ABSTRACT
Cell division presents a challenge for eukaryotic cells: how can chromosomes effectively segregate within the confines of a membranous nuclear compartment? Different organisms have evolved diverse solutions by modulating the degree of nuclear compartmentalization, ranging from complete nuclear envelope breakdown to complete maintenance of nuclear compartmentalization via nuclear envelope expansion. Many intermediate forms exist between these extremes, suggesting that nuclear dynamics during cell division are surprisingly plastic. In this review, we highlight the evolutionary diversity of nuclear divisions, focusing on two defining characteristics: (1) chromosome compartmentalization and (2) nucleocytoplasmic transport. Further, we highlight recent evidence that nuclear behavior during division can vary within different cellular contexts in the same organism. The variation observed within and between organisms underscores the dynamic evolution of nuclear divisions tailored to specific contexts and cellular requirements. In-depth investigation of diverse nuclear divisions will enhance our understanding of the nucleus, both in physiological and pathological states.
KEYWORDS: Cell division, meiosis, mitosis, nuclear compartmentalization, nuclear envelope, nuclear pore complex, nucleocytoplasmic transport
Introduction
Nuclear compartmentalization, the separation of nuclear material from the rest of the cell, is vital for eukaryotic cellular homeostasis. Nuclear components are physically separated from the cell cytoplasm by a double membrane termed the nuclear envelope (NE). Nuclear access is gated by nuclear pore complexes (NPCs), conserved supramolecular structures composed of over thirty different subunits named nucleoporins that are embedded in the NE (reviewed in [1]). NPCs prevent the passive diffusion of large proteins between the nucleus and cytoplasm, with structural nucleoporins scaffolding channel nucleoporins to form a selective permeability barrier [2–4]. Specialized proteins called karyopherins bind cargo to mediate directional transport through NPCs. Karyopherin movement and substrate binding is determined by the Ran gradient, the asymmetric distribution of GTP- and GDP-bound Ran, which controls cargo directionality (reviewed in [5]). Dysregulation of nuclear integrity or transport is associated with aging and neurodegenerative diseases, such as amyotrophic lateral sclerosis, Huntington’s disease, and frontotemporal dementia (covered in more detail in [6,7]). An improved understanding of nuclear structure is necessary to understand how it is altered in disease states and, ultimately, to develop new therapeutic approaches.
During cell division, the NE is drastically remodeled to accommodate chromosomal segregation, offering a context to better understand the regulation of nuclear structure and integrity. Diverse organisms have evolved unique forms of nuclear remodeling (reviewed in [8]). Many organisms, including some fungi and protozoan parasites, undergo a ‘closed’ mitosis, whereby the NE remains intact and nuclear transport occurs throughout the cell division. Microtubule organizing centers (MTOCs), which arrange microtubules (MTs) and form the mitotic spindle, are embedded into the NE to enable closed division. Closed division offers the cell extensive chromatin protection from the contents of the cytoplasm but comes at the energetic cost of rapidly expanding the intact NE to accommodate a growing spindle [9]. Instead of closed divisions, metazoans and many plants undergo an ‘open’ mitosis, whereby the NE is disassembled, and nuclear transport is abolished until chromosome division is complete. In NE breakdown (NEBD), the NE is destabilized and fenestrated by the phosphorylation and disassembly of both NPCs and nuclear lamina. This destabilization allows for dynein-dependent tearing of the NE to facilitate NE remodeling (reviewed in [10]). Open divisions allow extranuclear MTOCs access to chromosomes but pose the challenge of keeping chromatin distinct from other cytoplasmic components.
Notably, despite its fundamental importance to the eukaryotic cell, the nucleus exhibits remarkable evolutionary plasticity during cell divisions. There have been many transitions between ‘open’ and ‘closed’ forms of divisions [11], even on relatively short evolutionary timescales between closely related species. Further, the continued study of mitosis in different organisms has made it clear that nuclear dynamics exist on a spectrum rather than on a binary [8, 12–16]. The integrity of the nuclear envelope and the disruption of nuclear transport are often differentially regulated in these intermediates, suggesting that they are separable traits that may be responding to distinct selective pressures. Even within the same organism, it has recently become appreciated that different developmental contexts employ different modes of nuclear division, for example during the somatic and gametic divisions in Saccharomyces cerevisiae [17] and in various cell types in Drosophila melanogaster [18–20]. Further investigation into the mechanisms and functional consequences of these transitions in nuclear division promises to provide insight into how different evolutionary pressures shape nuclear properties and function.
