Abstract
Several studies have found that the promoter CpG island is frequently methylated in gastric cancer. The CpG island methylator phenotype (CIMP) defines concordant methylation of multiple promoter CpG island loci in a subset of gastric cancer. However, the relationship between CIMP and lymph node metastasis in gastric cancer is unknown. Our study aimed to characterize the role of CIMP in lymph node metastasis. Clinical specimens from 120 patients were analyzed and PCR was used to detect the methylation status of five genes (ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1). We measured the level of mRNA for the five genes by real‐time RT‐PCR. Microsatellite instability and Helicobacter pylori infection status were assayed by capillary electrophoresis and real‐time PCR, respectively. DNA methylation in the five genes was correlated with low expression of the respective mRNA. With CIMP as the dependent variable, CIMP‐high gastric cancer tended to show more distant lymph node metastasis, higher pathologic tumor classification, more pathologic metastasis, and higher pathologic TNM status. Microsatellite instability and H. pylori status were not significant predictors of prognosis. CIMP‐high gastric cancer showed significantly worse survival compared with that of CIMP‐low/CIMP‐negative gastric cancer (P < 0.001). Our results show that there is an association between CIMP status and lymph node metastasis in gastric cancer and CIMP‐high was an independent prognostic factor. (Cancer Sci 2012; 103: 73–79)
Gastric cancer is the second most common cause of global cancer mortality, accounting for >700 000 deaths annually.( 1 ) Despite a steady decline in global incidence, gastric cancer still causes prominent morbidity and mortality in China. The clinical outcome of surgery in combination with chemotherapies largely depends on the stage of the gastric cancer. Although the molecular mechanisms of gastric cancer carcinogenesis remain unclear, epigenetic alteration through promoter methylation is known to play an important role in the development of this cancer that inhibit the expression of tumor suppressor genes. Currently, DNA methylation markers have been used in early detection, prognosis, and prediction of response to cancer therapy.( 2 , 3 )
DNA methylation has been studied extensively in gastric cancer.( 4 , 5 , 6 , 7 ) However, most studies have focused on aberrant methylation in a single gene. Because methylated genes rarely occur singly, and more often in groups, the concept of a CpG island methylator phenotype (CIMP) in gastric and colorectal cancer was introduced,( 2 ) in which five to seven methylation‐sensitive genes were included for evaluating the methylation status in cancer and for correlating the CIMP with tumor risk and prevention. The CIMP was defined as a subset of malignancies that show widespread hypermethylation of multiple promoter CpG island loci. Several scientists have used their own CIMP marker panels for the determination of CIMP status, however, producing some inconsistent results.( 8 , 9 , 10 , 11 , 12 )
In this study we tried to evaluate the role of hypermethylation of multiple tumor‐related genes such as ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1 in gastric cancer. ALX4 methylation frequently occurs in colorectal cancer tissue as well as in patients’ serum.( 13 ) Aberrant methylation of the TMEFF2 gene inhibits the transforming growth factor β signaling pathway, and the gene plays an important role in human cancers.( 14 , 15 , 16 , 17 , 18 , 19 ) Using gene knockdown under in vitro conditions, it is thought that CHCHD10 is involved in oxidative phosphorylation and plays an important role in complex IV activity.( 20 ) NPR1 is regarded as a major natriuretic peptide receptor and its activation produces the second messenger cGMP, which plays a key role in maintaining blood pressure and cardiovascular homeostasis. It has been shown that NPR1 methylation is associated with survival in colorectal cancer( 21 ) and hepatocellular carcinoma.( 22 ) IGFBP3 is the main carrier of insulin‐like growth factors (IGFs) in the circulation, where this complex regulates the biologic function of IGFs.( 23 ) Hypermethylation of the IGFBP3 promoter is a frequent phenomenon and strongly associated with prognosis of non‐small‐cell lung cancer and ovarian cancer.( 24 , 25 )
In this study, we investigated the prevalence of the methylation of ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1 among gastric cancer tissues and its relation to various clinicopathological characteristics. In addition, to clarify the characteristics and their underlying mechanisms of gastric tumors with hypermethylation of the five genes, we also measured levels of expression of mRNAs for these genes by real‐time RT‐PCR and evaluated microsatellite instability (MSI) and Helicobacter pylori status. Finally, we evaluated the prognostic significance of CIMP status in gastric carcinomas.
Materials and Methods
Tissue samples. One hundred and twenty samples were obtained from patients with newly diagnosed primary tumor at the Gastric Cancer Center, First Affiliated Hospital, Sun Yet‐sen University (Guangzhou, China) between December 2003 and August 2009. These samples were from primary surgery, and the patients did not receive previous chemotherapy. Tumor sampling was carried out specifically for in vitro testing and was approved by the Ethical Research Committee, Sun Yet‐sen University. Tumor samples were transferred from the operating room to the laboratory within 30 min and stored in liquid nitrogen for later use. As a control, endoscopic gastric biopsies from 10 patients with chronic gastritis also were included.
