Abstract
The human cytomegalovirus (HCMV) major immediate-early (MIE) genes, encoding IE1 p72 and IE2 p86, are activated by a complex enhancer region (base positions -65 to -550) that operates in a cell type- and differentiation-dependent manner. The expression of MIE genes is required for HCMV replication. Previous studies analyzing functions of MIE promoter-enhancer segments suggest that the distal enhancer region variably modifies MIE promoter activity, depending on cell type, stimuli, or state of differentiation. To further understand the mechanism by which the MIE promoter is regulated, we constructed and analyzed several different recombinant HCMVs that lack the distal enhancer region (-300 to -582, -640, or -1108). In human fibroblasts, the HCMVs without the distal enhancer replicate normally at high multiplicity of infection (MOI) but replicate poorly at low MOI in comparison to wild-type virus (WT) or HCMVs that lack the neighboring upstream unique region and modulator (-582 or -640 to -1108). The growth aberrancy was normalized after restoring the distal enhancer in a virus lacking this region. For HCMVs without a distal enhancer, the impairment in replication at low MOI corresponds to a deficiency in production of MIE RNAs compared to WT or virus lacking the unique region and modulator. An underproduction of viral US3 RNA was also evident at low MOI. Whether lower production of IE1 p72 and IE2 p86 causes a reduction in expression of the immediate-early (IE) class US3 gene remains to be determined. We conclude that the MIE distal enhancer region possesses a mechanism for augmenting viral IE gene expression and genome replication at low MOI, but this regulatory function is unnecessary at high MOI.
Human cytomegalovirus (HCMV) replicates in many cell types, including endothelial, smooth muscle, fibroblast, hepatocyte, neuronal, glial, and macrophage cells (reviewed in reference 45). It replicates poorly or negligibly in lymphocytes, neutrophils, and certain embryonal cells (11, 12, 25, 44, 45), and it resides latently in monocytes and their precursors (16, 21, 22, 33, 34, 42, 46, 52, 53, 57). The mechanisms that govern HCMV replication or latency are poorly understood. Products of the HCMV major immediate-early (MIE) genes (e.g., IE1 p72 and IE2 p86) are required for viral replication (15, 18, 36, 38, 47). They are not expressed in latently or certain nonpermissively infected cell types (21, 22, 25, 33, 37, 42, 53). Hence, the regulation of their expression may be a pivotal step in controlling viral replication.
The MIE regulatory region controls transcription of its genes through interplay of both positive and negative cis-acting elements. The enhancer component of the MIE regulatory region contains many of these cis-acting elements. The enhancer's boundaries are inexact but are often considered to span base positions -65 to -550 with respect to the +1 start site of MIE RNAs (32). A variety of cellular and viral proteins interact with the enhancer's cis-acting elements to regulate activity of MIE promoter segments when assayed in transfection, in vitro, or transgenic animal studies (reviewed in references 32 and 35). Their roles in regulating the MIE enhancer-promoter during viral infection are not yet defined. The MIE enhancer's activity varies with availability or activity of the cellular or viral proteins that act on this region. For instance, enhancer activity is stimulated by several cellular transcription factors, including NF-κB/rel, CREB/ATF, AP1, retinoic acid receptor, SP-1, serum response factor and ELK-1 (3, 6, 32, 35). These transcription factors bind to sites located throughout the enhancer, and many of them bind to multiple sites. Their amounts or activities vary greatly depending on cell type, state of cellular differentiation, or stimulation of signal transduction pathways. Viral proteins, like pp71 and IE1 p72, can also bolster enhancer activity (8, 26, 29, 48). In contrast to positive regulation, enhancer function can be repressed by the cellular transcription factor YY1 (23, 27, 43). This factor binds to three sites located in the distal one-third of the enhancer. These sites are important in repressing enhancer function in undifferentiated monocytic and embryonal cells. The binding of cellular protein Gfi-1 to two cognate sites in the enhancer may also confer repression in certain cell types (58). Thus, enhancer activity is likely a combined result of complex mechanisms involving multiple and diverse transcription factors and signal transduction pathways.
HCMV latency in monocytes and their precursors is distinguished from productive viral infection by the lack of transcription from the +1 start site of the MIE promoter. However, viral RNAs colinear with the MIE genes are detectable in latently infected monocyte progenitors but not monocytes (16, 21, 22, 42, 53). These latency-specific (LS) RNAs originate from base position -292 or -356 of the MIE regulatory region and encode a protein (UL126/ORF94) of unknown function (22). The mechanism by which the LS and MIE promoters are differentially regulated is unknown. These findings further emphasize the complexity of the enhancer, which does not activate the MIE promoter during viral latency yet gives rise to LS RNAs.
Whether the entire enhancer region is needed for regulation of the MIE genes during a productive viral infection is unclear. Removal of the distal half of the enhancer does not reduce activity of MIE promoter segments in transfected fibroblasts as well as differentiated THP-1 and NTera-2 cells (26, 28, 50; J. L. Meier and M. F. Stinski, unpublished data). However, this same deletion increases activity of MIE promoter segments in transfected undifferentiated THP-1 and NTera-2 cells because of removal of YY1 binding sites (23, 27, 41, 43). In mice having a chromosomal insertion of the HCMV MIE enhancer-promoter and adjoining transgene, the exclusion of distal enhancer (-304 to -550) from the insert alters the cell types in which the transgene is expressed (5, 24). Thus, the distal enhancer may possess a regulatory function that is dependent on cell type, context of chromatin, or both. Whether the distal enhancer controls MIE or LS gene expression during viral latency is unknown, although deletion of this region en bloc would likely disrupt the transcriptional initiation of LS RNAs.
To extend our understanding of the regulatory mechanisms governing MIE gene expression and viral replication, we constructed an assortment of HCMVs lacking the MIE distal enhancer. In this report, we demonstrate that deletion of the MIE distal enhancer impairs HCMV replication at low multiplicity of infection (MOI) but not high MOI. This growth defect corresponds to an underproduction of MIE RNAs, which does not occur at high MOI. The MIE enhancer deletion also impairs production of viral US3 RNAs at low MOI but not high MOI. Hence, the deletion alters expression of two viral immediate-early (IE) class genes that are widely separated in the viral genome. Possible explanations for this finding are discussed.
MATERIALS AND METHODS
Cells and virus.
Primary human foreskin fibroblast (HFF) cells were grown in Eagle's minimal essential medium supplemented with 10% newborn bovine serum as described previously (31). These same culture conditions were applied to the growth of fibroblasts with hypoxanthine-guanine phosphoribosyltransferase (HGPRT) deficiency (GM02291), which were obtained from Coriell Institute for Medical Research (Coriell Cell Repositories, Camden, N.J.). HCMV strain Towne and recombinant viruses were propagated on HFF cells, using an MOI of 10−3 to 10−6 PFU/cell. Supernatants of infected cells were passed through a 0.45-μM-pore-size filter to yield cell-free virus stocks. Virus absorption was carried out for 1 to 1.5 h.
Viral growth curves and DNA replication rates were determined in HFF cells (between passages 5 and 10) in six-well culture plates. After virus absorption for 1.5 h, cells were washed with Hanks' balanced salt solution without calcium and magnesium (HBSS) and exposed 1 min to citrate buffer (50 mM sodium citrate, 4 mM KCl) of pH 3 to inactivate extracellular virus (39). Infected cells were washed twice more with HBSS prior to addition of growth medium. Input virus titer was determined by standard plaque assay on subconfluent HFF cells as described previously (31). However, this assay was inadequate for determining input titers of recombinant viruses that inefficiently form plaques. Therefore, titers of these viruses were determined by comparison to known titers of replication-competent viruses, using the following two methods. First, serial twofold dilutions of viral stocks were compared in abilities to induce cytopathic effect (CPE) in HFF cells at 24 h postinfection (hpi), as an indirect measure of relative viral titers. Second, the relative amount of viral DNA in HFF cells at 4 hpi, which precedes onset of viral DNA synthesis, was determined by Southern blot analysis as detailed below. These two methods were highly concordant in determination of viral titers. At the indicated times postinfection, cells were sacrificed for either viral growth curves or DNA replication studies. For viral growth curves, the infected cells were washed with HBSS, scraped into 1 ml of Eagle's minimal essential medium containing 10% newborn bovine serum, and stored at −70°C. In parallel, samples of each time point were thawed, sonicated (2 min), and centrifuged (1,000 × g for 5 min), and supernatants were subjected to plaque assay on subconfluent HFF cells. For viral DNA replication studies, the infected cells were washed with 1× phosphate-buffered saline (PBS) and DNA was prepared and analyzed as described below.
Plasmids.
