ABSTRACT
The DNA in bacterial viruses collectively contains a rich, yet relatively underexplored, chemical diversity of nucleobases beyond the canonical adenine, guanine, cytosine, and thymine. Herein, we review what is known about the genetic and biochemical basis for the biosynthesis of complex DNA modifications, also called DNA hypermodifications, in the DNA of tailed bacteriophages infecting Escherichia coli and Salmonella enterica. These modifications, and their diversification, likely arose out of the evolutionary arms race between bacteriophages and their cellular hosts. Despite their apparent diversity in chemical structure, the syntheses of various hypermodified bases share some common themes. Hypermodifications form through virus-directed synthesis of noncanonical deoxyribonucleotide triphosphates, direct modification DNA, or a combination of both. Hypermodification enzymes are often encoded in modular operons reminiscent of biosynthetic gene clusters observed in natural product biosynthesis. The study of phage-hypermodified DNA provides an exciting opportunity to expand what is known about the enzyme-catalyzed chemistry of nucleic acids and will yield new tools for the manipulation and interrogation of DNA.
KEYWORDS: DNA synthesis, hypermodified bases, nucleotide metabolism, nucleotides, oligonucleotides
The tailed double-stranded DNA (dsDNA) viruses of bacteria (Caudovirales) contain the largest diversity of naturally occurring noncanonical deoxynucleotides anywhere in the biological world. To date, more than 21 modified nucleotides have been observed in the DNA of bacteriophages (1), and many more modifications await discovery (2). Beyond the well-understood and relatively simple methyltransferase-catalyzed modifications to DNA at N-6 in adenosine, as well as N-4 and C-5 in cytosine, each of the bases in phage DNA can harbor other modifications that are complex both in their chemical structures and in their biosynthesis pathways (3). These modifications deriving from sugars, amino acids, and deazapurines mirror the diversity of hypermodifications found in tRNAs (4). It should come as no surprise then that the microcosm of phages infecting Escherichia coli and Salmonella species reflects this diversity as a whole, and among the phages of EcoSal can be found examples of hypermodifications to each of the four canonical bases of DNA.
THE CHEMICAL LANDSCAPE OF DNA MODIFICATION
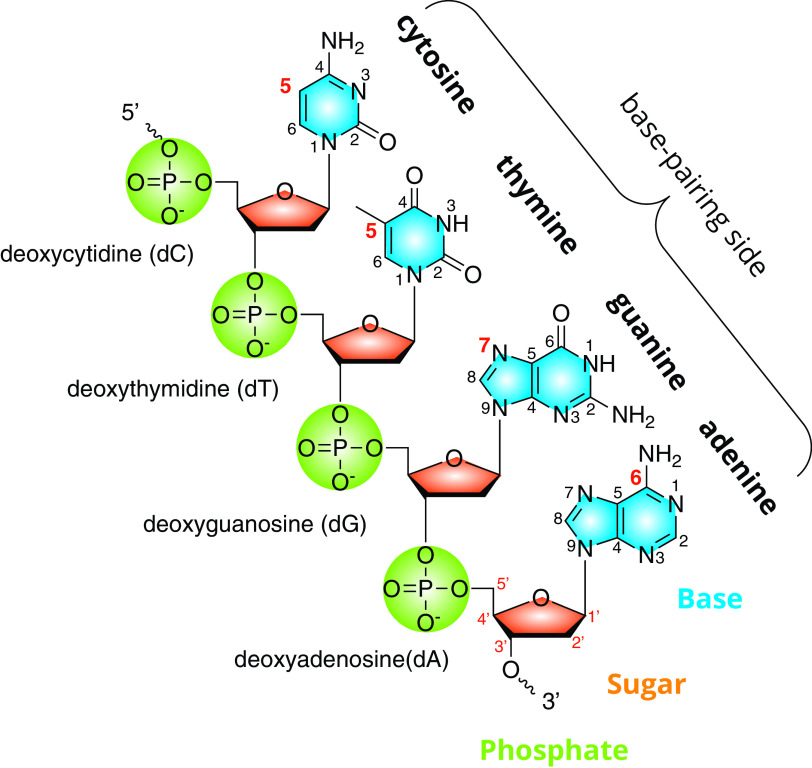
Similar to its cellular hosts, the DNA of the Caudovirales is a complementary double-stranded heteropolymer of nucleotides, each composed of a phosphate, a 2′-deoxyribose, and one of four bases, including adenine, guanine, cytosine, and thymine, as shown in Fig. 1. The nucleotide forms of the bases (i.e., with one or more phosphates), as well as the corresponding nucleoside (i.e., the base and sugar alone) are adenosine (A), guanosine (G), cytidine (C), and thymidine (T). Where clarity is required, these single-letter abbreviations are preceded by a lowercase “d” (abbreviation for 2′-deoxyribose) in order to distinguish them from their RNA counterparts. The nucleobase portion of the nucleotide is the primary site of DNA modification. The nucleobases of DNA consist of the heterocyclic purines (adenine and guanine) and the pyrimidines (cytosine and thymine). Abbreviations for modified bases often indicate the atom at the point of attachment between the modifying substituent and the base heterocycle according to the commonly accepted numbering scheme (e.g., 5mC for 5-methylcytidine; see also Fig. 1). A similar numbering scheme exists for the atoms of the sugar backbone, where each number is followed by a prime symbol (e.g., 5′-PO4 and 3′-OH) to differentiate them from the atoms of the base.
FIG 1.
DNA and its substituents. A schematic representation of a single strand of DNA composed of four different nucleotides showing the component parts phosphate, 2′-deoxyribose, and nucleobase. The four nucleotides are linked via a phosphodiester bond between the 3′ and 5′ carbon of the adjacent base. The atoms of the nucleobase heterocycles, as well as the deoxyribose, are numbered according to accepted chemical convention.
The nucleobase portion of the DNA carries out several functions. Principally, it supplies the information for templated synthesis of a complementary strand during replication and transcription. DNA is maintained in the dsDNA state via hydrogen bonding between the bases of antiparallel strands across the Watson-Crick interface. Modifications and adducts along this edge of the base potentially interfere with base pairing. Therefore, there are limited portions of the DNA base that can tolerate modification, which may partly explain the greater diversity of modifications seen in tRNA where regions of the molecule are not required to conform to Watson-Crick bonding geometries. To date, the sites of hypermodification seen in phages of Enterobacteriaceae are at position N-7 of dG, N-6 of dA, and C-5 of both dC and dT, and examples of these are shown in Fig. 2. Biological modifications to the phosphate backbone of DNA do occur as phosphorothioates (5), where a nonbridging oxygen is replaced with sulfur, but these have not yet been observed in bacteriophages. Modifications to the sugar portion of nucleotides occur in RNA in the form of methylation at the 2′ OH of the ribose (6), but the 2′-deoxyribose of DNA lacks this acceptor group, and the 3′ oxygen is occupied as part of the phosphodiester bond linking adjacent nucleotides, so one would not expect modifications to the sugar portion of nucleotides in DNA to occur.
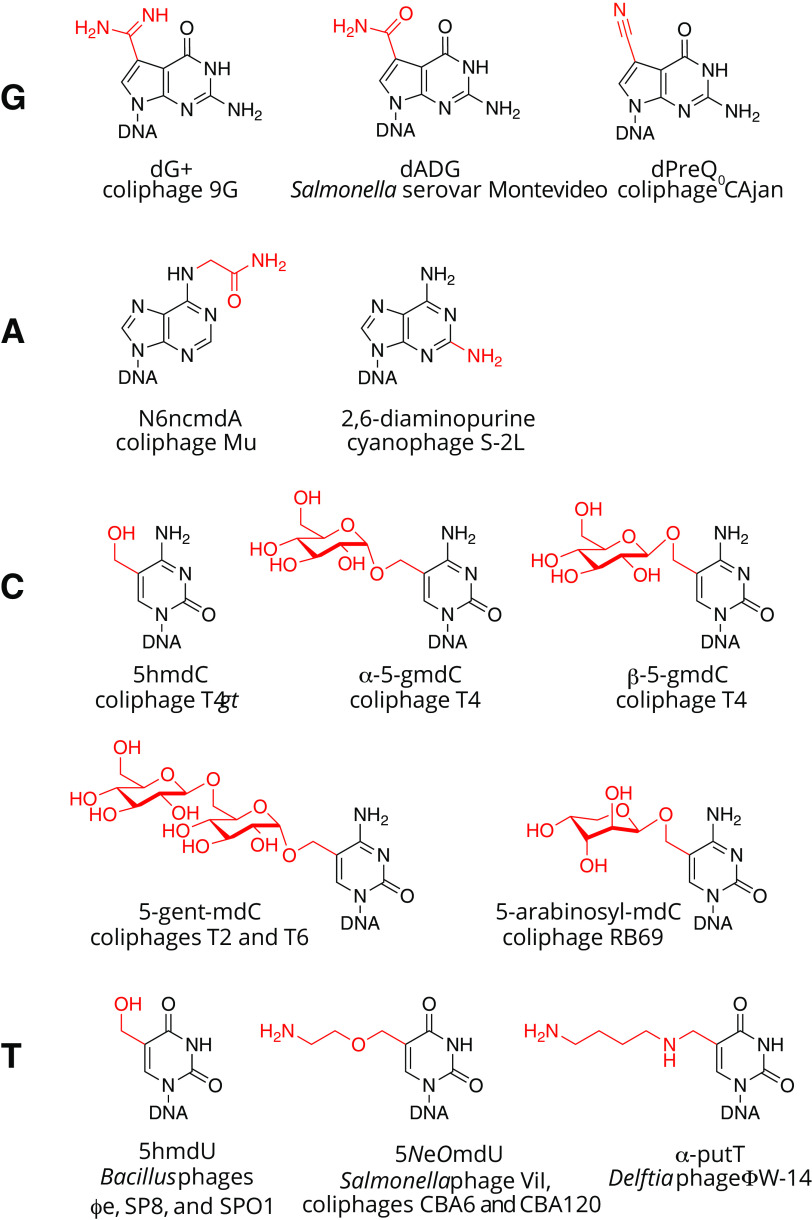
FIG 2.