DNA extraction and sodium bisulfite modification. Genomic DNA was isolated from gastric cancer specimens in liquid nitrogen using using a DNeasy Tissue Kit (Qiagen, Valencia, CA, USA). Bisulfite treatment of DNA was carried out with an EpiTect bisulfite kit (Qiagen) according to the manufacturer’s protocol. Bisulfite‐treated DNA was used as a template in subsequent MethyLight PCR analyses.
Methylation analysis of multiple genes. After genomic DNA was treated with sodium bisulfite, the methylation levels of five genes (ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1, as well as β‐actin as an internal marker) was analyzed using MethyLight PCR. Real‐time PCR‐based DNA methylation assay (MethyLight assay) was validated and carried out as described earlier.( 26 , 27 , 28 ) We used a percentage of methylated reference (PMR) cut‐off value of 4 to define positive versus negative methylation and to determine DNA methylation frequencies (PMR > 4) for each CpG island locus.
An aliquot of 2 μL was amplified with a primer set along with the TaqMan probe specific to methylated sequences. All PCR experiments were carried out in a volume of 25 μL using the EpiTect MethyLight PCR kit (Qiagen) with 96‐well plates and an ABI 7500 Sequence Detector (Applied Biosystems, Foster City, CA, USA). The primer and probe sequences are shown in Table 1. Each reaction contained 12.5 μL Master Mix for methylation‐specific real‐time PCR analysis, 0.5 μM each primer, and 2.0 μL bisulfite‐treated DNA in a total volume of 25 μL at 95°C for 10 min, followed by 45 cycles of 94°C for 30 s, and 60°C for 45 s in ALX4, TMEFF2, CHCHD10, IGFBP3, NPR1, and β‐actin. The latter was used to normalize for input DNA. These experiments were carried out in triplicate and the mean value was then calculated. Every PCR experiment included serial dilutions of a positive control for construction of the calibration curve, a positive and a negative DNA sample, and water blanks. CpGenome Universally Methylated DNA (Chemicon, Temecula, CA, USA) was used as a positive control for methylation, and CpGenome Universal Unmethylated DNA (Chemicon) was used as a negative control. The methylation value of target genes in the specimens was determined as the relative methylation ratio (methylation level of target gene/β‐actin in sample)/(methylation level of target gene/β‐actin in positive control DNA).
Table 1.
Primers and probes for DNA methylation and mRNA detection
| Gene | Status | Primer and probe sequence | Length (bp) |
|---|---|---|---|
| ALX4 | Methylation | 5′‐TTAGGTATGAATGTTGAGATTTGCG‐3′ | 83 |
| 5′‐CTACGACACCGAACTATAATAAACG‐3′ | |||
| 5′‐FAM‐TTATTGCGAGTCGTCGGTCGTTGTTATGG‐BHQ1‐3′ | |||
| TMEFF2 | Methylation | 5′‐GTTATCGTCGTCGTTTTTGTTGTC‐3′ | 87 |
| 5′‐GACTTCCGAAAAACACAAAATCG‐3′ | |||
| 5′‐FAM‐CGCGGGATGTTTAGTAGTTCGTTGTTCGG‐BHQ1‐3′ | |||
| CHCHD10 | Methylation | 5′‐AGGTTTCGTTCGGGGTTTCG‐3′ | 88 |
| 5′‐AAACGACGACAACGATACTATCG‐3′ | |||
| 5′‐FAM‐ACAAATACCGCAACGCTTATCACAACCGA‐BHQ1‐3′ | |||
| IGFBP3 | Methylation | 5′‐GTTTCGGGCGTGAGTACGA‐3′ | 119 |
| 5′‐GAATCGACGCAAACACGACTAC‐3′ | |||
| 5′‐FAM‐TCGGTTGTTTAGGGCGAAGTACGGG‐BHQ1‐3′ | |||
| NPR1 | Methylation | 5′‐GCGGGTAATTTGACGGTAGTCG‐3′ | 126 |
| 5′‐CAACAAATCGAAACGCGCCTTC‐3′ | |||
| 5′‐FAM‐AAACCAACTCCACGACGAATCCCACGC‐BHQ1‐3′ | |||
| ALX4 | mRNA | 5′‐GGGAACAGCTGGCCATGA‐3′ | 95 |
| 5′‐AAAACGCTCCCGCTTCCT‐3′ | |||
| 5′‐FAM‐CCCGCGTGCAGGTCTGGTTCC‐BHQ1‐3′ | |||
| TMEFF2 | mRNA | 5′‐CTGCTTTCCCTACCTCCTTAAG‐3′ | 110 |
| 5′‐TTTACAGGTGTTGGTGTCACAG‐3′ | |||
| 5′‐FAM‐ACTGCCAAACGCCCACCGG‐BHQ1‐3′ | |||
| CHCHD10 | mRNA | 5′‐TCCTGCACCCACCTCTACC‐3′ | 81 |
| 5′‐CCTCACTTCCAATCCCAGCTA‐3′ | |||
| 5′‐FAM‐CGCCGACAGCCAGACCACAAC‐BHQ1‐3′ | |||
| IGFBP3 | mRNA | 5′‐TGATACAACTGTGGCCATGACT‐3′ | 105 |
| 5′‐TCCCTGAGCCTGACTTTGC‐3′ | |||
| 5′‐FAM‐CTCTCCCGGAGGCCAAACCCA‐BHQ1‐3′ | |||
| NPR1 | mRNA | 5′‐ATACCTGAAAATTGATAGCA‐3′ | 105 |
| 5′‐GTCCCATTGTAGTTCAGTA‐3′ | |||
| 5′‐FAM‐AACCCTGAAGGCACCATTCT‐BHQ1‐3′ |
RNA isolation and real‐time RT‐PCR for determining mRNA expression. Total RNA was extracted from gastric cancer specimens using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Nucleic acid concentrations were determined using RiboGreen (Molecular Probes, Eugene, OR, USA). The RNA was stored at −80°C until further use.
