Abstract
Introduction:
Prenatal and early-life dog exposure has been linked to reduced childhood allergy and asthma. A potential mechanism includes altered early immune development in response to changes in the gut microbiome among dog-exposed infants. We thus sought to determine whether infants born into homes with indoor dog(s) exhibit altered gut microbiome development.
Methods:
Pregnant women living in homes with dogs or in pet-free homes were recruited in southeast Michigan. Infant stool samples were collected at intervals between 1 week and 18 months after birth and microbiome was assessed using 16S ribosomal sequencing. Perinatal maternal vaginal/rectal swabs and stool samples were sequenced from a limited number of mothers. Mixed effect adjusted models were used to assess stool microbial community trajectories comparing infants from dog-keeping versus pet-free homes with adjustment for relevant covariates.
Results:
Infant gut microbial composition among vaginally born babies became less similar to the maternal vaginal/rectal microbiota and more similar to the maternal gut microbiota with age-related accumulation of bacterial species with advancing age. Stool samples from dog-exposed infants were microbially more diverse (p = .041) through age 18 months with enhanced diversity most apparent between 3 and 6 months of age. Statistically significant effects of dog exposure on β-diversity metrics were restricted to formula-fed children. Across the sample collection period, dog exposure was associated with Fusobacterium genera enrichment, as well as enrichment of Collinsella, Ruminococcus, Clostridaceae and Lachnospiraceae OTUs.
Conclusion:
Prenatal/early-life dog exposure is associated with an altered gut microbiome during infancy and supports a potential mechanism explaining lessened atopy and asthma risk. Further research directly linking specific dog-attributable changes in the infant gut microbiome to the risk of allergic disorders is needed.
Keywords: allergy, dog, environmental microbes, gut microbiota, infant, microbial trajectory, prenatal
GRAPHICAL ABSTRACT
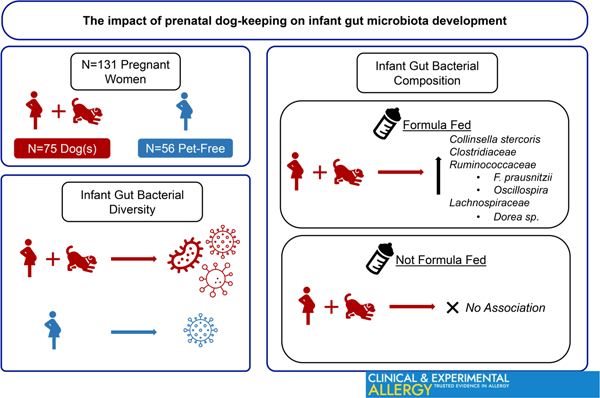
Prenatal dog-keeping associates with lower rates of childhood allergic disorders. The gut microbiome was analysed longitudinally for 18 months among 75 infants whose mothers kept indoor dogs during pregnancy and 56 infants whose mother’s homes were pet-free. Dog-keeping was associated with increased gut bacterial diversity. Among formula-fed children, dog keeping was associated with altered microbial composition indicating enrichment of Collinsella stercoris. Ruminococcacea, Clostridiaceae and Lachnospiraceae. These data support a potential microbial mechanism for associations between dog keeping and decreased allergy risk.
1 |. INTRODUCTION
Vertical transfer of maternal microbes, together with environmental microbial exposures, shape the initial bacterial communities of the neonatal gastrointestinal tract.1–3 These inceptive microbes regulate immune function4,5 and influence trajectories of gut microbiome maturation during this critical period of development.6 Birth cohort studies have provided evidence that early-life gut microbiome perturbations, and subsequent metabolic dysfunction, is associated with increased risk of atopy and asthma in childhood.7–10 We previously reported that gut microbial profiles associated with protection against atopy and asthma were more likely among babies with prenatal and early life exposure to dogs.7,8
Indoor pet exposure during pregnancy and early childhood is associated with lower total immunoglobulin E (IgE) levels, lessened allergen sensitization, childhood wheeze and lower risk of allergic disorders including asthma.11–21Household pets, particularly dogs, influence household environmental microbes22–26 and contribute to variance in infant stool microbiota.27
These observations are consistent with potential gut microbiome-related mechanisms linking pets to lowered risk of allergy-related disorders. However, reports directly linking indoor pet keeping to culture-independent alterations of the early-life gut microbiome are lacking. This study compares longitudinal gut microbiota taxonomic developmental trajectories among infants living in homes with indoor dogs versus pet-free homes, permitting analyses as to whether dog exposure influences initial gut microbiome development.
2 |. METHODS
2.1 |. Study population
The Microbes, Asthma, Allergy and Pets (MAAP) birth cohort includes 141 maternal–child pairs from southeastern Michigan enrolled to evaluate potential differences in the early-life gut microbiome between children living in homes with indoor dogs versus those living in pet-free homes. Pregnant women ages 18 through 49 years, receiving healthcare from Henry Ford Health (HFHS) obstetricians with planned delivery at selected HFHS hospitals, who intended to remain in southeast Michigan for at least 2 years post-partum were recruited during their second or third trimester of pregnancy between January 2014 and August 2016. An enrolment requirement included either: (1) living with a dog(s) kept indoors at least 12 h daily for at least 6 months prior to pregnancy with plans to keep the dog for the study’s duration or (2) living in furred pet-free homes for at least 2 years prior to pregnancy with no plans to obtain pets or have contact with pets for more than 4 h weekly for the study’s duration.
Women were required to communicate in English sufficient to provide written informed consent. The study was approved by the Henry Ford Hospital IRB.
2.2 |. Data collection
Women were interviewed prenatally and at 1 week, 6 months, and 18 months post-delivery. Questionnaires included: household demographics, tobacco smoke exposure, medication use, parental allergic history, pet keeping and questions related to their offspring’s health. Mothers completed dietary calendars to track weekly changes to their infant’s diet (breastfeeding, formula feeding and solid food introduction). When calendars were incomplete or missing, dietary information was utilized from questionnaires. Formula feeding included any formula use since the previous maternal contact. Breastfeeding duration was calculated as the total number of weeks of breastfeeding (whether exclusive or partial). First introduction of solid food was defined as the first week of life that any type of solid food was introduced. Pregnancy and delivery information were obtained from electronic health records. Allergen sensitization at 18 months was defined as sIgE ≥ .35 ku/L to at least one of following allergens: ragweed, dust mite, grass, cat, dog or mould mixture.
2.3 |. Specimen collection
Maternal vaginal/rectal swabs (Epicentre Catch-All™) were collected by their obstetrician within 6 weeks before delivery or on the day of delivery using the standard process for Group B Streptococcus screening. Swabs were stored in RNAlater at 4°C for at least 24 h, and then transferred to −80°C.
Maternal stool was collected during the last trimester and at approximately 1 week, 6 months and 18 months post-partum.
Infant stool was collected at approximately 1 week, 1 month, 3 months, 6 months and 18 months after birth.
Soiled diapers and maternal stool specimens were transported or mailed to the laboratory in insulated containers with ice packs and then stored at −80°C.
