethane (at −186 °C) using an FEI Vitrobot Mark IV robotic plunge freezing device.
Data acquisition for cryo-EM was performed on a 200 kV Talos Arctica (FEI, Thermo Fisher Scientific) transmission electron microscope equipped with a Falcon 3EC direct electron detector (FEI, Thermo Fisher Scientific). Imaging for both sCAA and fCAA-Dutch samples were performed as movies (100 fractions) per micrograph in electron counting mode at a nominal magnification of 92,000×, corresponding to a detector pixel size of 1.12 Å/pixel. Micrographs for sCAA samples were collected with under focus values varying between 0.8 and 1.4 μm and for the fCAA-Dutch samples between 0.6 and 1.4 μm. The total exposure dose for each micrograph for sCAA and fCAA-Dutch samples were 49.7 e−/Å2 and 56.92 e−/Å2 respectively. The parameters for cryo-EM acquisition and helical reconstruction can be found in Table 1 (Supporting Information) and the atomic model validation statistics can be found in Table 2 (Supporting Information).
2.8. Morphology assay
Motion-corrected micrographs (as detailed below) were converted to JPEG format using the mrc2tif program from the IMOD package (Kremer et al., 1996). A Gaussian blur was applied, and the contrast was enhanced for better visualization using ImageJ software. For all fibrils clearly distinguishable, i.e., unobstructed by intra-fibrillar interactions, both the width and cross-over distances at multiple points along each fibril were measured. Using an in-home written MATLAB script, average width and cross-over distance per fibril was calculated and plotted. The script can be found on GitHub (https://github.com/) under elliotcrooks/fibril.git.
3. Results
3.1. Extraction and isolation of Aβ fibrils from patients
Vascular deposits containing Aβ fibrils were extracted from the cerebral vasculature of two patients, diagnosed with sCAA or fCAA-Dutch. In contrast with other studies, we used laser capture microdissection to specifically target vascular amyloid (Irizarry et al., 2021). To further purify the samples and increase the length and number of Aβ fibrils, we employed templated fibril growth using wild-type Aβ40, which has been shown to propagate fibril conformation (Ghosh et al., 2018; Irizarry et al., 2021). Templated growth was verified using FTIR spectroscopy (Fig. S1) as described previously (Irizarry et al., 2021).
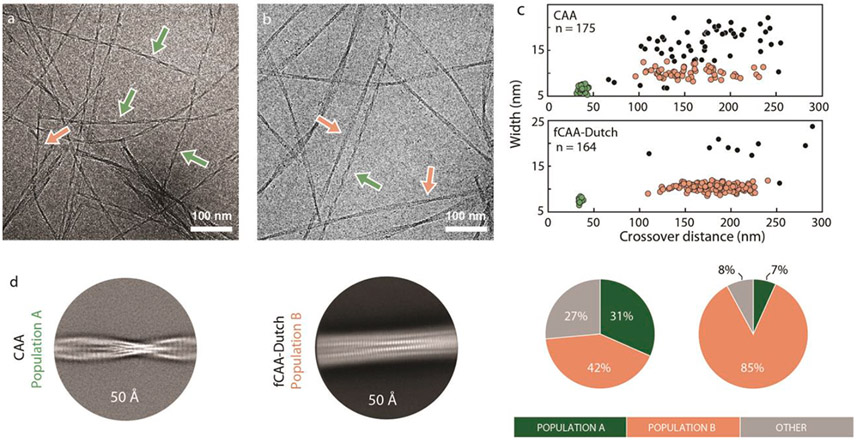
Morphology analyses were first performed on the cryo-EM datasets to characterize fibril homogeneity (Fig. 1 and Fig. S2). The Aβ peptide can exhibit extensive conformational variability that can hinder helical reconstruction in cryo-EM, and prior knowledge of the range of helical characteristics facilitates the separation of distinct fibril polymorphs. Morphology analyses revealed two primary fibril populations. The first population of fibrils (population A) exhibited narrow widths of ≈6.4 ± 0.9 nm and short crossover distances of ≈37 ± 3 nm. The second population of fibrils (population B) exhibited widths of ≈1O nm and a highly variable crossover distance averaging around 175 nm.
Fig. 1.
Two populations of Aβ fibrils in sporadic and familial Dutch vascular amyloid. Representative cryo-EM micrographs from Aβ fibrils derived from the (a) sCAA and (b) fCAA-Dutch patients. Arrows highlight examples of different fibril populations. (c) Morphology plots for each patient along with the distributions of each population within the cryo-EM micrographs. Each dot represents the average crossover-distance and width of a single fibril with colors corresponding to the arrows and different populations in panel (a). Two primary fibril clusters are observed: a highly twisted homogeneous population A (green), abundant in the sCAA case, and a more heterogeneous and less-twisted population B (red), predominant in the fCAA-Dutch case. We note that the second population primarily exhibits variability in crossover distance but not in width, suggesting that the number of peptides in each cross-β unit is maintained along the fibril but the conformation may vary. The highly twisted fibril populations in both cases resulted in cryo-EM volumes with similar backbone traces. (d) 2D class averages from helical reconstruction reflect the difference in fibril morphology found in samples derived from sCAA and fCAA patients. (left) The highly twisted fibrils abundant in the sample seeded from the sCAA patient (in green in panel c) – named population A – have an average crossover distance of ≈37 nm. (right) The major fibril morphology in the sample seeded from the fCAA patient (red in panel c) exhibit a greater and more variable crossover distance.
The highly twisted fibril morphology was previously observed in Aβ fibrils purified from the leptomeningeal vasculature of a CAA patient (Kollmer et al., 2019), and has been detected in vitro using gold nanoparticles (Cendrowska et al., 2020). The widths and longer crossover distances of population B, however, are not unique, being similar to most in vitro Aβ fibrils as well as fibrils seeded off parenchymal plaques of AD patients (Ghosh et al., 2021a). Based on the morphology analyses, the highly twisted population Aβ fibrils were found to be abundant in the sCAA patient, while population B was the predominant fibril morphology in the fCAA-Dutch patient sample (Fig. S2). Both populations were targeted for helical reconstruction. It is important to note that the morphologies present in populations A and B are observed when we seed to further generations (Irizarry et al., 2021) suggesting that the elongation rates of these two fibril forms are similar.
Parallel seeding experiments in the presence of Aβ40-Dutch monomer was also undertaken using seeds of sCAA and fCAA-Dutch fibrils to template fibril growth. The Aβ40-Dutch monomer has a higher rate of fibril formation compared to the Aβ40-WT monomer, and consequently has a higher propensity to self-nucleate. However, as with Aβ40-WT, self-nucleation can be minimized at 22–25 °C (Rajpoot et al., 2022).3
For sCAA seeds used to template fibril growth with the Aβ40-Dutch monomer, we replicate the morphology plots of the sCAA-case using Aβ40-WT monomer (Fig. S3). Both populations A and B are observed with a heterogeneous distribution of population B. When we used fCAA seeds to template fibril growth with the Aβ40-Dutch monomer, we replicated the morphology plots of the fCAA case using Aβ40-WT monomer (Fig. S3). In this case, there is a shift of population A to population B and the distribution of population B is more homogeneous. These observations indicate that both the Aβ40-WT and Aβ40-Dutch monomers replicate the existing structures in the fibril seeds. The lack of glutamate at position 22 in the E22Q mutant that is present in the fCAA-Dutch seeds appears to favor the population B structure. In the context of fCAA Dutch mutation, affected individuals possess one copy of the APP gene with a mutation and one copy with the normal sequence. The similar morphology plots using either the Aβ40-WT monomer or the Aβ40-Dutch monomer indicate that both Aβ peptides add to existing fibrils in vascular amyloid.