To analyze expression of the five genes, we carried out real‐time RT‐PCR as described previously.( 29 ) The primer and probe sequences are shown in Table 1. The mRNA expression level was detected by real‐time one‐step RT‐PCR using TaqMan probe specific for the five genes. Real‐time one‐step RT‐PCR was carried out on an ABI 7500 (Applied Biosystems) using standard 25 μL Universal PCR Master Mix on 1–2 μg total RNA. Reaction conditions were 50°C for 15 min, 95°C for 8 min, 40 cycles at 94°C for 30 s, with an annealing temperature of 55°C for 45 s. No‐template control was included in each assay. β‐actin was used as an endogenous control and vehicle control was used as a calibrator. Each sample was run in triplicate. The comparative threshold cycle method was used to calculate the relative changes in the expression of five genes. The relative change of gene expression was calculated using the following formula: fold change in gene expression, 2−ΔΔCt = 2−[ΔCt (tumor samples) − ΔCt (vehicle control)], where ΔCt = Ct (detected gene) – Ct (β‐actin) and Ct represents the threshold cycle number.
Microsatellite instability and H. pylori status. The MSI status was determined using a consensus panel of five reference microsatellite markers (BAT25, BAT26, D2S123, D3S546, and D17S250) by a previously described method.( 12 , 30 ) When no marker was altered the tumors were defined as microsatellite stable. When only one marker was altered, the tumors were defined as low MSI. When two or more markers were altered those were defined as high MSI.
Helicobacter pylori infection was analyzed by detecting urease A gene of H. pylori genomic DNA in the gastric cancer mucosa using real‐time PCR. The H. pylori specific primers and probe were: forward primer, 5′‐ATGAAGTGGGTATTGAAGCGAT‐3′, reverse primer, 5′‐TTAAGAACAACTCACCAGGAACTA‐3′, and probe, 5′‐FAM‐CCTCAATAGGGGTATGCACGG‐BHQ1‐3′.
Statistical analysis. All clinicopathologic variables were used as categorical variables. Differences in continuous variables between two groups were evaluated by Student’s t‐test, and differences in categorical variables were evaluated by the chi square ‐test. Associations between clinicopathological parameters and CIMP status were analyzed using Pearson’s chi square‐test and Fisher’s exact test. Disease‐free survival was measured from the date of resection of gastric cancer to the date of event or the last follow‐up date before 11 May 2011. Event was defined as recurrence, death due to any cause, or development of a second primary gastric cancer. The Kaplan–Meier method was used to calculate and display disease‐free survival curves, and the log–rank test was used to determine differences among all groups. The Cox proportional hazards regression method was used to determine independent prognostic factors. All statistical tests were done using the spss software package, version 17 (SPSS, Chicago, IL, USA). All P‐values were two‐sided, and P < 0.05 was considered statistically significant.
Results
Gene methylation and CIMP status in gastric cancer. We studied 120 patients based on sample availability. Mean age was 58 years (range, 25–86 years), 80 patients were male (66.7%). The clinicopathologic features of the patients analyzed by CIMP are summarized in Table 2.
Table 2.