2.4 |. 16S ribosomal RNA Sequencing
2.4.1 |. DNA extraction
Punch biopsies (4 mm) (VWR International) were used to transfer ~0.3 g of frozen stool to Lysing Matrix E tubes (LME; MP Biomedicals). Vaginal/rectal swabs in RNAlater were thawed on ice and transferred to LME tubes. RNAlater was transferred into a sterile tube and centrifuged at 16,000 × g for 5 min at 4°C. Pellets were re-suspended using cetyltrimethylammonium bromide (CTAB) and transferred to the LME tube containing the swab. All prepped LME tubes were stored at −80°C. Genomic DNA was extracted using a modified CTAB buffer protocol.7
2.5 |. Sequencing preparation, data processing and quality control
Sequencing preparation methods are described in the supplemental material. Following preparation, paired-end sequences were merged using flasH (v1.2.11), demultiplexed by barcode. Reads that contained two or more unexpected errors were discarded. Unique sequences were clustered at 97% identity into operational taxonomic units (OTU) and chimeras removed using USEARCH. USEARCH was used to map all raw quality filtered reads to the unique sequence OTUs at 97% identity. Sequences were aligned using PyNast and the alignment was filtered to remove gaps. A bacterial phylogenetic tree was built using FastTree. The OTU table was filtered to remove unaligned sequences and taxonomy was assigned using Greengenes database (v13_5). NTCs were assessed to determine potential contamination. OTUs present in over half of NTCs were removed from samples. For OTUs present in less than half of NTCs, the highest read count was determined and subtracted from samples. To denoise the OTU table, taxa with less than 1/1000 of a percent of the total read count across all samples were removed. To normalize read depth across samples, data were rarefied 100 times to a read depth of 60,503 reads, using a multiple rarefaction algorithm.7 This depth was chosen because it was a high rarefying depth that resulted in little sample loss (stool N = 36, swabs N = 4) and Procrustes analysis indicated the distances in this rarefied matrix were closer (lowest M2 values) to the non-rarefied matrix compared to other rarefying depths. For timing of stool sample collection and mean number of stool samples contributed per infant, refer to Table S1 and Table 2.
TABLE 2.
Characteristics of the overall MAAP cohort and those with infant stool microbiota measurements.
| Covariate | Statistic | Level | Overall cohort (N = 141) | Has baby stool microbiota (N = 131) | Indoor dog(s) during pregnancy |
p-Value | |
|---|---|---|---|---|---|---|---|
| (N = 131) | |||||||
| No N = 56 | Yes N = 75 | ||||||
| African American mothera | N (Col %) | No | 112 (79.4%) | 107 (81.7%) | 40 (71.4%) | 67 (89.3%) | .009b |
| N (Col %) | Yes | 29 (20.6%) | 24 (18.3%) | 16 (28.6%) | 8 (10.7%) | ||
| Maternal-reported marital status | N (Col %) | No | 19 (13.5%) | 16 (12.2%) | 6 (10.7%) | 10 (13.3%) | .651b |
| N (Col %) | Yes | 122 (86.5%) | 115 (87.8%) | 50 (89.3%) | 65 (86.7%) | ||
| Household income | N (Col %) | <$40 K | 21 (14.9%) | 20 (15.3%) | 14 (25%) | 6 (8%) | .006b |
| N (Col %) | $40 K–$80 K | 33 (23.4%) | 32 (24.4%) | 7 (12.5%) | 25 (33.3%) | ||
| N (Col %) | >$80 K | 64 (45.4%) | 60 (45.8%) | 28 (50%) | 32 (42.7%) | ||
| N (Col %) | Refused/do not know | 23 (16.3%) | 19 (14.5%) | 7 (12.5%) | 12 (16%) | ||
| Highest level of maternal education | N (Col %) | HS diploma or less | 22 (15.7%) | 19 (14.6%) | 8 (14.3%) | 11 (14.9%) | .493b |
| N (Col %) | Some college, Associate’s degree or tech school | 40 (28.6%) | 37 (28.5%) | 13 (23.2%) | 24 (32.4%) | ||
| N (Col %) | Bachelor’s degree | 40 (28.6%) | 38 (29.2%) | 20 (35.7%) | 18 (24.3%) | ||
| N (Col %) | Advanced degree | 38 (27.1%) | 36 (27.7%) | 15 (26.8%) | 21 (28.4%) | ||
| Lives in single family home | N (Col %) | No | 30 (21.7%) | 27 (20.9%) | 20 (35.7%) | 7 (9.6%) | <.001b |
| N (Col %) | Yes | 108 (78.3%) | 102 (79.1%) | 36 (64.3%) | 66 (90.4%) | ||
| First-born child | N (Col %) | No | 90 (63.8%) | 82 (62.6%) | 40 (71.4%) | 42 (56%) | .071b |
| N (Col %) | Yes | 51 (36.2%) | 49 (37.4%) | 16 (28.6%) | 33 (44%) | ||
| Mode of delivery | N (Col %) | Vaginal | 96 (68.1%) | 89 (67.9%) | 41 (73.2%) | 48 (64%) | .264b |
| N (Col %) | C-section | 45 (31.9%) | 42 (32.1%) | 15 (26.8%) | 27 (36%) | ||
| Mother ever diagnosed with eczema | N (Col %) | No | 122 (86.5%) | 112 (85.5%) | 47 (83.9%) | 65 (86.7%) | .66b |
| N (Col %) | Yes | 19 (13.5%) | 19 (14.5%) | 9 (16.1%) | 10 (13.3%) | ||
| Mother ever diagnosed with asthma | N (Col %) | No | 95 (67.4%) | 90 (68.7%) | 37 (66.1%) | 53 (70.7%) | .575b |
| N (Col %) | Yes | 46 (32.6%) | 41 (31.3%) | 19 (33.9%) | 22 (29.3%) | ||
| Mother tobacco smoker at pre-delivery | N (Col %) | No | 131 (93.6%) | 122 (93.8%) | 55 (98.2%) | 67 (90.5%) | .137c |
| N (Col %) | Yes | 9 (6.4%) | 8 (6.2%) | 1 (1.8%) | 7 (9.5%) | ||
| Household tobacco smoke at pre-delivery | N (Col %) | No | 125 (91.9%) | 117 (92.1%) | 53 (96.4%) | 64 (88.9%) | .185c |
| N (Col %) | Yes | 11 (8.1%) | 10 (7.9%) | 2 (3.6%) | 8 (11.1%) | ||
| Child sex | N (Col %) | Male | 68 (48.6%) | 63 (48.1%) | 24 (42.9%) | 39 (52%) | .30b |
| N (Col %) | Female | 72 (51.4%) | 68 (51.9%) | 32 (57.1%) | 36 (48%) | ||
| Antibiotic use in first 6 months of life | N (Col %) | No | 115 (89.8%) | 114 (89.8%) | 47 (88.7%) | 67 (90.5%) | .733b |
| N (Col %) | Yes | 13 (10.2%) | 13 (10.2%) | 6 (11.3%) | 7 (9.5%) | ||
| Antifungal use in first 6 months of life | N (Col %) | No | 121 (94.5%) | 120 (94.5%) | 50 (94.3%) | 70 (94.6%) | .99c |
| N (Col %) | Yes | 7 (5.5%) | 7 (5.5%) | 3 (5.7%) | 4 (5.4%) | ||
| Maternal age at birth (years) | N | 141 | 131 | 56 | 75 | .952d | |
| Mean | 31 | 31.05 | 31.02 | 31.07 | |||
| SD | 4.98 | 4.92 | 4.68 | 5.13 | |||
| Number of previous pregnancies | N | 141 | 131 | 56 | 75 | .072 e | |
| Mean | 1.52 | 1.51 | 1.73 | 1.36 | |||
| SD | 1.32 | 1.35 | 1.31 | 1.36 | |||
| Number of previous live births | N | 141 | 131 | 56 | 75 | .093 e | |
| Mean | 0.98 | 0.96 | 1.11 | 0.85 | |||
| SD | 0.99 | 1 | 1 | 0.98 | |||
| Number of children in the Home at pre-delivery | N | 141 | 131 | 56 | 75 | .555 e | |
| Mean | 0.99 | 0.99 | 1.04 | 0.96 | |||
| SD | 1.1 | 1.11 | 1.11 | 1.12 | |||
| Breastfeeding duration (weeks) | N | 125 | 124 | 51 | 73 | .26 e | |
| Mean | 30.63 | 30.69 | 33.8 | 28.51 | |||
| SD | 24.46 | 24.55 | 25.47 | 23.83 | |||
| Formula feeding duration (weeks) | N | 127 | 126 | 52 | 74 | .065 e | |
| Mean | 30.00 | 30.01 | 25.06 | 33.49 | |||
| SD | 22.88 | 22.98 | 23.30 | 22.25 | |||
| Formula feeding at 1 week | N (Col %) | No | 90 (67.2%) | 88 (67.2%) | 41 (73.2%) | 47 (62.7%) | .203b |
| N (Col %) | Yes | 44 (32.8%) | 43 (32.8%) | 15 (26.8%) | 28 (37.3%) | ||
| Formula feeding at 1 month | N (Col %) | No | 83 (66.9%) | 82 (66.7%) | 38 (74.5%) | 44 (61.1%) | .120b |
| N (Col %) | Yes | 41 (33.1%) | 41 (33.3%) | 13 (25.5%) | 28 (38.9%) | ||
| Formula feeding at 3 months | N (Col %) | No | 73 (58.4%) | 72 (58.1%) | 32 (61.5%) | 40 (55.6%) | .505b |
| N (Col %) | Yes | 52 (41.6%) | 52 (41.9%) | 20 (38.5%) | 32 (44.4%) | ||
| Formula feeding at 6 months | N (Col %) | No | 53 (44.9%) | 53 (45.3%) | 21 (43.8%) | 32 (46.4%) | .779b |
| N (Col %) | Yes | 65 (55.1%) | 64 (54.7%) | 27 (56.3%) | 37 (53.6%) | ||
| Formula feeding at 18 months | N (Col %) | No | 106 (96.4%) | 106 (96.4%) | 41 (95.3%) | 65 (97%) | .643c |
| N (Col %) | Yes | 4 (3.6%) | 4 (3.6%) | 2 (4.7%) | 2 (3.0%) | ||
| First solid food introduction (weeks) | N | 123 | 122 | 50 | 72 | .074d | |
| Mean | 20.72 | 20.75 | 21.92 | 19.94 | |||
| SD | 6 | 6.01 | 4.43 | 6.82 | |||
| Number of sequenced baby stool samples | N | 131 | 131 | 56 | 75 | .506e | |
| Mean | 4.03 | 4.03 | 3.91 | 4.12 | |||
| SD | 1.03 | 1.03 | 1.18 | 0.90 | |||
If multiracial with at least one part Black, assigned to Black category.