Examples of base hypermodifications observed in bacteriophage DNA.
THE ROLES OF HYPERMODIFIED DNA
Although we choose here to focus on the biochemical and genetic aspects of the synthesis of hypermodified bases, these modifications undoubtedly affect viral fitness. Hypermodified bases in bacteriophage DNA evolved, in part, under the selection pressure of an arms race between viruses and their hosts. Nucleobase hypermodifications render viral DNA resistant to cleavage by the endonuclease components of restriction modification (RM) systems of their cellular hosts (7–11), the most common system of defense in bacteria. In response, cellular hosts respond to the “armor” of hypermodified DNA through the evolution of modification-dependent endonucleases, such as the glucosylmethylcytidine-targeting type IV restriction endonuclease GmrSD (12, 13), encoded by a subset of E. coli strains (and discussed in further detail below). Other analogous networks of measure/countermeasures are sure to be found in the phages of E. coli and Salmonella. An understanding of the biochemical and genetic basis for the synthesis of DNA hypermodifications complements our understanding of their biological roles and can also provide the basis for predicting which phages contain hypermodified bases in their DNA.
Base modifications may also have more subtle functions affecting viral fitness. Modified bases contribute to DNA polymers with physicochemical properties different from their canonical counterparts. For example, the cyanophage S-2L fully replaces its adenines with 2-aminoadenine (14–16). This extra amino group at the 2 position of the purine ring can hydrogen bond with normally unpaired keto-oxygen at position 2 of thymine. The result is an A-T triple hydrogen bond and a DNA polymer having a melting temperature (Tm) much higher than would be predicted from a canonical DNA of equal GC content. DNA of the Delftia phage ΦW-14 contains a putrescine moiety attached to the 5 position of thymidine (α-putT) (17). Under physiological conditions, the free amino group of the putrescine adduct is likely positively charged and may counteract the negative charge of the phosphate DNA backbone. Experiments measuring DNA length and capsid volume indicate that the DNA of this phage is packed at a 25% higher density in the capsid than phages with canonical DNA (18). Finally, modification-dependent gene regulation could be an additional function potentially provided by modified bases as suggested by phage transcription factor TF1-specific binding of 5-hydroxymethyl-2′-deoxyuridine (5hmU) DNA in Bacillus phage SPO1 (19).
HOW DNA IS MODIFIED
Bacteriophages use a range of mechanisms operating before and/or after DNA replication to synthesize DNA containing modified bases (3, 20, 21). A range of bases can be completely or partially modified. Some phages have one base completely substituted for another. Other phages replicate their DNA with canonical bases but modify a subset of them just prior to encapsidation. Still others completely replace one kind of nucleotide with one containing a noncanonical base, and these noncanonical bases are further modified postreplicatively. Many of these pathways of modification rely on a combination of host and phage-encoded functions such that the phage can be viewed as a metabolic engineer subverting the host metabolism for its own ends with brutal efficiency.
THE HYPERMODIFIED BASES
Guanosine. (i) Discovery of deazaguanine modifications in DNA.
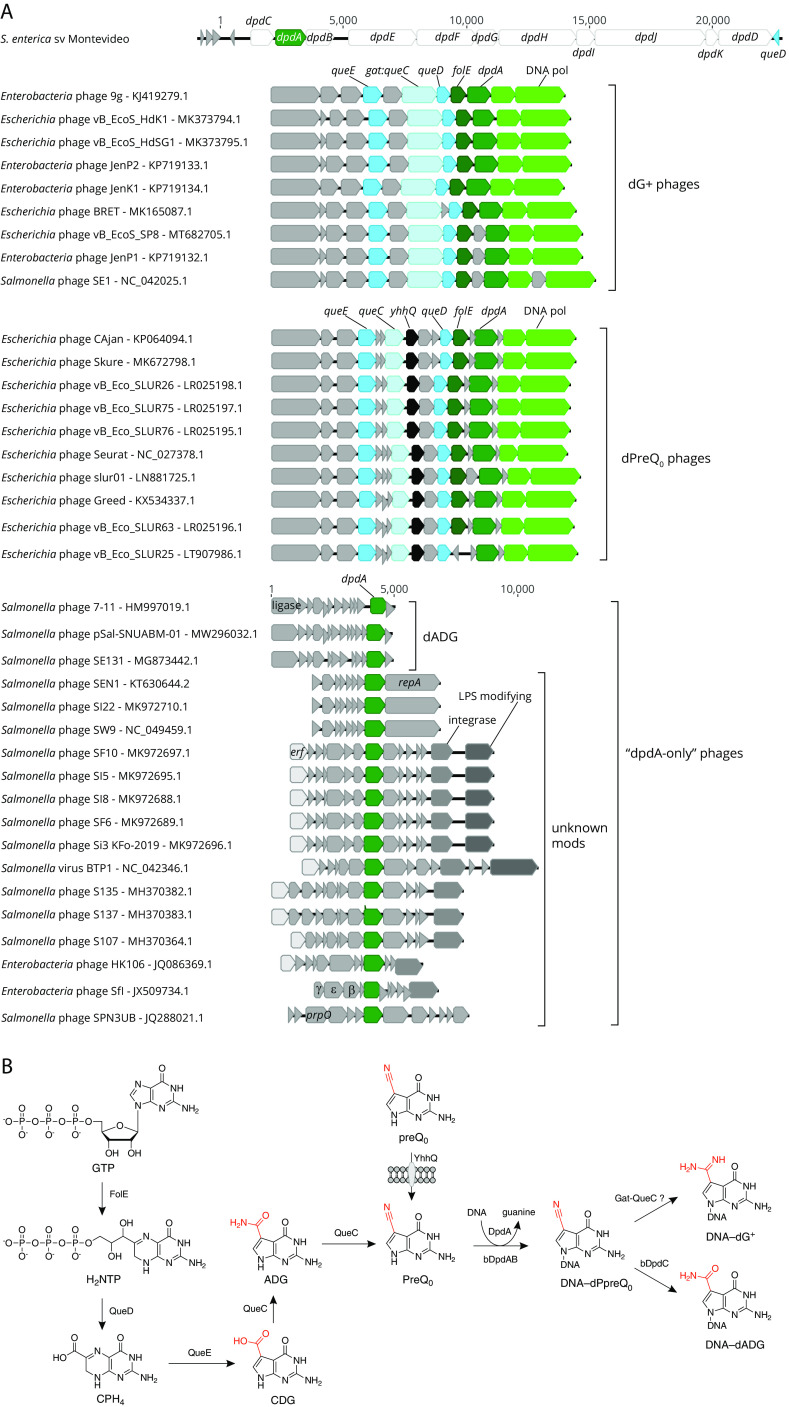
Deazaguanine derivatives are well-known modifications of tRNA molecules. In bacteria and eukaryotes, queuosine (Q) is found at position 34 of tRNAs harboring GUN anticodons (4). In archaea, archaeosine (G+) is found at position 14 or 15 of numerous tRNAs (4). The discovery of paralogs of tRNA-guanine transglycosylase (TGT), the signature enzyme of Q and G+ synthesis pathways, clustering with DNA processing genes in bacteria and phage genomes suggested that deazaguanine derivatives could also be found in DNA (22). These TGT paralogs were initially detected in E. coli E22 and S. enterica subsp. enterica serovar Montevideo (22). Small amounts of 7-amido-7-deazaguanine (ADG; 0.04% Gs) and even fewer amounts of 7-cyano-7-deazaguanine (preQ0; 0.00025% Gs) were detected, in the genomic DNA of S. Montevideo, both being precursors of the Q and G+ tRNA modification (4). The presence of these two modifications was associated with a cluster of 11 genes (Fig. 3A). Indeed, the deletion of this whole cluster in S. Montevideo led to the disappearance of both dADG and dPreQ0. This cluster of genes was renamed dpd, for deazapurine in DNA, with DpdA being the paralog of TGT. This cluster was detected in 44 E. coli and 80 S. enterica genomes at the time of the analysis, most of them of the Montevideo serovar, and one Tennessee (22). It was later shown that only 3 of the 11 genes were required for the synthesis of the dADG modification, including dpdA, dpdB, and dpdC (23). Another homolog of TGT/DpdA was found encoded in Escherichia phage 9g, strongly clustering with homologs of genes involved in G+ synthesis and DNA-processing enzymes (22) (Fig. 3A). Further analysis showed that 25% of the guanines in DNA were replaced by the G+ base in this phage (8, 22). Additional analyses of phage genomes later revealed that phages harbored 4 different deazaguanine modifications, including preQ0, ADG, G+, and 7-aminomethyl-7-deazaguanine (preQ1) (7, 24). It was shown that DpdA alone was sufficient for dPreQ0 modification in vivo, but the addition of Gat-QueC was required to obtain dG+ (7). Based on genome sequences, additional phages of Escherichia and Salmonella are predicted to be modified with deazaguanines (7, 25).
FIG 3.