Clinicopathologic features of gastric carcinomas with CpG island methylator phenotype (CIMP) status
| Characteristic | CIMP‐H | CIMP‐L | CIMP‐N | P† |
|---|---|---|---|---|
| No. of patients | 18 | 94 | 8 | |
| Age group | ||||
| <60 years | 9 | 45 | 3 | 0.890 |
| ≥60 years | 9 | 49 | 5 | |
| Sex | ||||
| Male | 10 | 65 | 5 | 0.516 |
| Female | 8 | 29 | 3 | |
| Gross | ||||
| Borrmann I | 1 | 7 | 0 | 0.186 |
| Borrmann II | 4 | 30 | 3 | |
| Borrmann III | 8 | 56 | 4 | |
| Borrmann IV | 5 | 5 | 1 | |
| Borrmann V | 0 | 6 | 0 | |
| Histologic grade | ||||
| G1 Well differentiated | 1 | 4 | 1 | 0.658 |
| G2 Moderately differentiated | 7 | 32 | 2 | |
| G3 Poorly differentiated | 10 | 58 | 5 | |
| G4 Undifferentiated | 0 | |||
| Lymph node metastasis | ||||
| Negative | 0 | 24 | 4 | <0.001 |
| <3 cm from tumor | 5 | 41 | 4 | |
| ≥3 cm from tumor | 13 | 29 | 0 | |
| Pathologic tumor classification | ||||
| pT1 | 0 | 8 | 5 | <0.001 |
| pT2 | 1 | 10 | 2 | |
| pT3 | 7 | 60 | 1 | |
| pT4 | 10 | 16 | 0 | |
| Pathologic lymph node status | ||||
| pNo | 3 | 33 | 6 | 0.229 |
| pN1 | 10 | 36 | 2 | |
| pN2 | 3 | 15 | 0 | |
| pN3 | 2 | 10 | 0 | |
| Pathologic metastasis status | ||||
| pM0 | 9 | 83 | 8 | 0.001 |
| pM1 | 9 | 11 | 0 | |
| Stage (pTNM) | ||||
| Stage IA | 0 | 5 | 5 | <0.001 |
| Stage IB | 1 | 11 | 2 | |
| Stage II | 1 | 15 | 0 | |
| Stage IIIA | 1 | 24 | 1 | |
| Stage IIIB | 1 | 12 | 0 | |
| Stage IV | 14 | 27 | 0 | |
| MSI | ||||
| MSI‐H | 1 | 16 | 2 | 0.244 |
| MSI‐L | 2 | 21 | 0 | |
| MSI‐N | 15 | 57 | 6 | |
| Helicobacter pylori | ||||
| Positive | 4 | 16 | 1 | 0.902 |
| Negative | 14 | 78 | 7 | |
H, high; L, low; MSI, microsatellite instability; N, negative.
†Statistical significance determined using Pearson’s χ2‐test and Fisher’s exact test.
The methylation status of 120 gastric cancer samples in five DNA methylation markers was detected by MethyLight technology and PMR values were calculated for each sample and MethyLight reaction. Bisulfite genomic DNA sequencing of representative methylated PCR products of each of the five genes showed that all cytosines at non‐CpG sites were converted to thymine (representative result shown in Fig. 1A).
Figure 1.
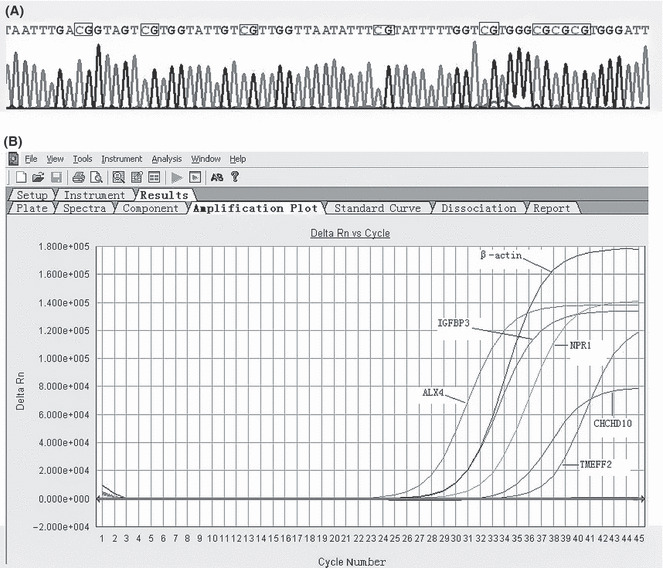
(A) Sequencing analysis of methylated PCR products of the NPR1 gene. All CpG sites were methylated and C to T transition was observed by bisulfite modification. To indicate methylation status, the wild‐type CpG sites were squared. Methylated CpG sites appear as CG. (B) Methylation analysis of five genes in gastric cancer by MethyLight PCR. The gene studied is indicated.
The methylation frequencies of the five genes analyzed were 62.5% for ALX4, 70% for TMEFF2, 39.2% for CHCHD10, 58.3% for IGFBP3, and 42.5% for NPR1. The average number of methylated genes per tumor was 2.73. The results indicated that hypermethylation of these loci is a common event in gastric cancer. Representative results of MethyLight of ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1 are shown in Figure 1B. We confirmed that DNA methylation of each gene was correlated with low expression of the respective mRNA (Table 3).
