Abstract
The BNLF-1 open reading frame of Epstein-Barr virus (EBV) encodes two related proteins, latent membrane protein-1 (LMP-1) and lytic LMP-1 (lyLMP-1). LMP-1 is a latent protein required for immortalization of human B cells by EBV, whereas lyLMP-1 is expressed during the lytic cycle and is found in the EBV virion. We show here that, in contrast to LMP-1, lyLMP-1 is stable, with a half-life of >20 h in tetradecanoyl phorbol acetate- and butyrate-treated B95-8 cells. Although lyLMP-1 itself has negligible effects on NF-κB activity, it inhibits NF-κB activation by LMP-1 in a dose-dependent manner. The lyLMP-1 protein does not oligomerize with LMP-1, and the negative effect of lyLMP-1 on NF-κB activation by LMP-1 does not result from lyLMP-1-mediated disruption of LMP-1 oligomers. Modulation of LMP-1-activated signaling pathways is the first identified biological activity associated with lyLMP-1, and this activity may contribute to the progression of EBV's lytic cycle.
Epstein-Barr virus (EBV), a ubiquitous human herpesvirus causally associated with several human tumors (23), infects resting B cells and establishes a latent infection resulting in unlimited proliferation. Latent membrane protein-1 (LMP-1) is an essential viral membrane protein required for immortalization by EBV and acts by regulating key cell signaling pathways. Although EBV-infected B cells rarely enter the lytic cycle and release virus (24, 30, 33), certain EBV-positive B-cell lines can be induced to release infectious progeny through treatment with agents such as tetradecanoyl phorbol acetate (TPA) and sodium butyrate (10, 34, 35). Lytic cycle entry results in temporally regulated expression of the majority of the viral genome (∼100 open reading frames [ORFs]) (2, 32).
One late lytic cycle promoter, EDL1A, lies within the LMP-1 gene and drives the expression of a transcript encoding a predicted ORF corresponding to an amino-terminally truncated form of LMP-1 (11). A protein in infected cells of molecular weight predicted by this ORF has been termed lytic LMP-1 (lyLMP-1) because of its expression during EBV's lytic cycle (1, 3, 7, 11, 29). The lyLMP-1 ORF begins at methionine 129 of the LMP-1 sequence and continues through the fifth and sixth transmembrane domains and entire carboxy terminus. lyLMP-1 shares none of LMP-1's known biological or biochemical properties (19, 31), and little is known about lyLMP-1's function in the infected cell. We reported previously that lyLMP-1 is a component of the EBV virion and proposed a function in the initial stages of infection and/or during the lytic cycle (7). Its sequence identity with LMP-1 suggests that lyLMP-1 may interact with LMP-1 itself or with effectors of LMP-1 signaling. We have begun to characterize the biochemical and biological properties of lyLMP-1, with the goal of understanding its role in the virus life cycle, and have tested the hypothesis that lyLMP-1 affects the ability of full-length LMP-1 to activate cell signaling pathways.
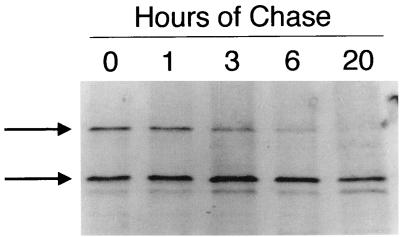
De novo synthesis of lyLMP-1 in TPA- and butyrate-induced B95-8 cells.