Genes and biosynthesis of deazaguanosine DNA. (A) Subgenomic regions depicting biosynthetic gene clusters that lead to dG+, dPreQ0, or dADG for bacteria-modified DNA are shown. (B) Biosynthetic pathways leading to deazaguanosine modifications in DNA are shown. The phage genomes can be classified into the following 3 groups: phages encoding the full pathway to dPreQ0 (dPreQ0 phages) or to dG+ (dG+ phages), and phages encoding only DpdA (“dpdA-only” phages). In addition to encoded biosynthetic functions discussed in the text, landmark genes are indicated, such as DNA polymerase (green), integrase, erf (essential for recombination function), a lambda-like beta/exo/gam cassette, and lipopolysaccharide (LPS)-modifying genes (gtrC-like).
(ii) Path to preQ0.
All sequenced E. coli and S. enterica genomes encode the entire de novo pathway, leading to the insertion of Q in tRNAs (7). One of the Q pathway intermediates is preQ0, which is also used by DpdA to modify DNA. Four enzymes are required to produce preQ0 from GTP, as diagrammed in Fig. 3B (4). Some phages encode all of these enzymes as well, such as Escherichia phages 9g and CAjan (Fig. 3A) (7, 22). The first step of preQ0 synthesis is shared with that of tetrahydrofolate (THF) (26). GTP cyclohydrolase I (FolE) catalyzes the formation of dihydroneopterin triphosphate (H2NTP) from GTP, releasing formic acid (26, 27). This enzyme is a tunnel-fold (T-fold) enzyme, using a Zn2+ cation cofactor (28). The first dedicated step of the preQ0 synthesis pathway is the hydrolysis of H2NTP into 6-carboxyl-5,5,7,8-tetrahydropterin (CPH4), catalyzed by CPH4 synthase (QueD) (29). This enzyme is also a T-fold protein and a member of the COG0720 family that contains close homologs involved in THF and BH4 synthesis that are difficult to differentiate based on sequence similarity alone (30). CPH4 is then transformed into 7-carboxy-7-deazaguanine (CDG) by CDG synthase (QueE). This enzyme is a member of the radical S-adenosyl-l-methionine (SAM) superfamily (31). The carboxyl group of CDG is then replaced by a nitrile group to form preQ0. This reaction is catalyzed by preQ0 synthase (QueC) consuming two ATP molecules and going through an ADG intermediate (32, 33). E. coli and S. enterica also encode a specific transporter of preQ0 and preQ1 (another intermediate of the Q pathway) called YhhQ (Fig. 3B) (33). Some phages also encode this transporter, such as Escherichia phage CAjan (Fig. 3A) (7).
(iii) DpdA is the central protein of this DNA modification pathway.
DpdA is a transglycosylase that replaces guanine in DNA by preQ0, similar to its TGT homolog in tRNA (7, 23, 34). This is the first step, and for some phages the last, in modification of DNA by deazaguanines. Both the bacterial and phage DpdA proteins can modify plasmid DNA with preQ0 if this base is available (7, 23). No structure is available to date, but it has been shown that DpdA shares essential amino acids with TGT, notably involved in substrate specificity and binding and catalytic activity (7, 22). These conserved amino acids were confirmed by modeling DpdA from Escherichia phage CAjan (34). It also seems that in this model, DpdA would have difficulty fully binding to a double-stranded DNA molecule, and a conformational change or a partial melting of the DNA would be required (34). The sequence specificity for Escherichia coli CAjan DpdA has also been determined. This enzyme recognizes two short sequences, GA and GGC (34). The sequence specificity of the bacterial DpdA is still to be determined. Though it can bind DNA by itself, it requires DpdB to modify DNA (23). DpdB is a distant homolog of DndB (22), a regulator involved in the sequence specificity of the phosphorothioate DNA modification in Streptomyces lividans (35). This indicates that DpdB could play a major role in the sequence recognition of the bacterial DNA modification system.
(iv) Beyond preQ0.
Though DpdA inserts preQ0 in DNA, it is not always the final modification observed (Fig. 3B). For example, ADG is present in S. Montevideo. Although, ADG is an intermediate in the synthesis of preQ0 from CDG by QueC, it was shown that ADG observed in S. Montevideo DNA is produced by the oxidation of preQ0 already inserted in DNA, a reaction catalyzed by DpdC (23). It is to be noted that ADG was also found in phage genomes, including Salmonella phage 7-11, that encode only for a DpdA (Fig. 3A) (7). The biosynthetic pathway has yet to be fully determined. Another variation is found in Escherichia phage 9g that, similar to archaeal tRNAs, harbors a G+ base. Indeed, phage 9g encodes a homolog of a fusion between a glutamine amidotransferase class II (GAT) domain and QueC previously shown to be involved in G+ synthesis in archaea (22, 36). It was shown that dG+ in DNA can be produced when both phage 9g DpdA and Gat-QueC were produced in an E. coli already producing the preQ0 base (7). It was also found that halovirus HVTV-1 DNA is modified with preQ1, most likely by the activity of the homolog of QueF encoded in HVTV-1 genome (7). Thus far, no phages of E. coli or S. enterica were found to encode a homolog of QueF, making it unlikely to find preQ1 in the phages of these species.
(v) Role of deazaguanine modifications.
For the bacterial dpd system, the main hypothesis so far is that it is a restriction modification system. Indeed, unmodified plasmids transform with a lesser efficiency than ADG-modified plasmids (∼600-fold difference) in S. Montevideo encoding the dpd cluster (22). No difference in transformation efficiency was observed when the recipient strain lacked the dpd cluster. The dpdD-J genes are not required for the modification synthesis, and it was only possible to delete the genes involved in the ADG modification in a dpdD mutant (23). The deletion of dpdD, dpdE, dpdG, dpdI, dpdJ, or dpdK, but not dpdF or dpdH, resulted in a high efficiency of transformation of unmodified plasmids. These phenotypes were complemented by expression in trans of the deleted gene, suggesting that all of these genes are involved in restriction (23).
To date, no restriction enzyme linked to the presence of deazaguanines was identified in phages. However, Escherichia phages modified with deazaguanines were found to be resistant to various degrees to a large set of restrictions enzymes, but each modification confers resistance to a different set of enzymes. For example, Escherichia phage 9g, modified by G+, is completely resistant to EcoRI and SwaI, but Escherichia phage CAjan, modified by preQ0, is not (7, 8, 37).
Adenosine. (i) An enigmatic adenosine derivative in phage Mu.
To date, the only known postreplicatively hypermodified adenine found in DNA is α-N-(9-,3D-2′-deoxyribofuranosylpurin-6-yl)glycinamide of E. coli bacteriophage Mu, shown in Fig. 2. Originally abbreviated as dA'x, this modification is also known as ncm6dA (short for N-6-aminocarboxymethyl-2′-deoxyadenosine) to more closely follow naming conventions for RNA hypermodifications. Approximately 15% of adenine bases are thus modified in phage Mu virion DNA (38) and are known to confer resistance to restriction endonuclease systems in vivo (39) and in vitro (40). Mutants defective in the protective function were mapped to the Mu mom gene (modification of Mu) encoding the Mom enzyme, whose expression has been shown to be subject to an unusual combination of epigenetic, transcriptional, and translational control mechanisms (41).
Despite extensive investigations into the complexities of mom expression, the mechanism by which the Mom enzyme synthesizes ncm6dA is not well understood, though interesting clues have been found. Using a combination of sensitive homology detection and structural modeling, Kaminska and Bujnicki revealed that Mom is a member of the GCN5-related N-acetyltransferase (GNAT) superfamily (42). They further pointed out that the original work on the structure of the Mu-modified adenosine did not distinguish between the currently accepted structure (as presented in Fig. 2) and a structural isomer consisting of a glycinamide group appended to N-6 of adenine. However, a coenzyme A (CoA) cofactor carrying a glycinamide group has never been observed, and later work by Karambelkar and coworkers, using collisional-induced dissociation-mass spectrometry (CID-MS), supports the accepted structure of ncm6dA (43). In that work, Karambelkar et al. also showed that recombinant expression of the Mom enzyme alone in E. coli was sufficient to convert a subset of adenines to the hypermodified form. Although they were unable to reconstitute the adenosine modification in vitro, purified Mom was shown to bind iron with micromolar affinity and also to bind acetyl-CoA. Attempting to reconcile these observations, they proposed a modification pathway drawing elements from mono-iron/alpha-ketoglutarate (Fe/αKG)-dependent dioxygenases and the proposed modification pathway of the Elongator complex, a multisubunit tRNA-modifying enzyme that uses acetyl-CoA to synthesize 5-aminocarboxymethyluridine (ncm5U). Key differences between ncm6dA and ncm5U are attachment of the modifying group at N-6 in adenine versus C-5 in uracil, respectively, and in the tRNA modification, after acetyl transfer, there is an amination step, presumably carried out by a subunit in the Elongator complex. Currently, the identity of the group donor(s) in the Mom enzyme DNA modification reaction and how an acetyl group is aminated by the same enzyme are unknown.
(ii) A-T triple hydrogen bonds in the DNA of the Salmonella phage PMBT28?
Recent papers published back-to-back in the journal Science detail the elucidation of the biosynthesis and polymerization of 2-amino-2′-deoxyadenosine (aka 2,6-diaminopurine, N2dA, or “dZ”) (15, 16, 44). N2dA DNA is resistant to a variety of restriction endonucleases targeting dA-containing sequences in vitro (45). The groups of Suwen Zhao and Pierre Kaminski independently demonstrated the two-step biosynthesis of N2dA in a pathway mirroring the cellular biosynthesis of adenosine ribonucleotides. The first step of N2dA biosynthesis is catalyzed by a phage-encoded enzyme, PurZ, a homolog of cellular adenylosuccinate synthetase (PurA). PurA catalyzes the formation of succinyladenosine monophosphate from XMP, glutamate, and ATP. A second cellular enzyme, adenylosuccinate lyase (PurB), cleaves a fumarate moiety from succinyladenosine monophosphate to yield AMP. Similarly, PurZ can form succinyl deoxyguanosine monophosphate from dGMP, glutamate, and ATP. Cleavage of a fumarate moiety from this nucleotide by the host-encoded PurB enzyme produces N2dAMP, which is, in turn, phosphorylated by phage- and host-encoded nucleotide kinases producing N2dATP for utilization by phage-encoded DNA polymerases. The Zhao group demonstrated an active recombinant PurZ could be derived from the genome sequence of the Salmonella phage PMBT28 (GenPept accession no. AUZ95522.1), suggesting the native DNA of this virus also contains the triple hydrogen bond-forming N2dA nucleotide.