3.2. Structure determination
We used cryo-EM to determine the structure of vascular amyloid fibrils seeded from both patients. From the two datasets, we obtained three cryo-EM maps (Figs. S4-S6). The population A structure was solved from data of the sCAA patient to a resolution of 2.9 Å (Fig. 2a and Fig. S4). The population B structure was determined from data of the fCAA-Dutch patient to a resolution of 3.1 Å (Fig. 2b and Fig. S6). The high degree of morphological heterogeneity and low particle number of population B in the sCAA patient sample precluded successful helical reconstruction. The low number of fibrils corresponding to population A in the fCAA-Dutch patient sample also hindered high-resolution reconstruction. However, using the population A structure as an initial model, we found that the same structure is likely present in the fibril pool from the fCAA-Dutch patient (Fig. S5).
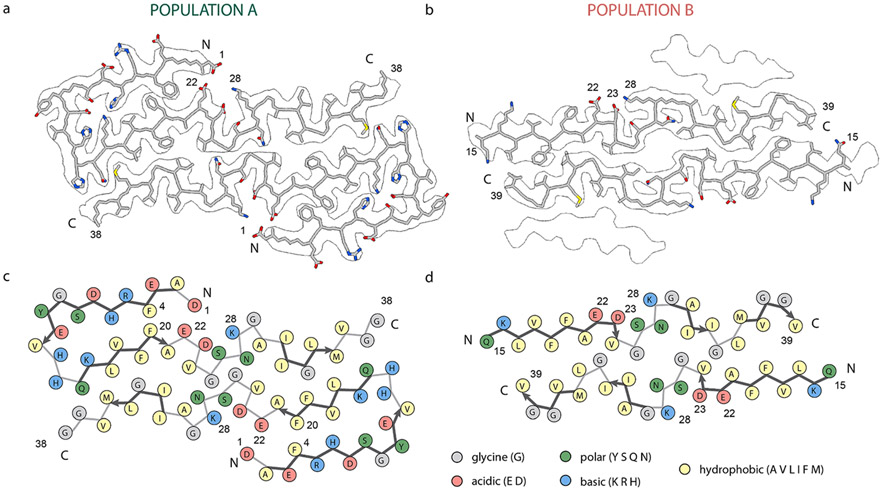
Fig. 2.
Ciyo-EM Aβ40 structures from vascular amyloid of CAA patients. (a) Structure of population A fibrils. Nearly the entire Aβ40 monomer fits within the 2.9 Å cryo-EM density with well-defined positions for uncharged side chains. Population A is composed of two protofilaments with an ordered N-terminal section and intermolecular hydrophobic interactions, with a β-strand from residues Q15 to A21. Reconstruction did not result in defined densities for the last two amino acids, V39 and V40. (b) Structure of population B fibrils within the outline of the cryo-EM density. Population B is composed of four layers. The 3.1 Å cryo-EM density identified a backbone fold with three β-strands composing each protofilament. The N-terminal region was not modeled due to a lack of resolvable density. (c) Schematic representation of population A with each residue colored according to its electrostatic property, as indicated in the legend. The two hydrophobic segments L17–A21 and A30–V36 interact in both structures but in an arrangement not found in many in vitro structures, suggesting a different mechanism of fibril formation. Multiple charged residue pairs are present in population A, both solvent-exposed and inside the fibril core. (d) Schematic representation of population B with each residue colored according to its electrostatic property as indicated in the legend. Similar to population A, hydrophobic segments L17–A21 and A30–V36 form intermolecular interactions. In this structure, charged residues do not form numerous interactions to stabilize the conformation.
The structure of population A is comprised of two cross β-units mediated by intermolecular hydrophobic interactions (Fig. 2a). In contrast with in vitro fibrils, the N-terminus has an ordered fold comprised of two β-strands mediated via several electrostatic interactions. The backbone fold of structure A is reminiscent of the conformation proposed by Kollmer et al. (2019) and Yang et al. (2023b) (Fig. S7). Our model, however, differs in the form of a one-register shift from the structure proposed by Kollmer et al., which we validate below.
The structure of population B fibrils is also formed from two cross β-units in the inner layers, but differs from population A in that the N-terminal segment remains disordered such that no defined density is visible prior to residue 14 (Fig. 2c). As in population A, the two hydrophobic segments interact in an inter-molecular fashion between protofilaments, distinguishing these structures from in vitro fibrils (Fig. 2d). We also found this population to exhibit a similar conformation to parenchymal-seeded Aβ40 fibrils determined by Ghosh et al. (Ghosh et al., 2021a) from an AD patient (Fig. S7).
3.3. Solid-state NMR spectroscopy as a probe of fibril structure and polymorphism
The structures of populations A and B were found to contain several elements typically not observed in in vitro fibrils, arguing that templated growth was able to capture aspects of fibril forms present in vascular deposits. Our structures also exhibit similarities and differences with Aβ fibril models previously developed from CAA and AD brain tissue. We used solid-state NMR spectroscopy to independently probe for specific structural features and assess polymorphism within the entire fibril population. We made use of several 13C labeled peptides, the first incorporating 13C labels at 1–13C F4, 2–13C L17, ring–13C F19, ring–13C F20, 2–13C G33, 5–13C M35, and 1–13C G38 (Figs. S8 and S9).
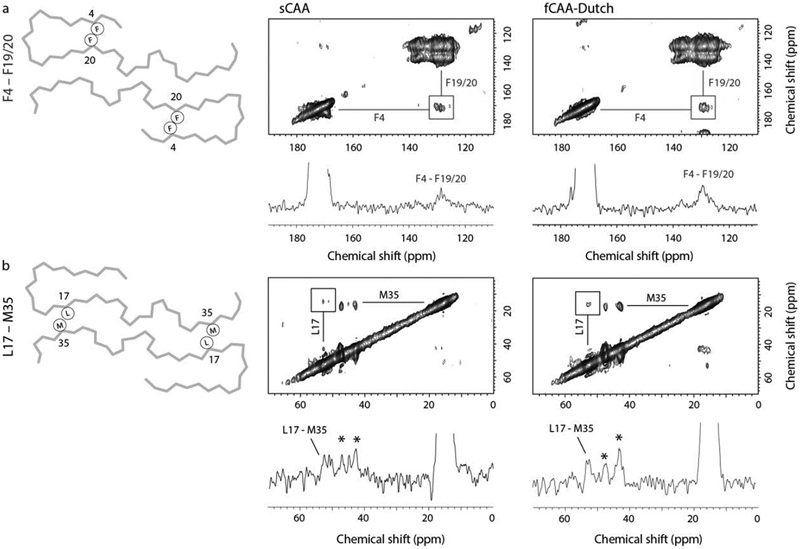
We first investigated the unique two-stranded β-sheet fold of the N-terminus in population A by probing for a close F4-F20 contact (Fig. 3a), which is absent from in vitro fibrillar Aβ structures as well as published models of Aβ40 fibrils seeded from parenchymal amyloid (Ghosh et al., 2021a; Lu et al., 2013) but present in ex vivo CAA fibrils (Kollmer et al., 2019). Evaluation of the resulting DARR NMR spectra reveals the presence of an F4-F20 cross-peak in samples from both the sCAA and the fCAA-Dutch individuals where population A fibrils are present, thus supporting the existence of the N-terminal fold as determined via cryo-EM. The high resolution from this population also indicated a clear register shift starting at position 26 when compared to the structure proposed by Kollmer et al. (2019), resulting in a 180° flip of the C-terminal segment. We probed the L17-M35 contact specific to our model and observe a cross-peak (Fig. 3b) in both brain-derived samples, supporting the register shift and therefore the change in C-terminal orientation.