Western blot analysis of permissive EBV-positive B-cell lines induced to enter the lytic cycle often reveals a ladder of LMP-1-immunoreactive proteins migrating with lower apparent molecular weights than does LMP-1. Detection of this LMP-1 ladder of bands depends upon the amount of LMP-1 expressed in the cell (reference 7 and unpublished observations). Whether these LMP-1-related proteins are derived from proteolysis of LMP-1 (before or after cell lysis) or from de novo translation initiating at internal methionines in the LMP-1 ORF has been difficult to ascertain (18). The 45-kDa LMP-1-immunoreactive protein, detected in induced cells, migrates with a molecular mass consistent with that reported for the migration of lyLMP-1 (1, 7) as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
To confirm that the 45-kDa lyLMP-1 protein arises from de novo translation of the EDL1A transcript, we asked if a precursor-product relationship existed between LMP-1 and the 45-kDa protein. B95-8 cells were cultured in 20 ng of TPA per ml and 3.5 mM sodium butyrate for 48 h and starved in methionine-free medium for 1 h, and then 1 mCi of Tran [35S]methionine (ICN) per ml was added for an additional 20 min. Following the pulse, cells were grown in RPMI 1640–10% calf serum for up to 20 h. Cell pellets were solubilized in 5× radioimmunoprecipitation assay [RIPA] buffer (100 mM Tris [pH 8.0], 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% deoxycholate) and sonicated, and LMP-1 proteins were immunoprecipitated as described previously (21). Boiled samples were precleared with protein G-agarose beads (Boehringer Mannheim), incubated with polyclonal LMP-1 antiserum, and precipitated with protein G-agarose beads. The LMP-1 antiserum was raised in rabbits against an LMP-1 carboxy terminus–glutathione S-transferase fusion protein. Immunoprecipitates were resolved by SDS-PAGE and analyzed by autoradiography (Fig. 1). If the 45-kDa lyLMP-1 protein arises de novo from its own transcript, then it should be detectable by autoradiography in cells labeled with [35S]methionine for short periods without subsequent chase. [35S]methionine-labeled 45-kDa lyLMP-1 was readily detectable after the 30-min pulse and persisted, with a calculated half-life of at least 20 h (Fig. 1). In contrast to lyLMP-1, 35S-labeled LMP-1 decreased over time and was undetectable 20 h following the pulse. Consistent with its reported rapid turnover in other cells (1, 20, 21), the half-life of LMP-1 in TPA- and butyrate-treated B95-8 cells was between 3 and 6 h. It is unlikely that radiolabeled lyLMP-1 results from LMP-1 degradation since a band comigrating with lyLMP-1 was not seen in 35S-labeled uninduced B95-8 cells, which express primarily LMP-1 (not shown). Also, unlike the relationship between the loss of LMP-1 and the appearance of the ∼40-kDa LMP-1 immunoreactive protein, there was no precursor-product relationship between LMP-1 and the 45-kDa lyLMP-1 protein (Fig. 1). These results provide experimental evidence for lyLMP-1's de novo translation from the EDL1A transcript in induced B95-8 cells. The difference in half-lives of the two LMP-1 proteins suggests distinct turnover mechanisms and is consistent with previous results demonstrating a correlation between LMP-1's biological activity and rapid turnover (21). The long half-life of lyLMP-1 is consistent with our observation that, once carried into the infected cell with the virion, the protein remains detectable for ∼48 h, in the absence of de novo protein synthesis (7). The persistence of lyLMP-1 protein in the cell early after infection may be important for regulation of cellular signaling pathways involved in establishment of immortalization.
FIG. 1.
The half-life of the lyLMP-1 protein in B95-8 cells is >20 h. Forty-eight hours after induction with TPA and butyrate, B95-8 cells were starved in methionine-free medium for 1 h, pulsed with [35S]methionine for 20 min, and chased in RPMI 1640 plus 10% bovine calf serum (R10C) medium for the times indicated. LMP-1 proteins were immunoprecipitated from cell lysates with affinity-purified anti-LMP-1 antiserum raised against LMP-1's carboxy terminus. Immunoprecipitates were resolved by SDS-PAGE and visualized by autoradiography. The hours of chase are the times prior to harvest following a 30-min pulse with [35S]methionine; the upper and lower arrows indicate the migrations of the 62-kDa full-length and 45-kDa lyLMP-1 proteins, respectively. Molecular mass markers are not shown.