Table 3.
Relationship between DNA methylation and mRNA expression in gastric cancer
| Gene | Methylation status | No. cases | mRNA level† | P‡ |
|---|---|---|---|---|
| ALX4 | M | 75 | 0.20 ± 0.03§ | 0.017 |
| U | 45 | 2.48 ± 0.12 | ||
| TMEFF2 | M | 84 | 1.14 ± 0.11 | 0.001 |
| U | 36 | 8.26 ± 2.04 | ||
| CHCHD10 | M | 47 | 0.04 ± 0.01 | 0.005 |
| U | 73 | 0.89 ± 0.05 | ||
| IGFBP3 | M | 70 | 0.13 ± 0.02 | 0.005 |
| U | 50 | 0.74 ± 0.14 | ||
| NPR1 | M | 51 | 0.05 ± 0.03 | 0.005 |
| U | 69 | 0.48 ± 0.08 |
†Mean values and standard errors for all gastric cancer samples including those that are methylated (M) and unmethylated (U).
‡Statistical significance determined using the Mann–Whitney U‐test.
§Units are arbitrary, and we calculated the respective mRNA expression level by standardization with 1 μg total RNA of NCI‐N87 gastric cancer cells, taken as 1.0.
For descriptive purposes, CIMP status was classified as: CIMP‐negative (CIMP‐N) if none of the evaluated genes were methylated; CIMP‐low (CIMP‐L) if fewer than four genes were methylated; and CIMP‐high (CIMP‐H) if four or more genes were methylated. Based on this classification, concordant methylation of multiple genes (CIMP‐H) was present in 15% (18 of 120) of tumors, CIMP‐L in 78.3% (94 of 120), and CIMP‐N in 6.7% (8 of 120) (Table 2). There were no differences in age, sex, tumor gross, histologic grade, pathologic lymph node status, MSI, or H. pylori infection status among the three groups. However, when three subtypes were correlated with lymph node metastasis, pathologic tumor classification, pathologic metastasis status, and stage (pTNM), CIMP‐H gastric cancers tended to show more distant metastasis and higher tumor stage (Table 2).
Microsatellite instability and H. pylori infection. Representative examples of MSI in gastric cancer are shown in Figure 2. The prevalence of MSI‐high (MSI‐H) was 15.8% (19 of 120), MSI‐low (MSI‐L) was 19.2% (23 of 120), and MSI‐stable was 65% (78 of 120) in gastric carcinomas. There was no statistical difference between MSI status in tumors with evaluated clinicopathologic features, including age, sex, tumor histology, and pathologic stage (data not shown).
Figure 2.
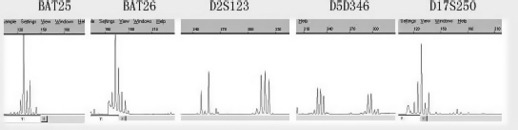
Representative examples of microsatellite instability in gastric carcinoma. A microsatellite instability‐high tumor had allelic shifts in two microsatellite markers (D2S123 and D5S346).
Helicobacter pylori was detected in 21 of 120 tumors (17.5%) using real‐time quantitative PCR. No differences were found in the number of methylated genes between H. pylori‐negative patients and H. pylori‐positive patients (data not shown).
Survival analysis. In univariate analysis, Borrmann stage (P < 0.001), lymph node metastasis (P < 0.001), tumor stage (pTNM) (P < 0.001), and CIMP‐H (P < 0.001) were statistically significant predictors for overall survival (Table 4). Histologic grade, MSI, and H. pylori infection status were not significant prognostic factors. By Kaplan–Meier survival analysis and the log–rank test, patients who had CIMP‐H gastric tumors showed significantly worse survival than patients with CIMP‐L/CIMP‐N tumors (Fig. 3).
Table 4.