Fig. 3.
Probing brain-derived Aβ40 fibrils for population A with solid-state NMR. Two-dimensional 13C DARR NMR measurements of fibrils derived from sCAA and fCAA-Dutch patients. The fibrils were produced via templated growth of brain amyloid using specifically 13C-labeled Aβ40 to probe inter-residue contacts modeled in the cryo-EM structures. (a) F4-F20 interaction. A scheme of the backbone trace of population A (left) highlighting the close intramolecular contact of F4 and F20. Region of the 2D DARR NMR spectrum (right) exhibiting cross peaks between 1-13C F4 and ring-13C-F20 (or F19). The 1D row extracted from the 2D spectrum passes through the diagonal resonance of 1-13C F4 and highlights the cross peak between 1-13C F4 and ring-13C-F20 (or F19). The 13C labeling scheme chosen for these measurements generally provides well-resolved resonances such that cross-peaks can be assigned to specific residue contacts. One exception is that the F19 and F20 13C labels overlap but are predicted to interact with either G33 or F4, respectively. The presence of the F4-F19/20 cross peak (square) is consistent with the close distance between F4 and F20 in the cryo-EM structure in both the sCAA and fCAA-Dutch patient samples. We also observe a cross peak between F19 and G33 (not shown), which is consistent with the population A structure. Note that the cross peak intensities in DARR NMR spectra are often not symmetric (see Ohashi and Takegoshi (2006)). (b) L17-M35 interaction. A scheme of the backbone trace of population A (left) highlights the close intermolecular contact of L17 and M35, while the 2D DARR NMR spectrum (right) exhibits cross peaks between 2-13C L17 and 5-13C M35. The 1D row extracted from the 2D spectrum passes through the diagonal resonance of 5-13C M35 and highlights the cross peak between 5-13C M35 and 2-13C L17. Cross peaks (asterisks) are also observed between 5-13C M35 and 2-13C G33, residues that are close together in the Aβ sequence. However, on the basis of the two structures presented here, these cross peaks may arise from the β-hairpins populating the outer densities in population B (see below). The population A structure indicates a register shift with respect to a previously published similar structure, leading to a flip of the C-terminal segment. This shift results in a close proximity between L17 and M35 consistent with the presence of observed cross-peaks in the DARR NMR spectra of the sCAA and fCAA-Dutch patient fibrils.
The structure of population B is identical to fibrils seeded from parenchymal deposits of an AD patient (Ghosh et al., 2021a) in that residues F19 and L34 are in close contact in the intermolecular interface (Fig. 2d). An F19-L34 interaction is also present in many structures of in vitro fibrils, also in an intermolecular fashion due to staggering of the individual monomers in the fibril, but with both hydrophobic residues in the interior of the fibril core. Using a second 13C-labeled Aβ peptide (ring–13C F19, 2-13C G33, U-13C L34, and 5-13C M35), NMR measurements confirm the presence of a close contact between the residues F19 and L34 (Fig 4a). To investigate the intermolecular nature of this contact in structure B, we performed isotope dilution experiments and observe a decreased normalized cross-peak intensity consistent with the structure of population B (Fig 4b).
The outer densities observed in the map of population B mirror findings from Ghosh et al. (2021a) who assigned these to β-hairpins. Although our map of population B suggests similar β-hairpins, the densities are too poorly defined to conclusively place these in this work. However, layering of these individual peptides atop the inner layers suggests a close intermolecular contact between residues F19 and M35, a contact that no other structure predicts. We observed the emergence of a cross-peak between these residues, consistent with β-hairpins populating these outer densities (Fig. S8).
3.4. The D1-E22-D23-K28 electrostatic cluster is a key structural element
The N-terminal structure in population A is unusual for Aβ fibrils. Many of the in vitro Aβ40 fibrils that have generated start with monomeric Aβ40 containing an unstructured N-terminus in solution. However, we have previously shown that the N-terminus adopts a two-stranded β-hairpin structure in C55 in membrane bilayers (Hu et al., 2017). C55 contains the extracellular and transmembrane domains of C99 and releases Aβ upon proteolysis by γ-secretase (Hu et al., 2017). As a result, monomeric Aβ40 originates from membranes and not from solution, suggesting that the starting structure for fibril formation in vivo is different than in vitro. This observation of an ordered N-terminus in membrane-embedded Aβ raises the possibility that the D1-E22-D23-K28 electrostatic cluster is a key element for both C99 and Aβ fibrils.
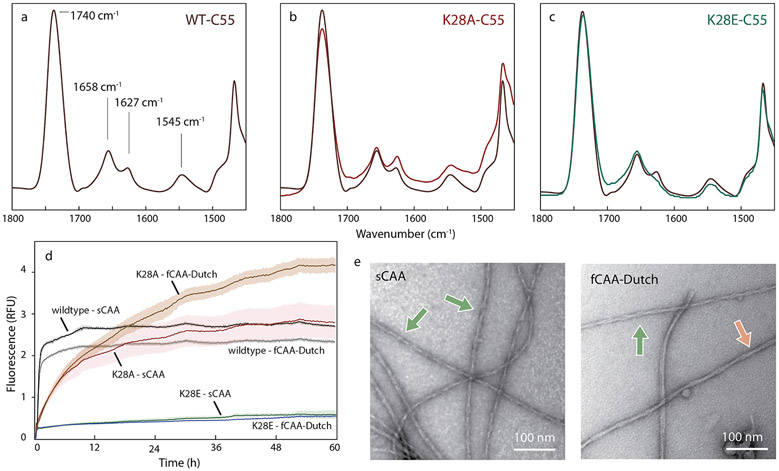
Our previous studies on C55 and C99 indicate that substitutions in the KLVFF and YEV sequences strongly disrupted the N-terminal β-hairpin and resulted in increased Aβ production (Hu et al., 2017). Mutation of E22-D23, however, only slightly influenced the N-terminal β-hairpin structure and subsequent proteolysis (Hu et al., 2017), in contrast with K28 mutations that have been shown to influence γ-secretase processing. Specifically, K28A strongly decreased total Aβ production (Kukar et al., 2011), while K28E increased total Aβ (Petit et al., 2019). To test whether the N-terminal β-hairpin structure in C55 extends to K28, we carried out FTIR measurements on C55 and the two K28 mutants, K28A and K28E. The amide I region in the FTIR spectrum is sensitive to secondary structure and contains two strong vibrational bands in wild-type C55: a 1655 cm−1 band, corresponding to the TM helix, and a 1627 cm−1 band, diagnostic of an N-terminal β-hairpin.
The K28A and K28E substitutions in C55 exhibit distinct behaviors, reflected primarily by changes in the intensity of the 1627 cm−1 band (Fig 5a-c). This band becomes more pronounced in the K28A mutant indicating a stabilized N-terminal β-hairpin, but is lost in K28E-C55, consistent with unraveling the N-terminal structure. These changes argue that the interactions that stabilize the N-terminal β-hairpin in C55 extend to the D1-E22-D23-K28 electrostatic cluster.
Fig. 5.