Calculated by Chi-square test.
Calculated by Fisher’s exact test.
Calculated by ANOVA.
Calculated by Kruskal–Wallis test.
2.6 |. Statistical analysis
Statistical significance for main and interaction effects was set at p < .05. ANOVA and chi-square tests were used to compare maternal and child characteristics across groups by prenatal indoor dog keeping (yes/no). Metrics describing α-diversity were calculated using the R packages vegan and picante. Mixed effect models were used to fit stool α-diversity trajectories. Exact age at stool collection was used rather than targeted collection time (Figure S1). Best-fitting shape (linear vs. quadratic) was determined by Bayesian information criterion (BIC). Effect modification by age at time of stool sample collection, child sex, mode of delivery, household income and formula feeding were selected a priori for evaluation. The effect of dog keeping was assessed before and after adjusting for pre-specified potentially confounding covariates: maternal reported race, household income, maternal age at birth, mode of delivery (vaginal vs. caesarean-section), child sex and whether child was firstborn.
β-diversity metrics were calculated using the R packages phyloseq and vegan. Principal co-ordinates analysis (PCoA) was performed using the R package labdsv on baby stool samples with each β-diversity metric. The first through fourth principal co-ordinates were extracted and used as outcomes in mixed effect models to assess compositional differences by dog exposure, before and after covariate adjustment, as described previously for α-diversity. Compositional differences in cross-sectional data sets were assessed using PERMANOVA, before and after covariate adjustment.
Differences in OTU and genera abundance trajectories by age and prenatal indoor dog exposure were fit using generalized linear mixed effect models with the R package glmmADMB. Each taxon was first modelled with the random subject effect and a time covariate only (continuous, actual age at sample collection); negative binomial versus zero-inflated negative binomial models were compared for best fit using BIC (unless only one of the two models were able to be estimated; if neither model was able to be estimated, results were not examined). Upon determining the best-fitting model, main effects of dog exposure and interaction effects with time were obtained (with subgroup effects reported for significant interactions) with p-values rate-adjusted for false discovery (pFDR <.05 considered significant). All models testing dog effects were adjusted for the pre-specified confounding covariates. Taxa tested included only those present in ≧10% of baby stool samples. For OTU models, interaction effects were added for potentially effect modifying covariates, as suggested by β-diversity results. All analyses were performed using R version 3.6.1 or SAS version 9.4.
3 |. RESULTS
3.1 |. Cohort description and participant samples
Of 141 children at 18 months of age (81 dog/60 pet-free), seven were sensitized to common inhalants (four dog vs. three pet-free), seven had parentally-reported, doctor diagnosed asthma (five dog vs. two pet-free), and 35 had reported eczema/atopic dermatitis by 18-months (24 dog vs. 11 pet-free). A total of 660 fecal samples across 134 maternal–child pairs were successfully sequenced; 131 offspring (93%) had at least one infant stool sample successfully sequenced. The number of samples successfully sequenced during each collection period are displayed in Table 1.
TABLE 1.
Description of 660 samples across 134 maternal–child pairs with microbiome data.
| Subject | Sample | Sample collection time (Number sent for sequencing) | Observed collection time in days, with respect to baby date of birth (N = sequenced samples) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Min | Q1 | Median | Mean | Q3 | Max | |||
| Mother | Stool | Pregnancy (36) | 35 | −65 | −27 | −20 | −21 | −10 | −1 |
| 1 Week (33) | 32 | 6 | 13 | 22 | 24 | 27 | 95 | ||
| 6 Months (21) | 21 | 163 | 175 | 188 | 188 | 193 | 258 | ||
| 18 Months (2) | 2 | 532 | 534 | 535 | 535 | 536 | 538 | ||
| Vaginal/rectal swab | Pre-delivery (52) | 17 | −45 | −25 | −21 | −20 | −10 | −1 | |
| Delivery (102) | 25 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Baby | Stool | 1 Week (119) | 101 | 4 | 7 | 8 | 8 | 9 | 14 |
| 1 Month (122) | 106 | 18 | 27 | 31 | 35 | 38 | 79 | ||
| 3 Months (119) | 108 | 81 | 90 | 95 | 99 | 99 | 166 | ||
| 6 Months (123) | 114 | 153 | 173 | 181 | 193 | 194 | 332 | ||
| 18 Months (103) | 99 | 516 | 532 | 544 | 560 | 566 | 720 | ||
The analysed 131 children were similar to the full cohort and 57% of the samples were from participants residing with a dog (Table 2). The contribution of at least one infant stool sample (chi-square p = .87) and the number of samples sequenced per child did not differ by dog exposure (Kruskal–Wallis p = .506). Several factors were significantly associated with dog ownership: Mothers with dogs were less likely to self-identify as African American (10.7% vs. 28.6%, p = .009) and more likely to have mid-level household incomes ($40,000–$80,000; p = .006), and live in single family homes (90.4% vs. 64.3%, p < .001).
3.2 |. Infant gut microbiota development over advancing chronological age
Previous studies indicate that infant gut microbiota composition changes with advancing chronological age.8,28 We first examined relationships between gut microbiota development and infant age. The gut microbiota exhibited increasing bacterial richness and diversity and age-related compositional changes over the first 18 months of life (Figure 1A and Figure 2). Using vaginal/rectal swab and maternal stool microbiota profiles for reference, we noted that infant gut microbiota were similar to maternal vaginal/rectal microbiota at delivery, but became increasingly similar to the maternal gut microbiome with advancing age (Figure 1A [all samples] and Figure 1B [vaginally born infants with paired maternal samples]), indicating compositional maturation.
FIGURE 1.
