Abstract
The coronavirus Avian infectious bronchitis virus (IBV) employs polyprotein processing as a strategy to express its gene products. Previously we identified the first cleavage event as proteolysis at the Gly673-Gly674 dipeptide bond mediated by the first papain-like proteinase domain (PLPD-1) to release an 87-kDa mature protein. In this report, we demonstrate a novel cleavage activity of PLPD-1. Expression, deletion, and mutagenesis studies showed that the product encoded between nucleotides 2548 and 8865 was further cleaved by PLPD-1 at the Gly2265-Gly2266 dipeptide bond to release an N-terminal 195-kDa and a C-terminal 41-kDa cleavage product. Characterization of the cleavage activity revealed that the proteinase is active on this scissile bond when expressed in vitro in rabbit reticulocyte lysates and can act on the same substrate in trans when expressed in intact cells. Both the N- and C-terminal cleavage products were detected in virus-infected cells and were found to be physically associated. Glycosidase digestion and site-directed mutagenesis studies of the 41-kDa protein demonstrated that it is modified by N-linked glycosylation at the Asn2313 residue encoded by nucleotides 7465 to 7467. By using a region-specific antiserum raised against the IBV sequence encoded by nucleotides 8865 to 9786, we also demonstrated that a 33-kDa protein, representing the 3C-like proteinase (3CLP), was specifically immunoprecipitated from the virus-infected cells. Site-directed mutagenesis and expression studies showed that a previously predicted cleavage site (Q2583-G2584) located within the 41-kDa protein-encoding region was not utilized by 3CLP, supporting the conclusion that the 41-kDa protein is a mature viral product.
Avian infectious bronchitis virus (IBV), the prototype of the family of enveloped viruses Coronaviridae, is now categorized under the newly established order Nidovirales, which comprises the families Coronaviridae and Arteriviridae. The name Nidovirales came from the Latin nidus, for nest, referring to the 3′-coterminal “nested” set of subgenomic mRNAs produced during viral infection (33). Two overlapping open reading frames (ORFs), 1a and 1b, are located at the 5′-end unique region of the genome and are expected to encode polyproteins of 441 and 300 kDa, respectively (2). By a unique frameshifting mechanism, the downstream ORF 1b is expressed as a fusion protein of 741 kDa with ORF 1a (3, 4). The 441-kDa 1a and 741-kDa 1a/1b polyproteins are proteolytically processed into smaller mature products required for RNA synthesis and other aspects of viral replication.
Nucleotide sequence analyses of coronaviruses from three different antigenic groups have predicted the presence of three proteinases (2, 7, 9, 10, 16), namely, two overlapping papain-like proteinase domains (PLPDs) and a picornavirus 3C-like proteinase domain (3CLP). To date, both PLPD-1 and 3CLP have been demonstrated to be active, and their catalytic properties have been characterized. PLPD-1 is involved in proteolytic cleavage of the N-terminal region of the 1a and 1a/1b polyproteins. For mouse hepatitis virus (MHV), it was shown that PLPD-1 mediates cleavage at Gly247-Val248 and Ala832-Gly833 dipeptide bonds to release p28 and p65, respectively (1, 6, 13). The Cys1137 and His1288 residues were defined as the catalytic dyad of this proteinase (1, 6, 13). For human coronavirus (HCV) 229E, it was reported that PLPD-1 is required for releasing p9 upon cleavage at the Gly111-Asn112 dipeptide bond, and the Cys1137 and His1205 residues were identified as its catalytic dyad (11). No experimental evidence has so far shown that PLPD-2 is functionally active for both viruses. Multiple proteinase domains have also been described for arteriviruses. For example, nsp1, nsp2, and nsp4 of equine arteritis virus were shown to have proteolytic activities (32).
In our previous reports, IBV PLPD-1 was shown to be responsible for cleavage of the 1a and 1a/1b polyproteins at the Gly673-Gly674 dipeptide bond to release an 87-kDa N-terminal cleavage product (17, 21). The Cys1274 and His1437 residues were revealed to be the catalytic dyad of this proteinase (17). In this paper, we report yet another cleavage event performed by PLPD-1. Our results show that PLPD-1 is responsible for the cleavage of the product encoded by nucleotides 2548 to 8865 (which is located between the 87-kDa protein and 3CLP) (Fig. 1) to release a 195-kDa N-terminal product and a 41-kDa C-terminal product. Both cleavage products were identified in IBV-infected cells. Systematic deletion and mutagenesis studies map the second cleavage site to the Gly2265-Gly2266 scissile bond. No further cleavage activity mediated by either PLPD-1 or 3CLP was found to occur before the region encoding 3CLP, suggesting that both the 195- and 41-kDa proteins may be mature cleavage products.
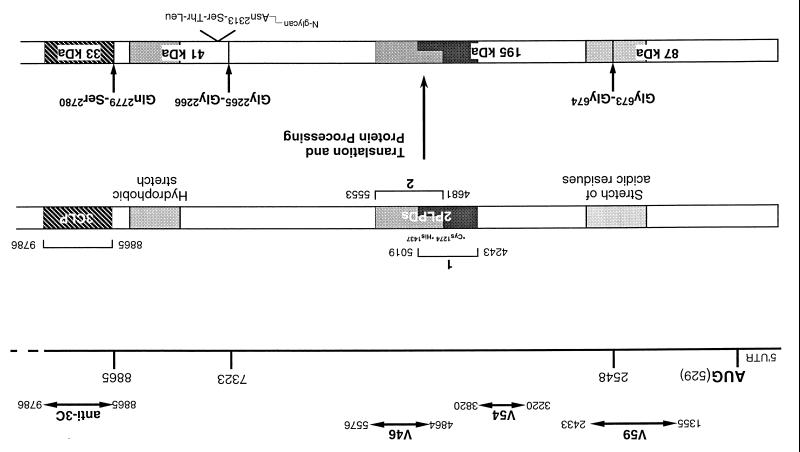
FIG. 1.
Diagram of the structure of the 5′-terminal 9-kb region of ORF 1a, illustrating the location of the 87-kDa protein, the 195-kDa protein with the two putative overlapping PLPDs, the 41-kDa glycoprotein, and 3CLP. The Gly673↓Gly674, Gly2265↓Gly2266, and Gln2779↓Ser2780 scissile bonds, the Asn2313 N-linked glycosylation site, and the catalytic residues of PLPD-1 (Cys1274 and His1437) are indicated. Also shown are the IBV sequence recognized by antisera V59, V54, V46, and anti-3C and the stretches of acidic and hydrophobic residues encoded by ORF1a.
Characterization of the two cleavage products showed that the 195-kDa, PLPD-1-containing protein possesses both cis- and trans-cleavage activities at the Gly2265-Gly2266 dipeptide bond when expressed in intact cells. Only cis-cleavage activity was detected when the protein was expressed in vitro in rabbit reticulocyte lysates. The 41-kDa protein was characterized as an N-linked glycoprotein, and mutagenesis studies confirmed that the putative Asn2313 residue is the actual N-linked glycosylation site.
MATERIALS AND METHODS
Virus and cells.
The egg-adapted Beaudette strain of IBV (ATCC VR-22) was obtained from the American Type Culture Collection and was adapted to Vero cells as described previously (23). Virus stocks were prepared by infecting Vero cells at a multiplicity of infection of approximately 0.1 PFU/cell and incubating them at 37°C in 5% CO2 for 48 h. The virus titer was determined by plaque assay on Vero cells.
Vero cells and Cos-7 cells were grown at 37°C in 5% CO2 and maintained in Glasgow's modified Eagle's medium (GMEM) supplemented with 10% fetal calf serum.
Transient expression of IBV sequence in Cos-7 cells by using a vaccinia virus-T7 expression system.
IBV sequences were placed under the control of a T7 promoter and transiently expressed in mammalian cells, using the system described by Fuerst et al. (8). Briefly, 60 to 80% confluent monolayers of Cos-7 cells grown on 35-mm-diameter dishes (Falcon) were infected at 10 PFU/cell with a recombinant vaccinia virus (vTF7-3) which expresses bacteriophage T7 RNA polymerase. The cells were then transfected with 2.5 μg of plasmid DNA (purified by Qiagen plasmid Midi kits) mixed with DOTAP liposomal transfection reagent according to the instructions of the manufacturer (Boehringer Mannheim). The transfection efficiency achieved by this method was about 25 to 35%, as determined by monitoring the expression of a reporter plasmid encoding green fluorescent protein from jellyfish Aequorea victoria (5). After incubation at 37°C in 5% CO2 for 5 h, the cells were washed twice with methionine-free medium and labeled with 25 μCi of [35S]methionine per ml. The radiolabeled cells were then harvested at 18 h posttransfection.