Cytidine. (i) Sugar-coated DNA.
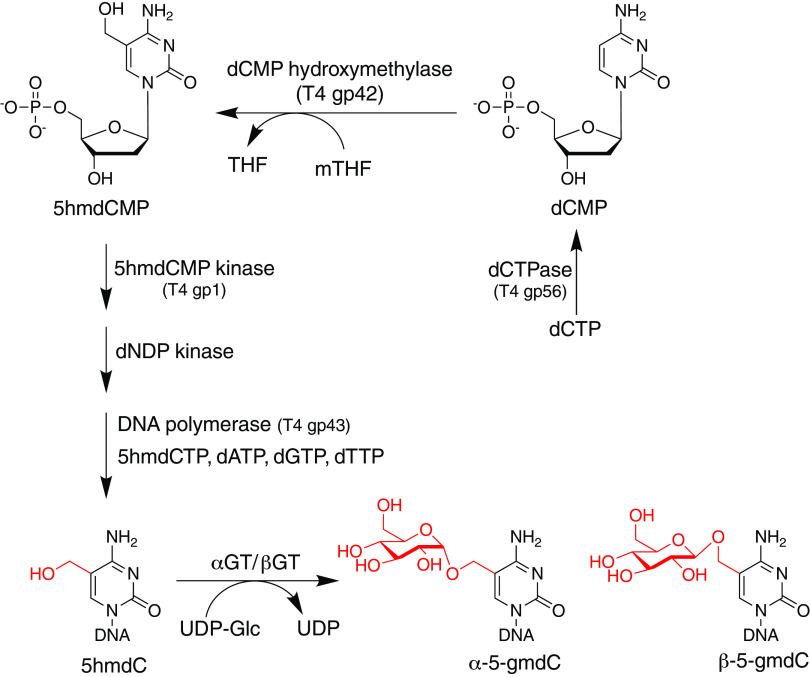
T4 and related T4-like phages contain sugar-modified hydroxymethylcytosines fully replacing cytosines. Of the total C-coding nucleotides in phage T4, approximately 70% are 5-(α-d-glucosyl)oxymethylcytosine (α-5gmC), and 30% are 5-(β-d-glucosyl)oxymethylcytosine (β-5gmC) (46). The two isomers of glucosylmethylcytosine differ in the stereochemistry at the anomeric carbon where the glucose moiety is attached to the methyl group at 5C via an ether linkage (46). Phage T6 additionally contains cytosines modified with a disaccharide, 5-(α-gentiobiosyl)oxymethylcytosine (α-5gentmC) (46). For Escherichia phage RB69, cytidines are analogously glycosylated, but evidence for arabinose as the modifying sugar has been reported (47). The major steps of the 5-glucosylmethylcytosine biosynthesis in phage T4 are shown in Fig. 4 and discussed in further detail below. Genome diagrams of other phages of E. coli and Salmonella predicted to have similarly modified cytosines are shown in Fig. 5. Although there is currently no available structure for glucosylmethylcytidine in the context of a DNA polymer, the attachment site at position 5 within the pyrimidine ring suggests the polar hexose moieties line DNA's major groove and likely alter both the steric topography as well as hydration patterns along the polymer. The differences between steric accessibility as well as altered solvation between canonical DNA versus glucosylated DNA almost certainly account for the extraordinary resistance of this modified DNA to cleavage by restriction endonucleases (11).
FIG 4.
Biosynthetic pathway of glucosylated methylcytosine.
FIG 5.

Genome maps of T4 and related phages. Schematic representation with open reading frames indicated as gray or colored boxes. Landmark genes such as rIIA (black) and DNA polymerase (green) are shown. Phage genes involved in DNA modification are also indicated, including dCMP hydroxymethylase (yellow) and DNA glucosyltransferases (brown). Note: many of the sequences obtained from GenBank were circularized in silico and the origin renumbered in order to follow the historical T4 convention of having rIIA as the first gene, albeit in reverse-complement orientation.
(ii) The biosynthesis of 5-glucosylmethylcytidine.
The biosynthesis of sugar-modified methylcytosines in the T4 and T4-related phages, summarized in Fig. 4, occurs in two stages, resulting from a combination of pre- and postreplicative mechanisms. The first stage is the synthesis of a phage DNA precursor containing 5-hydroxymethyl-2′-dCTP (5hmdCTP) completely replacing the dCTP pool. During lytic development, phage-encoded and host-encoded enzymes work in concert to eliminate dCTP from the nucleotide pool and produce its analog, 5hmdCTP from 5-hydroxymethyl-2′-dCMP (dCMP). During T4 phage DNA replication, the phage-encoded DNA polymerase (T4 gp43) incorporates 5hmdC into newly synthesized DNA. Following DNA replication but preceding packaging of the phage DNA into the capsid, the hydroxyl moieties accept a glucose from sugar carrier UDP glucose (UDP-Glu) in reactions catalyzed by phage-encoded DNA glucosyltransferases (48).
The major steps in the prereplicative stage are shown in Fig. 4. The T4 dCTPase (T4 gp56) converts dCTP to dCMP (49). dCMP is converted to 5hmdCMP by T4 gp42 (50), a thymidylate-synthase homolog (51), using 5,10-methylenetetrahydrofolate (mTHF) as a carbon donor (52). The amino acid residues of T4 gp42 dictating dCMP versus dUMP specificity have been determined, and residues correlating with resolution of a product intermediate to the hydroxymethyl form have also been identified, making it possible, in principle, to predict the synthesis of 5hmdCMP from sequence alone (3). The 5hmdCMP is converted to the diphosphate form by a phage-encoded dNMP kinase (T4 gp1) and is subsequently phosphorylated to the triphosphate form by E. coli dNDP kinase. After 5hmdC is polymerized into DNA by the T4 DNA polymerase (gp43), two DNA transferases, α-glucosyltransferase (AGT) and β-glucosyltransferase (BGT), convert 5hmC to their α- and β-glucosyloxymethylcytosine derivatives. Note that even though the hypermodified bases are glucosylated hydroxymethylcytosines, we use the term glucosylmethyl (5 gm) rather than frequently used glucosylhydroxymethyl (5ghm) since the bridging oxygen between m5C and the sugar is not protonated. Although both T4 enzymes glucosylating 5hmC belong to the GT-B fold glycosyltransferase family, BGT is an inverting transferase, and AGT is retaining (53).
(iii) The biology of T4 glucosyl-methylcytosine.
In addition to a detailed understanding of the genetic and biochemical basis of 5gmdC biosynthesis, much is known about the role DNA hypermodification plays in the conflict between T4 and its host. In vitro, T4 genomic DNA is highly resistant to cleavage by restriction endonucleases (10, 11). In vivo, glucosylated cytosines provide protection against the McrBC E. coli restriction system. McrBC is a type IV restriction endonuclease with a substrate dependence on 5hmdC and, to a lesser degree, 5-methyldeoxycytidine (5mdC). The genes encoding McrBC (54) were discovered as the locus of resistance to infection by phage T4 mutants deficient in glucosylation (called T4gt) and were originally called rgl (for restricts glucoseless) (55). Although DNA glucosylation provides protective functions to the phage, E. coli has evolved genome defense systems specifically targeting glucosylated bases. GmrSD, encoded by a subset of E. coli strains, is a type IV restriction endonuclease largely dependent on glucosylated bases for cleavage activity (12). Phage T4 has evolved an additional layer of countermeasure in the form of IPI* (13), a small DNA-mimicking protein (56) injected along with DNA into the host at the time of infection. Less clear is the effect of 5gmC on the activity of E. coli adaptive immunity systems, such as CRISPR/Cas. An initial study investigating this question (57) suggested that Cas9 endonuclease could not restrict T4, but subsequent papers revealed a more complex picture of partial resistance to CRISPR/Cas systems (58–60). The importance of glucosylated cytosine in genome defense is further supported by the T4 Arn protein, a protein mimic of DNA targeting E. coli McrBC, a restriction endonuclease targeting 5hmC in the absence of glucosylation (61, 62). Thus, T4 encodes a backup system in the event of the loss of glucosylation. Additional epigenetic regulatory functions for 5gmC are indicated by the finding that the T4 transcriptional activator MotA binds much more tightly to DNA containing 5gmC than its canonical counterpart, suggesting that the T4 modification aids in T4 expression (63).
Thymidine. (i) 5-NeOmdU, a complex modification in widespread phages of enterobacteria.