The derivation of pΔMSVgpt has been detailed previously (31). This pGEM-4Z-based plasmid (Promega, Madison, Wis.) contains the ScaI-SalI fragment (HCMV AD169 nucleotides [nt] 172864 to 176219 [7]) of HCMV Towne strain, in which the BsrGI-MluI segment (nt 174365 to 174833) was replaced with the basal simian virus 40 (SV40) early promoter (−138 to +57), guanine phosphoribosyltransferase (gpt) open reading frame (ORF), and SV40 early intron and polyadenylation signal. Plasmid pIE1EM has been detailed previously (31) and contains the HCMV BamHI-SalI fragment (nt 170970 to 176219). The SphI fragment (nt 173560 to 176219) of pIE1EM was subcloned into an SphI site of pGEM-4Z to produce p4EM. To construct pUS3α, the HindIII-BamHI fragment of pMal-C2 (31) was inserted into corresponding sites in pGEM-4Z and the HindIII-AccI segment of US3 ORF was deleted; the vector HindIII and AccI ends were blunted with T4 polymerase prior to their ligation. pSV71 was constructed by inserting the XbaI fragment of pCMV71 (26), which contains the HCMV pp71 ORF, into the BamHI site of pSG5 (Pharmacia, Piscataway, N.J.); the XbaI and BamHI ends were blunted with Klenow enzyme prior to ligation.
Plasmid pΔ-300/-1108SVgpt was derived from pΔMSVgpt by deletion of Sau96I-BsrGI fragment (nt 174034 to 174371), and the Sau96I and BsrGI ends were blunted with Klenow enzyme before ligation. The HindIII-BamHI segment in each of the pΔ-300/-1108SVgpt and pΔMSVgpt plasmids that contains the gpt ORF and SV40 intron and poly(A) site was replaced with the HindIII-BamHI segment of phGFP-S65T (Clontech, Palo Alto, Calif.) that contains the green fluorescent protein (gfp) ORF and SV40 intron and poly(A) site, to produce pΔ-300/-1108SVgfp and pΔ-640/-1108SVgfp, respectively. pΔ-300/-640SVgpt was constructed by replacing the corresponding BamHI (blunted with T4 polymerase)-SalI segment of pΔ-300/-1108SVgfp with a corresponding BsrGI (blunted with T4 polymerase)-SalI fragment of pIE1EM.
Plasmids pΔ-300/-1108Egfp and pΔ-640/-1108Egfp were derived from pΔ-300/-1108SVgfp and pΔ-640/-1108SVgfp, respectively, by replacing the entire basal early SV40 promoter with an adenovirus E1b TATA box. This was accomplished by removing the SV40 promoter as a BglII-HindIII fragment and replacing it with a synthetic duplex oligonucleotide (5′-GATCTGGGTATATAATGGATCCCGGG-3′) having compatible BglII and HindIII ends. This oligonucleotide contains the 8-bp E1b TATA box (underlined) flanked by downstream SmaI and BamHI sites, which are functionally inactive in in vitro transcription and transfection studies (31, 40). pΔ-582/-1108Egfp was derived from pΔ-640/-1108Egfp by deleting the SpeI-BglII segment and religating the remaining plasmid after blunting its ends with Klenow enzyme.
Plasmid p1.6 was constructed by subcloning an HCMV EcoRI-BamHI fragment (nt 185496 to 187110) of pMSDT-D (55) into corresponding sites of pGEM-4Z. This fragment contains predicted HCMV ORFs IRL3 and IRL4 (7).
HCMV recombination.
Recombinant HCMVs rΔ-300/-640SVgfp, rΔ-640/-1108SVgfp, rΔ-300/-1108SVgfp, rSVgfp, rΔ-300/-1108Egfp, rΔ-582/-1108Egfp, and rΔ-640/-1108Egfp were derived from rΔMSVgpt (r2 clone), which was described previously (31). In the parent rΔMSVgpt virus, the BsrGI-MluI segment (-640 to -1110) of the MIE regulatory region containing the modulator was replaced with the basal SV40 early promoter (-138 to +57), bacterial gpt ORF, and SV40 early intron and polyadenylation signal. This virus was used for homologous recombination with the plasmids listed below. The resultant recombinant viruses lack gpt, which allows for their selection in HGPRT-deficient fibroblasts and 6-thioguanine (50 μg/ml) as described by Greaves et al. (14). Viral DNA was prepared for recombination as described previously (31). Subconfluent HFF cells (100-mm-diameter dish) were cotransfected with viral DNA (30 to 50 μg) and the indicated plasmid (5 or 10 μg), using the DNA-calcium phosphate coprecipitation method of Graham and van der Eb (13). The rΔ-300/-640SVgfp, rΔ-640/-1108SVgfp, rΔ-300/-1108SVgfp, rΔ-300/-1108Egfp, rΔ-582/-1108Egfp, and rΔ-640/-1108Egfp viruses were made by using the pΔ-300/-640SVgfp, pΔ-640/-1108SVgfp, pΔ-300/-1108SVgfp, pΔ-300/-1108MEgfp, p-582/-1108Egfp, and p-640/-1108Egfp plasmids, respectively. Viruses arising from the cotransfections were used to infect HGPRT-deficient fibroblasts at an MOI of 0.3 to 0.5. Viral plaques were picked and transferred to HFF cells in 12-well dishes as described previously (31). Once these infected cells reached 100% CPE, the growth medium containing virus was removed and stored at −70°C. Infected cells were washed with PBS, and the cell-associated viral DNA was isolated and subjected to restriction endonuclease and Southern blot analyses as described previously (31). All recombinant viruses were subjected to at least two rounds of plaque isolation. Replication-impaired viruses were plaque purified on HGPRT-deficient fibroblasts, whereas the other viruses were plaque purified on HFF cells. Genomes of plaque-purified viruses were again scrutinized by restriction endonuclease and Southern blot analyses. Two or three recombinant virus clones were obtained from independent transfection-recombination procedures to control for spurious genomic mutations.
The rSVgfp virus was a by-product of homologous recombination between the rΔMSVgpt viral DNA and plasmid pΔ-300/-640SVgfp. Recombination occurring within the 200-bp SV40 early promoter and upstream of the modulator would produce an HCMV having an SV40 early transcription unit containing the gfp ORF located between the MIE enhancer and modulator at base position -640. Restriction endonuclease and Southern blot analyses confirmed such a structural arrangement in rSVgfp.
The rΔ21MSVgfp, which was derived from rΔMSVgpt, was reverted to wild-type HCMV (recombinant wild type [rWT]) by homologous recombination in HFF cells. Plasmids pIE1EM (5 or 10 μg) and pSV71 (5 μg) and viral rΔ21MSVgfp DNA (20 to 50 μg) were cotransfected into subconfluent HFF cells by the DNA-calcium phosphate coprecipitation method (13). The HCMV pp71 protein expressed from pSV71 greatly enhanced replication of transfected rΔ21MSVgfp DNA, which is consistent with the known function of pp71 (4). The resultant viruses were applied to HFF cells at an MOI of 0.05 to enrich for rWT, because rΔ21MSVgfp replicates very poorly at low MOI in comparison to WT. Serial 10-fold dilutions of the enriched viral stock (106 PFU/ml) were applied to HFF cells in 12-well dishes (1 ml/well) and overlaid with 0.5% agarose. Viral plaques developed at 10−4 dilution with a growth rate and plaque size consistent with those of WT. These viral plaques were picked, expanded, and shown by Southern blot analysis to contain rWT. Two rWT clones that were derived from separate transfection-recombination procedures were subjected to further plaque purification and analysis. Unlike the WT stock of 106 PFU/ml, the rΔ21MSVgfp stock of 106 PFU/ml failed to form plaques at 10−5 dilution and produced only a few small plaques at 10−4 dilution. We continuously monitored the rΔ21MSVgfp stock for contaminating WT, which should greatly increase in amount with successive passages at an MOI of 0.001. Despite more than 12 passages of rΔ21MSVgfp at an MOI of 0.001, we found no contaminating WT by Southern blot analysis or the plaque assay procedure described above.
DNA analysis.
HCMV genomic DNA was isolated as described previously (31). All viral genomes were digested with each of enzymes EcoRI, PstI, and BamHI and fractionated on a 0.5% agarose gel. The gels were stained with ethidium bromide, and the restriction enzyme fragment profiles were photographed and scrutinized. The gels were then subjected to Southern blot analysis and autoradiography as described previously (31). Hybridization was carried out at 68°C for 4 to 12 h; a high-stringency wash was performed at 55 to 65°C in 0.1X SSPE (1X SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% sodium dodecyl sulfate. Distal enhancer- and modulator-specific probes were generated from multiprime 32P-labeled NdeI-BsrG I (nt 170953 to 174371) and BsrGI-MluI (nt 174371 to 174836) fragments, respectively. The gfp probe was derived as a HindIII-BsrGI fragment of phGFP-S65T. Stripping of probes from blots was achieved by boiling the Nytran in 0.2% sodium dodecyl sulfate. DNA fragments used as probes were gel purified prior to radiolabeling.