This review provides an overview of the evolutionary diversity of eukaryotic nuclear division. We use (1) the means of chromosome compartmentalization and (2) the maintenance of nucleocytoplasmic transport as linked but distinct properties to categorize and understand nuclear divisions. We further describe what is currently known about the mechanisms driving this evolutionary variation. Finally, we highlight recently characterized examples of diverse nuclear divisions occurring within the same organism. Understanding the pressures driving different nuclear divisions promises to provide unique insights into nuclear dynamics and function and has the potential to shed light on its dysfunction in disease contexts.
Protecting chromosomes: keeping the chromatin distinct amidst division
The nucleus encloses chromatin in a physical compartment to allow for both the separation of transcription and translation and the physical protection of the genetic material. If chromatin is aberrantly removed from the nucleus, the results can be catastrophic [21]. This begs the question: how do cells keep chromatin safe during division?
Different organisms have evolved divergent mechanisms to compartmentalize chromatin, shielding it from the rest of the cellular environment. Closed divisions, which have been studied largely in the model fungi Saccharomyces cerevisiae (budding yeast) and Schizosaccharomyces pombe (fission yeast), allow for chromatin compartmentalization by keeping the NE intact throughout division (Figure 1, ‘Closed’). This physically separates chromatin and other nuclear material from interacting with large cytoplasmic elements such as organelles. The NE can also serve to separate different mitotic spindles in multinucleated states, such as during the multinucleated hyphal growth of the fungus Ashbya gossypii [22]. In cell divisions where the nuclear envelope persists, chromosome contacts with the nuclear periphery often need to be reorganized to facilitate proper division of chromosomes [23–26]. In S. pombe, the chromosome contacts may themselves play a role in chromatin division, as it has been demonstrated that chromatin division can still occur in the absence of spindle MTs [27]. The nuclear envelope can therefore serve as both a protective barrier and an active participant in closed nuclear divisions.
Figure 1.
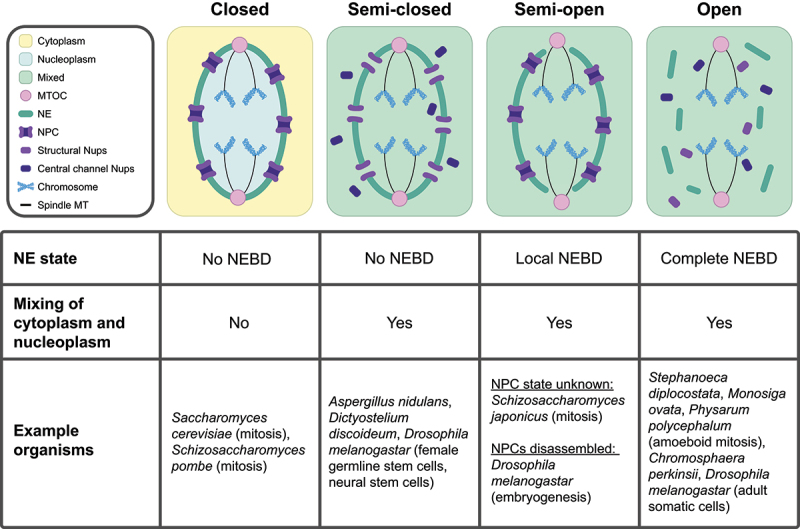
Nuclear divisions in diverse organisms utilize distinct nuclear envelope (NE) remodeling and nucleocytoplasmic mixing strategies. (Top) Visual representation of different types of nuclear division. From left to right: intact NE with no nucleocytoplasmic mixing (referred to as ‘Closed’), largely intact NE with nucleocytoplasmic mixing (referred to as ‘Semi-closed’), partially disrupted NE with nucleocytoplasmic mixing (referred to as ‘Semi-open’), and disrupted NE with nucleocytoplasmic mixing (referred to as ‘Open’). (Bottom) Table summarizing the role of the NE in compartmentalizing nuclear components. Abbreviations: NPC (Nuclear Pore Complex), Nups (Nucleoporins), NE (Nuclear Envelope), NEBD (Nuclear Envelope Breakdown), MT (Microtubule), and MTOC (Microtubule Organizing Center).