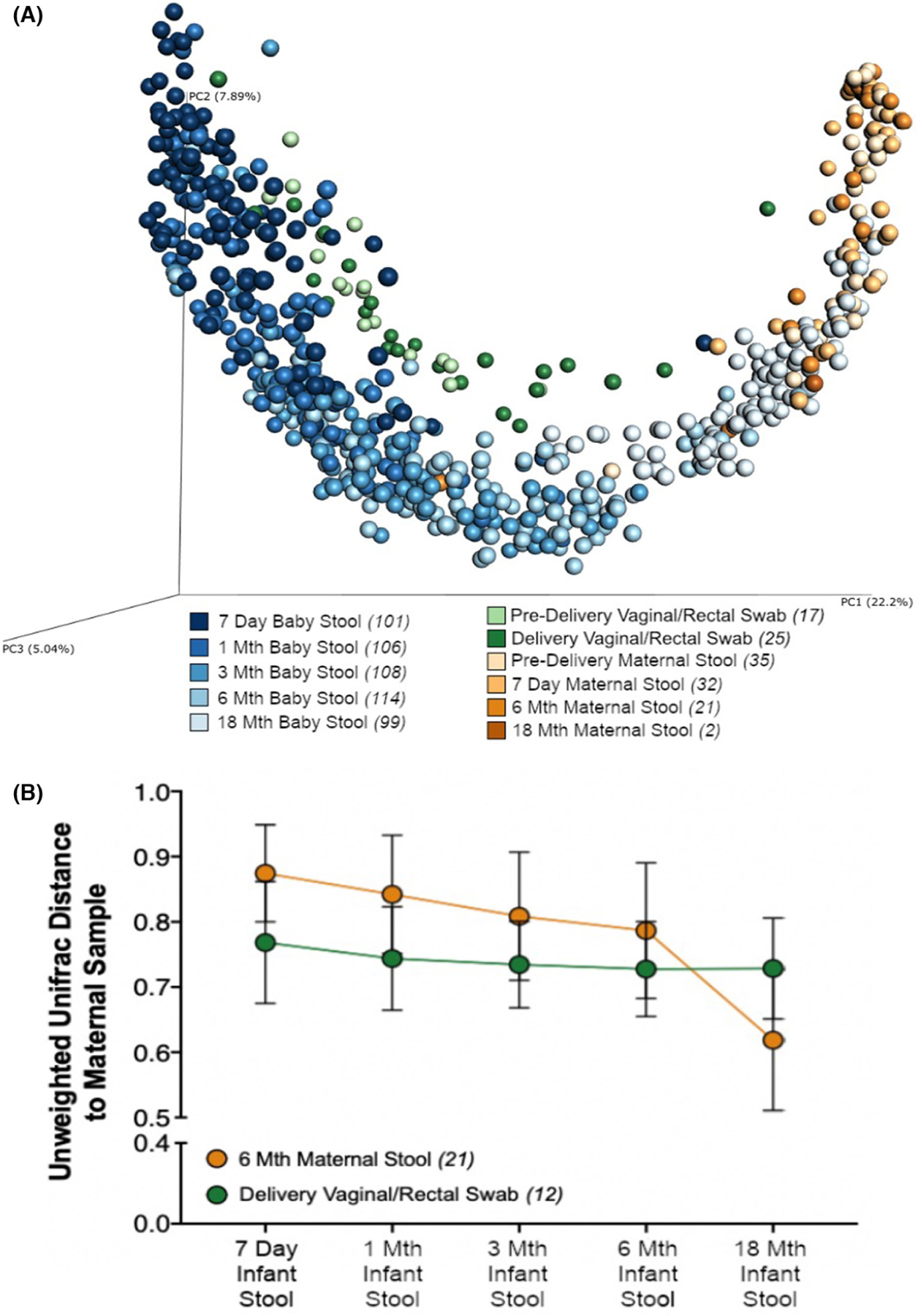
PCoA based on an unweighted Unifrac dissimilarity matrix showing bacterial community composition of infant stool samples (7 days and 1, 2, 6 and 18 months), maternal stool samples (pre-delivery, 7 days, 6 and 18 months), and maternal vaginal/rectal swabs (pre-delivery and delivery) (A). Mean unweighted Unifrac distance of delivery maternal vaginal/rectal swabs or 6 months maternal stool to paired, vaginally born infant stool samples at each time point (B).
FIGURE 2.
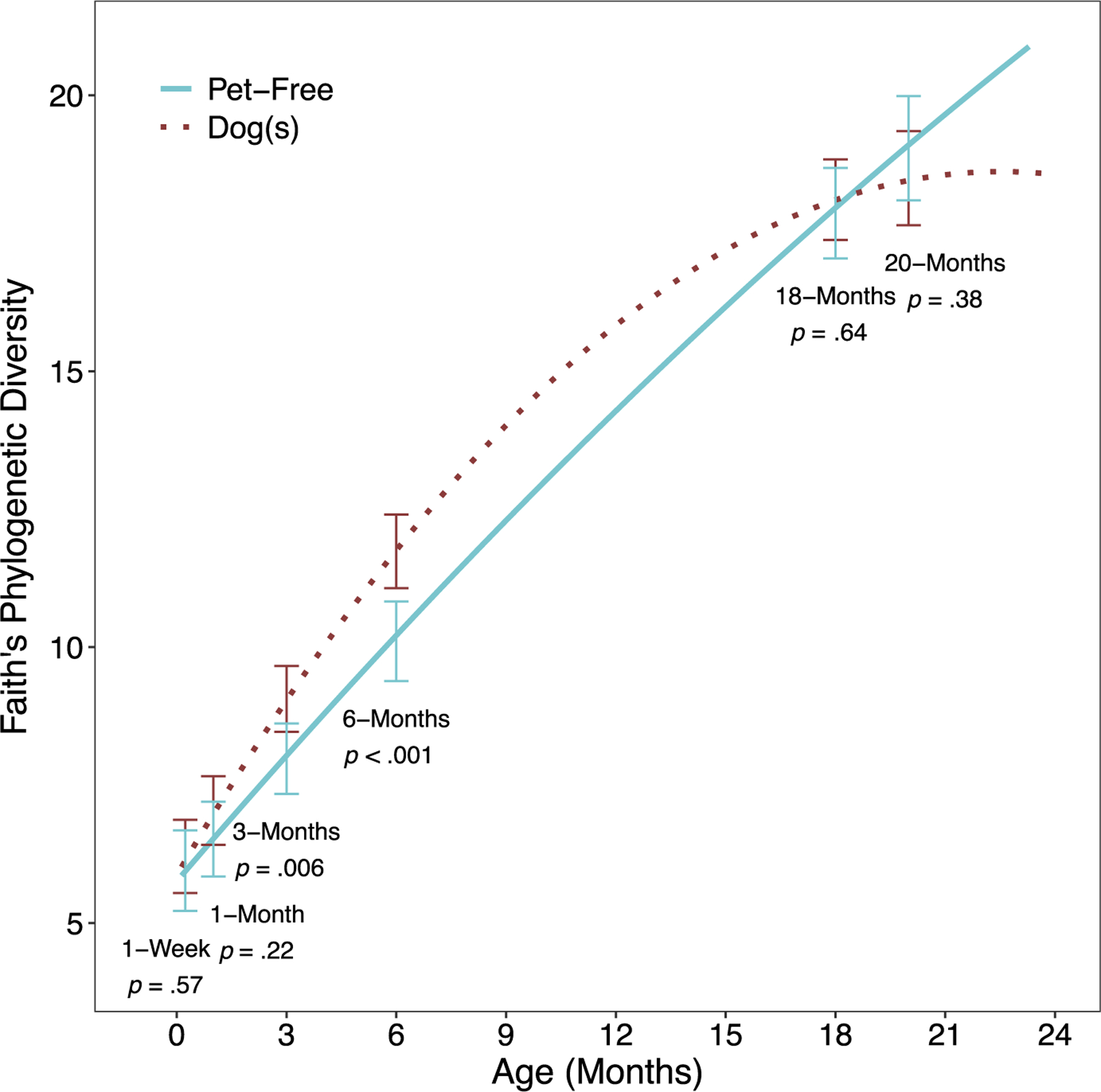
Fitted Faith’s phylogenetic diversity trajectories by prenatal indoor dogs. Model adjusts for maternal race, household income, maternal age at birth, mode of delivery, child sex and first-born child, and includes a term for the dog × time interaction, as this was statistically significant (p = .012).