PCR.
Complementary DNA templates for PCR were prepared from purified IBV virion RNA by using a first-strand cDNA synthesis kit (Boehringer Mannheim). Amplification of template DNAs with appropriate primers were performed with Pfu DNA polymerase (Stratagene) under the standard buffer conditions with 2 mM MgCl2. The reaction conditions used were 30 cycles of 95°C for 45 s, x°C for 45 s, and 72°C for x min. The annealing temperature (x°C) and the extension time (x min) were subjected to adjustments according to the melting temperature of the primers used and the length of PCR fragments synthesized.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by two rounds of PCR and two pairs of primers, as previously described (22).
Radioimmunoprecipitation.
IBV-infected Vero cells and plasmid-transfected Cos-7 cells were lysed with radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS]) and precleared by centrifugation at 12,000 rpm for 5 min at 4°C in a microcentrifuge. Immunoprecipitation with region-specific rabbit polyclonal and anti-T7 monoclonal antibodies (Novagen) were carried out as previously described (19).
Cell-free transcription and translation.
Plasmid DNAs were expressed in rabbit reticulocyte lysates by using a transcription-coupled translation (TnT) system according to the instructions of the manufacturer (Promega). Briefly, 1 μg of plasmid DNA was included in a 50-μl reaction mix, in the presence or absence of 5 μl of canine pancreatic microsomal membranes. The reaction mix was incubated at 30°C for 90 min in the presence of 50 μCi of [35S]methionine per ml.
SDS-PAGE.
Polyacrylamide gel electrophoresis (PAGE) of viral polypeptides was performed on SDS–12.5% polyacrylamide gels (15). The 35S-labeled polypeptides were detected by autoradiography of the dried gels.
Endo H treatment of immunoprecipitated proteins.
Endoglycosidase H (endo H) treatment (Boehringer Mannheim) of samples was performed as described by Machamer et al. (24). Briefly, the immunoprecipitated viral proteins were released from the Sepharose beads with immobilized Staphylococcus protein A by boiling in phosphate-buffered saline for 4 min. The supernatant was transferred to a fresh tube containing an equal volume of 2× endo H buffer (50 mM sodium citrate, 0.2% SDS, 100 mM 2-mercaptoethanol [pH 5.5]) and enzyme (40 U/mg) and incubated overnight at room temperature.
Construction of plasmids.
Plasmid pORF1a3, which covers nucleotides 2545 to 9786, was constructed by three steps of cloning. First, a reverse transcription (RT)-PCR fragment covering nucleotides 6870 to 9786 was generated from viral RNA, using primer XIANG-35 for RT and primers LN-1 and XIANG-35 for PCR. This fragment was digested with NcoI and BamHI, which were introduced by the two PCR primers, and ligated into NcoI- and BamHI-digested pKTO (19), giving rise to pIBV1a7. Second, an RT-PCR fragment covering nucleotides 3827 to 8885 was made from viral RNA, using primer LN-6 for RT and primers AD-2 and LN-6 for PCR. This fragment was digested with NcoI and SnaBI, which digest the IBV sequence at nucleotide positions 3831 and 7999, respectively, and ligated into NcoI- and SnaBI-digested pIBV1a7, giving rise to pIBV1a6(ext). Finally, a PCR fragment covering nucleotides 2548 to 5759 was generated using pIBV1a2 (17) as a template and LKP-14 and AD-3 as primers. This fragment was digested with BamHI and MluI and ligated into BglII- and MluI-digested pIBV1a6(ext), giving rise to pORF1a3. BamHI site was introduced by primer LKP-14. MluI digests the IBV sequence at nucleotide position 3996, and BglII cuts pIBV1a6(ext) at the position 16 nucleotides upstream of the IBV sequence. The construction and nucleotide sequence of pORF1a3 were confirmed by automated sequencing.
Two constructs were generated from pORF1a3. Plasmid pORF1a3Δ1 was made by deletion of the 3CLP-encoding region (nucleotides 8866 to 9786). This was achieved by replacing the SnaBI- and BamHI-fragment (nucleotides 7999 to 9786) from pORF1a3 with an SnaBI- and BamHI-digested PCR fragment covering nucleotides 7999 to 8865, resulting in the deletion of nucleotides 8866 to 9786. The PCR fragment was generated with primers SLKP-2 and LN-6. SnaBI digests the IBV sequence at nucleotide 7999, and the BamHI site was introduced by primer LN-6. Plasmid pORF1a3(M) contains a deletion of nucleotides 7999 to 8693 and a mutation of the nucleophilic Cys2922 residue to an Ala. This plasmid was constructed by ligation of an StuI- and BamHI-digested PCR fragment into SnaBI- and BamHI-digested pORF1a3. The PCR fragment was generated using pIBV14Δ1C2922-A (20) as the template and LDX-44 and XIANG-35 as primers. A single base (T) insertion was introduced immediately downstream of the StuI site by primer LDX-44 to keep the ligated IBV sequence in frame.
Plasmid pORF1a3Δ2 was constructed by two steps of cloning. A PstI-digested fragment covering nucleotides 4858 to 6919 was cloned into PstI-digested pIBV1a1 (17, 21), giving rise to pIBV1a3. The region coding for the 87-kDa protein (nucleotides 529 to 2545) was then deleted as described for pORF1a3, giving rise to pORF1a3Δ2.
Two constructs, pORF1a3Δ3 and pORF1a4, based on pORF1a3Δ2 were made using similar strategies. Plasmid pORF1a3Δ3, which contains nucleotides 2545 to 7324 and a UAA stop codon at the end of the viral sequence, was constructed by inserting a BstEII- and SmaI-digested PCR fragment into BstEII- and SmaI-digested pORF1a3Δ2. The PCR primers used to amplify the region between nucleotides 6174 and 7324 were XHY-7 and LKP-29. BstEII cuts the IBV sequence at nucleotide position 6452, and the SmaI site was introduced behind the UAA codon by primer LKP-29. Plasmid pORF1a4 was constructed by cloning a 1,500-bp, BstEII- and SmaI-digested PCR fragment, generated with primers XHY-7 and LKP-28, into BstEII- and SmaI-digested pORF1a3Δ2. This construct contains the IBV sequence starting from nucleotide 2548 and ending at nucleotide 7952. A termination codon (UAA) was inserted at the end of the viral sequence by primer LKP-28.
Plasmid pORF1a4Δ1 was made by deletion of nucleotides 4010 to 4619 from pORF1a4. This was achieved by replacing the BglII/StuI fragment (nucleotides 3369 to 4619) with a PCR fragment covering nucleotides 3369 to 4010. BglII and StuI digest the IBV sequences at nucleotides 3369 and 4619, respectively. The PCR fragment was made using pORF1a4 as the template and T7 and LKP-30 as primers. An StuI site was introduced at nucleotide position 4010 with primer LKP-30. Plasmid pORF1a4Δ2 was made by deletion of nucleotides 5026 to 5362 from pORF1a4. This was achieved by cloning an XbaI- and SmaI-digested PCR fragment covering nucleotides 5349 to 7965 into SpeI- and SmaI-digested pORF1a4. The XbaI and SmaI sites were introduced into the PCR fragment with primers LKP-31 and LKP-28, respectively. SpeI digests the IBV sequence at nucleotide 5026, and SmaI digests pORF1a4 at the position six nucleotides downstream of the viral sequence. Plasmid pORF1a4C1274-S covers nucleotides 2545 to 7952, with an alteration of the putative catalytic residue Cys1274 to a Ser. This construct was made by inserting an MluI- and SpeI-digested DNA fragment (nucleotides 3997 to 5027) from pIBV1a2Δ4C1274-S (16) into MluI- and SpeI-digested pORF1a4Δ1.