An electrostatic cluster mediates central interactions for both structures. Probing the D1-E22-D23-K28 cluster in C55 and brain-derived Aβ40 fibrils, (a–c) FTIR spectra of (a) wild-type C55, (b) K28A C55 and (c) K28E C55 reconstituted into DMPC:DMPG bilayers. The acyl chain C=0 vibration of the lipids is observed at ~ 1740 cm−1. The amide I region is between 1600 and 1700 cm−1 and the amide II region is between 1500–1560 cm−1. The band at ~1658 cm−, corresponds to the ™ α-helix, and the band at ~1627–1630 cm−1 corresponds to the N-terminal β-sheet. The K28A mutation increases the ~1630 cm−1 band relative to the 1658 cm−1 band, while the K28E mutation results in a decrease. (d) Templated growth upon G3 brain-derived fibrils was assessed by thioflavin T fluorescence using wild-type Aβ40, and the K28A and K28E mutants. For comparison, positive controls are shown (black and grey traces). The study was performed in triplicate and the results averaged, with noise representing standard deviation. The Aβ40 monomer containing the K28A mutation resulted in template growth for both brain-derived samples, but the initial fluorescence increase is slower than the positive control. The Aβ40 monomer containing the K28E mutation was not able to add to the brain-derived fibrils. (e) Representative negative stain TEM micrographs for the K28A Aβ40 mutant that successfully templated, as shown in (d), upon the sCAA (top) and fCAA-Dutch (bottom) patient samples. We observe fibrils in all cases, with helical characteristics similar to population A (green arrow) or population B (red arrow).
We next investigated whether the K28 mutations influence the ability of the Aβ40 monomers to template off the brain-seeded fibrils. In the population A fibril structure, the positively charged amine of K28 interacts with the negatively charged carboxylate of D23 in the neighboring monomer within the fibril (Fig 6a). The cryo-EM map also reveals a second intermolecular electrostatic interaction between D1 and K28 (Fig. 6a). In this case, the K28 side chain on monomer n interacts with the D1 side chain of monomer n + 2 (Fig. 6b). This stagger may drive fibril elongation by acting as an anchor, locking in the added monomer. In contrast to population A, the N-terminus in the population B structure is not resolved (Fig. 2d). Here, K28 solely interacts with the negative carboxylate of D23 and no longer interacts with D1 (Fig. 6a). The D23-K28 interaction is intra-molecular and consequently the monomeric stagger observed for population A fibrils is absent, suggestive of a different mechanism of fibril elongation (Fig. 6c).
Fig. 6.
D1-E22-D23-K28 cluster in populations A and B. (a) Top view of the D1-E22-D23-K28 cluster in population A (left) and population B (right). In population A, charged residue pairs are brought into close proximity by the N-terminal fold, resulting in electrostatic interactions that stabilize and tether the N-terminus in position. Contrary to population A, the linear conformation of the inner layers in population B does not allow many electrostatic interactions. However, the D23–K28 salt bridge remains and is now able to be fully involved. (b) Ribbon view of population A highlights the out-of-plane displacement of a monomer in red up to 10 Å. (c) (left) Cartoon representation of population A with three individual monomers highlighting interactions between monomers (n), (n + 1), and (n + 2). Residues of interest are highlighted (colors represent different monomers). The N-terminus of monomer (n) (green) is displaced above the monomer plane and interacts with K28 two monomers above (n + 2). Similarly, D23 interacts with K28 on the monomer (n + 1). (right) In contrast, the same residues of the electrostatic cluster are interacting in an intermolecular fashion in population B, leading to the absence of stagger and inter-molecular stabilizing interactions.
We compared thioflavin T fluorescence measurements (Fig. 5d) to assess the ability of monomers of wild-type Aβ40 and the K28Α-Aβ40 and K28E-Aβ40 mutants to undergo templated growth upon the brain-seeded fibrils. Wild-type Aβ40 efficiently polymerizes upon fibril seeds from the sCAA and the fCAA-Dutch patients (Fig. 5d and Fig. S1). In contrast, the efficiency of K28A monomer addition to fibril seeds is reduced. TEM imaging of the K28A fibrils (Fig. 5e) indicate the presence of abundant, highly twisted fibrils, indicating that retaining the NH+3–E22 interaction may be sufficient to induce population A. In contrast to K28A, Aβ40-K28E monomers do not add to the ex vivo fibrils consistent with disruption of the D1-E22-D23-K28 cluster and loss of N-terminal structure.
4. Discussion
We investigated the structure of Aβ40 fibrils seeded from vascular deposits of patients diagnosed with sCAA or (E22Q) fCAA. Vascular amyloid deposits were extracted from brain tissue sections with laser capture microdissection and used for templated fibril growth. Using cryo-EM and NMR spectroscopy, we describe two structures, identified as populations A and B, which are markedly different from in vitro fibril structures. These share a common fibril core but differ in the fold of the N-terminal sequence. The structure of population A contains a well-folded N-terminus that is unique to brain-derived vascular fibrils and has not yet been replicated under solution conditions. We suggest that the structured N-terminus may originate from the two-stranded β-hairpin structure of the N-terminus associated with membrane bilayers in C99, the Aβ precursor. Population B fibrils have a disordered N-terminus and are favored by the familial Dutch mutation. They are also observed in association with AD parenchymal deposits (Ghosh et al., 2021a). We discuss below the potential importance of a structured N-terminus of Aβ40 fibrils and the central role of the D1-E22-D23-K28 electrostatic cluster in guiding fibril formation in the human brain.
4.1. Implications of a structured N-terminus in Aβ40 fibrils
The Aβ40 fibrils comprising population A appeared morphologically homogeneous and allowed cryo-EM helical reconstruction to a resolution of 2.9 Å (Fig. S4). The resulting structure exhibits parallel, inregister β-sheet and a well-folded N-terminal sequence. A similar conformation of the N-terminus was identified by Kollmer et al. (2019) and Yang et al. (2023a) in fibrils isolated from leptomeningeal deposits of a sCAA patient. Coinciding structures in independent studies supports templated growth as a viable means of fibril expansion, with the possibility of utilizing FTIR as an accessible method of assessing the success of this method. In addition, the independent observation of similar N-terminal conformations in separate individuals, despite different extraction and purification methods, argues that the population A fibrils are a common feature of vascular amyloid.
The formation of a two-stranded β-sheet by the hydrophilic N-terminus stands in contrast with in vitro structures of wild-type Aβ40 and Aβ42. C99 also adopts a structured N-terminus prior to cleavage (Hu et al., 2017). In C99 (and C55) an N-terminal β-hairpin interacts with the surface of the lipid bilayer, while the hydrophobic residues downstream of K28 form a membrane-spanning α-helix. Upon cleavage and release of Aβ from γ-secretase the N-terminal two-stranded β-hairpin would need to convert to cros-β-sheet structure en route to population A fibrils and the hydrophobic C-terminus would need to associate with the hydrophobic L17-V18-F19-F20-A21 sequence via inter-molecular interactions as observed in the sCAA and fCAA-Dutch fibrils described here. It will be important to determine if either population A or B can be prepared in vitro by altering the solution environment in a way that favors intramolecular interactions of the N-terminus and inter-molecular interactions of the C-terminus.
Several studies suggest that a structured N-terminus may be important in Aβ40 fibrils in human brain. The N-terminal Aβ(1–28) sequence can form fibrils in solution with a parallel, in-register geometry (Mikros et al., 2001) that are stabilized by electrostatic interactions (Fraser et al., 1991) indicating that deletion of the C-terminal hydrophobic stretch may allow the N-terminus to fold independently. Intrahippocampal injections of Aβ(1–28) into rats resulted in cognitive deficits (Alvarez et al., 1997), emphasizing the importance of the N-terminus in disease progression.