We next sought to identify bacterial genera related to chronological age. Of the 57 genera with a prevalence of >10%, 48 (84%) were significantly associated with infant age at the time of sample collection. Of these 48 genera, 29 increased and 19 decreased in relative abundance with advancing age (Figure S2, Table S3, pFDR < .05). Twelve genera found to have the largest absolute effect size associated with advancing age are shown in Figure S2. Eleven of these genera increased in relative abundance with age and Staphylococcus decreased in relative abundance. These findings are consistent with previous observations of infant gut microbiota development over the first year of life and with age-related accumulation of bacterial species over this developmental period.2,7,9,29
3.3 |. Dog-ownership and gut microbial α-diversity metrics
We examined the relationship between dog ownership and infant gut microbiota richness, evenness and diversity trajectories. For richness and evenness trajectories, linear modelling was the best fit (Table S2). The overall dog effect was not statistically significant prior to covariate adjustment (Table 3, p = .094 and p = .54 respectively) nor following full adjustment (p = .080, p = .89). Interactions between dog exposure and the pre-specified effect modifiers were non-significant for richness and evenness (Table S4).
TABLE 3.
Association between dog exposure and infant gut α-diversity trajectories, before and after covariate adjustment.
| Model | N | β a | SE | p-Value | Percent change in unadjusted β |
|---|---|---|---|---|---|
| Richness (Linear model) | |||||
| Unadjusted | 131 | 10.0 | 6.0 | .094 | N/A |
| Adjusted for maternal race | 131 | 11.6 | 6.1 | .058 | 16.0% |
| Adjusted for household income | 131 | 7.0 | 6.1 | .25 | −30.0% |
| Adjusted for maternal age at birth | 131 | 9.7 | 5.8 | .098 | −3.0% |
| Adjusted for mode of delivery | 131 | 11.2 | 5.9 | .059 | 12.0% |
| Adjusted for child sex | 131 | 10.4 | 6.0 | .082 | 4.0% |
| Adjusted for first-born child | 131 | 10.4 | 6.0 | .088 | 4.0% |
| Fully adjusted (all of the above) | 131 | 11.1 | 6.3 | .080 | 11.0% |
| Evenness (Linear model) | |||||
| Unadjusted | 131 | 0.007 | 0.0122 | .54 | N/A |
| Adjusted for maternal race | 131 | 0.0082 | 0.0126 | .52 | 9.3% |
| Adjusted for household income | 131 | −0.001 | 0.0128 | .94 | −113.3% |
| Adjusted for maternal age at birth | 131 | 0.007 | 0.0122 | .56 | −6.7% |
| Adjusted for mode of delivery | 131 | 0.0089 | 0.0122 | .47 | 18.7% |
| Adjusted for child sex | 131 | 0.0084 | 0.0122 | .49 | 12.0% |
| Adjusted for first-born child | 131 | 0.006 | 0.0123 | .63 | −20.0% |
| Fully adjusted (all of the above) | 131 | 0.002 | 0.0136 | .89 | −74.7% |
| Faith’s phylogenetic diversity (Quadratic model) | |||||
| Unadjusted | 131 | 0.65 | 0.36 | .071 | N/A |
| Adjusted for maternal race | 131 | 0.73 | 0.37 | .050 | 12.3% |
| Adjusted for household income | 131 | 0.53 | 0.37 | .15 | −18.5% |
| Adjusted for maternal age at birth | 131 | 0.63 | 0.35 | .074 | −3.1% |
| Adjusted for mode of delivery | 131 | 0.72 | 0.35 | .042 | 10.8% |
| Adjusted for child sex | 131 | 0.68 | 0.36 | .061 | 4.6% |
| Adjusted for first-born child | 131 | 0.69 | 0.36 | .058 | 6.2% |
| Fully adjusted (all of the above) | 131 | 0.78 | 0.38 | .041 | 20.0% |
Average difference in specified α-diversity metric (comparing dog-exposed vs. pet-free), throughout early life, based on linear mixed effect models.
A quadratic trajectory model best-fit Faith’s phylogenetic diversity (Table S2). There were significant overall dog effects in the fully adjusted model, where diversity was on average 0.78 units higher in dog-exposed children across the observation period (Table 3; p = .041). Covariates including maternal race and delivery mode magnified the effect size of dog exposure, while household income and maternal age at birth diminished the effect size. However, the final multivariable model demonstrated that full covariate adjustment resulted in a 20% increase in effect size. A statistically significant dog exposure by time interaction was identified for diversity (Table S4; p = .012 and Figure 2). While phylogenetic diversity was greater in dog-exposed children for most of the trajectory, the curves were most divergent at 3 and 6 months (3 months: β = 1.08, p = .006; 6-months: β = 1.63, p < .001). By 18 months, the effect greatly diminished (β = 0.24, p = .64). Due to sparse data, estimates beyond 18 months should be interpreted with caution.
Assessment of potential confounding or effect modification by cat exposure was not possible as only dog-containing homes were allowed to have cats. We did examine the effect of cats on α-diversity trajectories from the 19 dog-containing homes that also contained a cat. Cats did not affect evenness (β (SE) = 0.024 (0.018), p-value = .19), while non-significant trends were observed for richness (β (SE) = 18.1 (9.2), p-value = .051) and phylogenetic diversity (β (SE) = 1.09 (0.56), p-value = .055).
3.4 |. Dog ownership and gut microbiota composition
Several cross-sectional associations of gut microbial composition (β-diversity) to indoor dogs were observed as listed in Table S5. However, to best understand how dog exposure may impact gut microbiota composition over time, we examined β-diversity trajectories using principal co-ordinates. The first principal co-ordinate of unweighted UniFrac was positively correlated with richness: r = .69 at 1 week, r = .80 at 1 month, r = .88 at 3 months, r = .89 at 6 months and r = .80 at 18 months. Although dog exposure was associated with overall compositional structure in some unadjusted models, effects were non-significant after covariate adjustment (Table S6). Effects were not time-dependent (Table S4; all dog × time interaction p ≥ .058). Other effect modifiers were non-significant except for formula-feeding on the first principal co-ordinate of unweighted UniFrac (interaction p = .036) and Canberra (interaction p = .039) distances. Specifically, the effect of dog ownership on β-diversity trajectory was apparent among formula-fed children only (Figure 3).
FIGURE 3.