Univariate and multivariate survival analysis in gastric cancer (n = 120)
| Variables | No. patients | No. deaths | Univariate analysis† | Multivariate analysis‡ | |
|---|---|---|---|---|---|
| χ2 (P) | Hazard ratio (95% CI) | P | |||
| Age group | |||||
| <60 years | 57 | 23 | 0.136 (0.712) | ||
| ≥60 years | 63 | 25 | |||
| Gross | |||||
| Borrmann I + II | 35 | 7 | 20.158 (<0.001) | Reference | |
| Borrmann III | 68 | 28 | 1.612 (0.688–3.779) | 0.272 | |
| Borrmann IV + V | 17 | 13 | 3.587 (1.339–9.612) | 0.011 | |
| Histologic grade | |||||
| G1 + G2 | 47 | 18 | 0.049 (0.825) | ||
| G3 | 73 | 30 | |||
| Lymph node metastasis | |||||
| Negative | 28 | 5 | 22.345 (<0.001) | Reference | |
| <3 cm from tumor | 50 | 17 | 0.949 (0.322–2.793) | 0.924 | |
| ≥3 cm from tumor | 42 | 26 | 1.438 (0.489–4.225) | 0.509 | |
| Stage (pTNM) | |||||
| Stage IA + IB | 24 | 1 | 44.676 (<0.001) | Reference | |
| Stage II | 16 | 3 | 2.583 (0.257–25.978) | 0.420 | |
| Stage IIIA | 26 | 9 | 10.476 (1.266–86.700) | 0.029 | |
| Stage IIIB | 13 | 5 | 11.715 (1.337–102.668) | 0.026 | |
| Stage IV | 41 | 30 | 22.182 (2.75–178.940) | 0.004 | |
| CIMP | |||||
| N + L | 102 | 32 | 91.108 (<0.001) | Reference | |
| H | 18 | 16 | 12.688 (5.249–30.671) | <0.001 | |
| MSI | |||||
| MSI‐N | 78 | 34 | 3.608 (0.058) | NA | NA |
| MSI‐L | 23 | 12 | NA | NA | |
| MSI‐H | 19 | 2 | NA | NA | |
| Helicobacter pylori | |||||
| Negative | 99 | 38 | 1.287 (0.257) | NA | NA |
| Positive | 21 | 10 | NA | NA | |
CI, confidence interval; CIMP, CpG island methylator phenotype; H, high; L, low; MSI, microsatellite instability; N, negative; NA, not applicable; Reference, represented in the contrast matrix as a row of zeros in multivariate analysis.
†Log–rank test. ‡Cox’s proportional hazards model.
Figure 3.
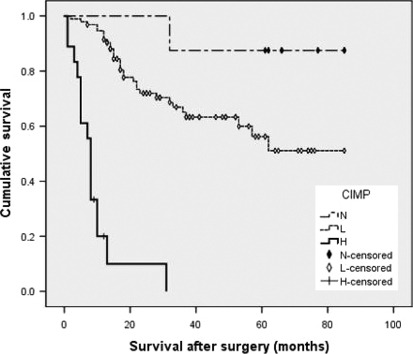
Kaplan–Meier survival curves in gastric cancers (n = 120) according to CpG island methylator phenotype status. H, high; L, low; N, negative.
Borrmann stage, lymph node metastasis, stage (pTNM), and CIMP were included in multivariate Cox regression analysis (Table 4). Patients with CIMP‐H gastric tumors tended to have worse survival than patients with CIMP‐L/CIMP‐N gastric tumors and the difference was significant (P < 0.001).
Discussion
The relationship between CIMP and tumor pathology is unclear. It seems reasonable that the biologic functions of each associated gene are also simultaneously silenced because DNA methylation is associated with low expression of the respective mRNA. Among the five marker genes used in the present study, ALX4 and TMEFF2 were reported as methylated in our previous colorectal cancer research by multiplex MethyLight assay.( 28 ) The methylation of ALX4 and TMEFF2 is regarded as an early event during tumor carcinogenesis.( 13 , 16 ) IGFBP‐3 expression may be protective against the development of gastric adenocarcinoma by preventing the formation of intestinal metaplasia and improve the prognosis of gastric cancer.( 31 ) The relationship between the methylation of CHCHD10 and NPR1 and gastric cancer clinical outcome is not clear so far. In this study, CIMP‐H including CHCHD10 and NPR1 methylation with lymph node metastasis, pathologic tumor classification, pathologic metastasis status, and pathologic TNM status show significant difference. In particular, patients with CIMP‐H show more distant lymph node metastasis, which can be explained by the fact that the methylation of ALX4 and TMEFF2 is an early event, the methylation of IGFBP‐3, CHCHD10, and NPR1 may be a later event associated with the lymph node metastasis. A project designed to clarify the concrete role of CHCHD10 and NPR1 methylation in lymph node metastasis of gastric cancer is underway.
Furthermore, most previous studies of methylation in gastric cancer focus on the prognostic significance of methylation of a single gene.( 4 , 5 , 6 , 7 ) The current research about the clinical outcome of CIMP‐positive gastric cancer is controversial.( 2 , 9 , 11 , 24 ) Most previous reports show that CIMP was related with better prognosis, but was not an independent prognostic factor on multivariable analysis, except in the report published by Seog‐Yun Park et al. ( 11 ) The present study shows that CIMP was closely associated with poor prognosis of gastric cancer patients and CIMP‐H was an independent prognostic marker. The differences might be attributed to two reasons. One is the different CIMP marker panels. Most CIMP reports used MINT1, MINT2, MINT12, MINT25, and MINT31 as the CIMP marker panel, but the CIMP marker panel in this study is ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1. The other is the different methods used to analyse the status and level of target gene methylation. Compared with combined bisulfite restriction analysis, methylation‐specific PCR, and quantitative methylation‐specific PCR, MethyLight assay is more accurate and more sensitive. However, the number of markers used in the CIMP panel in this study (five) was small, and the number of cases (120) was too few, therefore, it is necessary to launch a large‐scale study including more CIMP markers to attest the detailed clinicopathological features of tumors with CIMP.