The Salmonella phage Vi1 and related phages CBA6 and CBA120 of E. coli O157:H7 were shown to contain a hypermodified thymidine, 5-(2-aminoethoxy)methyluridine (5-NeOmdU) (64). 5-NeOmdU joins a growing list of phage-synthesized thymidine hypermodifications that includes 5-aminoethyl-2′-deoxyuridine (5-NedU) in the Pseudomonas phage M6 (64), 5-glutamylthymidine (α-glu-T) in the Bacillus phage SP10 (65), and 5-putrescinylthymdine (α-put-T) in the Delftia phage ΦW-14 (17, 66). The chemical structures of 5-NeOmdU and α-put-T are shown in Fig. 2. 5-NeOmdU had not been previously observed in any phage DNA, despite the Vi1 phage having been in culture since at least 1936 (67). Salmonella phage Vi1 originated in culture as part of a phage-typing scheme being specific for the Vi antigen of Salmonella typhi (68, 69), which was later shown to be chemically identical to the O157 antigen of pathogenic E. coli (70). The intense interest in isolating phages specific to E. coli O157 as biocontrol agents, and the subsequent sequencing of their genomes, has resulted in an abundance of Vi1-like virus sequences populating GenBank and the establishment of the essentially identical Viunalikevirus and Kuttervirus genera (71, 72). A similar scenario has developed with the sequencing of phages specific to Salmonella enterica serovar Enteriditis strains. Genome maps of E. coli and Salmonella phages from these genera are shown in Fig. 6 and reveal a remarkable degree of synteny.
FIG 6.
Genome maps of Vi1 and related phages. Schematic representation with open reading frames indicated as gray or colored boxes. Landmark genes such as rIIA (black) and DNA polymerase (green) are shown. Genes involved in thymidine hypermodification are also indicated, including dUMP hydroxymethylase (yellow), 5-hmdU DNA kinases (pink), and aGPT-PPlases (blue). Note: many of the sequences obtained from GenBank were circularized in silico and the origin renumbered in order to follow the historical T4 convention of having rIIA as the first gene, albeit in reverse-complement orientation. Applying this convention to the genomes of Vi1 and related phages reveals a noteworthy degree of synteny among geographically diverse phage genomes.
(ii) Hypermodification occurs through mechanisms acting before and after DNA replication.
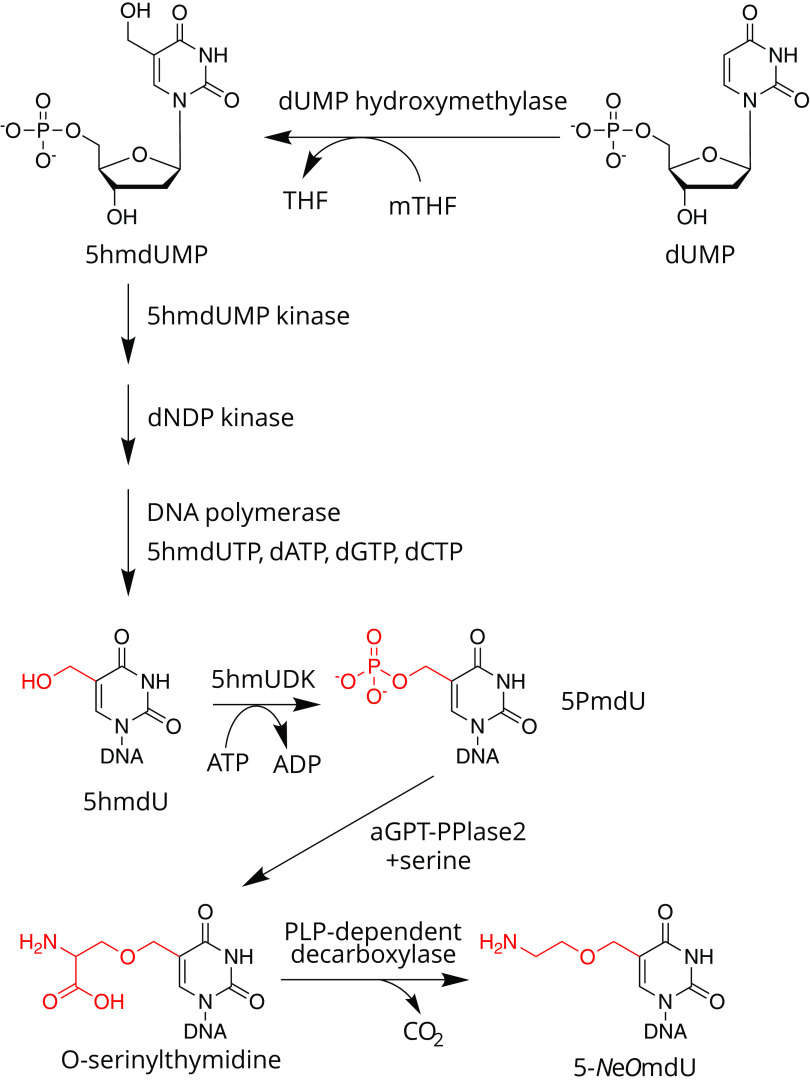
The Vi1-like phages, together with ΦW-14 and SP10, share a metabolic program in common with the 5hmU phages of Bacillus, a broad family that includes SPO1, SP8, and ϕe (73–75). The 5hmU phages use an analogous strategy to T4 to synthesize DNA containing 5hmU, fully replacing thymine, as illustrated in Fig. 7. Phage-infected cells express a dTTPase and synthesize 5-hydroxymethyl-2′-deoxyuridine monophosphate (5hmdUMP) from 2′-deoxyuridine monophosphate (dUMP) via dUMP hydroxymethylase (76), a variant of the classical thymidylate synthase (77). This enzyme transfers a single carbon unit from the C-1 donor 5,10-methylene tetrahydrofolate (mTHF) to the C-5 of the uracil ring, forming an exocyclic methylene intermediate. In the canonical thymidylate synthase, this exocyclic methylene is reduced to methyl via hydride transfer from tetrahydrofolate. But in the reaction catalyzed by dUMP hydroxymethylase, the methylene is instead hydroxylated by nucleophilic attack of a water molecule (78). This noncanonical nucleotide then feeds into the deoxynucleotide triphosphate pool for incorporation into DNA by the phage-encoded DNA polymerase. In the hmdU phages, this modification likely serves as protection against host-encoded endonucleases. From a chemical perspective, the installation of a nucleophilic hydroxyl group at the 5-methyl of thymidine serves as a reactive “handle” upon which additional molecules can be attached and sculpted. Phages Vi1, ΦW-14, and SP10 build on top of this 5hmU in two ways. First, a subset of the 5hmU nucleotides is further modified to form the hypermodified base. Second, the remaining 5hmU nucleotides that have not been hypermodified are converted to thymidine (65, 79–81). Following DNA modification, DNA is packaged into capsids as part of the virion morphogenetic pathway leading up to lysis.
FIG 7.
Biosynthetic pathway of 5-aminoethoxymethyl-2′-deoxyuridine (5-NeOmdU)-modified DNA. Biosynthesis of 5-NeOmdU begins at the nucleotide pool level with the formation of 5hmdUMP by a phage-encoded thymidylate synthase homolog. After conversion to the triphosphate form and incorporation into DNA polymer, 5-hmU is phosphorylated by a phage-encoded nucleobase kinase (5-HMUDK) producing 5PmdU. The phosphate of this intermediate is displaced by serine in a reaction catalyzed by aGPT-Pplase2 to produce O-serinylthymidine. Decarboxylation of O-serT yields the mature modification, 5-NeOmdU. 5hmU nucleobases not converted to 5-NeOmdU are instead converted to thymdine by an as-yet-unknown 5hmU reductase.
In work spanning the late 1960s to 1984, the laboratories of Heman Witmer at the University of Chicago and Tony Warren at the University of British Columbia in Canada laid the groundwork for understanding the postreplicative steps of thymidine hypermodification. Using a combination of conditional mutants, isotopic radiolabeling, and other techniques, including thin-layer chromatography and small-molecule ion-exchange chromatography, both groups were able to show that hypermodification in SP10 and ΦW-14 proceeded using the same chemical intermediate, at the time identified as pyrophosphorylated thy (65, 82). Both groups proposed that the pyrophosphate was needed to activate the C-5 methyl group and serve as a leaving group for either a substitution reaction with an incoming adduct or for reduction to thymidine. However, the enzyme activities downstream of this pyrophosphorylation step were not fully understood during this time, nor were the genes known.
Clues to the genetic basis of thymidine hypermodification in Vi1 and related phages came from computational biological investigations by Aravind and colleagues (83, 84). Using a combination of highly sensitive homology detection together with observations from genome context and genetic conservation across genomes, they deduced genes predicted to be involved in the biosynthesis of hypermodified thymidines in the Delftia phage ΦW-14 and the Bacillus phage SP10, including a 5hmU kinase predicted to synthesize phosphorylated 5hmU previously seen as an intermediate in the hypermodification process of these two phages. Using mixtures of recombinant crude extracts derived from cultures expressing predicted hypermodification enzymes, Lee et al. were able to reconstitute the conversion of 5hmU to 5-NeOmdU in vitro (64). Further biochemical characterization of the 5hmU kinase did not lead to formation of a pyrophosphorylated thymidine (85). Nonetheless, monophosphorylated thymidine (PmdU) was shown to be chemically competent to serve as a substrate in the hypermodification reaction (85).
Using recombinant lysate-catalyzed hypermodification reactions in various combinations, Lee and coworkers were able to work out what enzymes were necessary and sufficient to hypermodify 5hmU (85). This hypermodification pathway is summarized in Fig. 7. After phosphorylation of approximately 40% of 5hmdU residues by a kinase (Vi1 gp67), a second enzyme transfers a serine-5-methyl group in a nucleophilic substitution reaction between a free serine side chain hydroxyl and the 5-methyl group. This enzyme, Vi1gp247, is predicted to contain a DNA-glycosylase fold belonging to the helix-hairpin-helix family, despite not having a glycosylase activity. Following serinylation, the α-C-carboxy group is removed via decarboxylation by Vi1 gp226, a pyridoxyl-5′-phosphate (PLP) cofactor-dependent DNA decarboxylase, leading to the mature modification consisting of an amino ethylhydroxy group attached to the 5-methyl group by an ethoxy linkage.