For analysis of HCMV DNA replication, uninfected or infected HFF cell DNA was isolated at the indicated times postinfection by methods described previously (31). Lambda DNA (2 μg) was added to each sample after cell lysis, but before proteolysis and phenol-chloroform extraction, to control for sample-to-sample variation in processing, endonuclease digestion, and loading. The purified sample DNA was cut with HindIII until the internal lambda DNA control was completely digested (4 to 18 h). Restricted DNA fragments were fractionated electrophoretically on a 0.5% agarose gel and subjected to Southern blot analysis. The 1.6-kbp BamHI-HindIII fragment of plasmid p1.6 (termed, T probe) was used to probe HCMV genomic termini containing terminal repeat long (TRL) or inverted repeat long (IRL). Full-length lambda DNA was 32P labeled for use in probing lambda DNA fragments. Methods of probe labeling, hybridization, washes, autoradiography, and probe stripping were as described above. Hybridization signals were quantitated by image acquisition analysis (Hewlett Packard Instant Imager).
RNA analysis.
After virus absorption for 1.5 h, residual extracellular virus was removed and inactivated by the citiric acid (pH 3.0) wash procedure noted above. Whole cell RNA from uninfected or HCMV-infected HFF cells was isolated by the method of Chomczynski and Sacchi (9). The RNase protection (RNP) assay was performed as described previously (31). Antisense IE1 and actin 32P-labeled riboprobes were generated as described previously (31). Antisense MIE and US3 32P-labeled riboprobes were generated from templates p4EM and pUS3α, respectively, using T7 polymerase. p4EM and pUS3α were linearized with SpeI and EcoRI, respectively. The MIE riboprobe spans +171 to −582, which includes the enhancer, exon 1, and part of intron 1. This probe is predicted to protect 170-nt unspliced and 120-nt spliced MIE RNA products. It was shown previously that a probe spanning this region also protects an alternative spliced MIE RNA product of approximately 140 nt (49). The hybridization of riboprobe(s) to the RNA sample (20 or 25 μg) was performed overnight at 52°C, and the resultant hybrid sample was digested with 150 or 200 U of RNase T1 (Boehringer Mannheim, Indianapolis, Ind.) at 37°C for 1 h. Protected products were analyzed on 6% polyacrylamide-urea gels.
Quantitative competitive PCR (QC-PCR).
HFF cells in six-well culture plates were washed three times in HBSS at the completion of virus adsorption and then placed in growth medium. WT-, rΔ-582/-1108Egfp-, rΔ-300/-1108Egfp-, and mock-infected cells were processed in parallel. At 5 hpi, infected cells were washed with PBS and DNA within them was isolated as described above. The purified DNA was digested with HindIII in order to reduce sample viscosity, thereby minimizing error in DNA sampling and quantitation. The restricted DNA fragments were purified by standard phenol-chloroform and chloroform extraction and sodium acetate-ethanol precipitation and then resuspended in TE (Tris-HCl [pH 8.0], 1 mM EDTA) buffer at a concentration of 150 ng/μl, as determined spectrophotometrically. For each of these DNA samples, four 4-μl (600 ng of DNA) aliquots were analyzed in parallel by PCR (total volume, 50 μl) in the presence of 10, 3.0, 1.0, or 0.3 pg of a 1,765-bp fragment of IE1 cDNA. The PCR conditions have been report previously (21, 31). PCR amplification of genomic or copy IE1 DNA by using primers IEP3C and IE4BII generates unspliced and spliced PCR products of 387 and 217 bp, respectively. PCR amplification was done for 35 cycles at 94°C for 1 min, 62°C for 1 min, and 72°C for 2 min.
RESULTS
Construction of an HCMV without the MIE distal enhancer region.
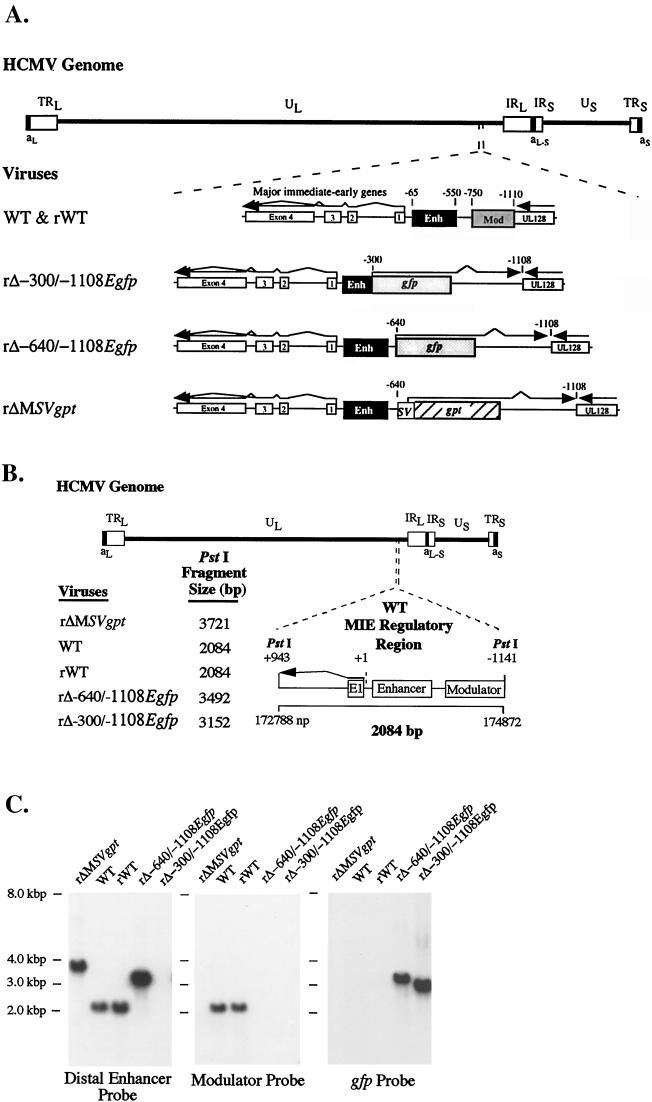
A set of recombinant HCMVs was assembled to determine the function of the MIE promoter's distal enhancer (Fig. 1A). These viruses were derived from the previously reported recombinant virus, rΔMSVgpt (31). This parental virus has an SV40 early kinetic class transcription unit containing the bacterial gpt ORF in place of the deleted MIE modulator region at base positions -640 to -1108 with respect to the MIE RNA cap site. rΔMSVgpt was shown previously not to differ appreciably from WT in its ability to transcribe the MIE genes at an MOI of 1 to 5. On the basis of design, rΔMSVgpt enabled selective mutagenesis of the MIE regulatory region by homologous recombination in conjunction with the elimination of gpt. Such construction afforded dominant selection of resultant recombinant viruses in HGPRT-deficient fibroblasts exposed to 6-thioguanine (14). Replacement of gpt with the gfp ORF further assisted in the selection of growth-defective recombinant viruses. The minimal SV40 early promoter lacking an enhancer was chosen to express the gfp gene because it does not appreciably alter MIE promoter activity when located at -640 in the MIE regulatory region (31).
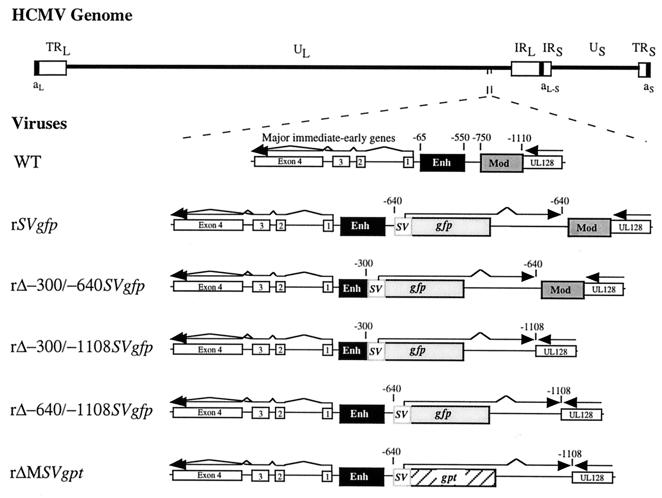
FIG. 1.
Schematic diagram of recombinant HCMVs containing mutations of the MIE regulatory region. HCMV genome and its unique long (UL) and short (US), internal repeat long (IRL) and short (IRS), terminal repeat long (TRL) and short (TRS), and a-sequence components are depicted. Locations of MIE regulatory region, MIE gene exons 1 to 4 (open boxes), and putative UL128 gene (open box) within UL component are shown. The MIE regulatory region of the WT is composed of proximal promoter (+1 to −64), enhancer (Enh; -65 to -550), unique region (-551 to -749), and modulator (Mod; -750 to -1140). Numerical base positions are assigned relative to start site of MIE RNAs. Recombinant HCMVs rΔ-300/-640SVgfp, rΔ-300/-1108SVgfp, rΔ-640/-1108SVgfp, and rSVgfp were derived from rΔMSVgpt (29). rΔ-300/-640SVgfp, rΔ-300/-1108SVgfp, and rΔ-640/-1108SVgfp have deletions from -300 to -640, -300 to -1108, and -640 to -1108, respectively. An enhancerless SV40 early promoter (−138 to +57), gfp ORF, and SV40 early intron and polyadenylation signal were inserted at the site of deletion. rSVgfp has the same insertion at -640 but has no deletion. rΔMSVgpt has a deletion of -640 to -1108 and insertion of the SV40 early transcription unit containing the gpt ORF.