The observation of N-terminal β-sheet secondary structure provides an explanation to a long-standing question: Why are the α-cleavage products Aβ(17–40) and Aβ(17–42) non-amyloidogenic even though they contain both the Leu17–Ala21 and Ala30–Ala42 hydrophobic sequences? Our findings suggest that the N-terminus may be an important component of amyloid deposition and disease progression. Naturally occurring mutations offer evidence that N-terminal structure may influence phenotype. For instance, while the protective A2T mutation (Jonsson et al., 2012) decreases the hydrophobicity of the N-terminus, the A2V mutation (Di Fede et al., 2009) increases the amyloidogenesis and neurotoxicity of the mutated Aβ peptide. The increased hydrophobic character of the N-terminus increases the likelihood of β-sheet formation in the region, as evidenced computationally by reduced disorder and increased β-hairpin population in A2V-Aβ(1–28) (Nguyen et al., 2014). Furthermore, while Aβ42 is extensively modified post-translationally in the N-terminal region, Aβ40 exhibits significantly lower levels of modifications (Brinkmalm et al., 2019), indicative of the importance of the N-terminus in fibril and plaque formation in AD, and possibly in CAA.
Proteomics studies of patient brain extracts provide further evidence of an N-terminal fold in vivo (Brinkmalm et al., 2019). Aβ40-Aβ40 dimers, with D1-E22 as the primary cross-linkage, were found in patients with extensive levels of parenchymal Aβ40. These two residues interact in population A, and the formation of this crosslink may be explained by the co-localization of lysyl oxidase in CAA-affected vessels (Wilhelmus et al., 2013).
4.2. The D1-E22-D23-K28 electrostatic cluster guides fibril assembly
The D1-E22-D23-K28 electrostatic cluster is an element common to both populations A and B. A bent backbone in this region was previously identified both in solution (Paravastu et al., 2008; Qiang et al., 2012) and in some brain-derived fibrils (Ghosh et al., 2021a), with a D23–K28 salt bridge present in many polymorphs (Paravastu et al., 2008; Qiang et al., 2012). In fact, the pioneering work of Tycko and others (Sciarretta et al., 2005) on the influence of D23 and K28 in the fibril formation of Aβ40-WT provides insights into the importance of these residues in nucleating fibril structure. One of the key observations made in this work is that the N-terminal β-hairpin in the C55 substrate is influenced by the mutation of K28. The K28E mutation, which disrupts this structure, results in a dramatic increase in APP processing (Petit et al., 2019), similar to what was previously observed for mutation of the LVFF sequence (Hu et al., 2017). This observation provides additional evidence that the structured N-terminus in C55 and C99 is inhibitory and agrees with the abundance of shorter peptides resulting from α-secretase cleavage compared to the Aβ peptides resulting from β-secretase cleavage.
The D1-E22-D23-K28 electrostatic cluster also provides an explanation as to why the structure of brain-derived Aβ40 fibrils differ from fibrils formed in vitro. Upon fibril formation, there appears to be a competition between the N- and C-termini to complex with the central region of the peptide. This idea has been proposed by Maji et al. (2009), who found that a single D1Y substitution significantly altered Aβ fibril assembly. The observation of a similar D1-E22-D23-K28 cluster in populations A and B suggests that loss of stabilizing D1-K28 and D1-E22 interactions and unraveling of the N-terminal fold in population A may be essential for exposing the electrostatic cluster and allowing binding of the β-hairpin structures in population B.
Our studies on fibrils seeded from a fCAA-Dutch patient suggest that rather than generating new fibril forms, familial mutations may instead alter the ratio of different fibril populations. It should be noted, however, that we used the wild-type Aβ40 monomer to expand the fCAA-Dutch patient sample for cryo-EM structure determination. The condition was chosen because the Aβ40-Dutch mutant peptide rapidly forms fibrils in solution, increasing the likelihood of forming de novo or self-nucleated fibrils. We have found that the wild-type Aβ40 and Aβ40-Dutch monomers are able to cross seed at comparable rates on fCAA-Dutch and sCAA fibrils, respectively. 1 As a result, using only the Aβ40 monomer for addition to fibril seeds eliminates self-nucleated fibrils, which do not form under our conditions for template growth (Irizarry et al., 2021). As discussed below, we might expect a further shift in the population ratios toward population B in the fCAA-Dutch case due to the higher propensity of the Aβ40-Dutch monomer to form β-hairpin structures that associate with the fibril surface in population B fibrils. When utilizing Aβ40-Dutch monomers at a lower temperature to expand tissue extracted from the fCAA-Dutch sample, we note a marked shift from population A to population B (Fig S7) when compared to Aβ40 expanded from sCAA tissue (Fig. 1). The implication, therefore, is that the E22Q substitution promotes a rapid expansion of population B fibrils, potentially more apt to interact with each other and with the cellular micro-environment, accelerating vascular inflammation and damage.
4.3. Origins of fibril polymorphism in vitro and in vivo
The differences between brain-derived and in vitro fibril structures suggest an importance of the cellular environment in fibril formation (Cendrowska et al., 2020). We discussed above the role of the existing N-terminal structure in C99 for guiding fibril formation in a cellular environment. Population A fibrils have well-defined widths and crossover lengths, while population B fibrils exhibit much greater morphological variability. This structural polymorphism may originate from β-hairpins associated with the outer densities as described by Ghosh et al. (2021a), or from the association of metal ions or proteins to the N-terminus in cellular environments. There is a wide range of data supporting the rapid formation of β-hairpin intermediates on the pathway to Aβ fibrils (Sandberg et al., 2010), providing a potential explanation for their inclusion in population B. Monomeric Aβ spontaneously forms β-hairpins (Rajpoot et al., 2022). However, we previously found that the Aβ40-Dutch peptide, and other fCAA mutants at positions E22 and D23, form β-hairpin structures much more rapidly than wild-type Aβ40 (Rajpoot et al., 2022). The peptides may then layer upon pre-formed fibrils having the linear structure of population B. Moreover, the β-hairpins on the fibril surface may be responsible for the formation of secondary nucleation sites as the Dutch (E22Q) mutation, along with other fCAA mutations, increase secondary nucleation in vitro (Törnquist et al., 2018).
The differences in the N-terminal structures of populations A and B may also impact the ability to bind metal ions and other proteins, and result in fibril polymorphism. The N-terminus is generally regarded as a binding site for metal ions, where His6, His13, and His14 are known sites of coordination (Crooks et al., 2020). These residues would only be available for coordinating metal ions in population B, which exhibits an unstructured N-terminus that coats the fibrils in a fuzzy, interactive sheath. Metal binding occurs in both vascular and parenchymal amyloid consistent with population B in both types of amyloids. While this study did not include metal binding experiments, future investigations, incorporating higher resolution structural analyses, could prove pivotal in unveiling the specific binding site of the metal ion. In a similar fashion, an unstructured N-terminus might facilitate the interaction of other known binding partners such as clusterin and apolipoproteins (Hondius et al., 2018). Exploring these interactions in subsequent studies may contribute to a more comprehensive understanding of the molecular mechanisms involved in amyloid formation and its potential implications in neurodegenerative diseases.