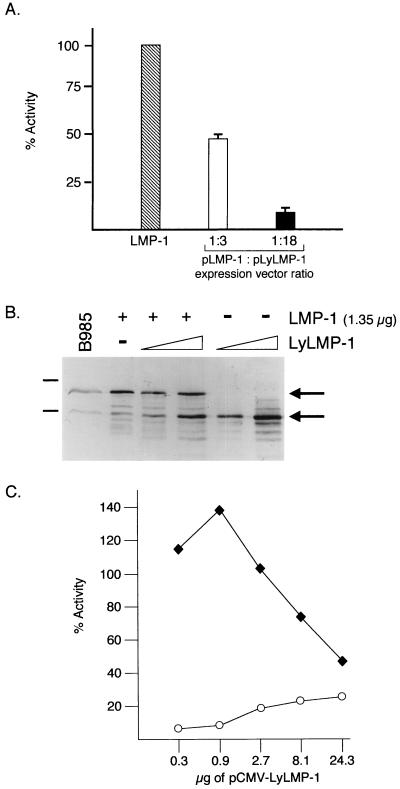
Inhibition of LMP-1-stimulated NF-κB activity by lyLMP-1.
The high levels of lyLMP-1 expressed during lytic cycle progression in EBV-infected cells may affect the function of LMP-1 and thereby contribute to the disruption of latency. Expression of LMP-1 in certain cell lines results in upregulation of NF-κB activity (20- to 50-fold) (9, 12, 26). Tumor necrosis factor receptor-associated factor (TRAF) binding to LMP-1's carboxy terminus is required for NF-κB activation (4, 14), and LMP-1 oligomerization is proposed to be required for both TRAF binding and NF-κB activation (8, 27). To determine whether expression of lyLMP-1 could affect LMP-1 activity, we coexpressed the two proteins in the human embryonic kidney cell line 293 and assayed for NF-κB activity (Fig. 2). 293 cells were electroporated (Bio-Rad Gene Pulser) with a luciferase reporter driven by the minimal fos promoter containing three upstream κB binding sites (p1242 [26]), pRSV-lacZ, and a constant amount of pCMV-LMP-1, together with increasing amounts of pCMV-lyLMP-1. pCMV-lyLMP-1 and pCMV-LMP-1 are pCDNA3-based expression vectors encoding the lyLMP-1 and LMP-1 ORFs, respectively, under the control of the cytomegalovirus promoter. Consistent with previous results, lyLMP-1 was deficient, relative to LMP-1, in its ability to activate NF-κB (Fig. 2A and C) (12, 26). Coexpression of lyLMP-1 and LMP-1 resulted in the inhibition of LMP-1-stimulated NF-κB activity. Maximal inhibition (∼90%) of LMP-1-stimulated NF-κB activity by lyLMP-1 occurred when there was an ∼18-fold excess of input pCMV-lyLMP-1 relative to the input pCMV-LMP-1 (Fig. 2A and C). This ratio of input DNA resulted in approximately equivalent expression levels of the two LMP-1 proteins (Fig. 2B). LMP-1-stimulated NF-κB activity was inhibited dose dependently by coexpression of lyLMP-1 (Fig. 2C). These results demonstrate that lyLMP-1 can negatively regulate LMP-1 signaling in 293 cells. Consistent with lyLMP-1's potential to negatively regulate LMP-1 signaling in virus-infected cells is the finding that the ratio of lyLMP-1 to LMP-1 required to block LMP-1 signaling in 293 cells (Fig. 2B) is roughly equivalent to the ratio of LMP-1 proteins in induced B95-8 cells (1, 7).
FIG. 2.