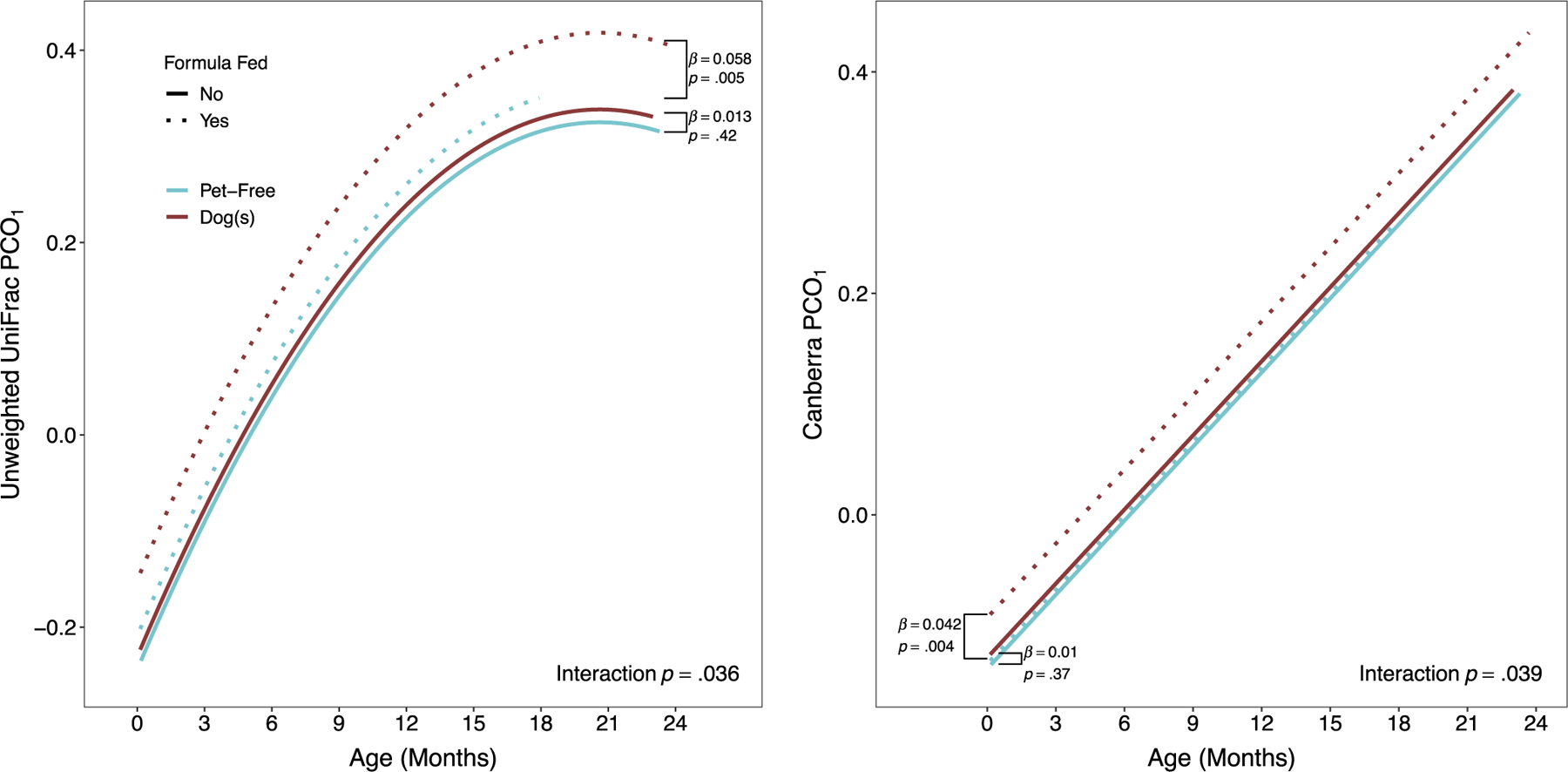
Trajectories of unweighted UniFrac and Canberra metrics (first principal co-ordinate), by prenatal dog exposure and type of feeding. Brackets show the difference between dog-exposed and unexposed children, among children who are formula fed (dotted lines) versus not (solid lines).
3.5 |. Indoor dogs and specific taxa
We next identified gut bacterial taxa differing between infants living with indoor dogs versus in pet-free homes. Among 120 genera, 57 were detected in >10% of samples. Following covariate adjustment, one genus, Fusobacterium (Figure 4), was significantly enriched among children living with dogs.
FIGURE 4.
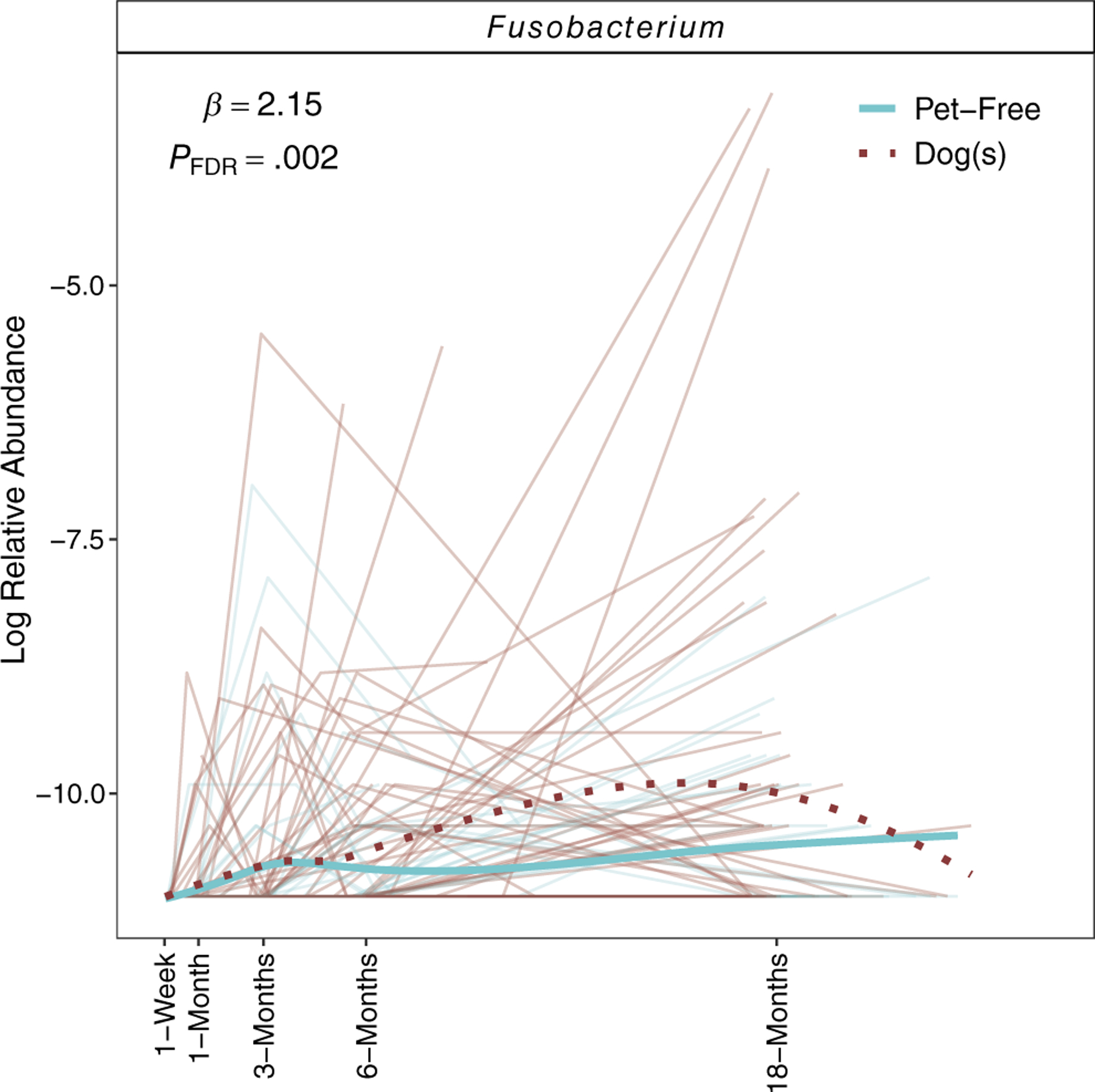
Fusobacterium was the only genus significantly associated with prenatal dog exposure (pFDR < .05), after covariate adjustment. Estimates shown represent the difference in log-transformed mean abundance, comparing infants living with dogs versus pet-free homes.
Of 1286 individual OTUs detected, 439 were prevalent in >10% of samples. After covariate adjustment, four were significantly related to living with a dog (Figure S3; Table S7). Specifically, Collinsella stercoris (OTU 93), Ruminococcus (OTU 1822) Lachnospiraceae (OTU 575) and Clostridiaceae (OTU 1099) were enriched in dog-exposed children.