The following four deletion and five mutation plasmids were constructed using two rounds of PCR (22). The primers used for the second round of PCR were XHY-7 and LKP-28. The PCR fragments (covering nucleotides 6097 to 7975) generated were digested with BstEII (which cuts at nucleotide 6451) and SmaI and then ligated into BstEII- and SmaI-digested pORF1a3Δ2; the resulting plasmids cover nucleotides 2545 to 7952 with desired deletions or mutations. The four deletion constructs, pORF1a4ΔG2265-G2266, pORF1a4ΔG2265, pORF1a4ΔG2246-A2247, and pORF1a4ΔG2237-V2238 (which contain deletions of nucleotides coding for Gly2265-Gly2266, Gly2265, Gly2246-Ala2247, and Gly2237-Val2238, respectively), were made using deletion primer pairs LKP-28dGGU and LKP-28dGGD, LKP-28dG1U and LKP-28dG1D, LKP-28dGAU and LKP-28dGAD, and LKP-28dGVU and LKP-28dGVD, respectively. The five mutants, pORF1a4A2264-N, pORF1a4G2266-N, pORF1a4G2265-N, pORF1a4G2265-A, and pORF1a4N2313-Q (which contain substitution mutations of A2264-N, G2266-N, G2265-N, G2265-A, and N2313-Q, respectively), were constructed using mutagenesis primer pairs LKP-28A-NU and LKP-28A-ND, LKP-28G2-NU and LKP-28G2-ND, LKP-28G1-NU and LKP-28G1-ND, LKP-28G1-AU and LKP-28G1-AD for pORF1a4G2265-A, and LKP-28N-QU and LKP-28N-QD, respectively.
Plasmid pP41, which covers nucleotides 7324 to 8865, was made by cloning a BamHI- and SmaI-digested PCR fragment into BglII- and SmaI-digested pKT0. The PCR fragment was generated using primer pair LKP-32 and LKP-33. An artificial AUG start codon and a UAA stop codon were introduced with the upstream and downstream primers, respectively.
Three plasmids were constructed based on pIBV1a7. Plasmid pIBV1a7Δ1 was made by digesting pIBV1a7 with SnaBI and BamHI (nucleotides 7999 to 9786) and ligated into EcoRV- and BamHI-digested pKT0, resulting in the deletion of nucleotides 6870 to 7999 from pIBV1a7. Plasmid pIBV1a7Δ1Q2779-E, which contains the Gln2779-to-Glu mutation, was created by two rounds of PCR with pIBV1a7Δ1 as the template. The primers used to introduce the Gln2779-to-Glu mutation are LN-10 and LN-11. Plasmid pIBV1a7Δ2Q2779-E was made by deletion of nucleotides 7999 to 8277 from pIBV1a7Δ1Q2779-E. The primers used for generating the PCR fragment are LN-5 and XIANG-35.
The sequences of all primers used are listed in Table 1. All mutants and deletion constructs were selected and confirmed by automated sequencing.
TABLE 1.
Oligonucleotides used for PCR Amplification and site-directed mutagenesis
| Name | Nucleotide sequence (5′-3′)a | Position |
|---|---|---|
| T7 | TAATACGACTCACTATAGGG | |
| AD-2 | CAGCTGCCATGGACAACAACTTGCATGAGA | 3827–3856 |
| AD-3 | AATTAGGATCCTAGCTAGCGACAACACTCTT | 5759–5740 |
| XHY-7 | GTGGCAGGATCCGTTATTATTTGC | 6097–6120 |
| LKP-14 | GGTTTGCGGATCCATGGGTAAGACTGTCAC | 2532–2561 |
| LDX-4 | TCTGTTTGCAAGTTACATCG | 4060–4041 |
| SLKP-2 | GGTGTTTATATTTAACAGCCAG | 7832–7853 |
| LN-6 | ACCAGAGGATCCTATTGTAATCTACT | 8885–8854 |
| LN-1 | GTTATACCATGGATAAAGTGTGTAGC | 6865–6885 |
| LN-5 | TTTCAACCATGGGTGTTTTTAAAGCT | 8272–8292 |
| LN-10 | GTTAGTAGATTAGAGTCTGGTTTTAAG | 8852–8877 |
| LN-11 | CAGTTTCTTAAAACCAGACTCTAATCT | 8857–8883 |
| LN-54 | AAGGCAGGTGGATCCGTATTGTTAGCGGC | 7315–7338 |
| LDX-44 | ATTTGAGGCCTTACCTTTCAGCG | 8685–8702 |
| XIANG-35 | TTAACAGGATCCTATTGTAATCTAAC | 9879–9855 |
| LKP-28 | GTGGAACTATCCCGGGTTAACCATTGGGTTG | 7975–7945 |
| LKP-29 | GCCGCTCCCGGGTTAACCTGCCTTTTTC | 7338–7311 |
| LKP-28dGG | AGAGAAAAAGGCAATTGTTAGCGGCAC | 7308–7340 |
| LKP-28dG1 | AGAGAAAAAGGCAGGTATTGTTAGCGG | 7308–7337 |
| LKP-28dGA | CTTTATAACAAAGTCTAAACAAGTTATTG | 7248–7282 |
| LKP-28dGV | CTACTGTTAAGTCACGTTTCTTTATAAC | 7223–7256 |
| LKP-28A-N | GTAGAGAAAAAGAACGGTGGTATTGTTA | 7306–7333 |
| LKP-28G1-A | GAGAAAAAGGCAGCCGGTATTGTTAGCG | 7309–7336 |
| LKP-28G1-N | GAGAAAAAGGCAAACGGTATTGTTAGCG | 7309–7336 |
| LKP-28G2-N | GAAAAAGGCAGGTAACATTGTTAGCGGC | 7311–7338 |
| LKP-28N-Q | GTATGATGTACAGTCCACACTGCATGT | 7455–7481 |
| LKP-30 | CTTGGCTCAGGCCTAACAAAAGAACGCG | 4024–3997 |
| LKP-31 | GGTCATCCATCTAGATATAGTAAGTCTC | 5350–5377 |
| LKP-32 | GAGAAAGGATCCATGGGTATTGTTAGCG | 7309–7336 |
| LKP-33 | CAGTTTCCCCGGGCCTTATTGTAATCTAC | 8883–8855 |
Underlining denotes the initiation codon (ATG), termination codon (TAA), and a T insertion, respectively.
RESULTS
Expression of the IBV sequence from nucleotides 2545 to 9786.
In a previous report, we demonstrated that the first cleavage product of the 1a and 1a/1b polyproteins is an 87-kDa protein encoded by ORF 1a between nucleotides 529 and 2547. To study the processing events occurring downstream of the 87-kDa protein, plasmid pORF1a3 was constructed to cover nucleotides 2545 to 9786. As shown in Fig. 2a, both PLPDs and 3CLP were included in this construct. Transient expression of pORF1a3 in Cos-7 cells using the vaccinia virus-T7 expression system resulted in the immunoprecipitation of two polypeptides of approximately 33 and 195 kDa (Fig. 2b, lanes 2 and 4). The 33-kDa protein, which represents 3CLP (27), was immunoprecipitated with anti-3C antiserum (lane 2), and the 195-kDa protein was detected by immunoprecipitation with antiserum V46, which was raised against the IBV sequence encoded by nucleotides 4864 to 5576. To investigate if the 195-kDa protein represents the N-terminal cleavage product of polyprotein encoded by this construct, two deletion constructs, pORF1a3Δ1 and pORF1a4, were made. Plasmid pORF1a3Δ1 contains a C-terminal deletion of the IBV sequence coding for 3CLP (nucleotides 8866 to 9786), and pORF1a4 contains a UAA termination codon inserted at nucleotide position 7952 (Fig. 2a). Expression of these plasmids in Cos-7 cells resulted in the detection of a 195-kDa protein which comigrates with the 195-kDa protein expressed from pORF1a3 (Fig. 2b, lanes 4 to 6). Interestingly, a smeary band of approximately 41 kDa and several smaller protein species migrating around 20 to 25 kDa were detected by immunoprecipitation with V46 from the expression of pORF1a3Δ1 and pORF1a4, respectively (lanes 5 and 6). As the same 195-kDa protein was detected from all three constructs and the only difference among them lies in the 3′-terminal region, it was suggested that a novel cleavage event might occur, resulting in the release of the N-terminal 195-kDa protein and a C-terminal cleavage product. The detection of the potential C-terminal cleavage protein by antiserum V46 may indicate the interaction between the N- and C-terminal cleavage product; the reason for failing to detect the 41-kDa protein from the expression of pORF1a3 with antiserum V46 is unclear (lane 4).
FIG. 2.
(a) Diagram showing the IBV sequence contained in plasmids pORF1a3, pORF1a3Δ1, pORF1a4, and pORF1a3(M). (b) Analysis of transiently expressed ORF 1a products from cells transfected with pORF1a3(M), pORF1a3, pORF1a3Δ1, and pORF1a4. Plasmid DNAs were transiently expressed in Cos-7 cells, using the vaccinia virus-T7 expression system. Cells were labeled with [35S]methionine, lysates were prepared, and polypeptides were immunoprecipitated with anti-3C and V46. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons.