Maintaining an intact NE during cell division necessitates membrane expansion of the NE via phospholipid synthesis to accommodate the space required to segregate chromosomes [28,29]. The closely related fission yeasts Schizosaccharomyces pombe and Schizosaccharomyces japonicus demonstrate the importance of lipid synthesis in the control of nuclear division. S. pombe undergoes a closed mitosis and must shift its lipid production toward NE expansion during mitosis. It accomplishes this via cell-cycle dependent phosphorylation of lipin, the enzyme which converts precursor phosphatidic acid (PA) into storage-type lipids. Lipin phosphorylation causes its inactivation, allowing for PA to be processed into structural lipids used in NE membranes [30]. S. japonicus does not undergo a closed mitosis, instead partially disassembling the NE at anaphase (Figure 1, ‘Semi-open’) [28]. Notably, S. japonicus does not phosphorylate lipin during mitosis, resulting in the lack of NE membrane expansion [30]. Interestingly, while forced phosphorylation or deletion of lipin in S. japonicus does lead to NE membrane expansion, local disassembly of the NE still occurs with wild-type timing [30]. This demonstrates that, while control over lipid synthesis is necessary to facilitate closed divisions, it may not be the sole deciding factor as to whether NEBD occurs.
Similar to S. japonicus, other organisms undergo local NEBD but retain most of the NE (Figure 1, ‘Semi-open’). While the timing, location, and extent of partial NEBD can vary between organisms, they may share the functional similarity of retaining the NE as a tool of organization and protection during division. In some cases of partial NEBD, the maintenance of most of the NE may help to organize and protect chromosomes through divisions. This is thought to be particularly important for the separation of multiple dividing nuclei within the same cell [31]. For example, the single-celled parasite Giardia intestinalis is binucleate and, throughout division, the NE only locally disassembles at the poles of the nucleus to allow spindle MTs from extranuclear MTOCs to access chromatin (see ‘Semi-open’ in Figure 1) [32].
Interestingly, in other cases of partial NEBD, the remaining NE does not appear to protect the chromatin. The pathogenic corn fungus Ustilago maydis undergoes a unique form of open division with local NEBD [33]. Local NEBD occurs at the bud neck, the junction between the mother and newly forming daughter cell, and all chromosomes are pulled out of the NE and into the bud. The old NE collapses and, like mammalian mitosis, new NE forms after the chromosomes segregate. The mechanisms of protection during mitosis in U. maydis are currently not well characterized, but examples of chromatin protection during complete NEBD may provide clues.
Even in completely open divisions (Figure 1, ‘Open’), chromatin is never entirely exposed to the cytoplasm. In human cells, chromatin is coated with the protein Ki-67 for the duration of NEBD [34]. During chromosome segregation, Ki-67 allows individual chromosomes to segregate as discrete units instead of collapsing together [34]. At the end of anaphase, Ki-67 plays a role in both compacting chromosomes and acting as a surfactant, which is vital for the exclusion of cytoplasmic materials from the newly formed nuclear compartments [35]. The protein BAF then packages chromosomes together, providing a scaffold for the new NE [36,37]. Notably, BAF also condenses chromatin, which may play a role in forming a new nuclear compartment that excludes components that are not already associated with chromatin [37]. The physical separation of dividing chromatin from the endoplasmic reticulum (ER) is also essential to prevent chromosome mis-segregation [21], demonstrating the importance of cellular organization in mitosis. The endosomal sorting complex required for transport-III (ESCRT-III) is required for membrane sealing at the end of new NE formation [38,39], completing the compartmentalization of the newly formed nuclei. Metazoan cells provide insight into how an open division can still exclude cytoplasmic components from chromatin and newly formed nuclei during division.