(iii) Function of 5-NeOmdU.
It is not yet known what affect the 5-NeOmdU modification has on the physicochemical properties of DNA. However, the modification can serve as a steric inhibitor of endonuclease cleavage. In recent work by Flodman et al., it was shown that up to 60% of commercially available enzymes fail to cut DNA containing 5-NeOmdU in vitro (9). Given how widespread this modification is predicted to be in the Vi1-like phages, it seems likely that bacterial hosts would have evolved a 5-NeOmdU-dependent endonuclease targeting this modification. Although no epigenetic gene regulatory function has been assigned to hypermodified thymidines, 5hmdU has been shown to modulate gene expression in the Bacillus phage SPO1 (19, 86).
DNA HYPERMODIFICATION: OUTLOOK AND BEYOND
Base hypermodification and the intersection between the RNA and DNA worlds.
DNA and RNA modifications are related, as proteins involved in the synthesis of these modifications often evolved from one another. For example, DNA methyltransferases probably first originated from RNA-modifying enzymes (87). Methyltransferase played a crucial role in the appearance of DNA as we know it. Indeed, dT is a dU methylated in C-5 prior to polymerization by a thymidine synthase (88). Other phage DNA modifications are related to other RNA modifications. DpdA that inserts preQ0 in DNA is a close homolog of the archaeal TGT that inserts preQ0 at position 14 or 15 of the transfer RNA and then is further modified in archaeosine (4). In bacteria, TGT evolved to insert preQ1 at position 34 of GUN-anticodon tRNAs, which is then further modified in queuosine (4). Though 5hmC is also found in RNA from all domains of life (89), the mechanism of insertion is different from its DNA counterpart. Indeed, the hydroxyl group is added to a 5mC by the TET proteins (90, 91). Some TET proteins evolved to modify mammalian DNA from 5mC to 5hmC (92) that can be further oxidized in 5-formylcytosine and 5-carboxylcytosine (93). TET enzyme homologs called J-binding proteins (JBPs) also participate in forming 5hmU from T in mammalian DNA but do not further oxidize it (94). These DNA modifications seem to be involved in activation and repression of transcription (95). No 5hmU was identified in RNA (91).
Engineering chemical and biological functions through base hypermodification.
Given that canonical bases in regular nonmodified DNA exhibit only limited chemical functionality (thus contributing to the genetic stability of DNA), the diversification of modified base observed in bacteriophages evokes the technological potential of using these newly identified functional groups for manipulating DNA molecules. As shown in Fig. 2, various kinds of substituents amend the native bases, and those chemically active functional groups, such as hydroxyl or amino, are of particular interest, as they can serve as “handles” for further functionalization. For example, the modifications of Salmonella phage Vi1 and Delftia phage ΦW-14 each display a reactive primary amine on DNA. Lee and coworkers demonstrated the use of amine-reactive chemical reagents such as 3-(4-carboxybenzoyl)-quinoline-2-carboxaldehyde (CBQCA) and fluorescamine to specifically label the DNA with fluorescent dyes (64). DNA base-modifying enzymes, together with unnatural substrates, can be used to introduce other chemical handles. One successful example uses phage T4's beta-glucosyltransferase (BGT), which transfers a glucose moiety from the UDP glucose (UDP-Glu) to the hydroxymethyl group of 5hmdC in DNA. The laboratory of Chuan He demonstrated that BGT can take a nonnative substrate (UDP-6-N-3-Glu) and transfer the azide containing sugar to 5hmdC on DNA (96). This functionalization allowed downstream labeling of DNA with biotin via click chemistry and was used in the aforementioned study for mapping the epigenetic 5hmdC sites on DNA. Other DNA base-modifying enzymes could potentially be used in similar ways. For example, tRNA base-modifying enzyme tRNA guanine transglycosylase (TGT) swaps out the guanine base with 7-deazaguanine derivatives in situ to the RNA polymer and has been demonstrated to be capable of mediating the site-specific tRNA functionalization with many 7-deazaguanine analogs (97). The DNA-modifying TGT homolog, DpdA, similarly incorporates 7-deazaguanine derivatives into DNA, suggesting the possibility of using the DpdA to functionalize DNA molecules for labeling or altering DNA properties.
In addition to chemical manipulation of DNA, base hypermodification might be harnessed to alter biological properties of recombinant DNA and phage therapeutics. It was proposed that deazaguanine DNA modifications could be used to protect DNA from degradation upon entry into wild-type strains (international patent application no. PCT/US20/21886). Indeed, DpdA protein and preQ0 are the only two components required to modify mobile elements such as phages, transposons, or plasmids to shield them against the main system of defense from bacteria, restriction enzymes. DNA modified in this way could not only be used to increase the transformation efficiency of plasmids being moved into wild-type industrial bacterial strains but also potentially broaden the host range of phages used as antimicrobial therapies. Lastly, mutation and other manipulations of base hypermodification genes in phages might lead to a deeper understanding of genome conflict systems, including identification of other modification-dependent restriction endonucleases (98) and observing the interplay of modification versus other, less well-characterized gene clusters having antiphage activity (99).
Contributor Information
Peter R. Weigele, Email: weigele@neb.com.
Deborah Hinton, National Institutes of Health.
REFERENCES
- 1.Sood AJ, Viner C, Hoffman MM. 2019. DNAmod: the DNA modification database. J Cheminform 11:30. doi: 10.1186/s13321-019-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y-J, Weigele PR. 2021. Detection of modified bases in bacteriophage genomic DNA. Methods Mol Biol 2198:53–66. doi: 10.1007/978-1-0716-0876-0_5. [DOI] [PubMed] [Google Scholar]
- 3.Weigele P, Raleigh EA. 2016. Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev 116:12655–12687. doi: 10.1021/acs.chemrev.6b00114. [DOI] [PubMed] [Google Scholar]
- 4.Hutinet G, Swarjo MA, de Crécy-Lagard V. 2017. Deazaguanine derivatives, examples of crosstalk between RNA and DNA modification pathways. RNA Biol 14:1175–1184. doi: 10.1080/15476286.2016.1265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Jiang S, Deng Z, Dedon PC, Chen S. 2019. DNA phosphorothioate modification—a new multi-functional epigenetic system in bacteria. FEMS Microbiol Rev 43:109–122. doi: 10.1093/femsre/fuy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiss T, Jády BE. 2004. Functional characterization of 2’-O-methylation and pseudouridylation guide RNAs. Methods Mol Biol 265:393–408. doi: 10.1385/1-59259-775-0:393. [DOI] [PubMed] [Google Scholar]
- 7.Hutinet G, Kot W, Cui L, Hillebrand R, Balamkundu S, Gnanakalai S, Neelakandan R, Carstens AB, Lui CF, Tremblay D, Jacobs-Sera D, Sassanfar M, Lee Y-J, Weigele P, Moineau S, Hatfull GF, Dedon PC, Hansen LH, Crécy-Lagard V. 2019. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat Commun 10:5442. doi: 10.1038/s41467-019-13384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai R, Corrêa IR, Xu MY, Xu S-Y. 2017. Restriction and modification of deoxyarchaeosine (dG+)-containing phage 9 g DNA. Sci Rep 7:8348. doi: 10.1038/s41598-017-08864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flodman K, Tsai R, Xu MY, Corrêa IR, Copelas A, Lee Y-J, Xu M-Q, Weigele P, Xu S. 2019. Type II restriction of bacteriophage DNA with 5hmdU-derived base modifications. Front Microbiol 10:584. doi: 10.3389/fmicb.2019.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang LH, Farnet CM, Ehrlich KC, Ehrlich M. 1982. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res 10:1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flodman K, Corrêa IR, Dai N, Weigele P, Xu S. 2020. In vitro type II restriction of bacteriophage DNA with modified pyrimidines. Front Microbiol 11:604618. doi: 10.3389/fmicb.2020.604618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bair CLC, Black LWL. 2007. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol 366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bair CL, Rifat D, Black LW. 2007. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J Mol Biol 366:779–789. doi: 10.1016/j.jmb.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirnos MD, Khudyakov IY, Alexandrushkina NI, Vanyushin BF. 1977. 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 270:369–370. doi: 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, Xu X, Wei Y, Cheng Y, Guo Y, Khudyakov I, Liu F, He P, Song Z, Li Z, Gao Y, Ang EL, Zhao H, Zhang Y, Zhao S. 2021. A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science 372:512–516. doi: 10.1126/science.abe4882. [DOI] [PubMed] [Google Scholar]
- 16.Sleiman D, Garcia PS, Lagune M, Loc'h J, Haouz A, Taib N, Röthlisberger P, Gribaldo S, Marlière P, Kaminski PA. 2021. A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science 372:516–520. doi: 10.1126/science.abe6494. [DOI] [PubMed] [Google Scholar]
- 17.Kropinski AM, Bose RJ, Warren RA. 1973. 5-(4-Aminobutylaminomethyl)uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage ΦW-14. Biochemistry 12:151–157. doi: 10.1021/bi00725a025. [DOI] [PubMed] [Google Scholar]