Next, plasmid pORF1a3(M) was constructed and expressed. In this plasmid, the nucleophilic cysteine residue (Cys2922) of 3CLP was mutated to an alanine residue, so that the 3CLP activity was abolished (20). Meanwhile, nucleotides 7999 to 8693, a region which codes for hydrophobic amino acid stretches, were deleted to facilitate detection of the potential C-terminal cleavage products (Fig. 2a). Expression of this construct resulted in the detection of the 195-kDa protein and two smaller products of approximately 57.5 and 64 kDa (Fig. 2b, lanes 1 and 3). The 33-kDa protein was not observed (lane 1), confirming that the 3CLP activity was abolished by the Cys2922→Ala mutation. Once again, the detection of all three products by both anti-3C and V46 may suggest the interaction between the cleavage products, which will be addressed later in greater detail. These results imply that at least one more cleavage site exists in the 1a region between the 87-kDa protein and 3CLP.
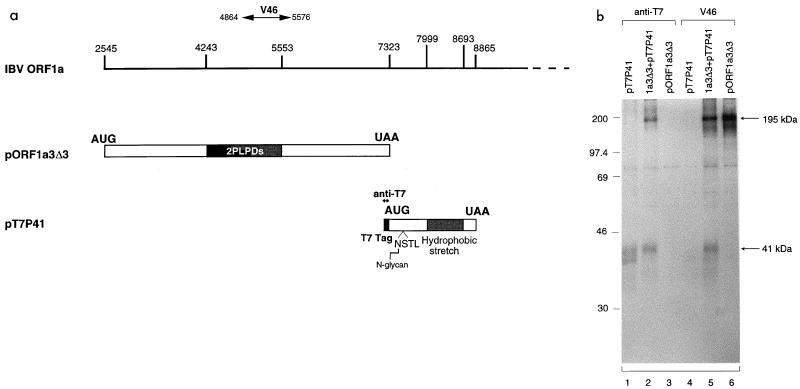
In vitro translation and processing of the polyprotein encoded by nucleotides 2545 to 8865.
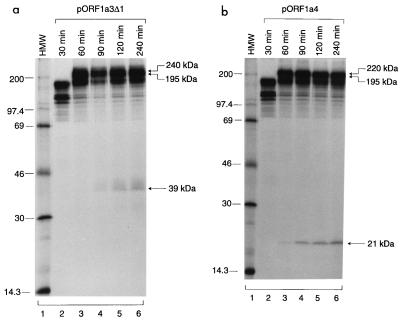
To confirm that a novel cleavage event did occur in the region between the 87-kDa protein and 3CLP and to investigate if the cleavage activity could be detected in vitro as described for MHV and HCV 229E (1, 6, 11, 13), plasmids pORF1a3Δ1 and pORF1a4 were expressed in in vitro time course experiments using the TnT reticulocyte lysate system. For each reaction, 10-fold unlabeled methionine was added to the reaction mixture after incubation for 90 min in the presence of 50 μCi of [35S]methionine per ml. A prolonged pulse time was required to allow labeling of the polypeptide towards the C terminus. As can be seen, expression of pORF1a3Δ1 led to the detection of a polypeptide with an apparent molecular mass of 39 kDa after incubation for 90 min; the product was increased toward the end of the time course (Fig. 3a). Similarly, expression of pORF1a4 led to the detection of a small protein of approximately 20 kDa (Fig. 3b). This product was first seen at the 60-min time point and was increased over time (Fig. 3b). The appearance of the 195-kDa protein, which was more clearly observed from gels with lighter exposure (data not shown), coincided with that of the 39- and 20-kDa proteins (Fig. 3). These results clearly showed the occurrence of a novel cleavage event, resulting in the release of the N-terminal 195-kDa protein and a C-terminal product. We noted that the C-terminal cleavage products detected in vitro migrate more rapidly than those detected in intact cells. This reflects the posttranslational modification of the cleavage products and will be addressed later.
FIG. 3.
Time course analysis of polypeptides synthesized in rabbit reticulocyte lysates from pORF1a3Δ1 (a) and pORF1a4 (b). A 10-fold excess of unlabeled methionine was added to the in vitro translation reaction mixture after incubation at 30°C for 90 min, and aliquots were taken from the reaction mixture after incubation for 30, 60, 90, 120, and 240 min. [35S]methionine-labeled translation products were separated on an SDS–15% polyacrylamide gel and detected by fluorography. HMW, high-molecular-mass markers (numbers indicate kilodaltons).
Identification of the proteinase domain required for the cleavage event and demonstration of its trans-cleavage activity.
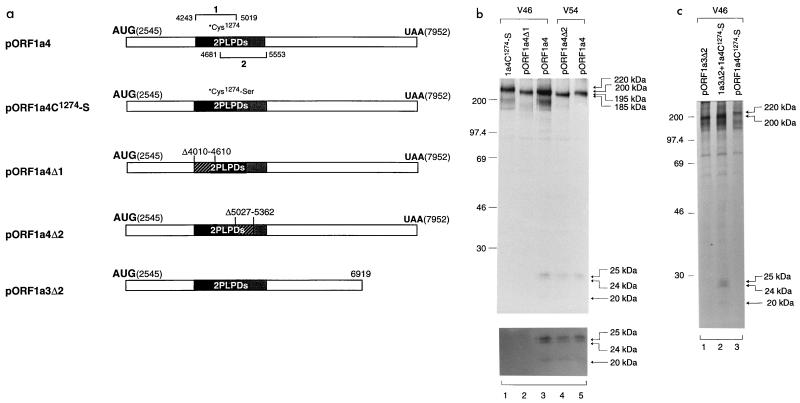
We previously demonstrated that PLPD-1 encoded between nucleotides 4243 and 5576 was responsible for the cleavage of the 87-kDa protein at the Gly673-Gly674 dipeptide bond (17), and the catalytic dyad for this activity was the Cys1274 and His1437 residues. We then used deletion and mutational analyses to test if either of the two PLPDs is involved in this novel cleavage activity. Two constructs, pORF1a4C1274-S and pORF1a4Δ1, were initially constructed and expressed. pORF1a4C1274-S is similar to pORF1a4 but contains a mutation of the putative catalytic residue Cys1274 to a Ser residue; pORF1a4Δ1 contains a deletion of nucleotides 4010 to 4610, which encode part of PLPD-1 including the catalytic residue Cys1274 (Fig. 4a). Expression of these constructs showed no detection of the cleavage product compared with pORF1a4 (Fig. 4b, lanes 1 to 3), suggesting that PLPD-1 may be involved in this cleavage event.
FIG. 4.
(a) Diagram showing the IBV sequences present in plasmids pORF1a4, pORF1a4C1274-S, pORF1a4Δ1, pORF1a4Δ2, and pORF1a3Δ2. (b) Analysis of transiently expressed ORF 1a products from transfection of pORF1a4, pORF1a4C1274-S, pORF1a4Δ1, and pORF1a4Δ2. Plasmid DNAs were transiently expressed in Cos-7 cells, using the vaccinia virus-T7 expression system. Cells were labeled with [35S]methionine, lysates were prepared, and polypeptides were immunoprecipitated with antisera V46 and V54. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. The upper panel was prepared from a gel exposed for 1 day, and the lower panel was prepared from the same gel exposed for 5 days. Numbers indicate molecular masses in kilodaltons. (c) trans-cleavage assay of PLPD-1 by coexpression of pORF1a3Δ2 and pORF1a4C1274-S. Cos-7 cells were transfected with either a single plasmid or two plasmids together and labeled with [35S]methionine. Lysates were prepared, and polypeptides were immunoprecipitated with antiserum V46. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons.
To support this conclusion, we constructed and expressed pORF1a4Δ2, a plasmid with a deletion of the second putative PLPD (nucleotides 5027 to 5362). Expression of this plasmid in Cos-7 cells led to the detection of the two cleavage products by region-specific antiserum V54 (Fig. 4b, lane 4). This antiserum was raised against the IBV sequence encoded by nucleotides 3220 to 3820. V54 could also precipitate the same cleavage products from cell lysate transfected with pORF1a4 (Fig. 4b, lane 5).