Within the context of open nuclear divisions, striking diversity exists in the timing of NEBD. Complete NEBD can occur anywhere from prophase to telophase, depending on the organism [15]. For example, the choanoflagellate Stephanoeca diplocostata fully disassembles its NE during prophase, whereas the choanoflagellate species Monosiga ovata fully disassembles its NE during anaphase [40]. Interestingly, M. ovata first locally disassembles its NE at the poles of the nucleus to allow spindle MT access to chromatin before it fully disassembles the NE [40]. This is strikingly similar to the pattern of partial disassembly at the nuclear poles, followed by complete NEBD at anaphase observed in Caenorhabditis elegans [41]. It is possible that delaying full NEBD until anaphase may provide a longer period of partial nuclear compartmentalization or organization and that eventual full NEBD may be necessitated by the constraints of lipid synthesis [30]. It is also possible that the timing of nuclear compartmentalization abolishment may play different roles in different contexts. Better understanding how and why the nuclear periphery exhibits such diversity in timing, location, and extent of disruption during nuclear divisions promises to provide new insight into the principles of nuclear organization.
Altering nucleocytoplasmic transport: changes to nuclear identity during cell division
While the state of the NE influences the integrity of the nucleocytoplasmic compartment during cell divisions, nucleocytoplasmic transport can be subject to additional regulation, especially within the context of ‘closed’ divisions. Canonical ‘closed’ divisions retain both an intact NE and the separation of cytoplasm and nucleoplasm (Figure 1, ‘Closed’). In addition to changes in lipid synthesis allowing for NE expansion, additional mechanisms are often in place to maintain compartmentalization of the nucleus from the cytoplasm. This has been best studied in the fission yeast S. pombe. Unlike S. cerevisiae, the spindle pole body (the MTOC) in S. pombe is not constitutively inserted into the NE, and the insertion of the spindle pole body into the NE necessitates local breakdown [42,43]. It has been shown that spindle pole body insertion and the location of insertion rely on chromatin association with the linker of nucleoskeleton and cytoskeleton (LINC) complex [44]. Importantly, the NEBD required for spindle pole body insertion is grommeted by the ESCRT machinery, allowing for the NEBD to be restricted and maintaining separation of nucleoplasm and cytoplasm [45]. Additionally, karyokinesis in S. pombe is facilitated at the end of mitosis by NPC disassembly at the midzone of the nuclear division, facilitating local NEBD in that region of the nucleus [46]. Similar to the grommet at spindle pole body insertion, the rest of the nuclear compartment is insulated from midzone breakdown by inner nuclear membrane protein Les1 [46]. These insulating factors, which enable continued separation of the nucleoplasm and cytoplasm, distinguish local breakdowns used in a closed division context from those in other types of partial NEBD.
Some divisions, assessed as ‘closed’ by the maintenance of an intact NE, still intermix their nucleoplasm and cytoplasm (Figure 1, ‘Semi-closed’). In some cases, the NPC is partially disassembled, leading to the mixing of the two compartments. The fungus Aspergillus nidulans retains a continuous NE throughout mitosis, but phosphorylation of several channel nucleoporins drives their dispersal, even as structural nucleoporins remain at the nuclear periphery [47]. The kinase NIMA facilitates both this partial NPC disassembly in A. nidulans [47] and complete NPC disassembly in mammalian open divisions (reviewed in [13], implying either convergent evolution or the existence of an ancestral nuclear remodeling event. The Ran gradient is therefore abolished, despite the presence of an intact NE. Interestingly, nucleoporins disassembled from the NPC can play alternate roles in mitosis. In the case of A. nidulans, nucleoporin Nup2 relocalizes from the NPC to chromatin, an association which is vital for the proper inheritance of the remaining NPCs into daughter cells [48]. It is currently unclear whether the function of nucleoporin disassembly from NPCs is to allow for alternate nucleoporin functions, to allow for nucleocytoplasmic intermixing, or both.
While A. nidulans is a particularly well-characterized example, there are other reported instances of partial NPC disassembly during closed mitotic divisions. In the slime mold Dictyostelium discoideum, partial NPC disassembly is observed, which corresponds to the timing of the dispersal of an NLS-containing reporter [49]. In U. maydis, channel and Y-complex nucleoporins disperse from the NPC during chromosome segregation into the bud, while the remaining structural nucleoporins remain associated with the soon-to-be discarded NE [50]. In this example, partial disassembly of NPCs occurs, despite the NE no longer functioning to compartmentalize chromosomes. It is possible that partial NPC disassembly may allow for nucleoporin turnover or may be necessary for nucleoporins that play critical mitotic roles when not associated with the NPC (such as in A. nidulans). Selective disassembly of nucleoporins without NEBD may also serve as an intermediate form of compartmentalization. In the starfish Asterina miniate oocytes, there are two stages of nuclear permeabilization [51]. In the first stage, nucleoporin disassembly leads to free diffusion of molecules ~40 nm in diameter, whereas in the second stage, local NEBD increases the permeability of the nucleus to allow for free diffusion of molecules ~100 nm in diameter [51].