4.4. Implications for structure-disease correlations in amyloidopathies
Structure-disease correlations have been identified in other amyloidogenic diseases, such as ALS (Cao et al., 2019) and Parkinson’s disease (Heise et al., 2005), but have not been established in the case of AD or CAA. The possibility that the difference between AD and CAA originates at the structural level is supported by multiple recent findings (Han et al., 2011; Rutgers et al., 2011; Scherpelz et al., 2021; Schrag et al., 2011). The results of this work add to this list; population A fibrils with an N-terminus comprised of two β-strands is unique to CAA. Our findings are supported by prior findings that N-terminal residues in vascular-derived fibrils are more ordered than in parenchyma-derived fibrils (Scherpelz et al., 2021). As such, this study provides a high-resolution structure as a potential pharmacological target to distinguish CAA from AD. Recently, a novel β-hairpin fold of the N-terminus of Aβ has been used to generate antibodies that reduce plaque formation in mouse of AD (Bakrania et al., 2022), suggesting that the cross β-structure in the N-terminus of population A may provide a unique structural signature for antibody development. To date, antibodies developed toward AD appear to target an unstructured N-terminus. For example, the epitopes of aducanumab and gantenerumab target residues 3–7 and 2–9, respectively (Arndt et al., 2018; Bohrmann et al., 2012). These would be exposed in population B fibrils. An unstructured N-terminus is likely recognized by lecanemab, a therapeutic antibody that has recently yielded significant improvements in reducing cognitive decline in a phase three clinical trial, although with significant side effects involving brain edema and hemorrhaging (van Dyck et al., 2023). The observation that population B fibrils are identical to fibrils found in parenchymal amyloid suggests that lecanemab binding to vascular amyloid may result in amyloid disaggregation and disruption of the blood vessel walls leading to the side effects observed during the clinical trials.
Finally, the differences in fibril populations between sCAA and fCAA-Dutch may be associated with the differences in clinical phenotype. The clinical presentation of fCAA-Dutch involves fewer ischemic strokes and more immune-active vascular amyloid deposits than sCAA (Kumar-Singh, 2009; van Broeckhoven et al., 1990; van Duinen et al., 1987). Cryo-EM structures have been determined of the Osaka (Δ22) (Schütz et al., 2015) and Arctic (E22G) (Yang et al., 2023b) mutations. In the Aβ40-Osaka structure (in vitro), the N-terminus bends around and Asp1 interacts with Lys28 of the neighboring peptide. It does not fold back and form a β-hairpin like structure as in population A. Hence, the structure appears to be somewhat between population A and other Aβ40 structures. The fibril structure of the Arctic mutant from human brain is also unusual with Lys28 interacting with Asp23 across the protofilament interface within the fibril (Yang et al., 2023b). Further structural studies of fibrils derived from vascular amyloid, in conjunction with proteomics studies to assess differences in metal and protein interactions, will be valuable in addressing the origins of the different phenotypes that have been described for CAA. In addition, model systems that replicate in vitro the fibril structures observed in human brain will be critical for understanding the mechanism(s) of fibril formation in cellular environments.
Supplementary Material
Acknowledgements
This work was supported through grants from the National Institutes of Health to SOS and WVN (AG-027317 and AG-3061775). Cryo-EM data were collected at the Stony Brook University cryo-EM center, which was established with National Institutes of Health Grant number S10 OD012272. We thank Drs. Sjors Scheres and Takanori Nakane for their useful help and suggestions with cryo-EM data processing, and Drs. Kent Thurber and Robert Tycko for their MATLAB scripts for optimizing particle alignment. SOS is grateful to Dr. Lucia Alvarez-Gutierrez and Matthias Koch for discussions and insights into the importance of the D1-E22-D23-K28 cluster in the processing of C99.
Footnotes
B. Irizarry et al. unpublished work.
CRediT authorship contribution statement
Ziao Fu: Writing – review & editing, Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization. Elliot J. Crooks: Writing – review & editing, Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization. Brandon A. Irizarry: Writing – review & editing, Investigation, Formal analysis, Data curation. Xiaoyue Zhu: Investigation, Data curation. Saikat Chowdhury: Writing – review & editing, Investigation, Formal analysis. William E. Van Nostrand: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization. Steven O. Smith: Writing – review & editing, Supervision, Project administration, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: William E. Van Nostrand, Steven O. Smith reports financial support was provided by National Institutes of Health. Co-author S.C. serves on the editoral board of the Journal of Structural Biology If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsb.2024.108092.
Data availability
Density maps for the sCAA (population A) and fCAA-Dutch (population B) samples were deposited in the EM Data Bank with reference IDs EMD-29036 and EMD-29037, respectively, and corresponding atomic models deposited in the Protein Data Bank (PDB) with reference IDs 8FF2 and 8FF3, respectively. The density map for population A in the fCAA-Dutch sample was deposited in the EM Data Bank with reference ID EMD-29038.
References
- Alvarez X, Miguel-Hidalgo J, Fernandez-Novoa L, Cacabelos R, 1997. Intrahippocampal injections of the beta-amyloid 1–28 fragment induces behavioral deficits in rats. Methods Find. Exp. Clin. Pharmacol 19, 471–479. [PubMed] [Google Scholar]
- Arndt JW, Qian F, Smith BA, Quan C, Kilambi KP, Bush MW, Walz T, Pepinsky RB, Bussière T, Hamann S, Cameron TO, Weinreb PH, 2018. Structural and kinetic basis for the selectivity of aducanumab for aggregated forms of amyloid-β. Sci. Rep 8, 6412. 10.1038/s41598-018-24501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakrania P, Hall G, Bouter Y, Bouter C, Beindorff N, Cowan R, Davies S, Price J, Mpamhanga C, Love E, Matthews D, Carr MD, Bayer TA, 2022. Discovery of a novel pseudo β-hairpin structure of N-truncated amyloid-β for use as a vaccine against Alzheimer’s disease. Mol. Psychiatry 27, 840–848. 10.1038/S41380-021-01385-7. [DOI] [PubMed] [Google Scholar]
- Bohrmann B, Baumann K, Benz J, Gerber F, Huber W, Knoflach F, Messer J, Oroszlan K, Rauchenberger R, Richter WF, Rothe C, Urban M, Bardroff M, Winter M, Nordstedt C, Loetscher H, 2012. Gantenerumab: a novel human anti-Aβ antibody demonstrates sustained cerebral amyloid-β binding and elicits cell-mediated removal of human amyloid-β. J. Alzheimer’s Disease 28, 49–69. 10.3233/JAD-2011-110977. [DOI] [PubMed] [Google Scholar]