It was reported that MHV PLPD-1 is able to act in trans at both Gly247-Val248 and Ala832-Gly833 dipeptide bonds to release p28 and p65, respectively (1, 6, 11, 13, 16). Recently, Herold et al. (11) reported that HCV 229E PLPD1 could also cleave the Gly111-Asn112 dipeptide bond in trans to release p9. We were interested to know whether this trans-cleavage activity could be demonstrated for IBV PLPD-1 at this cleavage event. Figure 4c shows the results obtained from a trans-cleavage assay. Immunoprecipitation of cell lysates prepared from cells transfected with pORF1a3Δ2, a plasmid containing the two overlapping PLPDs, resulted in the detection of a protein of approximately 200 kDa, representing the full-length product encoded by this construct (lane 3). Expression of pORF1a4C1274-S yielded the 220-kDa, full-length protein precursor (Fig. 4c, lane 1). Coexpression of pORF1a3Δ2 and pORF1a4C1274-S, however, resulted in the detection of both cleavage products, demonstrating the ability of the IBV PLPD-1 to act in a trans manner (Fig. 4c, lane 2).
Mapping of the second cleavage site to the Gly2265-Gly2266 scissile bond.
As shown in Fig. 3b, the actual C-terminal cleavage product of the pORF1a4-encoded product was approximately 20 kDa. This allows us to estimate that the cleavage site is located near nucleotide position 7300. By comparative analysis of the cleavage sites utilized by this and other viral papain-like proteinases (1, 6, 11, 13, 16), one of the following scissile bonds may be the cleavage site: Gly2265-Gly2266, Gly2246-Ala2247, or Gly2247-Val2248. Deletion and site-directed mutagenesis approaches were used to investigate this possibility.
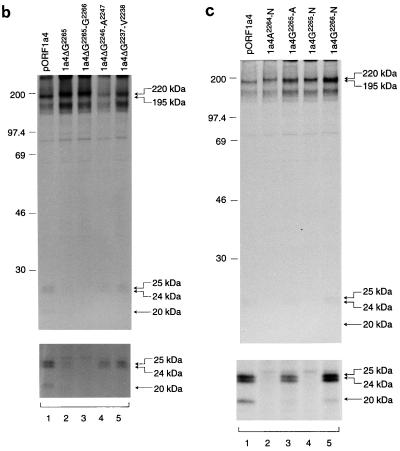
Four deletion mutants, pORF1a4ΔG2265-G2266, pORF1a4ΔG2265, pORF1a4ΔG2246-A2247, and pORF1a4ΔG2237-V2238, were first constructed based on pORF1a4. They contain deletions of Gly2265-Gly2266, Gly2265, Gly2246-Ala2247, and Gly2237-Val2238, respectively (Fig. 5a). Expression of these mutants showed that deletion of the Gly2265-Gly2266 residues resulted in dramatic inhibition of the cleavage activity; only the full-length 220-kDa protein was clearly detected (Fig. 5b, lane 3). However, deletion of either the Gly2246-Ala2247 or Gly2237-Val2238 had no obvious effect on the release of the 195-kDa and the C-terminal cleavage product (Fig. 5b, lanes 4 and 5). This implies that Gly2265-Gly2266 may be essential for this cleavage activity. To support this conclusion, deletion of the Gly2265 residue only showed that the cleavage activity was drastically inhibited (Fig. 5b, lane 2).
FIG. 5.
(a) Effects of substrate deletions and mutations on the second cleavage activity mediated by PLPD-1. The amino acid sequence flanking the Gly2265-Gly2266 dipeptide bond and the deletions and mutations introduced are outlined. (b) Effects of deletions of the Gly2265, Gly2265-Gly2266, Gly2246-Ala2247, and Gly2237-Val2238 residues on cleavage activity. Plasmid DNAs were transiently expressed in Cos-7 cells, using the vaccinia virus-T7 expression system. [35S]methionine-labeled cell lysates were prepared, and polypeptides were immunoprecipitated with antiserum V46. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. The upper panel was prepared from a gel exposed for 1 day, and the lower panel was prepared from the same gel exposed for 5 days. Numbers indicate molecular masses in kilodaltons. (c) Effects of mutations of the A2264-N, G2265-A, G2265-N, and G2266-N residues on cleavage activity. The mutants were expressed and analyzed as described for panel b. The upper panel was prepared from a gel exposed for 1 day, and the lower panel was prepared from the same gel exposed for 5 days.
Site-directed mutagenesis was then carried out to further test if the Gly2265-Gly2266 dipeptide bond was the scissile bond. The four point mutations created are depicted in Fig. 5a, and expression data are presented in Fig. 5c. As can be seen, mutation of Ala2264 to Asn (lane 2) and Gly2265 to Asn (lane 4) almost abolished cleavage activity, whereas substitution of Gly2265 with Ala (lane 3) and Gly2266 with Asn (lane 5) had no dramatic effect on cleavage activity (Fig. 5c). Taken together with the deletion data present in Fig. 5b, these results suggest strongly that IBV PLPD-1 may act on the Gly2265-Gly2266 dipeptide bond to release the 195- and 41-kDa proteins.
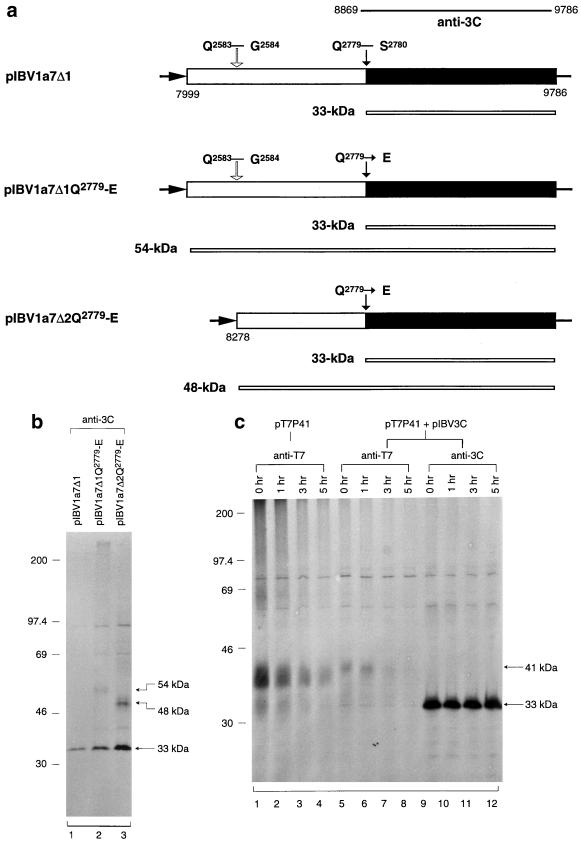
No cleavage at the previously predicted Q2583-G2584 dipeptide bond by 3CLP.
In a previous study using an in vitro expression and processing system, Tibbles et al. (35) showed that the two predicted Q-S dipeptide bonds (Q2779-S2780 and Q3086-S3087) may be the cleavage sites responsible for releasing 3CLP from the 1a and 1a/1b polyproteins. In addition, the Q2583-G2584 dipeptide bond encoded by nucleotides 8275 to 8280 was predicted to be a cleavage site of 3CLP (9). As this site is located in the 41-kDa protein-encoding region, mutagenesis studies were carried out to test if cleavage could occur at this position. As outlined in Fig. 6a, expression of pIBV1a7Δ1 would result in the detection of a 33-kDa protein representing 3CLP. Mutation of the Q2779 to an E was introduced to pIBV1a7Δ1 to generate plasmid pIBV1a7Δ1Q2779-E (Fig. 6a). A 54-kDa protein containing the 33-kDa protein and the upstream region would be immunoprecipitated by anti-3C antiserum (Fig. 6a). As can be seen, expression of pIBV1a7Δ1 resulted in the detection of the 33-kDa protein only (Fig. 6b, lane 1). The same product was also detected from expression of pIBV1a7Δ1Q2779-E (lane 2); in addition, a product of approximately 54-kDa was detected (lane 2). No other cleavage products were observed, suggesting that the Q2583-G2584 dipeptide bond may not be used by 3CLP. To explore this possibility further, plasmid pIBV1a7Δ2Q2779-E, which covers nucleotides 8278 to 9786, was constructed and expressed (Fig. 6a). As expected, expression of this construct resulted in the detection of the 33-kDa protein and a product of approximately 48 kDa (Fig. 6b, lane 3). The 48-kDa protein represents the full-length product encoded by this construct. If cleavage occurred at the Q2583-G2584 dipeptide bond, a product comigrating with the 48-kDa protein would be detected from the expression of pIBV1a7Δ1Q2779-E. These results support the conclusion that no cleavage occurs at this position.
FIG. 6.