Several studies have found that multiple gene methylation correlates with H. pylori infection in non‐neoplastic gastrointestinal tissues.( 32 , 33 ) However, several studies have shown that H. pylori infection induced the altered DNA methylation in gastric cancer.( 34 , 35 ) However, in this study, H. pylori infection was not a differential influence on CIMP including ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1 in gastric cancer.
Controversial results were reported by a different research group regarding the relationship between MSI, DNA replication errors, and clinical prognosis.( 12 , 36 , 37 ) In this study, MSI‐H was present in 15.8% of gastric cancers. There was no relationship between MSI status and clinicopathologic characters including CIMP. Compared with MSI‐L/MSI‐stable tumors, overall survival was slightly worse but without statistically significant difference (P = 0.058) in patients with MSI‐H tumors. The MSI status of gastric cancer in this study was not a significant predictor of prognosis. It is possible that CIMP in this study did not include DNA repair genes such as hMLH1, as has been reported previously.( 12 )
In conclusion, using the methylation profile of five unique genes (ALX4, TMEFF2, CHCHD10, IGFBP3, and NPR1) as marker genes, we found that CIMP‐H was associated with lymph node metastasis, pathologic tumor classification, pathologic metastasis status, and pathologic TNM status. CIMP‐H was an independent prognostic factor in gastric cancer. However, more studies are needed to validate the role of CIMP and elucidate its mechanism in gastric cancer.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81001085), the Key Project of the National Science Foundation of Guangdong Province (Grant No. 07117381), and the Young Teacher Cultivative Project of Sun Yat‐sen University (Grant No. 09ykpy49). Thanks also to Professor Zhou Luo‐Jin for her statistical guidance.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 2. Toyota M, Ahuja N, Suzuki H et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res 1999; 59: 5438–42. [PubMed] [Google Scholar]
- 3. Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128: 683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang XH, Zhang LH, Zhong XY et al. S100A6 overexpression is associated with poor prognosis and is epigenetically up‐regulated in gastric cancer. Am J Pathol 2010; 177: 586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim J, Min SY, Lee HE, Kim WH. Aberrant DNA methylation and tumor suppressive activity of the EBF3 gene in gastric carcinoma. Int J Cancer 2011; doi: 10.1002/ijc.26038 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 6. Yu J, Tao Q, Cheng YY et al. Promoter methylation of the Wnt/beta‐catenin signaling antagonist Dkk‐3 is associated with poor survival in gastric cancer. Cancer 2009; 115: 49–60. [DOI] [PubMed] [Google Scholar]
- 7. Al‐Moundhri MS, Al‐Nabhani M, Tarantini L, Baccarelli A, Rusiecki JA. The prognostic significance of whole blood global and specific DNA methylation levels in gastric adenocarcinoma. PLoS ONE 2010; 5: e15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Enomoto S, Maekita T, Tsukamoto T et al. Lack of association between CpG island methylator phenotype in human gastric cancers and methylation in their background non‐cancerous gastric mucosae. Cancer Sci 2007; 98: 1853–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular‐pathological prognostic factors of gastric cancer: a review. Gastric Cancer 2005; 8: 86–94. [DOI] [PubMed] [Google Scholar]
- 10. Kusano M, Toyota M, Suzuki H et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein‐Barr virus. Cancer 2006; 106: 1467–79. [DOI] [PubMed] [Google Scholar]
- 11. Park SY, Kook MC, Kim YW et al. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch 2010; 457: 415–22. [DOI] [PubMed] [Google Scholar]
- 12. An C, Choi IS, Yao JC et al. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res 2005; 11(2 Pt 1): 656–63. [PubMed] [Google Scholar]
- 13. Ebert MP, Model F, Mooney S et al. Aristaless‐like homeobox‐4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology 2006; 131: 1418–30. [DOI] [PubMed] [Google Scholar]
- 14. Suzuki H, Igarashi S, Nojima M et al. IGFBP7 is a p53‐responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 2010; 31: 342–9. [DOI] [PubMed] [Google Scholar]