For purpose of comparison, we made HCMVs with deletions of the distal enhancer alone (-300 to -640), distal enhancer and modulator (-300 to -1108), and modulator alone (-640 to -1108). These recombinant viruses were designated rΔ-300/-640SVgfp, rΔ-640/-1108SVgfp, and rΔ-300/-1108SVgfp, respectively. Each of these viruses has an SV40 early kinetic class transcription unit containing the gfp ORF that was inserted at the site of the deletion. To control for possible adventitious affects of the insertion, rSVgfp was added to the study set. This recombinant virus has no deletion in the MIE regulatory region but has the SV40 early transcription unit containing gfp inserted between the distal enhancer and modulator at -640.
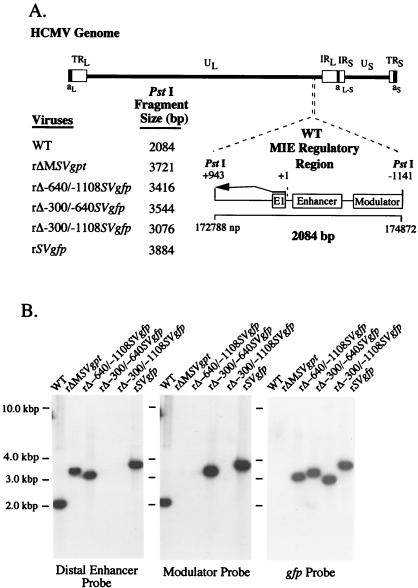
For each viral construct in the set, we randomly picked and studied two or three viral isolates from independent recombination procedures to control for spurious mutations. The genomes of these recombinant viruses were analyzed and compared to those of WT and rΔMSVgpt viruses with regard to restriction fragment length polymorphism (RFLP), as produced by three separate endonucleases (see Materials and Methods). The predicted sizes of PstI RFLPs are depicted in Fig. 2A. Findings of Southern blot analyses of PstI RFLPs are shown in Fig. 2B for a representative set of viral constructs. The sequential probing of the blot verifies a deletion of -300 to -640 in rΔ-300/-640SVgfp and rΔ-300/-1108SVgfp and deletion of -640 to -1108 in rΔ-640/-1108SVgfp and rΔ-300/-1108SVgfp. The gfp ORF is positioned correctly in rΔ-300/-640SVgfp rΔ-640/-1108SVgfp, rΔ-300/-1108SVgfp, and rSVgfp but is absent in WT and rΔMSVgpt. For each of the recombinant viruses, the PstI RFLP correctly matches its predicted size, and genomic instability was not detected. These findings indicate that the distal enhancer (-300 to -640) and combined distal enhancer and modulator (-300 to -1108) regions are deleted from HCMVs rΔ-300/-640SVgfp and rΔ-300/-1108SVgfp, respectively.
FIG. 2.
Structural analysis of rΔ-300/-640SVgfp, rΔ-300/-1108SVgfp, rΔ-640/-1108SVgfp, and rSVgfp. (A) Schematic diagram of HCMV genome and its components, as well as exploded view of the PstI fragment (+943 to -1141) containing the WT MIE regulatory region. Predicted sizes of PstI fragments of WT, rΔ-300/-640SVgfp, rΔ-300/-1108SVgfp, rΔ-640/-1108SVgfp, rSVgfp, and rΔMSVgpt are shown. Locations of exon 1, enhancer, and modulator are depicted. (B) Analyses of genomes of WT, rΔ-300/-640SVgfp, rΔ-300/-1108SVgfp, rΔ-640/-1108SVgfp, rSVgfp, and rΔMSVgpt. Viral DNAs were subjected to PstI digestion, agarose gel fractionation, Southern blot analysis, and autoradiography as described in Materials and Methods. The Southern blot was serially hybridized to 32P-labeled probes corresponding to the distal enhancer (-300 to -640), modulator (-640 to -1108), and gfp ORF (see Materials and Methods). Positions of accompanying size markers are provided.
An HCMV without the MIE distal enhancer region is replication impaired at low MOI but not high MOI.
In the early stages of creating HCMVs without the MIE distal enhancer (-300 to -640 or -1108), it became clear that these viral constructs produce substantially fewer plaques in HFF cells at low MOI (≤10−3) in comparison to WT. While the gfp expressed from these viruses permitted detection of a single infected cell among thousands of uninfected cells, we found neither individual nor small foci of fluorescent infected cells failing subsequently to form plaques via cell-to-cell spread. Despite their inefficiency in establishing initial infection, the viral plaques that formed slowly developed to yield 106 to 107 infectious extracellular viruses per ml of medium, which is comparable to that produced by WT (see Materials and Methods). The same viruses did not exhibit overt growth impairment at high MOIs such as 1 to 5.
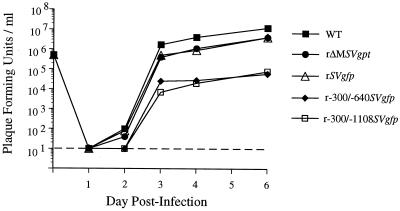
On the basis of these preliminary findings, we conducted a comparative study of viral replication rates after normalizing input viral titers by two independent methods that circumvent potentially deceptive plaque assay results. As detailed in Materials and Methods, titers of viral inocula were adjusted to yield equivalent amounts of input viral DNA in HFF cells at 4 hpi and to produce equivalent CPE at 24 hpi. There was high concordance among these two methods, enabling reliable determination of MOI for each virus relative to WT. Figure 3 shows single-step growth curves of WT, rΔMSVgpt, rSVgfp, rΔ-300/-640SVgfp, and rΔ-300/-1108SVgfp that were performed in parallel at an MOI of 1.0 in HFF cells. The amount of cell-associated virus produced at 1, 2, 3, 4, and 6 days postinfection (dpi) was measured by the standard viral plaque assay. The HCMVs lacking the distal enhancer, rΔ-300/-640SVgfp and rΔ-300/-1108SVgfp, register substantially less PFU per ml (−1.5 to −2.5 log10 at 3 to 6 dpi) than WT, rSVgfp, or rΔMSVgpt. Growth abnormalities of similar magnitude were seen with other rΔ-300/-640SVgfp and rΔ-300/-1108SVgfp constructs made from independent recombination procedures (data not shown). The abnormal single-step growth curves of rΔ-300/-640SVgfp and rΔ-300/-1108SVgfp could reflect impaired viral replication, assay bias, or both.
FIG. 3.
Single-step growth curves of WT, rΔMSVgpt, rSVgfp, rΔ-300/-640SVgfp, and rΔ-300/-1108SVgfp in HFF cells. In parallel, HFF cells were infected with these viruses at an MOI of 1. Cell-associated virus was prepared on 1, 2, 3, 4, and 6 dpi and subjected to standard plaque assay as detailed in Materials and Methods. PFU per milliliter of extract was determined on subconfluent HFF cells. The assay's lower limit of sensitivity was 10 PFU per ml (dashed line). Input viral titers at day 0 were not determined by plaque assay but by two other methods described in Materials and Methods and Results. Each growth curve represents the average of two independent experiments.
We examined whether rΔ-300/-640SVgfp and rΔ-300/-1108SVgfp differ from WT and rSVgfp in the abilities to replicate viral DNA in HFF cells at an MOI of 1.0 or 0.005. The infections were performed in parallel. Cell-associated DNA was isolated at various times after infection and subjected to HindIII digestion and Southern blot analysis. The blot was hybridized to a probe, termed T probe, which recognizes the RL regions flanking both ends of the long segment (L) of the HCMV genome (Fig. 4A). This method determines the proportion of RL regions that are not fused (free end) versus fused (fused end) to the short segment (S) of the viral genome. The unit-length linear viral genome is composed of an equimolar ratio of RL free and fused ends, although genomic isomerism yields L-S junction fragments of two sizes containing RL fused ends. Shortly after infection, the linear viral genomes form closed circles and large linear concatemers that lack RL free ends and possess only fused ends (30). Subsequently, genome cleavage and packaging regenerate RL free ends (30). Thus, serial analyses of RL free and fused ends during infection can reflect production of both replicative intermediates and cleaved or packaged genomes.