(a) Diagram showing the IBV sequence contained in plasmids pIBV1a7Δ1, pIBV1a7Δ1Q2779-E, and pIBV1a7Δ2Q2779-E. (b) Analysis of transiently expressed ORF 1a products from cells transfected with pIBV1a7Δ1, pIBV1a7Δ1Q2779-E, and pIBV1a7Δ2Q2779-E. Plasmid DNAs were transiently expressed in Cos-7 cells, using the vaccinia virus-T7 expression system. Cells were labeled with [35S]methionine, lysates were prepared, and polypeptides were immunoprecipitated with anti-3C. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons. (c) Time course analysis of the 41-kDa protein by coexpression of pT7P41 and pIBV3C. Cos-7 cells were transfected with either pT7P41 or pT7P41 and pIBV3C together and labeled with [35S]methionine for 4 h. The cells were washed three times with GMEM and were incubated in GMEM before they were harvested 0, 1, 3, and 5 h later. Lysates were prepared, and polypeptides were immunoprecipitated with antisera anti-T7 and anti-3C. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gels, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons.
To further address the possibility that cleavage may occur at the predicted Q2583-G2584 dipeptide bond, plasmid pT7P41 was constructed to tag an 11-amino-acid T7 tag (MASMTGGQQMG) to the N terminus of the 41-kDa protein. This construct was coexpressed in intact cells with pIBV3C (26) in time course experiments. As can be seen, both the 33-kDa and the T7-tagged 41-kDa proteins were efficiently detected at the beginning of the time course (Fig. 6c, lanes 5 to 12). The 33-kDa protein remained stable throughout the time course (lanes 9 to 12). However, the T7-tagged 41-kDa protein was rapidly decreased over time; only a trace amount of the protein was detected at the end of the time course (lanes 5 to 8). As no smaller product was detected, it is unlikely that the decrease in the detection of the T7 tagged 41-kDa protein is due to further cleavage of the 41-kDa protein. Instead, it may be caused by the instability of the 41-kDa protein. In support of this notion, rapid turnover of the 41-kDa protein was also observed when pT7P41 was expressed on its own (lanes 1 to 4).
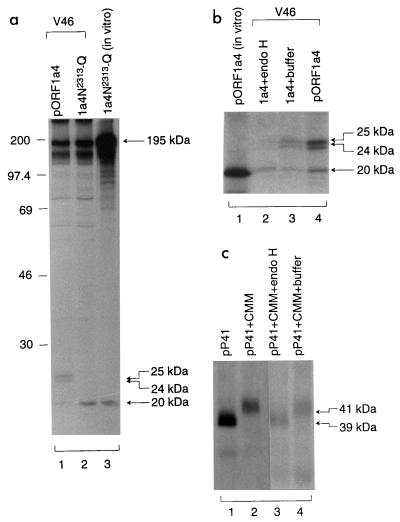
N-linked glycosylation of the 41-kDa protein.
As presented in Fig. 2b and 3b, migration of the C-terminal cleavage products was different when expressed in vitro and in intact cells, respectively, suggesting that they may be modified posttranslationally. Examination of the amino acid sequence indicates that a potential N-linked glycosylation site (Asn2313-Ser2314-Thr2315) is encoded by nucleotides 7465 to 7413. Two approaches were used to explore if glycosylation did occur at this position. First, the Asn2313 residue of the putative N-linked glycosylation site (Asn2313-Ser2314-Thr2315) was mutated to a Gln residue, giving rise to plasmid pORF1a4N2313-Q. When this plasmid was expressed in intact cells, only the 195- and 20-kDa cleavage products were detected (Fig. 7a, lane 2). Both proteins comigrated with the 195- and 20-kDa proteins yielded from the in vitro translation system (lanes 2 and 3). Next, the in vivo-generated products of pORF1a4 were subjected to digestion with endo H. Figure 7b shows that the two upper bands (24- and 25-kDa proteins) were sensitive to endo H treatment (lane 2); the resulting product comigrated with the protein generated from the in vitro-expressed 20-kDa protein (lanes 2 and 3). These results confirm that the C-terminal 20-kDa product is modified by N-linked glycosylation.
FIG. 7.
(a) Mutational analysis of the putative N-linked glycosylation site of the 41-kDa protein. Plasmids pORF1a4 and pORF1a4N2313-Q were transiently expressed in Cos-7 cells, using the vaccinia virus-T7 expression system. Cells were labeled with [35S]methionine, lysates were prepared, and polypeptides were immunoprecipitated with antiserum V46. Gel electrophoresis of the in vivo-synthesized polypeptides (lanes 1 and 2) together with the in vitro-synthesized products (lane 3) was performed on an SDS–12.5% polyacrylamide gel. Polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons. (b) Endo H treatment of the C-terminal cleavage products from expression of pORF1a4. Plasmid DNA was transfected into Cos-7 cells, using the vaccinia virus-T7 expression system. The [35S]methionine-labeled cell lysates were prepared, and polypeptides were immunoprecipitated with antiserum V46. The immunoprecipitated viral proteins were analyzed either by gel electrophoresis directly (lane 4) or after incubation with endo H (40 U/mg; lane 2). Incubation of the immunoprecipitated viral proteins with endo H buffer alone (lane 3) and the 20-kDa cleavage product obtained from in vitro expression of pORF1a4 (lane 1) were included as controls. The [35S]methionine-labeled polypeptides were subjected to gel electrophoresis on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. (c) Endo H treatment of the in vitro-synthesized 41-kDa protein. Plasmid pP41 was expressed in vitro, using the cell-free transcription-coupled translation system, in the presence or absence of 5 μl of canine pancreatic microsomal membranes. The [35S]methionine-labeled polypeptides were analyzed either by gel electrophoresis directly or after incubation with endo H (40 U/mg) or buffer only.
As the 20-kDa protein represents only a C-terminally truncated version of the 41-kDa cleavage product, plasmid pP41, which contains nucleotides 7324 to 8865 coding for the entire 41-kDa protein, was constructed to test if it could also be glycosylated. As shown in Fig. 7c, expression of this plasmid in vitro in rabbit reticulocyte lysates led to the detection of a product of approximately 39 kDa, representing the full-length product encoded by this construct (lane 1). In the presence of canine microsomal membranes, this product was translated as 41 kDa (lane 2). Treatment of the 41-kDa product with endo H reversed its size to 39 kDa (lane 3), confirming that it was also glycosylated.
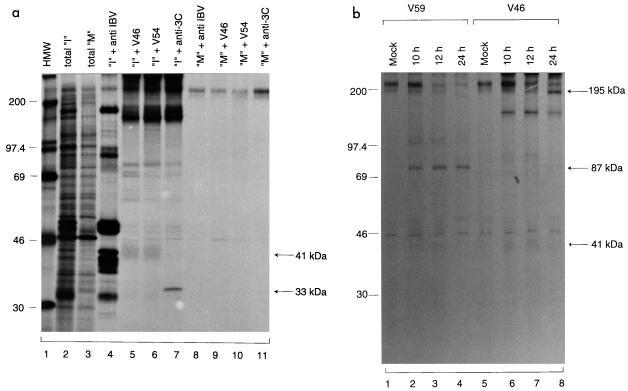
Interaction between the cleavage products.
During the course of this study, we used antiserum V46 (V54 in one case) to immunoprecipitate both the N- and C-terminal cleavage products. As V46 was raised against the IBV sequence encoded by nucleotides 4864 to 5576, the detection of both products by this antiserum may suggest that they are physically associated. To test the specificity of this antiserum and explore the potential interaction between the two products, plasmid pORF1a3Δ3, which encodes the 195-kDa protein, and pT7P41 were expressed (Fig. 8a). pT7P41 was used because attempts to raise antiserum against the 41-kDa protein were unsuccessful. As shown in Fig. 8b, when the two constructs were separately expressed, anti-T7 antiserum could precipitate only the 41-kDa protein encoded by pT7P41 (lane 1); no 195-kDa protein was detected (lane 3). Similarly, V46 could precipitate only the 195-kDa protein encoded by pORF1a3Δ3 (lane 6); no immunoprecipitation of the 41-kDa protein was observed (lane 4). However, coexpression of the two constructs led to precipitation of both the 195- and 41-kDa proteins by either antiserum (lanes 2 and 5). These results demonstrate that physical interactions of the two cleavage products may occur in intact cells, allowing us to coprecipitate the C-terminal cleavage product with a region-specific antiserum against the N-terminal cleavage product.
FIG. 8.