- 15. Young J, Biden KG, Simms LA et al. HPP1: a transmembrane protein‐encoding gene commonly methylated in colorectal polyps and cancers. Proc Natl Acad Sci USA 2001; 98: 265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gu P, Xing X, Tanzer M et al. Frequent loss of TIMP‐3 expression in progression of esophageal and gastric adenocarcinomas. Neoplasia 2008; 10: 563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geddert H, Kiel S, Iskender E et al. Correlation of hMLH1 and HPP1 hypermethylation in gastric, but not in esophageal and cardiac adenocarcinoma. Int J Cancer 2004; 110: 208–11. [DOI] [PubMed] [Google Scholar]
- 18. Brucher BL, Geddert H, Langner C et al. Hypermethylation of hMLH1, HPP1, p14(ARF), p16(INK4A) and APC in primary adenocarcinomas of the small bowel. Int J Cancer 2006; 119: 1298–302. [DOI] [PubMed] [Google Scholar]
- 19. Sabbioni S, Miotto E, Veronese A et al. Multigene methylation analysis of gastrointestinal tumors: TPEF emerges as a frequent tumor‐specific aberrantly methylated marker that can be detected in peripheral blood. Mol Diagn 2003; 7: 201–7. [DOI] [PubMed] [Google Scholar]
- 20. Martherus RS, Sluiter W, Timmer ED, VanHerle SJ, Smeets HJ, Ayoubi TA. Functional annotation of heart enriched mitochondrial genes GBAS and CHCHD10 through guilt by association. Biochem Biophys Res Commun 2010; 402: 203–8. [DOI] [PubMed] [Google Scholar]
- 21. Cavalieri D, Dolara P, Mini E et al. Analysis of gene expression profiles reveals novel correlations with the clinical course of colorectal cancer. Oncol Res 2007; 16: 535–48. [DOI] [PubMed] [Google Scholar]
- 22. Deng YB, Nagae G, Midorikawa Y et al. Identification of genes preferentially methylated in hepatitis C virus‐related hepatocellular carcinoma. Cancer Sci 2010; 101: 1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giovannucci E. Nutrition, insulin, insulin‐like growth factors and cancer. Horm Metab Res 2003; 35: 694–704. [DOI] [PubMed] [Google Scholar]
- 24. Jang TJ, Kim DI, Shin YM, Chang HK, Yang CH. p16(INK4a) Promoter hypermethylation of non‐tumorous tissue adjacent to gastric cancer is correlated with glandular atrophy and chronic inflammation. Int J Cancer 2001; 93: 629–34. [DOI] [PubMed] [Google Scholar]
- 25. Torng PL, Lin CW, Chan MW, Yang HW, Huang SC, Lin CT. Promoter methylation of IGFBP‐3 and p53 expression in ovarian endometrioid carcinoma. Mol Cancer 2009; 8: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ogino S, Cantor M, Kawasaki T et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut 2006; 55: 1000–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogino S, Kawasaki T, Brahmandam M et al. Precision and performance characteristics of bisulfite conversion and real‐time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn 2006; 8: 209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Q, Chen HY, Bai EQ et al. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet 2010; 202: 1–10. [DOI] [PubMed] [Google Scholar]
- 29. Zhang KL, Sun Y, Li Y et al. Increased frequency of CpG island methylator phenotype and CDH1 methylation in a gastric cancer high‐risk region of china. Transl Oncol 2008; 1: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boland CR, Thibodeau SN, Hamilton SR et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998; 58: 5248–57. [PubMed] [Google Scholar]
- 31. Zhang ZW, Newcomb PV, Moorghen M et al. Insulin‐like growth factor binding protein‐3: relationship to the development of gastric pre‐malignancy and gastric adenocarcinoma (United Kingdom). Cancer Causes Control 2004; 15: 211–8. [DOI] [PubMed] [Google Scholar]
- 32. Maekita T, Nakazawa K, Mihara M et al. High levels of aberrant DNA methylation in Helicobacter pylori‐infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 2006; 12: 989–95. [DOI] [PubMed] [Google Scholar]
- 33. Tahara T, Arisawa T, Shibata T et al. Increased number of methylated CpG islands correlates with Helicobacter pylori infection, histological and serological severity of chronic gastritis. Eur J Gastroenterol Hepatol 2009; 21: 613–9. [DOI] [PubMed] [Google Scholar]
- 34. Yan J, Zhang M, Zhang J, Chen X, Zhang X. Helicobacter pylori infection promotes methylation of WWOX gene in human gastric cancer. Biochem Biophys Res Commun 2010; 408: 99–102. [DOI] [PubMed] [Google Scholar]
- 35. Niwa T, Tsukamoto T, Toyoda T et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res 2010; 70: 1430–40. [DOI] [PubMed] [Google Scholar]
- 36. Seo HM, Chang YS, Joo SH et al. Clinicopathologic characteristics and outcomes of gastric cancers with the MSI‐H phenotype. J Surg Oncol 2009; 99: 143–7. [DOI] [PubMed] [Google Scholar]
- 37. Beghelli S, de Manzoni G, Barbi S et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery 2006; 139: 347–56. [DOI] [PubMed] [Google Scholar]