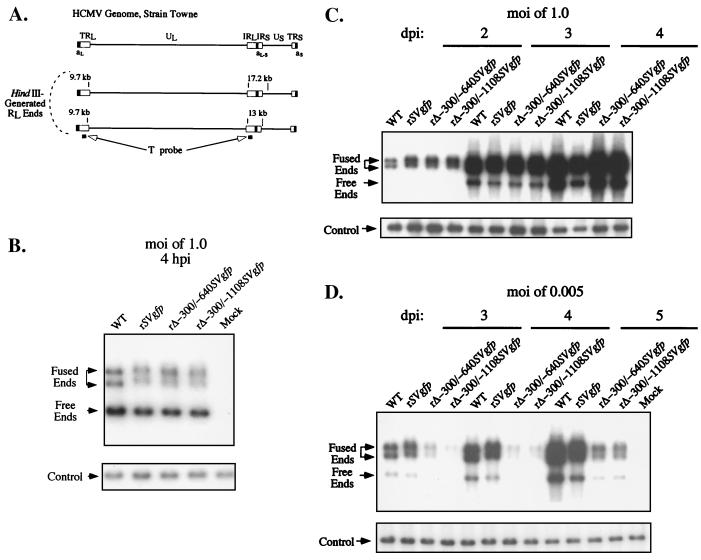
FIG. 4.
Analysis of DNA replication of WT, rSVgfp, rΔ-300/-640SVgfp, and rΔ-300/-1108SVgfp in HFF cells. (A) Schematic diagram of HindIII-generated RL-containing fragments and the T probe. Shown above is a diagram of the HCMV genome and its structural components as detailed in Fig. 1. Sizes of HindIII-generated RL-containing fragments are depicted for HCMV strain Towne. Genome isomerism yields L-S junction fragments of 17.2 and 13 kb containing RL fused ends, as well as 9.7-kb fragments representing RL free ends. Formation of circular or long concatemeric genomes results in conversion of RL free ends to RL fused ends, which are contained in 17.2- or 13-kb L-S junction fragments. Arrows point to regions of genome recognized by the T probe (black bars). (B) Analysis of viral genomes within HFF cells prior to viral DNA replication. In parallel, HFF cells were infected with WT, rSVgfp, rΔ-300/-640SVgfp, and rΔ-300/-1108SVgfp at an MOI of 1.0. Infected cells were lysed at 4 hpi and spiked with a constant amount (2 μg) of λ DNA for purpose of control of sample-to-sample variation (see Materials and Methods). Infected cell DNA was isolated, digested with HindIII, subjected to Southern blotting, hybridized to a 32P-labeled T probe, and visualized by autoradiography. Arrows point to positions of fragments containing RL fused ends (17.2 and 13 kb) and free ends (9.7 kb), as gauged by size markers not shown. The blot was stripped and rehybridized with 32P-labeled λ DNA (λ probe). The internal λ control to which the probe is hybridized is shown in the lower panel and denoted by an arrow. (C) Analysis of viral DNA replication in HFF cells at an MOI of 1.0. WT, rSVgfp, rΔ-300/-640SVgfp, and rΔ-300/-1108SVgfp infections were performed in parallel with those shown in panels B and D. Infected cell DNA was analyzed on 2, 3, and 4 dpi, using methods described for panel B. (D) Analysis of viral DNA replication in HFF cells at an MOI of 0.005. WT, rSVgfp, rΔ-300/-640SVgfp, and rΔ-300/-1108SVgfp infections were performed in parallel with those shown in panels B and C. Infected cell DNA was analyzed on 3, 4, and 5 dpi as described for panel B. Hybridization signals were quantitated by image acquisition analysis. Differences between WT or rSVgfp versus rΔ-300/-640SVgfp or rΔ-300/-1108SVgfp varied approximately 3- to 4.5-fold, 5- to 11.5-fold, and 6- to 10.5-fold on 3, 4, and 5 dpi, respectively.
We analyzed infected cell DNA of an MOI of 1 at 4 hpi, which precedes onset of viral DNA synthesis, to control for potential differences in input viral titers (Fig. 4B). Viral genome abundance and configuration resulting from replication was evaluated on 3 consecutive dpi. As shown in Fig. 4C, the rates of production and processing of viral genomes at an MOI of 1.0 were similar among rΔ-300/-640SVgfp, rΔ-300/-1108SVgfp, rSVgfp, and WT. A remarkably different finding emerged from an MOI of 0.005 (Fig. 4D). At this low MOI, the genomes of rΔ-300/-640SVgfp or rΔ-300/-1108SVgfp were produced in approximately 5- to 11-fold lesser amount than those produced by WT or rSVgfp on 4 or 5 dpi. The ratio of free to fused ends was not appreciably different among the viruses.
The combined findings suggest that a deletion of -300 to -640 of the MIE regulatory region impairs viral DNA replication at low MOI but not high MOI. The exhibited deficiency of these viruses in establishing initial infection at low MOI is consistent with this inference.
Validation of the MIE distal enhancer region's role in HCMV replication.
Two strategies were applied to substantiate the notion that deletion of the distal enhancer region (-300 to -640) impairs viral replication at low MOI but not high MOI. First, the deletion/insertion was repaired, with the objective of rescuing the WT phenotype. Second, we made another HCMV without the distal enhancer in which the 8-bp adenovirus Elb TATA box was inserted instead of the 200-bp SV40 promoter to control for possible confounding interactions of neighboring MIE and SV40 promoters.
rWT (Fig. 5A) was made from rΔ-300/-1108SVgfp by selection in low-MOI conditions for robustly replicating rWT, as described in Materials and Methods. Recombinant viruses rΔ-300/-1108Egfp and rΔ-640/-1108Egfp were derived from rΔMSVgpt (Fig. 5A). These viruses have deletions at -300 to -1108 and -640 to -1108 of the MIE regulatory region, respectively. The Elb TATA box, gfp ORF, and SV40 early intron and polyadenylation signal were inserted into the deletion. For each of the rWT, rΔ-300/-1108Egfp, and rΔ-640/-1108Egfp constructs, two or more isolates were derived from independent recombination procedures and fully analyzed. Their genomes were compared to those of WT and rΔMSVgpt by RFLP, using three separate restriction endonucleases. The predicted sizes of the PstI RFLPs are depicted in Fig. 5B. Findings of the Southern blot shown in Fig. 5C reveal that the PstI RFLPs of rWT, rΔ-300/-1108Egfp, and rΔ-640/-1108Egfp match the size predictions. The series of probes to which the blot was hybridized indicate that the insertions/deletions were correctly constructed to eliminate the distal enhancer (-300 to -640) in rΔ-300/-1108Egfp and the modulator (-640 to -1108) in rΔ-300/-1108Egfp and rΔ-640/-1108Egfp. These probes also confirm that rWT was fully restored to WT genotype.
FIG. 5.
Construction of rWT, rΔ-300/-1108Egfp, and rΔ-640/-1108Egfp. (A) Schematic diagram of WT, rWT, rΔ-300/-1108Egfp, rΔ-640/-1108Egfp, and rΔMSVgpt. The MIE regulatory region in relation to the HCMV genome is shown. rΔ-300/-1108Egfp and rΔ-640/-1108Egfp were derived from rΔMSVgpt and have deletions from -300 to -1108 and -640 to -1108 of the MIE regulatory region, respectively. The 8-bp adenovirus Elb TATA box, gfp ORF, and SV40 early intron and polyadenylation signal were inserted at the site of deletion. rWT was derived from rΔ-300/-1108SVgfp (Fig. 1). Enh, enhancer; Mod, modulator. (B) Depiction of predicted sizes of PstI fragments of rΔMSVgpt, WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp containing the MIE regulatory region. (C) Southern blot analysis of rΔMSVgpt, WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp genomes. Viral DNAs were subjected to PstI digestion and Southern blot analysis as described for Fig. 2B. The 32P-labeled probes recognize the distal enhancer (-300 to -640), modulator (-640 to -1108), or gfp ORF. Positions of accompanying size markers are provided.
We compared rates of genome replication of WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp in HFF cells at MOIs of 1.0 and 0.001. The DNA of infected cells was isolated at designated times after infection and subjected to HindIII digestion and Southern blot analysis. The viral RL-containing fragments were hybridized to the T probe depicted in Fig. 4A. Analysis of viral DNA within HFF cells at 4 hpi (MOI of 1.0) controlled for differences in input viral titers (Fig. 6A). On 2 and 3 dpi at an MOI of 1.0, the cohort produced similar amounts of genomic RL free and fused ends (Fig. 6B). On 4 and 5 dpi at an MOI of 0.001, the genomes of rΔ-300/-1108Egfp were not detected, in contrast to those of WT and rWT, in which the genomes were in abundance (Fig. 6C). Genomes of rΔ-640/-1108Egfp accumulated at a lower rate than that of WT or rWT but at a much greater rate than that of rΔ-300/-1108Egfp (Fig. 6C). We have also observed modest delays in replication of other HCMV recombinants devoid of a modulator, although this difference has been difficult to consistently demonstrate at the level of DNA replication (31; Meier and Stinski, unpublished data).
FIG. 6.