(a) Diagram showing the IBV sequences present in plasmids pORF1a3Δ3 and pT7P41. (b) Analysis of transiently expressed proteins from transfection of pORF1a3Δ3 and pT7P41. Plasmid DNAs were transiently expressed in Cos-7 cells, using the vaccinia virus-T7 expression system. Cells were labeled with [35S]methionine, lysates were prepared, and polypeptides were immunoprecipitated with anti-T7 or V46 antiserum. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons.
Identification of two novel gene products encoded by ORF1a in IBV-infected cells.
Confluent monolayers of Vero cells were infected with IBV at a multiplicity of infection of approximately 2 PFU/cell. The cells were labeled with [35S]methionine at 6 h postinfection (p.i.) for 4 h and then harvested (Fig. 9a). Immunoprecipitation of cell lysates with antisera V46 and V54 resulted in the detection of the 41-kDa protein from IBV-infected cells (Fig. 9a, lanes 5 and 6) but not from mock-infected cells (lanes 9 and 10). Meanwhile, immunoprecipitation of the same lysates with anti-3C antiserum led to the detection of the 33-kDa 3CLP in IBV-infected cells (lane 7). However, the expected 195-kDa protein was only marginally, if any, detected by either V46 or V54 (lanes 5 and 6).
FIG. 9.
(a) Detection of polypeptides encoded by ORF1a in IBV-infected Vero cells. IBV-infected (“I”) and mock-infected (“M”) Vero cell monolayers in 35-mm-diameter dishes were labeled with [35S]methionine for 4 h at 6 h p.i. Cell lysates were prepared, and polypeptides were either analyzed directly or immunoprecipitated with antisera anti-IBV, V46, V54, and anti-3C. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons. (b) Time course analysis of three ORF 1a-specific polypeptides in IBV-infected Vero cells. Cells were infected with IBV at a multiplicity of infection of approximately 2. After incubation in complete GMEM for 5.5 h, the cells were incubated in methionine-free medium for 0.5 h and were labeled with [35S]methionine (100 μCi/ml) for 4 h. The cells were then washed three times with GMEM and incubated in GMEM until harvested at 10, 12, and 24 h p.i. Lysates were prepared, and polypeptides were immunoprecipitated with antisera V59 and V46. Gel electrophoresis of the polypeptides was performed on an SDS–12.5% polyacrylamide gel, and polypeptides were detected by fluorography. Numbers indicate molecular masses in kilodaltons.
A time course experiment was then carried out to further investigate the processing and maturation of the three N-terminal cleavage products (87-, 195-, and 41-kDa proteins). Immunoprecipitation using antiserum V59 (17, 21) showed that the 87-kDa protein was detected from IBV-infected cells harvested at 10 h p.i. (Fig. 9b, lane 2). The protein remained stable over the time course (lanes 2 to 4). Immunoprecipitation of the same cell lysates with V46 showed that, once again, the 195-kDa protein was marginally detected at 10 h p.i. (lane 6). However, the protein level was markedly increased over the time course (lanes 6 to 8). The reason for this increase of the detection of the 195-kDa protein at the late stages of viral infection is uncertain. The 41-kDa product was first observed at 10 h p.i. but was gradually decreased over time (Fig. 9b, lanes 6 to 8), reflecting the instability of the protein. We also noted that a product of approximately 150 kDa was immunoprecipitated by V46, V54, and anti-3C. The identity of this product is not known, but we do not think that it is a specific viral polypeptide, as it could be immunoprecipitated by several region-specific antisera against other 1a as well as 1b regions (data not shown).
DISCUSSION
Data presented in this report demonstrate a novel cleavage activity of IBV PLPD-1. Based on sequence comparison, mutagenesis, and deletion studies, the Gly2265-Gly2266 dipeptide bond was defined as most likely the second cleavage site utilized by PLPD-1 to release a 195- and a 41-kDa protein.
It is generally believed that the sequence context determines the recognition of a cleavage site by a proteinase. The initial deletion studies present in this communication demonstrated that among the three double amino acid deletions (Gly2265-Gly2266, Gly2246-Ala2247, and Gly2237-Val2238), the only construct that abolished the cleavage activity was the Gly2265-Gly2266 deletion. These results essentially narrowed down the potential candidates for the cleavage sites to the Ala2264-Gly2265 [Val (position 5 {P5})-Glu(P4)-Lys (P3)-Lys (P2)-Ala (P1)-Gly (P1′)-Gly (P2′)] and Gly2265-Gly2266 [Glu (P5)-Lys (P4)-Lys (P3)-Ala (P2)-Gly (P1)-Gly (P1′)-Ile (P2′)] dipeptide bonds. Expression of the construct containing a single Gly (Gly2265 or Gly2266) deletion resulted in a drastic drop in the cleavage activity, indicating that Gly2265 may occupy P1. This is because deletion of the putative P1 site will shift all amino acid residues at P2 to P5 and therefore inhibit cleavage activity. For example, an Ala (P2) would occupy the P1 site and a Lys (P3) would be at P2, resulting in a nonconservative substitution of the nonpolar, uncharged Ala residue with the positively charged Lys at P2. This is consistent with the extensive mutagenesis studies reported for PLPD-1 of MHV and HCV 229E (1, 6, 11, 13), showing that the upstream sequences of a cleavage site play a more important role than the downstream sequences. Furthermore, the inhibitory effect observed is unlikely caused by the replacement of Gly with Ala, as proteolysis was permissive at Ala2265-Gly2266 when Gly2265 was mutated to an Ala. On the other hand, if Ala2264 is at P1, deletion of Gly2265 does not cause any change in P1 to P5 of the recognition site. As the next amino acid residue is Gly2266, deletion of Gly2265 shifts Gly2266 to P1′, effectively creating the same Ala-Gly dipeptide bond. No obvious inhibitory effect would be expected, because the amino acids at the P1′ and P2′ sites were less crucial to the cleavage efficiency of the substrate. In fact, mutation of the Gly2266 residue to Asn has little, if any, effect on the proteolytic event. It is therefore established that the Gly2265-Gly2266 dipeptide bond is most likely the second cleavage site of IBV PLPD-1.
The two cleavage activities of IBV PLPD-1 identified so far are on the Gly-Gly dipeptide bond. It is remarkable that the proteinase shows markedly different properties at the two positions. For the second cleavage event, IBV PLPD-1 behaves more like the PLPD-1 of other coronaviruses studied. For example, cleavage of the precursor at this position could occur both in intact cells and in an in vitro expression system, as seen for MHV and HCV 229E (1, 6, 11, 13). Interestingly, cleavage was not significantly affected when Gly2265 (P1) was mutated to an Ala, which creates an Ala-Gly scissile bond similar to the second cleavage site of MHV PLPD-1 (Ala832-Gly833) (1). In fact, mutation of the Ala832-Gly833 to Gly-Gly dipeptide did not affect the cleavage activity of MHV PLPD-1 (1). A mutation of Gly2266 at the P1′ site to Asn creates a dipeptide bond similar to the Gly111-Asn112 cleavage site utilized by HCV 229E to release the p9. This mutation was also permissive to proteolytic processing. It seems that as long as the dipeptide bond fit into consensus cleavage sites utilized by other coronavirus PLPDs, cleavage activity was not affected. IBV PLPD-1 could also act on the second cleavage site in a trans-cleavage manner, which would allow us to characterize the proteinase activity in more detail. Recently, Teng et al. (34) used the trans-cleavage activity of MHV PLPD-1 to define the temperature and minimum lengths of both substrate and enzyme required for optimal cleavage efficiency. By contrast, cleavage at the first Gly-Gly dipeptide bond (Gly673-Gly674) of IBV occurred only when both the substrate and enzyme were expressed in intact cells and no trans-cleavage activity was detected at the position (17, 21). It would be interesting to investigate whether these differences are dictated simply by the sequence context of the individual cleavage sites or, more sophisticatedly, by the overall folding and/or cellular compartmentalization of the substrates and the enzyme.
Furthermore, the Gly673-Gly674 and Gly2265-Gly2266 cleavage sites identified for IBV PLPD-1 are not exactly compatible with the general alignment model [P5-(R, K)XXX(G, A)↓(G, A, V)-P1′] proposed for coronavirus PLPD (13). For the cleavage sites utilized by PLPD-1 of MHV and HCV 229E (1, 6, 11, 13), P5 is occupied by a conserved basic amino acid, either an Arg or a Lys. But for IBV PLPD-1, instead of a basic amino acid residue, a Val for the first cleavage site and a Glu for the second cleavage site lie at this position. Interestingly, P3 of both IBV cleavage sites is occupied by a Lys. The significance of this residue to the two cleavage activities is under investigation.