- Brinkmalm G, Hong W, Wang Z, Liu W, O’Malley TT, Sun X, Frosch MP, Selkoe DJ, Portelius E, Zetterberg H, 2019. Identification of neurotoxic cross-linked amyloid β dimers in the Alzheimer’s brain. Brain 142, 1441–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Boyer DR, Sawaya MR, Ge P, Eisenberg DS, 2019. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat. Struct. Mol. Biol 26, 619–627. 10.1038/s41594-019-0248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendrowska U, Silva PJ, Ait-Bouziad N, Müller M, Guven ZP, Vieweg S, Chiki A, Radamaker L, Kumar ST, Fändrich M, 2020. Unraveling the complexity of amyloid polymorphism using gold nano particles and cryo-EM. Proc. Natl. Acad. Sci 117, 6866–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello C, Lernmin T, Stöhr J, Nick M, Wu Y, Maxwell AM, Watts JC, Caro CD, Oehler A, Keene CD, Bird TD, van Duinen SG, Lannfelt L, Ingelsson M, Graff C, Giles K, DeGrado WF, Prusiner SB, 2018. Structural heterogeneity and intersubject variability of Aβ in familial and sporadic Alzheimer’s disease. Proc. Natl. Acad. Sci 115, E782–E791. DOI: 10.1073/pnas.l714966115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks EJ, Irizarry BA, Ziliox M, Kawakami T, Victor T, Xu F, Hojo H, Chiu K, Simmerling C, Van Nostrand WE, Smith SO, Miller LM, 2020. Copper stabilizes antiparallel β-sheet fibrils of the amyloid β40 (Aβ40)-Iowa variant. J. Biol. Chem 295, 8914–8927. 10.1074/jbc.RA119.011955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fede G, Catania M, Morbin M, Rossi G, Suardi S, Mazzoleni G, Merlin M, Giovagnoli AR, Prioni S, Erbetta A, 2009. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 323, 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PE, Nguyen JT, Surewicz WK, Kirschner DA, 1991. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys. J 60, 1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh U, Thurber KR, Yau W-M, Tycko R, 2021a. Molecular structure of a prevalent amyloid-β fibril polymorph from Alzheimer’s disease brain tissue. Proc. Natl. Acad. Sci 118, e2023089118. DOI: 10.1073/pnas.2023089118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh U, Yau W-M, Collinge J, Tycko R, 2021b. Structural differences in amyloid-β fibrils from brains of nondemented elderly individuals and Alzheimer’s disease patients. Proc. Natl. Acad. Sci 118, e2111863118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh U, Yau W-M, Tycko R, 2018. Coexisting order and disorder within a common 40-residue amyloid-β fibril structure in Alzheimer’s disease brain tissue. Chem. Commun 54, 5070–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SM, Bacskai BJ, Hernandez-Guillamon M, Pruzin J, Sperling R, van Veluw SJ, 2020. Cerebral amyloid angiopathy and Alzheimer disease—one peptide, two pathways. Nat. Rev. Neurol 16, 30–42. 10.1038/S41582-019-0281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BH, Zhou M, Vellimana AK, Milner E, Kim DH, Greenberg JK, Chu W, Mach RH, Zipfel GJ, 2011. Resorufin analogs preferentially bind cerebrovascular amyloid: potential use as imaging ligands for cerebral amyloid angiopathy. Mol. Neurodegener 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M, 2005. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci 102, 15871–15876. DOI: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondius DC, Eigenhuis KN, Morrema THJ, van der Schors RC, van Nierop P, Bugiani M, Li KW, Hoozemans JJM, Smit AB, Rozemuller AJM, 2018. Proteomics analysis identifies new markers associated with capillary cerebral amyloid angiopathy in Alzheimer’s disease. Acta Neuropathol. Commun 6, 46. 10.1186/s40478-018-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Kienlen-Campard P, Tang T-C, Perrin F, Opsomer R, Decock M, Pan X, Octave J-N, Constantinescu SN, Smith SO, 2017. β-Sheet structure within the extracellular domain of C99 regulates amyloidogenic processing. Sci. Rep 7, 17159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry BA, Davis J, Zhu X, Boon BDC, Rozemuller AJM, Van Nostrand WE, Smith SO, 2021. Human cerebral vascular amyloid contains both antiparallel and parallel in-register Aβ40 fibrils. J. Biol. Chem 297 10.1016/j.jbc.2021.101259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, 2012. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99. [DOI] [PubMed] [Google Scholar]
- Kollmer M, Close W, Funk L, Rasmussen J, Bsoul A, Schierhorn A, Schmidt M, Sigurdson CJ, Jucker M, Fändrich M, 2019. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun 10, 4760. 10.1038/s41467-019-12683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR, 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Kukar TL, Ladd TB, Robertson P, Pintchovski SA, Moore B, Bann MA, Ren Z, Jansen-West K, Malphrus K, Eggert S, 2011. Lysine 624 of the amyloid precursor protein (APP) is a critical determinant of amyloid β peptide length: support for a sequential model of γ-seeretase intramembrane proteolysis and regulation by the amyloid β precursor protein (APP) juxtamembrane region. J. Biol. Chem 286, 39804–39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Singh S., 2009. Hereditary and sporadic forms of Aβ-cerebrovascular amyloidosis and relevant transgenic mouse models. Int. J. Mol. Sci 10, 1872–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnmer MP, Heneka MT, 2014. Truncated and modified amyloid-beta species. Alzheimer’s Res. Ther 6, 28. 10.1186/alzrt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Costantino I, Venugopalan N, Fischetti RF, Hyman BT, Frosch MP, Gomez-Isla T, Makowski L, 2016. Amyloid structure exhibits polymorphism on multiple length scales in human brain tissue. Sci. Rep 6, 33079. 10.1038/srep33079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J-X, Qiang W, Yau W-M, Schwieters CD, Meredith SC, Tycko R, 2013. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154, 1257–1268. 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji SK, Loo RRO, Inayathullah M, Spring SM, Vollers SS, Condron MM, Bitan G, Loo JA, Teplow DB, 2009. Amino acid position-specific contributions to amyloid β-protein oligomerization. J. Biol. Chem 284, 23580–23591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikros E, Benaki D, Humpfer E, Spraul M, Loukas S, Stassinopoulou CI, Pelecanou M, 2001. High-resolution NMR spectroscopy of the β-amyloid (1–28) fibril typical for Alzheimer’s disease. Angew. Chem. Int. Ed 40, 3603–3605. [DOI] [PubMed] [Google Scholar]
- Natté R, Maat-Schieman ML, Haan J, Bornebroek M, Roos RA, Van Duinen SG, 2001. Dementia in hereditary cerebral hemorrhage with amyloidosis-Dutch type is associated with cerebral amyloid angiopathy but is independent of plaques and neurofibrillary tangles. Ann. Neurol 50, 765–772. [DOI] [PubMed] [Google Scholar]
- Nguyen PH, Tarns B, Derreumaux P, 2014. Familial Alzheimer A2 V mutation reduces the intrinsic disorder and completely changes the free energy landscape of the Aβ1–28 monomer. J. Phys. Chem. B 118, 501–510. [DOI] [PubMed] [Google Scholar]
- Ohashi R, Takegoshi K, 2006. Asymmetric 13C–13C polarization transfer under dipolar-assisted rotational resonance in magic angle spinning NMR. J. Chem. Phys 125, 214503. [DOI] [PubMed] [Google Scholar]