- 18.Scraba DG, Bradley RD, Leyritz-Wills M, Warren RA. 1983. Bacteriophage ΦW-14: the contribution of covalently bound putrescine to DNA packing in the phage head. Virology 124:152–160. doi: 10.1016/0042-6822(83)90298-2. [DOI] [PubMed] [Google Scholar]
- 19.Sayre MH, Geiduschek EP. 1988. TF1, the bacteriophage SPO1-encoded type II DNA-binding protein, is essential for viral multiplication. J Virol 62:3455–3462. doi: 10.1128/JVI.62.9.3455-3462.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren R. 1980. Modified bases in bacteriophage DNAs. Annu Rev Microbiol 34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 21.Gommers-Ampt JH, Borst P. 1995. Hypermodified bases in DNA. FASEB J 9:1034–1042. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- 22.Thiaville JJ, Kellner SM, Yuan Y, Hutinet G, Thiaville PC, Jumpathong W, Mohapatra S, Brochier-Armanet C, Letarov AV, Hillebrand R, Malik CK, Rizzo CJ, Dedon PC, de Crécy-Lagard V. 2016. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci U S A 113:E1452–E1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y, Hutinet G, Valera JG, Hu J, Hillebrand R, Gustafson A, Iwata-Reuyl D, Dedon PC, de Crécy-Lagard V. 2018. Identification of the minimal bacterial 2′-deoxy-7-amido-7-deazaguanine synthesis machinery. Mol Microbiol 110:469–483. doi: 10.1111/mmi.14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crippen CS, Lee Y-J, Hutinet G, Shajahan A, Sacher JC, Azadi P, de Crécy-Lagard V, Weigele PR, Szymanski CM. 2019. Deoxyinosine and 7-deaza-2'-deoxyguanosine as carriers of genetic information in the DNA of Campylobacter viruses. J Virol 93:307–314. doi: 10.1128/JVI.01111-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngazoa-Kakou S, Shao Y, Rousseau GM, Addablah AA, Tremblay DM, Hutinet G, Lemire N, Plante P-L, Corbeil J, Koudou A, Soro BK, Coulibaly DN, Aoussi S, Dosso M, Moineau S. 2019. Complete genome sequence of Escherichia coli siphophage BRET. Microbiol Resour Announc 8:66–63. doi: 10.1128/MRA.01644-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips G, Yacoubi BE, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. 2008. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP cyclohydrolase I. J Bacteriol 190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Nakagawa N, Kuramitsu S, Yokoyama S, Masui R. 2005. Novel reaction mechanism of GTP cyclohydrolase I. High-resolution X-ray crystallography of Thermus thermophilus HB8 enzyme complexed with a transition state analogue, the 8-oxoguanine derivative. J Biochem 138:263–275. doi: 10.1093/jb/mvi120. [DOI] [PubMed] [Google Scholar]
- 28.Colloc'h N, Poupon A, Mornon J-P. 2000. Sequence and structural features of the T-fold, an original tunnelling building unit. Proteins 39:142–154. doi:. [DOI] [PubMed] [Google Scholar]
- 29.McCarty RM, Somogyi A, Bandarian V. 2009. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry 48:2301–2303. doi: 10.1021/bi9001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips G, Grochowski LL, Bonnett S, Xu H, Bailly M, Blaby-Haas C, Yacoubi BE, Iwata-Reuyl D, White RH, de Crécy-Lagard V. 2012. Functional promiscuity of the COG0720 family. ACS Chem Biol 7:197–209. doi: 10.1021/cb200329f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis JK, Bruender NA, Bandarian V. 2018. QueE: a radical SAM enzyme involved in the biosynthesis of 7-deazapurine containing natural products. Methods Enzymol 606:95–118. doi: 10.1016/bs.mie.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelp MT, Bandarian V. 2015. A single enzyme transforms a carboxylic acid into a nitrile through an amide intermediate. Angew Chem Int Ed Engl 54:10627–10629. doi: 10.1002/anie.201504505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zallot R, Yuan Y, de Crécy-Lagard V. 2017. The Escherichia coli COG1738 member YhhQ is involved in 7-cyanodeazaguanine (preQ0) transport. Biomolecules 7:12. doi: 10.3390/biom7010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kot W, Olsen NS, Nielsen TK, Hutinet G, de Crécy-Lagard V, Cui L, Dedon PC, Carstens AB, Moineau S, Swairjo MA, Hansen LH. 2020. Detection of preQ0 deazaguanine modifications in bacteriophage CAjan DNA using Nanopore sequencing reveals same hypermodification at two distinct DNA motifs. Nucleic Acids Res 48:10383–10396. doi: 10.1093/nar/gkaa735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang J, Wang Z, He X, Li J, Zhou X, Deng Z. 2007. DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res 35:2944–2954. doi: 10.1093/nar/gkm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips G, Swairjo MA, Gaston KW, Bailly M, Limbach PA, Iwata-Reuyl D, de Crécy-Lagard V. 2012. Diversity of archaeosine synthesis in Crenarchaeota. ACS Chem Biol 7:300–305. doi: 10.1021/cb200361w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulikov EE, Golomidova AK, Letarova MA, Kostryukova ES, Zelenin AS, Prokhorov NS, Letarov AV. 2014. Genomic sequencing and biological characteristics of a novel Escherichia coli bacteriophage 9g, a putative representative of a new Siphoviridae genus. Viruses 6:5077–5092. doi: 10.3390/v6125077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. 1983. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2’-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci U S A 80:7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toussaint A. 1976. The DNA modification function of temperate phage Mu-1. Virology 70:17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- 40.Allet B, Bukhari AI. 1975. Analysis of bacteriophage Mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol 92:529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- 41.Hattman S. 1999. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol Ther 84:367–388. doi: 10.1016/s0163-7258(99)00042-x. [DOI] [PubMed] [Google Scholar]
- 42.Kaminska KH, Bujnicki JM. 2008. Bacteriophage Mu Mom protein responsible for DNA modification is a new member of the acyltransferase superfamily. Cell Cycle 7:120–121. doi: 10.4161/cc.7.1.5158. [DOI] [PubMed] [Google Scholar]
- 43.Karambelkar S, Udupa S, Gowthami VN, Ramachandra SG, Swapna G, Nagaraja V. 2020. Emergence of a novel immune-evasion strategy from an ancestral protein fold in bacteriophage Mu. Nucleic Acids Res 48:5294–5305. doi: 10.1093/nar/gkaa319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pezo V, Jaziri F, Bourguignon P-Y, Louis D, Jacobs-Sera D, Rozenski J, Pochet S, Herdewijn P, Hatfull GF, Kaminski P-A, Marliere P. 2021. Noncanonical DNA polymerization by aminoadenine-based siphoviruses. Science 372:520–524. doi: 10.1126/science.abe6542. [DOI] [PubMed] [Google Scholar]
- 45.Szekeres M, Matveyev AV. 1987. Cleavage and sequence recognition of 2,6-diaminopurine-containing DNA by site-specific endonucleases. FEBS Lett 222:89–94. doi: 10.1016/0014-5793(87)80197-7. [DOI] [PubMed] [Google Scholar]
- 46.Lehman IR, Pratt EA. 1960. On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem 235:3254–3259. doi: 10.1016/S0021-9258(20)81347-7. [DOI] [PubMed] [Google Scholar]
- 47.Thomas JA, Orwenyo J, Wang L-X, Black LW. 2018. The odd “RB” phage-identification of arabinosylation as a new epigenetic modification of DNA in T4-like phage RB69. Viruses 10:313. doi: 10.3390/v10060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kornberg SR, Zimmerman SB, Kornberg A. 1961. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J Biol Chem 236:1487–1493. doi: 10.1016/S0021-9258(18)64202-4. [DOI] [PubMed] [Google Scholar]
- 49.Munro JL, Wieberg JS. 1968. Evidence that gene 56 of bacteriophage T4 is a structural gene for deoxycytidine triphosphatase. Virology 36:442–446. doi: 10.1016/0042-6822(68)90169-4. [DOI] [PubMed] [Google Scholar]
- 50.Epstein RH, Bolle A, Steinberg CM, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar RS, Susman M, Denhardt GH, Lielausis A. 1963. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harb Symp Quant Biol 28:375–394. doi: 10.1101/SQB.1963.028.01.053. [DOI] [Google Scholar]
- 51.Lamm N, Wang Y, Mathews CK, Rüger W. 1988. Deoxycytidylate hydroxymethylase gene of bacteriophage T4. Nucleotide sequence determination and over-expression of the gene. Eur J Biochem 172:553–563. doi: 10.1111/j.1432-1033.1988.tb13925.x. [DOI] [PubMed] [Google Scholar]
- 52.Flaks JG, Cohen SS. 1959. Virus-induced acquisition of metabolic function. I. Enzymatic formation of 5-hydroxymethyldeoxycytidylate. J Biol Chem 234:1501–1506. doi: 10.1016/S0021-9258(18)70038-0. [DOI] [PubMed] [Google Scholar]
- 53.Parker MJ, Lee Y-J, Weigele PR, Saleh L. 2020. 5-methylpyrimidines and their modifications in DNA, p 465–488. In Liu HW, Begley TP (ed), Comprehensive natural products III, vol. 5, Elsevier, Amsterdam, Netherlands. doi: 10.1016/B978-0-12-409547-2.14838-3. [DOI] [Google Scholar]