Despite extensive work highlighting diverse mechanisms by which nuclear permeability is disrupted, the functional significance of losing nuclear compartmentalization during cell divisions remains surprisingly poorly understood. It has been suggested that, in some instances, compartment mixing may be necessary to allow for a rapid influx of tubulin to enable rapid spindle assembly in mitosis [13] Mixing may also be a form of rapid regulation facilitated by the interaction of cytoplasmic and nuclear components that are usually kept separated. Others argue that intercompartmental mixing itself does not serve a function per se but is instead a consequence of other necessary nuclear remodeling events in mitosis [8]. For example, partial NEBD, where the integrity of the nuclear compartment is lost, may be a tradeoff to allow for spindle MT access to the nucleus from an extranuclear MTOC (such as in G. intestinalis) or to avoid costly membrane lipid production (such as in S. japonicus).
Different nuclear division strategies within the same organism
Recent studies have revealed that, even within the same organism, different forms of nuclear division can co-exist depending on the cellular context. Combined with the rapid evolvability of this trait [11], the co-existence of different modes of nuclear division within the same organism provides strong evidence that nuclear division strategies are adapted to specific cellular environments and purposes.
To date, there have been four different reported mechanisms of nuclear division in the fruit fly Drosophila melanogaster (Figure 2A). During embryonic development, the embryos are syncytial (multinucleated) and undergo a form of partial NEBD during nuclear division. The NPCs fully disassemble and the NE is locally disassembled at the nuclear poles to allow for spindles to access chromatin from extranuclear MTOCs [20]. These both leave the NE fenestrated but still surrounding the dividing chromosomes and therefore compartmentalizing different spindles [20]. Adult flies, however, exhibit different types of nuclear division based on cell type [18,19]. Most adult somatic cells in the fly undergo an open nuclear division, disassembling the nuclear lamina and the NE. Female germline stem cells undergo division somewhat reminiscent of A. nidulans, in which central channel and Y-complex nucleoporins, but not other structural nucleoporins, are disassembled [18]. Nucleocytoplasmic mixing is implied by the abolishment of fully intact NPCs but has not been directly tested. Unlike the fly embryos, the MTOCs (centrosomes) are inserted into the NE during division in these cells, such that the division likely occurs without NE fenestrations [18]. In contrast to partial disassembly in the germline stem cells, Drosophila neural stem cells fenestrate their NE through the full disassembly of NPCs but keep the nuclear lamina assembled [19]. The functional needs that drive the differences in nuclear dynamics observed between adult somatic cells and stem cells remain unknown. Given that the non-canonical cell divisions in Drosophila stem cells were only recently described, divergent nuclear behavior in different cell types may be more common in multicellular organisms than previously appreciated and merits further investigation.
Figure 2.