Analysis of DNA replication of WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp in HFF cells. (A) Analysis of viral genomes within HFF cells prior to viral DNA replication. HFF cells were infected in parallel with WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp at an MOI of 1.0. Infected cell DNA was isolated at 4 hpi, digested with HindIII, and subjected to Southern blotting using the 32P-labeled T or λ probes described in the legend to Fig. 4A. λ DNA served as an internal control, as detailed for Fig. 4A. Arrows point to positions of RL 9.7-kb free ends, 17.2- and 13-kb RL fused ends, and internal λ control. (B) Analysis of viral DNA replication in HFF cells at an MOI of 1.0. WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp infections were performed in parallel with those shown in panels A and C. Infected cell DNA was analyzed on 2 and 3 dpi as described for Fig. 4B. (C) Analysis of viral DNA replication in HFF cells at an MOI of 0.001. WT, rWT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp infections were performed in parallel with those shown in panels A and B. Infected cell DNA was analyzed on 4 and 5 dpi as described for Fig. 4B.
These data further support the notion that removal of the MIE distal enhancer region (-300 to -640) hinders viral DNA replication at low MOI but not high MOI.
An MIE distal enhancer deletion reduces expression of viral IE RNAs at low MOI but not high MOI.
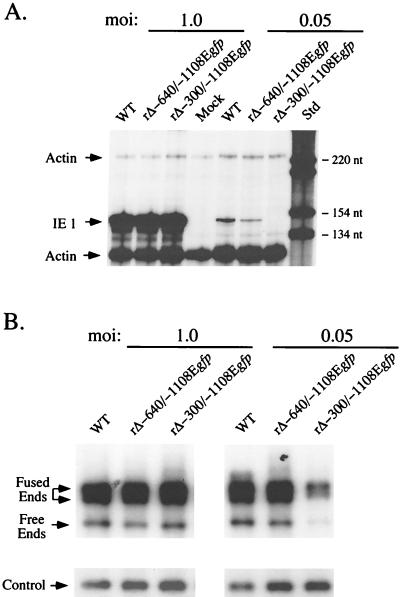
We compared WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp with regard to productivity of both IE1 RNA and viral DNA in HFF cells at MOIs of 1.0 and 0.05. Infections were performed in parallel. Infected cell DNA was isolated at 4 dpi and subjected to HindIII digestion and Southern blot analysis with the T probe (Fig. 4A). Whole cell RNA was isolated at 8 hpi, during peak production of IE1 RNA under these experimental conditions (see Materials and Methods). The relative amount of IE1 RNA was determined by RNP assay, using a riboprobe corresponding to exon 4 of IE1 (31). The concurrent analysis of cellular actin RNA controlled for sample-to-sample variation (31). Findings of the analyses are shown in Fig. 7. WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp produce equivalent amounts of IE1 RNA and viral DNA at an MOI of 1.0. In contrast, rΔ-300/-1108Egfp produces substantially less IE1 RNA and viral DNA at an MOI of 0.05 compared to levels for WT and rΔ-640/-1108Egfp. The difference in IE1 RNA amounts was not quantifiable because IE1 RNA of rΔ-300/-1108Egfp was below the limit of detection of the RNP assay. Free and fused RL regions of rΔ-300/-1108Egfp genomes were produced in 3- and 2.7-fold lesser amounts, respectively, than those of WT or rΔ-640/-1108Egfp. These findings suggest that deletion of the MIE distal enhancer region (-300 to -640) decreases production of both viral DNA and IE1 RNA at low MOI but not high MOI.
FIG. 7.
Analysis of productivity of IE1 RNA and viral DNA by WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp in HFF cells at high or low MOI. (A) Analysis of viral IE1 RNAs. HFF cells were infected in parallel with WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp at MOIs of 1.0 and 0.05. Total cellular RNA was harvested at 8 hpi. Equal amounts of RNAs (20 μg) were subjected to RNP assay, using both IE1- and actin-specific riboprobes, as described previously (17, 31, 56) (see Materials and Methods). RNA of uninfected HFF cells (Mock) served as a control. Positions of protected actin (230 and 122 nt) and IE1 (145 nt) RNAs (denoted by arrows), as well as selected size markers (Std), are shown. (B) Analysis of viral DNAs. HFF cells were infected with WT, rΔ-640/-1108Egfp, and rΔ-300/-1108Egfp at MOIs of 1.0 and 0.05. Infections were performed in parallel with those shown in panel A. Infected cell DNA was isolated at 4 dpi, digested with HindIII, and subjected to Southern blotting using the 32P-labeled T or λ probe described in the legend to Fig. 4A. λ DNA served as an internal control as detailed for Fig. 4A. Arrows indicate positions of 9.7-kb RL free ends, 17.2- and 13-kb RL fused ends, and internal λ control. At an MOI of 0.05, abundance of RL fused and free ends of rΔ-300/-1108Egfp differed by approximately 3- and 2.7-fold, respectively, from those of WT or rΔ-640/-1108Egfp.
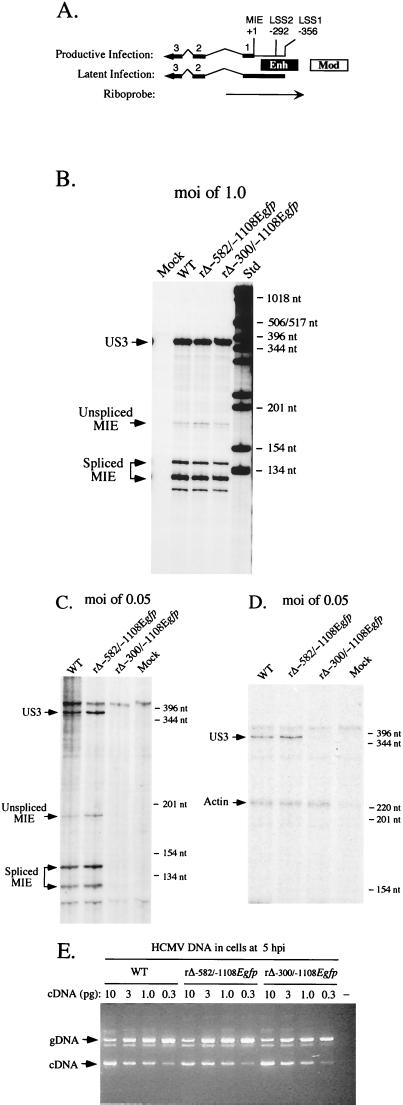
We examined whether deletion of the MIE distal enhancer specifically reduces transcription from the MIE promoter at low MOI. Using the RNP assay, we analyzed MIE RNA originating from the +1 start site of the MIE regulatory region. The riboprobe detects both unspliced and spliced MIE RNAs, which yield IE1 and IE2 RNAs via differential splicing events. This probe is also capable of detecting the overlapping latency-specific RNAs of UL126, should they be activated in HFF cells by the mutations (Fig. 8A). We also analyzed RNA produced by the viral IE US3 gene that is separated from the MIE genes by approximately 21 kbp. To more accurately delineate the MIE distal enhancer's role, we made and characterized another recombinant virus, rΔ-582/-1108Egfp (data not shown). This virus lacks -582 to -1108 of the MIE regulatory region and is therefore missing virtually the entire unique region, as well as the modulator. Like rΔ-300/-1108Egfp, this virus contains the E1b TATA box and gfp cassette at the site of the deletion. Thus, rΔ-300/-1108Egfp and rΔ-582/-1108Egfp differ only by whether they possess the MIE distal enhancer region (-300 to -582). The rΔ-582/-1108Egfp, as well as other HCMV constructs having deletions of -582 to -1108 (28), replicate at rates comparable to those of HCMVs lacking the modulator (-640 to -1108) (28; Meier and Stinski, unpublished data).
FIG. 8.