Activation of NF-κB by LMP-1 is inhibited by coexpression of lyLMP-1. 293 cells were electroporated with 1 μg of p1242 (luciferase reporter driven by the minimal fos promoter with three upstream κB sites), 1 μg of pRSV-lacZ, and 1.35 μg of pCMV-LMP-1, with and without increasing amounts of pCMV-lyLMP-1. Forty-eight hours following transfection, cells were harvested and samples were assayed for LMP-1 expression and NF-κB activity. Luciferase values were averaged for each sample and normalized to averaged β-galactosidase values to yield relative light units. The relative light units were averaged for each set of duplicate transfections. (A) LMP-1 activation of NF-κB activity in the presence of lyLMP-1. Data are expressed as percentages of LMP-1-stimulated NF-κB activity; the percent NF-κB activity attributed to lyLMP-1 alone (∼20%) was considered background and subtracted from the percent NF-κB activity when both proteins were coexpressed. Hatched bar, LMP-1 alone; open bar, 1.35 μg of pCMV-LMP-1 plus 4.05 μg of pCMV-lyLMP-1; filled bar, 1.35 μg of pCMV-LMP-1 plus 24.3 μg of pCMV-lyLMP-1. The ratios of the expression vectors (pLMP-1/pLyLMP-1) are indicated below the graph. Each error bar represents the standard error of mean of results from three experiments. The average fold induction for lyLMP-1 alone was 5, and that for LMP-1 alone was 25. (B) Western analysis of LMP-1 expression. Extracts of 2.5 × 103 cells from panel A were analyzed by Western blotting with anti-LMP-1 antiserum. +, 1.35 μg of pCMV-LMP-1; −, absence of the indicated LMP-1 expression vector; triangle, low (4.05 μg) and high (24.3 μg) amounts of introduced pCMV-lyLMP-1; B95-8, extract of 5 × 104 B95-8 cells included as a marker for migration of LMP-1 proteins; upper and lower arrows, migrations of full-length and lyLMP-1 proteins, respectively. Sixty-eight- and 45-kDa markers are indicated to the left of the blot. (C) Dose-dependent inhibition of LMP-1-stimulated NF-κB activity by lyLMP-1 expression. Data are expressed as percentages of LMP-1-stimulated NF-κB activity; activity from lyLMP-1 alone was not subtracted from LMP-1 values. Open circles, NF-κB activity in cells expressing lyLMP-1; filled squares, NF-κB activity in cells transfected with 1.35 μg of pCMV-LMP-1 and increasing amounts of pCMV-lyLMP-1.
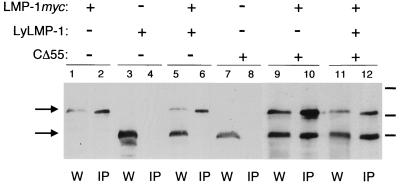
LyLMP-1 does not disrupt LMP-1 oligomerization.
LMP-1 is believed to activate signaling pathways as a constitutive TRAF-binding oligomer (4, 5, 13). Overexpression of lyLMP-1 (relative to the level of LMP-1) may interfere with LMP-1 signaling by disrupting LMP-1 oligomerization. High levels of lyLMP-1 may inhibit LMP-1 oligomerization by associating with LMP-1 and preventing formation of full-length LMP-1 oligomers. To explore the mechanism by which lyLMP-1 inhibits LMP-1 signaling, we determined if lyLMP-1 either oligomerizes with LMP-1 or alters LMP-1's ability to homo-oligomerize. To assess if lyLMP-1 associates with LMP-1, pCMV-lyLMP-1 and pCMV-LMP-1myc were cotransfected into 293 cells and 48 h later cell extracts prepared as described by Gires et al. (8) were assayed for oligomerization by coimmunoprecipitation with the anti-myc monoclonal antibody 9E10 (Santa Cruz Biochemicals) (Fig. 3). pCMV-LMP-1myc was constructed from pCMV-LMP-1 by insertion of the myc epitope tag (EQKLISEEDL) at LMP-1's carboxy terminus. pCMV-CΔ55 is an expression vector encoding a mutant LMP-1 lacking the last carboxy-terminal 55 amino acids and has been described previously (22). The NP-40 soluble fraction was precleared with protein G-agarose beads, and LMP-1 myc was immunoprecipitated from the precleared supernatant with anti-myc antibody (9E10; Santa Cruz). Complexes were recovered by incubation with protein G-agarose beads, washed with 1× RIPA buffer, resuspended in 4× SDS sample buffer, and analyzed by SDS-PAGE and Western blotting. lyLMP-1 was not detectable in LMP-1 myc immunoprecipitates, despite the large (≥5-fold) excess of lyLMP-1 protein relative to the level of LMP-1 myc protein (Fig. 3, lane 5). The inability of lyLMP-1 to coimmunoprecipitate with LMP-1 myc was not due to immunoprecipitation conditions or to the presence of the myc epitope tag at LMP-1's carboxy terminus since CΔ55, a mutant LMP-1 lacking the last carboxy-terminal 55 amino acids, interacted efficiently with LMP-1 myc (Fig. 3, lane 10). These results are consistent with the work of Gires et al., demonstrating a role for the amino terminus and transmembrane domains of LMP-1 in oligomerization (8). Importantly, CΔ55 was detected in LMP-1 myc immunoprecipitates from cells coexpressing LMP-1 myc and lyLMP-1, indicating that lyLMP-1 did not interfere with LMP-1's ability to oligomerize (Fig. 3, lane 12). These results demonstrate that lyLMP-1 does not associate with LMP-1 and that it does not prevent LMP-1 from forming homo-oligomers. It is unlikely, therefore, that lyLMP-1 inhibits LMP-1 signaling via disruption of LMP-1 oligomerization.