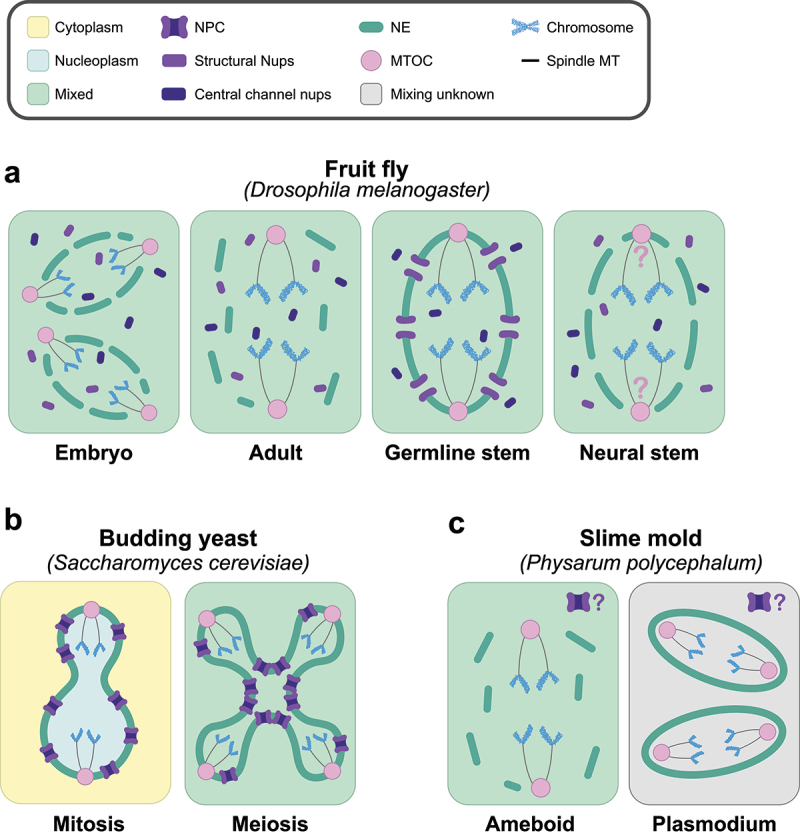
Distinct nuclear division strategies are used in different developmental contexts within the same organism. a. Cartoon representing the four currently identified types of nuclear division found in the fruit fly Drosophila melanogaster. From left to right: partial nuclear envelope breakdown (NEBD) in the embryo [20], open division in adult somatic cells, semi-closed division with partial NPC disassembly in female germline stem cells [18], and semi-closed division with complete NPC disassembly in neural stem cells [19]. The position of the MTOC (centrosome) in fly neural stem cells is currently unknown, represented with question marks. b. Cartoon representations of the budding yeast Saccharomyces cerevisiae nucleus undergoing closed mitosis (left) [52]; and semi-closed meiosis (right) [17,53]. c. Cartoon representations of the slime mold Physarum polycephalum nucleus undergoing an open division in the cell’s ameboid stage of development (left) and a closed division during the syncytial plasmodium form (right) [54]. It is currently unknown if the nuclear pore complexes (NPCs) disassemble in either mode of division, represented with question marks. Nuclear lamina is omitted for simplicity. MTOC (Microtubule Organizing Center).
Single-celled organisms also exhibit different types of nuclear division. For example, it has recently been shown that the yeasts S. cerevisiae and S. pombe undergo different modes of nuclear division in mitosis and meiosis (Figure 2B). Both undergo mitosis and meiosis I (MI) with no nucleocytoplasmic mixing, but exhibit compartment mixing during meiosis II (MII), a phenomenon called virtual nuclear envelope breakdown (vNEBD [53,55,56]. The nuclear envelope appears intact by electron microscopy (EM) [56]; [17]), but – at least in S. pombe – the Ran gradient appears to be abolished [55,56]. Intriguingly, there appears to be no evidence of central channel nucleoporin dispersal from the nuclear periphery in S. pombe [56], pointing to a novel mechanism of compartment abolishment. In S. cerevisiae, the channel nucleoporins become sequestered into a discrete nuclear compartment along with the core nucleoporins [17] [57]. Whether this NPC remodeling event contributes to vNEBD is currently unknown. Notably, these two yeasts are diverged by more than 400 million years [58], suggesting vNEBD may have an important and evolutionarily conserved function. As with other open and semi-open divisions, it has been suggested that compartment mixing is necessary for the timely disassembly of the MII spindle during S. pombe meiosis [59]. We speculate that vNEBD may have another unique role in meiosis: facilitating the largescale turnover of soluble nuclear proteins and contributing to cellular rejuvenation during the gametogenesis program [60].
The nuclear periphery undergoes additional meiosis-specific remodeling in budding yeast, exhibiting two distinct nuclear pore complex remodeling events. During meiosis I, the nuclear basket partially detaches from the NPC in a phosphorylation-driven event [57]. Although the functional significance of the detachment is currently unknown, basket detachment may allow for chromosomes to be decoupled from the nuclear periphery after extensive contacts are made during meiotic recombination, consistent with the genetic interactions observed between nuclear basket proteins and chromatin tethers in meiosis [61,62]. During meiosis II, NPCs are sequestered to a fifth nuclear compartment called the GUNC (gametogenesis uninherited nuclear compartment) and are ultimately destroyed [17]). Age-associated nuclear damage is also sequestered to this compartment, providing a means to turn over structures in the nuclear periphery even as the nuclear envelope remains intact. Intriguingly, only the nuclear basket detaches and returns to gametes during this remodeling event, implying that it may play an important role in re-organizing the nuclear periphery during this cell division.