3.6 |. Dog exposure in formula-fed infants and taxa trajectories
Noting significant interaction with formula feeding in the β-diversity trajectory models, we examined individual OTU trajectories contributing to this interaction. Seventy OTUs significantly contributed to the dog by formula-feeding interaction (interaction pFDR < .05; Table S8) with 60 exhibiting enrichment among formula-fed children. Of the four OTUs with a significant main effect of dog exposure (Table S7), Collinsella stercoris (OTU 93) exhibited a stronger effect of living with a dog in formula-fed versus non-formula-fed children (Table S8). Nearly all Ruminococcaceae and Lachnospiraceae OTUs having a significant dog exposure by formula-feeding interaction also showed greater enrichment with dog exposure among formula-fed children (Table S8). Many Ruminococcaceae OTUs were identified as Faecalibacterium prausnitzii or Oscillospira, while Lachnospiraceae OTUs included Dorea sp.
4 |. DISCUSSION
We observed 131 infants over the first 18 months of life that exhibited gut microbial developmental trajectories consistent with published studies describing increasing richness and progression towards and adult-like gut microbiome.2,30 These microbial developmental trajectories indicated that compared to infants living in homes without furred pets, those living with dogs had elevated gut bacterial phylogenetic diversity over the first 18 months of life. Early life represents a critical period of immunological development when low diversity or inappropriate assembly of the gut microbiota is associated with adverse respiratory health outcomes.31–33 Our results lend support to hypotheses linking dog exposure to decreased risks of atopy, allergy and asthma11,12,16,18,34,35 through an impact of dog exposure on the developing gut microbiome.
For phylogenetic diversity, we observed a statistically significant dog exposure by time interactions with the most robust associations at 3 and 6 months of age; a common period of complementary food introduction also known to influence the repertoire of local and circulating microbial metabolic products known to shape immune function and development of atopy and asthma.7,9,36–38 Whether diversification of the gut microbiota, specifically at this key point in development results in differences in the functional pathways or metabolic output of the gut microbiome or associated effects on immune function merits further study.
Several taxa enriched in infants living with dogs compared to pet-free homes included Collinsella stercoris, Ruminococcus sp., Lachnospiraceae sp. and Clostridiaceae sp. Gut Ruminococcus enrichment was also reported in 3–4-month-old infants exposed to furry pets.39 We previously reported Ruminococcaceae, Lachnospiraceae and Clostridiaceae enrichment in the gut of mice orally supplemented with house dust collected from dog-containing homes and resulting in microbiota restructuring associated with protection against allergic airway inflammation.40 This effect appeared mediated, in part, by alteration of the serum metabolome including increased concentrations of circulating anti-inflammatory polyunsaturated fatty acids that reduced bone marrow-derived dendritic cell activation and inflammatory cytokine production in vitro.41 Thus suggesting that dog exposure enriches gut microbes that can down-regulate responses to inflammatory stimuli.
The impact of dog cohabitation on gut microbiota was greatest among formula-fed infants, revealing enrichment of OTUs such as Collinsella stercoris (OTU 93) and Dorea sp. (OTUs 1015). Both have been identified in breast milk42 with Collinsella abundance highest in the gut of actively breastfeeding infants.2 Whereas breast milk contains bacterial communities43 capable of colonizing the infant gut44–47 and influencing community composition,2,47 formula fails to provide these microbial exposures. Perhaps, in the absence of breast milk, dogs provide an alternative source of environmental microbes for the developing gut microbiome of formula-fed infants. Supporting this concept, Dorea was found to be depleted in stool from dust mite-sensitized, 4–5-year-old children with allergic rhinitis compared to healthy controls with Dorea relative abundance negatively correlated to fecal IgE levels.48 Dorea is also underrepresented in the gut microbiota food-allergic children.49
Faecalibacterium prausnitzii was also enriched in the formula-fed infants living with a dog. F. prausnitzii is an abundant and key inhabitant of the human gut capable of blocking nuclear factor kB activation and enhancing anti-inflammatory, antigen-specific T cell proliferation and IL-10 expression.50,51 Its loss from the gut microbiota is characteristic of infants at high risk of developing allergies and asthma in childhood and in adults with chronic inflammatory conditions such as inflammatory bowel disease.7,8,52 Exposure of formula-fed infants to dogs in early life may serve to increase the presence of this species in early infancy.
Few studies have examined the relationship between household pets and the infant gut microbial composition and prior studies were limited by small sample size, lack of pet-free controls and/or examination at a single time point.39,53,54 Our inclusion of infants from pet-free homes allowed comparison of the impact of dog-associated exposures on early gut microbiota trajectories. Frequent longitudinal stool collection also allowed us to determine whether the impact of dog on the developing gut microbiome was associated with specific windows of microbial divergence while detailed surveys permitted adjustment for key, potentially confounding covariates. However, unmeasured, residual confounding is possible and should be further explored in larger studies. To the best of our knowledge, this study is the first to conduct longitudinal analyses examining the impact of dog exposure and the interaction of dog exposure and other early-life factors on infant gut microbiota trajectories.
Our study has several limitations. The limited cohort size and assessment of non-high-risk infants for less than 2 years limits the statistical power to directly assess whether the observed dog-attributable microbes are linked to differences in relevant allergic outcomes and also limits statistical power to assess potential associations among some subgroups. MAAP participants assessments at age 5–8 years are ongoing and may provide an opportunity to address some of these potential associations. Although low DNA yields from the vaginal/rectal swabs contributed to a high sequencing failure rate, this limitation did not impact analyses relating dog keeping to the infant gut microbiome since these samples were used solely as adult microbiota references to compare with the infant’s developing gut microbiome. Our study allowed dog-containing homes to also contain cats, whereas cats were not allowed in pet-free homes. This precluded the ability to examine confounding or effect modification by cat exposure. Although, cats within dog-containing homes did not significantly alter α-diversity trajectories, a trend for enhanced richness and phylogenetic diversity was observed. The possibility that cats, or other furred pets could have partially contributed to the altered gut microbiota of dog-exposed infants was not excluded. Finally, while fungi or viruses may play an important role as inhabitants of the gut microbiome and influence immune development, we were limited in our study to only 16 S ribosomal sequencing of gut bacteria.
5 |. CONCLUSIONS
Prenatal dog-keeping associates with altered infant gut microbial community trajectories including higher microbial diversity, particularly at 3 and 6 months of age. The strongest dog-associated compositional effects were observed among formula-fed infants and involved enhancement of key immunomodulatory gut bacteria. These data support ongoing hypotheses positing that associations of dog exposure and decreased allergy may be related to early-life gut microbiome development. Additional, appropriately scaled future studies are needed to definitively link dog-attributable changes in the infant gut microbiome to effects on immune development that causally explain lowered risk of allergy among dog-exposed infants.
Supplementary Material
Key Messages.
Prior studies indicate prenatal and early-life dog exposure associates with lower IgE production and allergy.
Infants of mothers that lived with dogs during pregnancy exhibit an altered gut microbiome.
An altered infant gut microbiome may explain associations of early-life dog exposure on allergy-related outcomes.
ACKNOWLEDGEMENTS
We acknowledge the MAAP cohort families for the substantial time and effort necessary to collect the many samples and questionnaires to complete this project. We also acknowledge the continued efforts of all our supporting MAAP research team members.
FUNDING INFORMATION
This study was supported by the US National Institutes of Health (Grant no. P01 AI089473) and the Fund for Henry Ford Hospital.
Fund for Henry Ford Hospital; National Institute of Allergy and Infectious Diseases
Footnotes
CONFLICT OF INTEREST STATEMENT
SVL is a consultant, sits on the board of directors and holds stock in Siolta Therapeutics Inc. She also consults for Solarea Bio. ARP, ARS, DF, SLH, KJ, BD, SF, GRW, KW, NWL, AML, DRO, CCJ and EMZ have no disclosures related to the submitted work to report.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The Raw sequence read data that support the findings of this study been submitted to the European Nucleotide Archive under accession number PRJEB50482. Additional supplementary and supporting data are available within this article and can be made available from the corresponding author [EMZ], upon reasonable request.