Analysis of both MIE and US3 RNAs produced by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp in HFF cells at high or low MOI. (A) Schematic diagram of MIE riboprobe. Position of MIE riboprobe (rightward arrow) in relation to +1 start site of MIE RNA is depicted. LS RNAs arise from two sites, LSS1 and LSS2 at base positions -356 and -292, respectively (21, 22). Both MIE and LS RNAs (leftward arrows) possess exons (thick lines; only exons 1, 2, 3, and L [UL126 ORF] are depicted) and introns (thin lines). The riboprobe extends from +171 to −582, spanning the enhancer (Enh), exon 1, and part of intron 1. It is predicted to protect unspliced and spliced MIE RNAs of 170 and 120 nt, respectively. It also can protect an alternatively spliced MIE RNA of ∼140 that was noted previously by Stenberg et al. (49). Mod, modulator. (B) Analysis of MIE and US3 RNAs in infected HFF cells at an MOI of 1.0. Cells were infected in parallel by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp. Total cellular RNA (20 μg) was isolated at 8 hpi and subjected to RNP assay, using both MIE- and US3-specific riboprobes (see Materials and Methods). RNA of uninfected HFF cells (Mock) served as a control. Positions of protected US3 (370 nt) and unspliced (170) and spliced (120 and 140 nt) MIE RNAs (denoted by arrows), as well as selected size markers (Std), are shown. (C) Analysis of MIE and US3 RNAs in infected HFF cells at an MOI of 0.05. Cells were infected in parallel by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp. Total cellular RNA (25 μg) was isolated at 8 hpi and subjected to RNP assay, using both MIE- and US3-specific riboprobes. (D) Analysis of US3 and actin RNAs in infected HFF cells at an MOI of 0.05. Total cellular RNA (25 μg) isolated from WT-, rΔ-582/-1108Egfp-, and rΔ-300/-1108Egfp-infected HFF cells and employed in studies shown in panel C was analyzed by RNP assay, using US3- and cellular actin-specific riboprobes. Positions of protected US3 (370 nt) and actin (230 nt) RNAs (denoted by arrows), as well as selected size markers, are shown. (E) Viral entry into HFF cells. HFF cells were infected at an MOI of 0.05 with WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp in parallel to those shown in panels C and D. Infected cells were thoroughly washed, and DNA was isolated at 5 hpi. This cell-associated DNA was then subjected to QC-PCR that amplifies a region spanning intron 3 of the HCMV IE1 gene (see Materials and Methods). Amplification of viral genomes (gDNA) and IE1 cDNA generates 387- and 217-bp products, respectively. Each QC-PCR mixture contained 600 ng of cell-associated DNA and the indicated amount (10, 3.0, 1.0, or 0.3 pg) of the 1,765-bp IE1 cDNA. QC-PCR products were fractionated on an agarose gel containing ethidium bromide. All steps were done in parallel. The negative control (−) contains uninfected cell DNA but no IE1 cDNA. Arrows indicate positions of 387 bp (gDNA) and 217 bp (cDNA) PCR products, as gauged by DNA size markers that are not shown.
Whole cell RNA was isolated at 8 hpi from HFF cells infected in parallel by WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp at MOIs of 1.0 and 0.05. The RNA samples were analyzed for both US3 and MIE RNAs. The findings of the analyses are shown in Fig. 8B and C. These findings reveal that WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp produce similar amounts of US3 and MIE RNAs at an MOI of 1.0. Furthermore, the amount of MIE RNA originating from the 1+ start site does not appreciably differ among the viruses, and there is no use of alternative or aberrant start sites despite the mutations in rΔ-582/-1108Egfp and rΔ-300/-1108Egfp. Infections conducted at an MOI of 0.05 yield strikingly different findings. Surprisingly, the 20-fold reduction in MOI greatly diminishes the amount of both US3 and MIE RNAs expressed from rΔ-300/-1108Egfp in comparison to WT and rΔ-582/-1108Egfp. This difference cannot be quantitated because both US3 and MIE RNAs are below the level of detection of the RNP assay. The same finding was also documented for another rΔ-300/-1108Egfp isolate that had been constructed separately (data not shown). Notably, the findings in Fig. 8C also reveal that deletion of the unique region and modulator from rΔ-582/-1108Egfp does not appreciably change the amount of US3 or MIE RNA produced relative to that of WT. We confirmed the comparable integrity of these RNA samples by subjecting them to RNP assay, using both US3- and actin-specific riboprobes. As shown in Fig. 8D, the findings of this assay reflect a reproducible diminution in amount of US3 RNA produced by rΔ-300/-1108Egfp.
We determined whether WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp were equivalent in their abilities to enter HFF cells at low MOI. Infections were performed at an MOI of 0.05 in parallel with those used in the RNA studies shown in Fig. 8C and D. The infected HFF cells were thoroughly washed, and DNA was isolated at 5 hpi, a time point well preceding that of viral DNA synthesis. Relative abundance of HCMV genomes was determined by QC-PCR that amplifies a portion of the viral IE1 gene. The findings of this analysis, as shown in Fig. 8E, indicate that the numbers of viral genomes that are cell associated at 5 hpi are comparable (<2-fold difference) for these three viruses. Thus, the impaired production of MIE and US3 RNA by rΔ-300/-1108Egfp is unlikely a result of deficient entry into the cell at low MOI.
DISCUSSION
We have reported previously that deletion of the modulator, -640 to -1108, of the MIE regulatory region has minimal effect on HCMV replication and negligible effect on MIE gene expression in infected HFF, Tera2, or THP-1 cells (31). Here, we extend our genomic deletion analysis to determine the function of the MIE distal enhancer. Of the various deletions made between base positions -300 to -1108 of the MIE regulatory region, only those that remove the distal enhancer (-300 to -582 or -640) markedly affect HCMV replication in HFF cells (Fig. 3, 4, 6, and 7). The distal enhancer's elimination substantially decreases rate of viral DNA replication at an MOI of ≤0.05 but not at an MOI of 1.0. The extent of this decrease is commensurate with the level of reduction in MOI (compare MOIs of 0.001 and 0.05 in Fig. 6 and 7). Cleavage and packaging of the viral genomes appear to progress normally. The virus's ineffectiveness at replicating its DNA at low MOI corresponds to an antecedent underproduction of viral IE RNAs, which involves both MIE and US3 genes (Fig. 7 and 8). Because the MIE gene products are required for viral genome replication (15, 18, 36, 38, 47), we surmise that their reduced expression likely hinders replication. Notably, the replication phenotype of the distal enhancer mutants closely resembles that of recombinant HCMVs lacking IE1 p72 (15, 36). We do not dismiss the possibility that other viral IE gene products (e.g., those encoded by UL36-38, UL115-119, or TRS1/IRS1) may also be reduced in amount to impede viral genome replication, although the US3 gene product is not needed for viral replication in HFF cells (reviewed in reference 10). At an MOI of 1.0, viral DNA replication progresses at a normal rate in association with normal levels of MIE and US3 RNAs, despite the distal enhancer's absence (Fig. 7). Thus, the MIE distal enhancer functions to augment viral IE gene expression and genome replication at low MOI, but this regulatory function is unnecessary at high MOI.
The following considerations substantiate the specificity of the deletion analysis in determining the distal enhancer's role in viral replication and IE gene expression. First, deletions of neighboring upstream regions, such as the unique region or modulator (-582 or -640 to -1108), cause little or no abnormality in production of viral genomes or MIE and US3 RNAs (Fig. 4 and 6 to 8). Second, the inserted marker gene does not confound the findings, since viral DNA replication is unaltered by the insertion itself (Fig. 4). Furthermore, replacement of the MIE distal enhancer with a reconfigured marker gene containing only the 8-bp E1b TATA box as its promoter generates phenotypic abnormalities comparable to that of other HCMVs lacking the distal enhancer (Fig. 6). Similar phenotypic abnormalities are exhibited by a recently constructed HCMV lacking the MIE distal enhancer (-300 to -582) but not possessing a marker gene insertion (data not shown). Last, placement of the distal enhancer in a virus missing this region restores replication kinetics to levels matching those of WT (Fig. 6). These cumulative findings indicate that the MIE enhancer's absence is the cause of impaired HCMV replication and IE gene expression at low MOI.
How does the MIE distal enhancer affect expression of two viral IE genes separated by approximately 21 kbp? Although we do not yet have the answer to this question, we have examined some of the possibilities and contemplated others. Foremost, we verify the reproducibility of this finding among several independently constructed HCMVs lacking the MIE distal enhancer (Fig. 7 and 8), including a recombinant HCMV in which the distal enhancer and unique region were replaced with heterologous DNA not containing a marker gene (data not shown). Three different types of assays were performed to ensure that this finding is not merely the result of difference in titer of infectious virus or ability of virus to penetrate HFF cells. Equivalence in viral entry is maintained even after dilution of viral inocula (Fig. 8E). The distal enhancer's ability to augment the steady-state amount of unspliced +1 MIE RNA at low MOI suggests that this region's functional role extends to the transcriptional regulation of the MIE genes (Fig. 8C). On this basis, we propose that the MIE distal enhancer's function is to increase or hasten transcription of the viral MIE genes at low MOI. The mechanism by which it does so is unclear. The distal enhancer does contain known binding sites for SRF, ELK-1, NF-κB/rel, CREB/ATF, SP1, YY1, and retinoic acid receptor (3, 6, 32, 35), but whether these and/or other cis-acting sites are relevant is speculative. We suspect that the MIE distal enhancer indirectly affects US3 RNA production through its control on expression of MIE gene products, IE1 p72 and IE2 p86. Whether IE1 p72 and IE2 p86 actually stimulate US3 gene expression during infection remains to be determined, but they are known or reputed to activate a wide variety of other viral and cellular genes (35, 47) and can stimulate US3 promoter activity in transient transfection assay (B. J. Biegalke, unpublished data). Of the RNAs traversing US3 at 4 to 8 hpi, the great majority originate from the US3 promoter, whereas a much lesser amount arise from upstream early/late genes (e.g., US6 and US7) (10, 20, 54). Because our US3 RNP assay does not distinguish among these different RNAs, the findings may not precisely reflect the MIE distal enhancer's effect on US3 promoter activity. Further studies, including analyses performed in the presence of protein synthesis inhibitors, are required in order to ascertain the mechanism by which the MIE distal enhancer deletion alters US3 RNA production, as well as determine whether the deletion affects other viral IE genes.