Cleavage at the Gly2265-Gly2266 dipeptide bond would result in the release of the N-terminal 195- and the C-terminal 41-kDa proteins. The 195-kDa protein is therefore the cleavage product immediately downstream of the 87-kDa protein. It contains PLPD-1 and the putative PLPD-2 (17, 21) and has a calculated molecular mass of 180 kDa (1,592 amino acid residues). However, the protein migrates more slowly on the SDS-PAGE system used, probably due to the presence of a stretch of negatively charged residues at its N terminus, as mentioned in our previous report (17). As the two Gly-Gly (Gly673-Gly674 and Gly2265-Gly2266) cleavage sites flank the 195-kDa protein, cleavage at these positions effectively results in autoprocessing of the 195-kDa protein. Autoprocessing is so far the only known function of the IBV 195-kDa protein. Characterization of the MHV and HCV PLPD-1 activities showed that the minimal sequences required for functionally active proteinase are 232 (between amino acids 1084 and 1316) and 422 (between amino acid residues 863 and 1285) amino acid residues, respectively (1, 11). More recently, Herold et al. (12) have demonstrated that the HCV PLPD-1 activity is dependent on a unique Zn2+-binding finger that connects the two domains of a papain-like fold (12). It is suggested that although the relatively small portion of the proteinase domain is contained in a large protein molecule, the proteinase domain may maintain a unique folding. We do not know whether the bulk of the molecule may play additional, independent functions.
The C-terminal cleavage product migrates at 41 kDa on SDS-PAGE, although it has a calculated molecular mass of 58 kDa (514 amino acids) and has undergone N-linked glycosylation. This is probably due to its high content of hydrophobic residues. No cleavage site at the position equivalent to the IBV Gly2265-Gly2266 dipeptide bond has been identified so far for other coronaviruses. The available evidence indicated that a potential cleavage site might exist at a similar position for MHV-JHM. Schiller et al. (29) have recently reported the identification of two cleavage products of 250 and 210 kDa, which may be encoded by regions corresponding to the IBV sequences coding for the 240- and 195-kDa proteins. Pulse-chase experiments further showed that the 250-kDa precursor may undergo proteolytic processing to release the 210-kDa protein and a protein of 40 kDa (29) strongly suggesting the existence of a cleavage site equivalent to the second cleavage site of IBV identified in this report. The 41-kDa glycoprotein (39 kDa when unglycosylated) would be the equivalent of the 40-kDa protein suggested by Schiller et al. (29).
The 41-kDa protein is the first coronavirus nonstructural protein demonstrated so far to be posttranslationally modified by N-linked glycosylation. Examination of the amino acid sequences showed that the equivalent regions of other coronaviruses also encode potential N-linked glycosylation sites (e.g., MHV contains two, transmissible gastroenteritis virus contains three, and HCV contains one) (7, 10, 16). It is suggested that this modification may be conserved among different groups of coronaviruses. When tagged to green fluorescent protein, the 41-kDa exhibits a staining pattern resembling that of the M protein, a protein shown to be localized to the Golgi apparatus, suggesting that the 41-kDa protein may be also localized to this subcellular compartment (data not shown). N-linked glycosylation of nonstructural proteins was also described for other viral systems. For example, the NS1 protein of flaviviruses was reported to be an N-linked glycoprotein (25).
It is intriguing that the 41-kDa protein is physically associated with the 195-kDa product via its N terminus. In EAV, it was shown that the second and third cleavage products of ORF 1a (corresponding to the 195- and 41-kDa proteins of IBV, respectively) tend to be coimmunoprecipitated (31). As the N-terminal half of the 195-kDa mature product was previously shown to be able to interact with the 87-kDa protein (17), it seems likely that the three products may form a complex. The prime role of this complex formation would be to sequester, by the action of the 41-kDa protein, the proteinase domain-containing 195-kDa protein and the 87-kDa protein to the membranous compartment where the IBV RNA replicates and the virion particles assemble (14). In two recent report, Shi et al. (30) and van der Meer et al. (37) have demonstrated the colocalization and membrane association of several gene 1 products with de novo-synthesized MHV-59 viral RNA in virus-infected cells. When the IBV counterparts of these products were separately expressed, distinct distribution patterns were observed for each protein (D. X. Liu, unpublished observations). It is suggested that the colocalization patterns reported by Shi et al. (30) may reflect the interactions between viral proteins. Such interactions between ORF 1a-encoded products have been demonstrated in arteriviruses (28, 36). They may facilitate the association of the replication complex with cellular membranous compartments (28, 36). Characterization of the multiprotein interaction observed in this study would provide significant insights into the subcellular translocation of coronavirus proteins and the involvement of nonstructural proteins in the viral replication cycle.
ACKNOWLEDGMENTS
We thank H. Y. Xu for technical assistance and S. Shen for help with photography.
This work was supported by a grant from the National Science and Technology Board of Singapore.
REFERENCES
- 1.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boursnell M E G, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 3.Brierley I, Boursnell M E G, Binns M M, Bilimoria B, Blok V C, Brown T D K, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Digard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for a RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutant of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 6.Dong S H, Baker S C. Determination of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herold J, Raabe T, Schelle-Prinz B, Siddell S G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold J, Gorbalenya A E, Thiel V, Schelle B, Siddell S G. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J Virol. 1998;72:910–918. doi: 10.1128/jvi.72.2.910-918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold J, Siddell S G, Gorbalenya A E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J Biol Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes S A, Bonilla P J, Weiss S R. Identification of the murine coronavirus p28 cleavage site. J Virol. 1995;69:809–813. doi: 10.1128/jvi.69.2.809-813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klumperman J, Locker J K, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, Monica N L, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim K P, Liu D X. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology. 1998;245:303–312. doi: 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D X, Cavanagh D, Green P, Inglis S C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D X, Brierly I, Tibbles K W, Brown T D K. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J Virol. 1994;68:5772–5780. doi: 10.1128/jvi.68.9.5772-5780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D X, Brown T D K. Characterization and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D X, Tibbles K W, Cavanagh D, Brown T D K, Brierly I. Identification, expression and processing of an 87 kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology. 1995;208:48–57. doi: 10.1006/viro.1995.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D X, Xu H Y, Brown T D K. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J Virol. 1997;71:1814–1820. doi: 10.1128/jvi.71.3.1814-1820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D X, Shen S, Xu H Y, Wang S F. Proteolytic mapping of the coronavirus infectious bronchitis virus 1b polyprotein: evidence for the presence of four cleavage sites of the 3C-like proteinase and identification of two novel cleavage products. Virology. 1998;246:288–297. doi: 10.1006/viro.1998.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machamer C E, Mentone S A, Rose J K, Farquhar M G. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc Natl Acad Sci USA. 1990;87:6944–6948. doi: 10.1073/pnas.87.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muylaert I R, Chambers T J, Galler R, Rice C M. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology. 1996;222:159–168. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- 26.Ng L F P, Liu D X. Identification of a 24 kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology. 1998;243:388–395. doi: 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng L F P, Liu D X. Further characterization of the coronavirus IBV ORF 1a products encoded by the 3C-like proteinase domain and the flanking regions. Adv Exp Med Biol. 1998;440:161–71. doi: 10.1007/978-1-4615-5331-1_21. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiller J J, Kanjanahaluethai A, Baker S C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF 1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi S T, Schiller J J, Kanjanahaluethai A, Baker S C, Oh J W, Lai M M C. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snijder E J, Wassenaar A L M, Spaan W J M. Proteolytic processing of the replicase ORF1a protein of the equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 33.Stern D F, Sefton B M. Coronavirus multiplication: localizations of genes for virion proteins on the avian infectious bronchitis virus genome. J Virol. 1984;50:22–29. doi: 10.1128/jvi.50.1.22-29.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng H, Pinon J D, Weiss S R. Expression of murine coronavirus recombination papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides. J Virol. 1999;73:2658–2666. doi: 10.1128/jvi.73.4.2658-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibbles K W, Brierley I, Cavanagh D, Brown T D K. Characterization in vitro of an autocatalytic processing activity associated with the predicted 3C-like proteinase domain of the coronavirus avian infectious bronchitis virus. J Virol. 1996;70:1923–1930. doi: 10.1128/jvi.70.3.1923-1930.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Meer Y, van Tol H G, Locker J K, Snijder E J. ORF1a-encoded replicase subunits are involved in membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J M, Locker J K. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]