- Paravastu AK, Leapman RD, Yau W-M, Tycko R, 2008. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc. Natl. Acad. Sci 105, 18349–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit D, Hitzenberger M, Lismont S, Zoltowska KM, Ryan NS, Mercken M, Bischoff F, Zacharias M, Chávez-Gutiérrez L, 2019. Extracellular interface between APP and Nicastrin regulates Aβ length and response to γ-secretase modulators. EMBO J. 38, e101494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau W-M, Luo Y, Mattson MP, Tycko R, 2012. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc. Natl. Acad. Sci 109, 4443–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau W-M, Lu JX, Collinge J, Tycko R, 2017. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 541, 217–221. 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpoot J, Crooks EJ, Irizarry BA, Amundson A, Van Nostrand WE, Smith SO, 2022. Insights into cerebral amyloid angiopathy type 1 and type 2 from comparisons of the fibrillar assembly and stability of the Aβ40-IOWA and Aβ40-Dutch peptides. Biochemistry 61, 1181–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J, Mahler J, Beschorner N, Kaeser SA, Häsler LM, Baumann F, Nyström S, Portelius E, Blennow K, Lashley T, Fox NC, Sepulveda-Falla D, Glatzel M, Oblak AL, Ghetti B, Nilsson KPR, Hammarström P, Staufenbiel M, Walker LC, Jucker M, 2017. Amyloid polymorphisms constitute distinct clouds of conformational variants in different etiological subtypes of Alzheimer’s disease. Proc. Natl. Acad. Sci 114, 13018–13023. 10.1073/pnas.1713215114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmer YD, van Veluw SJ, Greenberg SM, 2016. Ischemic brain injury in cerebral amyloid angiopathy. J. Cereb. Blood Flow Metab 36, 40–54. 10.1038/jcbfm.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgers KS, van Remoortere A, van Buchem MA, Verrips CT, Greenberg SM, Bacskai BJ, Frosch MP, van Duinen SG, Maat-Schieman ML, van der Maarel SM, 2011. Differential recognition of vascular and parenchymal beta amyloid deposition. Neurobiol. Aging 32, 1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A, Luheshi LM, Söllvander S, Pereira de Barros T, Macao B, Knowles TP, Biverstål H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, 2010. Stabilization of neurotoxic Alzheimer amyloid-β oligomers by protein engineering. Proc. Natl. Acad. Sci 107, 15595–15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherpelz KP, Wang S, Pytel P, Madhurapantula RS, Srivastava AK, Sachleben JR, Orgel J, Ishii Y, Meredith SC, 2021. Atomic-level differences between brain parenchymal- and cerebrovascular-seeded Aβ fibrils. Sci. Rep 11, 247. 10.1038/s41598-020-80042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag M, Crofton A, Zabel M, Jiffry A, Kirsch D, Dickson A, Mao XW, Vinters HV, Domaille DW, Chang CJ, 2011. Effect of cerebral amyloid angiopathy on brain iron, copper, and zinc in Alzheimer’s disease. J. Alzheimer’s Disease 24, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH, 2015. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew. Chem. Int. Ed 54, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighauser M, Shi Y, Tarutani A, Kametani F, Murzin AG, Ghetti B, Matsubara T, Tomita T, Ando T, Hasegawa K, 2020. Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC, 2005. Aβ40-Lactam (D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry 44, 6003–6014. [DOI] [PubMed] [Google Scholar]
- Seino Y, Nakamura T, Harada T, Nakahata N, Kawarabayashi T, Ueda T, Takatama M, Shoji M, 2021. Quantitative measurement of cerebrospinal fluid amyloid-β species by mass spectrometry. J. Alzheimer’s Disease 79, 573–584. 10.3233/JAD-200987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina S, La Joie R, Petersen C, Nolan AL, Cuevas D, Cosme C, Hepker M, Hwang J-H, Miller ZA, Huang EJ, Karydas AM, Grant H, Boxer AL, Gorno-Tempini ML, Rosen HJ, Kramer JH, Miller BL, Seeley WW, Rabinovici GD, Grinberg LT, 2021. Comorbid neuropathological diagnoses in early versus late-onset Alzheimer’s disease. Brain 144, 2186–2198. 10.1093/brain/awab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Iwatsubo T, Odaka A, Ishibashi Y, Kitada C, Ihara Y, 1994. High tissue content of soluble beta 1–40 is linked to cerebral amyloid angiopathy. Am. J. Pathol 145, 452. [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Montagne A, Sagare AP, Nation DA, Schneider LS, Chui HC, Harrington MG, Pa J, Law M, Wang DJ, 2019. Vascular dysfunction—the disregarded partner of Alzheimer’s disease. Alzheimer’s Dementia 15, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Arnold SE, Raible K, Brettschneider J, Xie SX, Grossman M, Monsell SE, Kukull WA, Trojanowski JQ, 2013. Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer’s Coordinating Centre. Brain 136, 2697–2706. 10.1093/brain/awtl88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törnquist M, Michaels TCT, Sanagavarapu K, Yang X, Meisl G, Cohen SIA, Knowles TPJ, Linse S, 2018. Secondary nucleation in amyloid formation. Chem. Commun 54, 8667–8684. 10.1039/C8CC02204F. [DOI] [PubMed] [Google Scholar]
- Tycko R., 2014. Physical and structural basis for polymorphism in amyloid fibrils. Protein Sci. 23, 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Broeckhoven C, Haan J, Bakker E, Hardy J, Van Hul W, Wehnert A, Vegter-Van der Vlis M, Roos R, 1990. Amyloid β protein precursor gene and hereditary cerebral hemorrhage with amyloidosis (Dutch). Science 248, 1120–1122. [DOI] [PubMed] [Google Scholar]
- Van Duinen SG, Castano EM, Prelli F, Bots G, Luyendijk W, Frangione B, 1987. Hereditary cerebral hemorrhage with amyloidosis in patients of Dutch origin is related to Alzheimer disease. Proc. Natl. Acad. Sci 84, 5991–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyck CH, Swanson CJ, Aisen P, Bateman RJ, Chen C, Gee M, Kanekiyo M, Li D, Reyderman L, Cohen S, Froelich L, Katayama S, Sabbagh M, Vellas B, Watson D, Dhadda S, Irizarry M, Kramer LD, Iwatsubo T, 2023. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med 388, 9–21. 10.1056/NEJMoa2212948. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe A, Xiao Y, Kobayashi N, Wang S, Scherpelz KP, Yamazaki T, Meredith SC, Ishii Y, 2021. Sensitivity-enhanced solid-state NMR detection of structural differences and unique polymorphs in pico-to nanomolar amounts of brain-derived and synthetic 42-residue amyloid-β fibrils. J. Am. Chem. Soc 143, 11462–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmus MM, Bol JG, van Duinen SG, Drukarch B, 2013. Extracellular matrix modulator lysyl oxidase colocalizes with amyloid-beta pathology in Alzheimer’s disease and hereditary cerebral hemorrhage with amyloidosis—Dutch type. Exp. Gerontol 48, 109–114. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ, 1999. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517. 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- Yang Y, Arseni D, Zhang W, Huang M, Lövestam S, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Ryskeldi-Falcon B, Scheres SHW, Goedert M, 2022. Cryo-EM structures of amyloid-β 42 filaments from human brains. Science 375, 167–172. 10.1126/science.abm7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Murzin AG, Peak-Chew S, Franco C, Garringer HJ, Newell KL, Ghetti B, Goedert M, Scheres SHW, 2023a. Cryo-EM structures of Aβ40 filaments from the leptomeninges of individuals with Alzheimer’s disease and cerebral amyloid angiopathy. Acta Neuropathol. Commun 11, 191. 10.1186/S40478-023-01694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang W, Murzin AG, Schweighauser M, Huang M, Lövestam S, Peak-Chew SY, Saito T, Saido TC, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordberg A, Goedert M, Scheres SHW, 2023b. Cryo-EM structures of amyloid-β filaments with the Arctic mutation (E22G) from human and mouse brains. Acta Neuropathol. 145, 325–333. 10.1007/s00401-022-02533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Density maps for the sCAA (population A) and fCAA-Dutch (population B) samples were deposited in the EM Data Bank with reference IDs EMD-29036 and EMD-29037, respectively, and corresponding atomic models deposited in the Protein Data Bank (PDB) with reference IDs 8FF2 and 8FF3, respectively. The density map for population A in the fCAA-Dutch sample was deposited in the EM Data Bank with reference ID EMD-29038.