Despite speculation that US3 gene expression is partly linked to the availability of IE1 p72 and IE2 p86, we do not discount the possibility of the MIE distal enhancer having an alternative role in regulating viral IE genes. It is conceivable that a distal enhancer deletion disrupts an unrecognized viral gene encoding a trans-acting factor. However, previous studies of this region have not reported the presence of RNAs colinear with the distal enhancer in a productive infection (1, 19, 49, 51). Nonetheless, viral RNAs of very low abundance emanate from this region on both DNA strands in infected HFF cells (data not shown). The many potential short ORFs that overlap this region do not share appreciable structural homology to other herpesvirus ORFs and are of unknown significance. Murine cytomegalovirus has putative ORFs (m124, m124.1, and m125) in its enhancer, but they do not appear to have a role in viral replication (2). The possibility of the distal enhancer possessing a cis-acting mechanism that can directly enhance or derepress expression of viral IE genes separated by kilobases is an intriguing notion but an unprecedented circumstance. We also cannot exclude the possibility of the distal enhancer possessing a cis-acting function that indirectly secures the timing or efficiency of IE transcriptional events. Such a paradigm might entail, for example, a cis-acting function that facilitates the uncoating of the viral genome, import of the viral genome to a preferred nuclear compartment, or organization of viral chromatin into a form that submits to transcription. For this set of possibilities, it is perplexing that their operations would manifest differently at low versus high MOI.
While the MIE distal enhancer's mechanism of action is unknown, its effect on viral MIE gene expression and genome replication is remarkable. The manner by which this 280 bp region importantly contributes to the HCMV life cycle merits further investigation.
ACKNOWLEDGMENTS
We are grateful to Mark F. Stinski for critical reading of the manuscript. We thank Mark Stinski and members of his laboratory for helpful discussions of this work.
This work was supported by the National Institutes of Health (grant AI-40130), American Cancer Society (Institutional Research Grant IN-122R), and March of Dimes (grant FY99-549). J.L.M. is a recipient of the Burroughs Wellcome Young Investigator Award of the Infectious Disease Society.
REFERENCES
- 1.Akrigg A, Wilkonson G W G, Oram J D. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 1985;2:107–121. doi: 10.1016/0168-1702(85)90242-4. [DOI] [PubMed] [Google Scholar]
- 2.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angulo A, Suto C, Heymen R A, Ghazal P. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol Endocrinol. 1996;10:781–793. doi: 10.1210/mend.10.7.8813719. [DOI] [PubMed] [Google Scholar]
- 4.Baldick C J, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskar J F, Smith P P, Ciment G S, Hoffman S, Tucker C, Tenney D J, Colberg-Poley A M, Nelson J A, Ghazal P. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J Virol. 1996;70:3215–3226. doi: 10.1128/jvi.70.5.3215-3226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan Y-J, Chiou C-J, Huang Q, Hayward G S. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness or primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J Virol. 1996;70:8590–8605. doi: 10.1128/jvi.70.12.8590-8605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee M S, Bankier S, Beck S, Bohni R, Brown C R, Horsnell T, Hutchisno III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 8.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Colberg-Poley A M. Functional roles of immediate early proteins encoded by the human cytomegalovirus UL36-38, UL115-119, TRS1/IRS1 and US3 loci. Intervirology. 1996;39:350–360. doi: 10.1159/000150506. [DOI] [PubMed] [Google Scholar]
- 11.Gonczol E, Andrews P W, Plotkin S A. Cytomegalovirus infection of human teratocarcinoma cells. J Gen Virol. 1985;66:509–515. doi: 10.1099/0022-1317-66-3-509. [DOI] [PubMed] [Google Scholar]
- 12.Gonczol E, Andrews P W, Plotkin S A. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science. 1984;224:159–161. doi: 10.1126/science.6322309. [DOI] [PubMed] [Google Scholar]
- 13.Graham F L, van der Eb A J. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the E. coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 15.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahn G, Knust E, Schmolla H, Sarre T, Nelson J A, McDougall T K, Fleckenstein B. Predominant immediate early transcripts of human cytomegalovirus AD169. J Virol. 1984;49:363–370. doi: 10.1128/jvi.49.2.363-370.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T R, Muzithras V P. Fine mapping of transcripts expressed from the US6 gene family of human cytomegalovirus strain AD169. J Virol. 1991;65:2024–2036. doi: 10.1128/jvi.65.4.2024-2036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo K J X, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothari S, Baillie J, Sissons J G, Sinclair J H. The 21bp repeat element of the human cytomegalovirus major immediate early enhancer is a negative regulator of gene expression in undifferentiated cells. Nucleic Acids Res. 1991;19:1767–1771. doi: 10.1093/nar/19.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kothary R, Barton S C, Franz T, Norris M L, Hettle S, Surani M A H. Unusual cell specific expression of a major human cytomegalovirus immediate early gene promoter-lacZ hybrid gene in transgenic mouse embryo. Mech Dev. 1991;35:25–31. doi: 10.1016/0925-4773(91)90038-8. [DOI] [PubMed] [Google Scholar]
- 25.LaFemina R, Hayward G S. Constitutive and retinoic acid-inducible expression of cytomegalovirus immediate-early genes in human teratocarcinoma cells. J Virol. 1986;58:434–440. doi: 10.1128/jvi.58.2.434-440.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu R, Baillie J, Sissons J G, Sinclair J H. The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in non-permissive cells. Nucleic Acids Res. 1994;22:2453–2459. doi: 10.1093/nar/22.13.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundquist C A, Meier J L, Stinski M F. A strong transcriptional negative regulatory region between the human cytomegalovirus UL127 gene and the major immediate-early enhancer. J Virol. 1999;73:9032–9052. doi: 10.1128/jvi.73.11.9039-9052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1505. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier J L, Stinski M F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier J L, Stinski M F. Regulation of cytomegalovirus immediate early genes. Intervirology. 1996;39:331–342. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 33.Mendelson M, Monard S, Sissons P, Sinclair J. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 34.Minton E J, Tysoe C, Sinclair J H, Sissons J G. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocarski E. Cytomegalovirus and its replication. In: Fields B N, editor. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2523. [Google Scholar]
- 36.Mocarski E S, Kemble G, Lyle J, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1 149aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson J A, Groudine M. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol Cell Biol. 1986;6:452–461. doi: 10.1128/mcb.6.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roller R J, Herold B C. Characterization of a BHK(TK−) cell clone resistant to postattachment entry by herpes simplex virus types 1 and 2. J Virol. 1997;71:5805–5813. doi: 10.1128/jvi.71.8.5805-5813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadowski I J, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 41.Shelbourn S L, Kothari S K, Sissons J G P, Sinclair J H. Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acid Res. 1989;17:9165–9171. doi: 10.1093/nar/17.22.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair J, Sissons P. Cytomegalovirus: latent and persistent infection of monocytes and macrophages. Intervirology. 1996;39:293–301. doi: 10.1159/000150501. [DOI] [PubMed] [Google Scholar]
- 43.Sinclair J H, Baillie J, Bryant L A, Taylor-Wiedeman J A, Sissons J G. Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J Gen Virol. 1992;73:433–435. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 44.Sinzger C, Grefte A, Plachter B, Gouw A S H, Hauw The T, Jahn G. Fibroblasts, epithelial cells, endothelial cells, and smooth muscle cells are the major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol. 1995;76:741–750. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 45.Sinzger C, Jahn G. Human cytomegalovirus cell tropism and pathogenesis. Intervirology. 1996;39:302–319. doi: 10.1159/000150502. [DOI] [PubMed] [Google Scholar]
- 46.Soderberg-Naucler N C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus from by allogenic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 47.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 48.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involve common and unique protein domains. J Virol. 1990;1990:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stinski M F, Roehr T J. Activation of the major immediate-early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J Virol. 1985;55:431–441. doi: 10.1128/jvi.55.2.431-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L C. Organization and expression of the immediate-early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 53.Taylor-Wiedeman J A, Sissons J G P, Sinclair J H. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenney D J, Santomenna L D, Goudie K B, Colberg-Poley A M. The human cytomegalovirus US3 immediate-early protein lacking the putative transmembrane domain regulates gene expression. Nucleic Acids Res. 1993;21:2931–2937. doi: 10.1093/nar/21.12.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomsen D R, Stinski M F. Cloning of the human cytomegalovirus genome as XbaI fragments. Gene. 1981;16:207–216. doi: 10.1016/0378-1119(81)90077-9. [DOI] [PubMed] [Google Scholar]
- 56.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate early gene (US3) by a silencer/enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Laer D, Meyer-Koenig U, Serr A, Finke J, Kanz L, Fauser A A, Neumann-Haefelin D, Brugger W, Hufert F T. Detection of cytomegalovirus DNA in CD34+ cells from blood and bone marrow. Blood. 1995;86:4086–4090. [PubMed] [Google Scholar]
- 58.Zweidler-McKay P A, Grimes H L, Flubacher M M, Tsichlis P N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]