FIG. 3.
lyLMP-1 does not interfere with LMP-1 oligomerization. 293 cells were electroporated (conditions were as described in the Fig. 1 legend) with 1.5 μg of pCMV-LMP-1myc, with or without either 25 μg of pCMV-lyLMP-1 or 5 μg of pCMV-CΔ55. Cells were transfected with equivalent amounts of the cytomegalovirus promoter (pcDNA3) and equivalent amounts of total DNA. Forty-eight hours following transfection, 5 × 106 cells were harvested for immunoprecipitation (IP) and the remaining cells were used for the whole-cell lysate sample (W). Lanes: W, 5 × 104 cell equivalents of whole-cell lysate; IP, 1.25 × 106 cell equivalents of immunoprecipitated samples. The presence (+) or absence (−) of the transfected expression vector is indicated above the blot. The upper and lower arrows indicate the migrations of the full-length LMP-1 and lyLMP-1 or CΔ55 proteins, respectively; 87-, 64-, and 52-kDa molecular mass markers are indicated by bars to right of the blot.
Immortalization of human B cells by EBV is complex and results from the expression of several viral gene products, one of which is LMP-1. Since expression of LMP-1 proteins is restricted to full-length LMP-1 in latently infected lymphoblastoid cell lines, we think it unlikely that lyLMP-1 plays a role in the maintenance of immortalization. This conclusion is supported by the observation that lymphoblastoid cell lines infected with recombinant EBV and expressing LMP-1 proliferate despite the presence in these cells of LMP-1 immunoreactive, smaller-molecular-weight proteins (15). LMP-1 can activate divergent signaling pathways (6, 16, 17, 25). Not all LMP-1-activated pathways are likely to be required for immortalization (16, 28), and overexpression of lyLMP-1 may not regulate all LMP-1-activated signaling.
The high level of expression of lyLMP-1 in virus-producing cells suggests a function during the lytic cycle, and the presence of lyLMP-1 protein in the EBV virion suggests a role shortly after infection. lyLMP-1 can negatively affect LMP-1 function in 293 cells without disrupting LMP-1 oligomerization. These observations are consistent with a model in which the lyLMP-1 protein may downregulate NF-κB activation in the late stages of lytic infection by negatively affecting the function of LMP-1. In addition, lyLMP-1 has the potential to regulate signaling in infected cells in an LMP-1-independent manner, i.e., upon virus entry prior to LMP-1 expression (7). Studies are in progress to determine the mechanism by which lyLMP-1 affects LMP-1-regulated pathways and to identify the role of lyLMP-1 in EBV's life cycle.
Acknowledgments
We thank Brad Olwin, Tin Tin Su, and members of the Martin laboratory for critical reading of the manuscript.
This work was supported by NIH CA-64610-06 and AI-01537-02 grants to J.M.M.