REFERENCES
- 1.Yassour M, Jason E, Hogstrom LJ, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 2018;24(1):146 154.e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015;17(5):690–703. [DOI] [PubMed] [Google Scholar]
- 3.Ferretti P, Pasolli E, Tett A, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018;24(1):133–145.e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rackaityte E, Lynch SV. The human microbiome in the 21(st) century. Nat Commun 2020;11(1):5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Constantinides MG, Link VM, Tamoutounour S, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366(6464):eaax6624. 10.1126/science.aax6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016;22(10):1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durack J, Kimes NE, Lin DL, et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by lactobacillus supplementation. Nat Commun 2018;9(1):707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015;7(307):307ra152. [DOI] [PubMed] [Google Scholar]
- 10.Johnson CC, Ownby DR. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res 2017;179:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aichbhaumik N, Zoratti EM, Strickler R, et al. Prenatal exposure to household pets influences fetal immunoglobulin E production. Clin Exp Allergy 2008;38(11):1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA 2002;288(8):963–972. [DOI] [PubMed] [Google Scholar]
- 13.Anyo G, Brunekreef B, de Meer G, Aarts F, Janssen NA, van Vliet P. Early, current and past pet ownership: associations with sensitization, bronchial responsiveness and allergic symptoms in school children. Clin Exp Allergy 2002;32(3):361–366. [DOI] [PubMed] [Google Scholar]
- 14.Svanes C, Jarvis D, Chinn S, Burney P. Childhood environment and adult atopy: results from the European Community respiratory health survey. J Allergy Clin Immunol 1999;103(3 Pt 1):415–420. [DOI] [PubMed] [Google Scholar]
- 15.Kerkhof M, Wijga AH, Brunekreef B, et al. Effects of pets on asthma development up to 8 years of age: the PIAMA study. Allergy 2009;64(8):1202–1208. [DOI] [PubMed] [Google Scholar]
- 16.Gern JE, Reardon CL, Hoffjan S, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol 2004;113(2):307–314. [DOI] [PubMed] [Google Scholar]
- 17.Wegienka G, Havstad S, Kim H, et al. Subgroup differences in the associations between dog exposure during the first year of life and early life allergic outcomes. Clin Exp Allergy 2017;47(1):97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havstad S, Wegienka G, Zoratti EM, et al. Effect of prenatal indoor pet exposure on the trajectory of total IgE levels in early childhood. J Allergy Clin Immunol 2011;128(4):880–885.e884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hesselmar B, Aberg N, Aberg B, Eriksson B, Björkstén B. Does early exposure to cat or dog protect against later allergy development? Clin Exp Allergy 1999;29(5):611–617. [DOI] [PubMed] [Google Scholar]
- 20.Eapen AA, Sitarik AR, Cheema G, et al. Effect of prenatal dog exposure on eczema development in early and late childhood. J Allergy Clin Immunol Pract 2022;10(12):3312–3314.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tutino M, Granell R, Curtin JA, et al. STELAR/UNICORN investigators dog ownership in infancy is protective for persistent wheeze in 17q21 asthma-risk carriers. J Allergy Clin Immunol 2022;151(2):423–430. [DOI] [PubMed] [Google Scholar]
- 22.Sitarik AR, Havstad S, Levin AM, et al. Dog introduction alters the home dust microbiota. Indoor Air 2018;28(4):539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimura KE, Johnson CC, Ownby DR, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol 2010;126(2):410–412, 412.e411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maier RM, Palmer MW, Andersen GL, et al. Environmental determinants of and impact on childhood asthma by the bacterial community in household dust. Appl Environ Microbiol 2010;76(8):2663–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettleson EM, Adhikari A, Vesper S, Coombs K, Indugula R, Reponen T. Key determinants of the fungal and bacterial microbiomes in homes. Environ Res 2015;138:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barberán A, Dunn RR, Reich BJ, et al. The ecology of microscopic life in household dust. Proc Biol Sci 2015;282(1814):20151139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin AM, Sitarik AR, Havstad SL, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep 2016;6:31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasparrini AJ, Wang B, Sun X, et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol 2019;4(12):2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402): 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renz H, Adkins BD, Bartfeld S, et al. The neonatal window of opportunity-early priming for life. J Allergy Clin Immunol 2018;141(4):1212–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352(6285):539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez FD, Guerra S. Early origins of asthma. Role of microbial Dysbiosis and metabolic dysfunction. Am J Respir Crit Care Med 2018;197(5):573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hesselmar B, Hicke-Roberts A, Lundell AC, et al. Pet-keeping in early life reduces the risk of allergy in a dose-dependent fashion. PLoS One. 2018;13(12):e0208472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fall T, Lundholm C, Örtqvist AK, et al. Early exposure to dogs and farm animals and the risk of childhood asthma. JAMA Pediatr 2015;169(11):e153219. [DOI] [PubMed] [Google Scholar]
- 36.Levan SR, Stamnes KA, Lin DL, et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol 2019;4(11):1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee-Sarwar KA, Lasky-Su J, Kelly RS, Litonjua AA, Weiss ST. Gut microbial-derived metabolomics of asthma. Metabolites. 2020;10(3):97. 10.3390/metabo10030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014;20(2):159–166. [DOI] [PubMed] [Google Scholar]
- 39.Tun HM, Konya T, Takaro TK, et al. Exposure to household furry pets influences the gut microbiota of infant at 3–4 months following various birth scenarios. Microbiome 2017;5(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimura KE, Demoor T, Rauch M, et al. House dust exposure mediates gut microbiome lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A 2014;111(2):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonseca W, Lucey K, Jang S, et al. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol 2017;10(6):1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr 2013;110(7):1253–1262. [DOI] [PubMed] [Google Scholar]
- 43.Togo A, Dufour JC, Lagier JC, Dubourg G, Raoult D, Million M. Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol 2019;14:623–641. [DOI] [PubMed] [Google Scholar]
- 44.Fehr K, Moossavi S, Sbihi H, et al. Breastmilk feeding practices are associated with the Co-occurrence of bacteria in Mothers’ Milk and the infant gut: the CHILD cohort study. Cell Host Microbe 2020;28(2):285–297.e284. [DOI] [PubMed] [Google Scholar]
- 45.Asnicar F, Manara S, Zolfo M, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017;2(1):e00164–16. 10.1128/mSystems.00164-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol 2014;16(9):2891–2904. [DOI] [PubMed] [Google Scholar]
- 47.Pannaraj PS, Li F, Cerini C, et al. Association between breast Milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 2017;171(7):647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu CY, Chan YL, Tsai MH, et al. Cross-talk between airway and gut microbiome links to IgE responses to house dust mites in childhood airway allergies. Sci Rep 2020;10(1):13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savage JH, Lee-Sarwar KA, Sordillo J, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018;73(1):145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breyner NM, Michon C, de Sousa CS, et al. Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-κB pathway. Front Microbiol 2017;8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossi O, van Berkel LA, Chain F, et al. Faecalibacterium prausnitzii A2–165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep 2016;6:18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pittayanon R, Lau JT, Leontiadis GI, et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020;158(4):930–946.e931. [DOI] [PubMed] [Google Scholar]
- 53.Azad MB, Konya T, Maughan H, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clini Immunol 2013;9(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konya T, Koster B, Maughan H, et al. Associations between bacterial communities of house dust and infant gut. Environ Res 2014;131:25–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Raw sequence read data that support the findings of this study been submitted to the European Nucleotide Archive under accession number PRJEB50482. Additional supplementary and supporting data are available within this article and can be made available from the corresponding author [EMZ], upon reasonable request.