- 54.Raleigh EA. 1992. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol Microbiol 6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 55.Revel HR. 1967. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology 31:688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- 56.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. 2008. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol 375:720–734. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yaung SJ, Esvelt KM, Church GM. 2014. CRISPR/Cas9-mediated phage resistance is not impeded by the DNA modifications of phage T4. PLoS One 9:e98811. doi: 10.1371/journal.pone.0098811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bryson AL, Hwang Y, Sherrill-Mix S, Wu GD, Lewis JD, Black L, Clark TA, Bushman FD. 2015. Covalent modification of bacteriophage T4 DNA inhibits CRISPR-Cas9. mBio 6:e00648-15. doi: 10.1128/mBio.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Dai L, Dong J, Chen C, Zhu J, Rao VB, Tao P. 2020. Covalent modifications of bacteriophage genome confer a degree of resistance to bacterial CRISPR systems. J Virol 94:e01630-20. doi: 10.1128/JVI.01630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vlot M, Houkes J, Lochs SJA, Swarts DC, Zheng P, Kunne T, Mohanraju P, Anders C, Jinek M, van der Oost J, Dickman MJ, Brouns SJJ. 2018. Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Res 46:873–885. doi: 10.1093/nar/gkx1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dharmalingam K, Revel HR, Goldberg EB. 1982. Physical mapping and cloning of bacteriophage T4 anti-restriction endonuclease gene. J Bacteriol 149:694–699. doi: 10.1128/jb.149.2.694-699.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho C-H, Wang H-C, Ko T-P, Chang Y-C, Wang AH-J. 2014. The T4 phage DNA mimic protein Arn inhibits the DNA binding activity of the bacterial histone-like protein H-NS*. J Biol Chem 289:27046–27054. doi: 10.1074/jbc.M114.590851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuypers MG, Robertson RM, Knipling L, Waddell MB, Moon K, Hinton DM, White SW. 2018. The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA. Nucleic Acids Res 46:5308–5318. doi: 10.1093/nar/gky292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee Y-J, Dai N, Walsh SE, Müller S, Fraser ME, Kauffman KM, Guan C, Corrêa IR, Weigele PR. 2018. Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc National Acad Sci 115:E3116–E3125. doi: 10.1073/pnas.1714812115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Witmer H. 1981. Synthesis of deoxythymidylate and the unusual deoxynucleotide in mature DNA of Bacillus subtilis bacteriophage SP10 occurs by postreplicational modification of 5-hydroxymethyldeoxyuridylate. J Virol 39:536–547. doi: 10.1128/JVI.39.2.536-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelln RA, Warren RA. 1973. Studies on the biosynthesis of alpha-putrescinylthymine in bacteriophage FW-14-infected Pseudomonas acidovorans. J Virol 12:1427–1433. doi: 10.1128/JVI.12.6.1427-1433.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Craigie J, Brandon KF. 1936. Bacteriophage specific for the O‐resistant V form of B. typhosus. J Pathol 43:233–248. doi: 10.1002/path.1700430202. [DOI] [Google Scholar]
- 68.Pickard D, Toribio AL, Petty NK, van Tonder A, Yu L, Goulding D, Barrell B, Rance R, Harris D, Wetter M, Wain J, Choudhary J, Thomson N, Dougan G. 2010. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J Bacteriol 192:5746–5754. doi: 10.1128/JB.00659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craigie J, Brandon KF. 1936. The identification of the V and W forms of B. typhosus and the occurrence of the V form in cases of typhoid fever and in carriers. J Pathol 43:249–260. doi: 10.1002/path.1700430203. [DOI] [Google Scholar]
- 70.Samuel G, Hogbin JP, Wang L, Reeves PR. 2004. Relationships of the Escherichia coli O157, O111, and O55 O-antigen gene clusters with those of Salmonella enterica and Citrobacter freundii, which express identical O antigens. J Bacteriol 186:6536–6543. doi: 10.1128/JB.186.19.6536-6543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matilla MA, Fang X, Salmond GPC. 2014. Viunalikeviruses are environmentally common agents of horizontal gene transfer in pathogens and biocontrol bacteria. ISME J 8:2143–2147. doi: 10.1038/ismej.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adriaenssens EM, Ackermann H-W, Anany H, Blasdel B, Connerton IF, Goulding D, Griffiths MW, Hooton SP, Kutter EM, Kropinski AM, Lee J-H, Maes M, Pickard D, Ryu S, Sepehrizadeh Z, Shahrbabak SS, Toribio AL, Lavigne R. 2012. A suggested new bacteriophage genus: “Viunalikevirus”. Arch Virol 157:2035–2046. doi: 10.1007/s00705-012-1360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, Pedulla ML. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 388:48–70. doi: 10.1016/j.jmb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kropinski A, Turner D, Nash J, Ackermann H-W, Lingohr E, Warren R, Ehrlich K, Ehrlich M. 2018. The sequence of two bacteriophages with hypermodified bases reveals novel phage-host interactions. Viruses 10:217–214. doi: 10.3390/v10050217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoet PP, Coene MM, Cocito CG. 1992. Replication cycle of Bacillus subtilis hydroxymethyluracil-containing phages. Annu Rev Microbiol 46:95–116. doi: 10.1146/annurev.mi.46.100192.000523. [DOI] [PubMed] [Google Scholar]
- 76.Schellenberger U, Livi LL, Santi DV. 1995. Cloning, expression, purification, and characterization of 2’-deoxyuridylate hydroxymethylase from phage SPO1. Protein Expr Purif 6:423–430. doi: 10.1006/prep.1995.1057. [DOI] [PubMed] [Google Scholar]
- 77.Wilhelm K, Rüger W. 1992. Deoxyuridylate-hydroxymethylase of bacteriophage SPO1. Virology 189:640–646. doi: 10.1016/0042-6822(92)90587-f. [DOI] [PubMed] [Google Scholar]
- 78.Fritz TA, Liu L, Finer-Moore JS, Stroud RM. 2002. Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access. Biochemistry 41:7021–7029. doi: 10.1021/bi012108c. [DOI] [PubMed] [Google Scholar]
- 79.Maltman KL, Neuhard J, Lewis HA, Warren RA. 1980. Synthesis of thymine and alpha-putrescinylthymine in bacteriophage ΦW-14-infected Pseudomonas acidovorans. J Virol 34:354–359. doi: 10.1128/JVI.34.2.354-359.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warren RA. 1981. Ordered distribution of α-putrescinylthymine in the DNA of bacteriophage ΦW-14. Curr Microbiol 6:185–188. doi: 10.1007/BF01642396. [DOI] [Google Scholar]
- 81.Witmer H, Wiatr C. 1985. Polymer-level synthesis of oxopyrimidine deoxynucleotides by Bacillus subtilis phage SP10: characterization of modification-defective mutants. J Virol 53:522–527. doi: 10.1128/JVI.53.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maltman KL, Neuhard J, Warren RA. 1981. 5-[(Hydroxymethyl)-O-pyrophosphoryl]uracil, an intermediate in the biosynthesis of alpha-putrescinylthymine in deoxyribonucleic acid of bacteriophage ΦW-14. Biochemistry 20:3586–3591. doi: 10.1021/bi00515a043. [DOI] [PubMed] [Google Scholar]
- 83.Iyer LM, Tahiliani M, Rao A, Aravind L. 2009. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iyer LM, Zhang D, Burroughs AM, Aravind L. 2013. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res 41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee Y-J, Dai N, Müller SI, Guan C, Parker MJ, Fraser ME, Walsh SE, Sridar J, Mulholland A, Nayak K, Sun Z, Lin Y-C, Comb DG, Marks K, Gonzalez R, Dowling DP, Bandarian V, Saleh L, Corrêa IR, Jr, Weigele PR. 2021. Pathways of thymidine hypermodification. Nucleic Acids Res. doi: 10.1093/nar/gkab781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grove A, Galeone A, Mayol L, Geiduschek EP. 1996. On the connection between inherent DNA flexure and preferred binding of hydroxymethyluracil-containing DNA by the type II DNA-binding protein TF1. J Mol Biol 260:196–206. doi: 10.1006/jmbi.1996.0392. [DOI] [PubMed] [Google Scholar]
- 87.Rana AK, Ankri S. 2016. Reviving the RNA world: an insight into the appearance of RNA methyltransferases. Front Genet 7:99–99. doi: 10.3389/fgene.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forterre P. 2006. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol 24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 89.Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F, Miska EA, Balasubramanian S. 2015. Formation and abundance of 5‐hydroxymethylcytosine in RNA. Chembiochem 16:752–755. doi: 10.1002/cbic.201500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin S-G, Niedernhofer LJ, Pfeifer GP, Xu G-L, Wang Y. 2014. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc 136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCown PJ, Ruszkowska A, Kunkler CN, Breger K, Hulewicz JP, Wang MC, Springer NA, Brown JA. 2020. Naturally occurring modified ribonucleosides. Wiley Interdiscip Rev RNA 11:e1595. doi: 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song C-X, Zhang K, He C, Xu G-L. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olinski R, Starczak M, Gackowski D. 2016. Enigmatic 5-hydroxymethyluracil: oxidatively modified base, epigenetic mark or both? Mutat Res Rev Mutat Res 767:59–66. doi: 10.1016/j.mrrev.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 95.Janoušková M, Vaníková Z, Nici F, Boháčová S, Vítovská D, Šanderová H, Hocek M, Krásný L. 2017. 5-(Hydroxymethyl)uracil and -cytosine as potential epigenetic marks enhancing or inhibiting transcription with bacterial RNA polymerase. Chem Commun (Camb) 53:13253–13255. doi: 10.1039/c7cc08053k. [DOI] [PubMed] [Google Scholar]
- 96.Song C, Sun Y, Dai Q, Lu X, Yu M, Yang C, He C. 2011. Detection of 5‐hydroxymethylcytosine in DNA by transferring a keto‐glucose by using T4 phage β‐glucosyltransferase. Chembiochem 12:1682–1685. doi: 10.1002/cbic.201100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alexander SC, Busby KN, Cole CM, Zhou CY, Devaraj NK. 2015. Site-specific covalent labeling of RNA by enzymatic transglycosylation. J Am Chem Soc 137:12756–12759. doi: 10.1021/jacs.5b07286. [DOI] [PubMed] [Google Scholar]
- 98.Lutz T, Flodman K, Copelas A, Czapinska H, Mabuchi M, Fomenkov A, He X, Bochtler M, Xu S. 2019. A protein architecture guided screen for modification dependent restriction endonucleases. Nucleic Acids Res 47:9761–9776. doi: 10.1093/nar/gkz755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120-18. doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]