REFERENCES
- 1.Baichwal V R, Sugden B. Posttranslational processing of an Epstein-Barr virus-encoded membrane protein expressed in cells transformed by Epstein-Barr virus. J Virol. 1987;61:866–875. doi: 10.1128/jvi.61.3.866-875.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggin M, Bodescot M, Perricaudet M, Farrell P. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J Virol. 1987;61:3120–3132. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boos H, Berger R, Kuklik-Roos C, Iftner T, Mueller-Lantzch N. Enhancement of Epstein-Barr virus membrane protein (LMP) expression by serum, TPA, or n-butyrate in latently infected Raji cells. Virology. 1987;159:161–165. doi: 10.1016/0042-6822(87)90360-6. [DOI] [PubMed] [Google Scholar]
- 4.Devergne O, McFarland E C, Mosialos G, Izumi K M, Ware C F, Kieff E. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eliopoulos A G, Young L S. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16:1713–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 7.Erickson K D, Martin J M. Early detection of the lytic LMP-1 protein in EBV-infected B-cells suggests its presence in the virion. Virology. 1997;234:1–13. doi: 10.1006/viro.1997.8638. [DOI] [PubMed] [Google Scholar]
- 8.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammerskjold M, Simurda M. Epstein-Barr latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF-κB activity. J Virol. 1992;66:6496–6501. doi: 10.1128/jvi.66.11.6496-6501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudewentz J, Bornkamm G V, zur Hausen H. Effect of the diterpene ester TPA on Epstein-Barr virus antigen and DNA synthesis in producer and nonproducer cells. Virology. 1980;100:175–178. doi: 10.1016/0042-6822(80)90563-2. [DOI] [PubMed] [Google Scholar]
- 11.Hudson G S, Farrell P J, Barrell B G. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J Virol. 1985;53:528–535. doi: 10.1128/jvi.53.2.528-535.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huen D S, Henderson S A, Croom-Carter D, Rowe M. The Epstein-Barr virus latent membrane protein (LMP1) mediates activation of NFκB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 13.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchili R, Kieff E, Mosialos G. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebowitz D, Kopan R, Fuchs E, Sample J, Kieff E. An Epstein-Barr virus transforming protein associates with vimentin in lymphocytes. Mol Cell Biol. 1987;7:2299–2308. doi: 10.1128/mcb.7.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann K P, Staunton D, Thorley-Lawson D. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann K P, Thorley-Lawson D. Posttranslational processing of the Epstein-Barr virus-encoded p63/LMP protein. J Virol. 1987;61:2100–2108. doi: 10.1128/jvi.61.7.2100-2108.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin J M, Sugden B. The LMP onco-protein resembles activated receptors in its properties of turnover. Cell Growth Differ. 1991;2:653–660. [PubMed] [Google Scholar]
- 22.Martin J M, Sugden B. Transformation by the LMP onco-protein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J Virol. 1991;65:3246–3258. doi: 10.1128/jvi.65.6.3246-3258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller G, editor. Epstein-Barr virus. 1st ed. New York, N.Y: Raven Press; 1985. [Google Scholar]
- 24.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller W E, Mosialos G, Kieff E, Raab-Traub N. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 28.Roberts M L, Cooper N R. Activation of a Ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology. 1998;240:93–99. doi: 10.1006/viro.1997.8901. [DOI] [PubMed] [Google Scholar]
- 29.Rowe M, Evans H S, Young L S, Hennessy K, Kieff E, Rickinson A B. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J Gen Virol. 1987;68:1575–1586. doi: 10.1099/0022-1317-68-6-1575. [DOI] [PubMed] [Google Scholar]
- 30.Sugden B, Phelps M, Domoradzki J. Epstein-Barr virus DNA is amplified in transformed lymphocytes. J Virol. 1979;31:590–595. doi: 10.1128/jvi.31.3.590-595.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Liebowitz D, Kieff E. The truncated form of the Epstein-Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J Virol. 1988;62:2337–2346. doi: 10.1128/jvi.62.7.2337-2346.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel R, Miller G. Major EB virus-specific cytoplasmic transcripts in a cellular clone of the HR-1 Burkitt's lymphoma line during latency and after induction of virus replicative cycle by phorbol esters. Virology. 1983;125:287–298. doi: 10.1016/0042-6822(83)90202-7. [DOI] [PubMed] [Google Scholar]
- 33.Wilson G, Miller G. Recovery of Epstein-Barr virus from nonproducer neonatal human lymphoid cell transformants. Virology. 1979;95:351–358. doi: 10.1016/0042-6822(79)90490-2. [DOI] [PubMed] [Google Scholar]
- 34.zur Hausen H, Bornkamm G W, Schmidt R, Hecker E. Tumor initiators and promoters in the induction of Epstein-Barr virus. Proc Natl Acad Sci USA. 1979;76:782–785. doi: 10.1073/pnas.76.2.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.zur Hausen H, O'Neill J, Freese U K. Persisting oncogenic herpesvirus induced by the tumour promoter TPA. Nature. 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]