Diverse eukaryotes provide additional novel forms of meiotic nuclear remodeling. In the ciliate Tetrahymena thermophila, the somatic macronucleus (MAC) and germline micronucleus (MIC) co-exist in the same cytoplasm. These nuclei are differentiated at least in part by nucleus-specific nucleoporins that facilitate distinct transport profiles [63,64]. The deposition of these nucleus-specific nucleoporins – and the conferral of their transport properties – occurs during meiosis, suggesting that nuclear remodeling during cell divisions can confer disparate nuclear identities [64]. Collectively, the meiosis-specific nuclear remodeling observed in diverse eukaryotes – flies, ciliates, and yeast – suggest that meiosis imposes unique requirements on the nucleus compared to mitosis.
Cell division switching is also observed in the context of other life-cycle transitions. In the slime mold Physarum polycephalum, different nuclear division strategies are observed in different developmental stages [54] Figure 2C). In its uninucleate amoeboid form, P. polycephalum undergoes complete NEBD; in its syncytial (multinucleated) plasmodium form, P. polycephalum switches to a closed mitosis strategy [54]. As additional organisms are studied in greater depth, more examples of different nuclear division strategies employed by the same organism are likely to be uncovered.
Conclusions and future directions
There is considerable diversity in the ways that different organisms approach nuclear division. Chromosome compartmentalization can be achieved by complete or partial NE maintenance in closed and intermediate divisions or by chromatin-binding proteins (e.g., Ki-67 or BAF in human cells) in open cell divisions. Nuclear compartmentalization can be maintained by the NE and other insulating factors in closed divisions; alternatively, it can be disrupted by diverse mechanisms, ranging from partial NPC disassembly to NE disruption, in semi-closed, semi-open, and open divisions. Even within the same organism, different types of nuclear division have been observed at different life cycle stages and in different cell types, implying that different division dynamics may be driven by unique demands. Future research should seek to characterize additional tractable relatives in clades that exhibit diverse nuclear divisions (e.g., fungi) to provide insight into how and why by which these transitions occur.
Despite striking differences between distinct nuclear division strategies, they often use the same molecular tools in different ways. This may enable the same organism to switch nuclear division types across both cell types and evolutionary space. For example, the closed mitotic division of S. pombe relies on local NEBD mediated by the disassembly of NPCs, a small-scale version of how NEBD occurs during open divisions [46]. The partial NPC disassembly events observed in A. nidulans mitosis and S. cerevisiae meiosis are phosphorylation-driven events mediated by the same kinases (NIMA and Polo kinase, respectively) involved in metazoan NPC assembly [47,57]. The ESCRT-III complex similarly plays a role in sealing the NE at the end of mitosis in both metazoan open divisions [38,39] and in the closed division of S. pombe [45]. Future comparative cell biology will provide further insight into the molecular mechanisms that underpin the evolution of nuclear organization during division.
Current understanding of nuclear division in many non-model organisms relies primarily or solely on EM data [15,65]. EM images can provide information about the state of the NE and location of a MTOC but fail to fully capture the nuances of how nuclear compartmentalization changes throughout nuclear division. Organisms annotated as conducting closed nuclear divisions based on EM data alone may or may not undergo nucleocytoplasmic mixing during division. Partial disassembly or remodeling of NPCs is similarly not easily observed in EM images. Local NEBD can also be protected from nucleocytoplasmic leaking, such as spindle pole body insertion in S. pombe, necessitating studies that directly assay nucleocytoplasmic compartmentalization. For example, nuclear compartmentalization has not been directly assayed in G. intestinalis, which undergoes local NEBD to accommodate mitotic spindle access. Historically, many organisms have been difficult to study in-depth at the cellular level due to the difficulty of genetic modification. With the widespread implementation of CRISPR-Cas9 editing systems, it is more feasible than ever before to study a diverse array of non-model eukaryotic organisms [66]. Future studies – and reexamination of older studies – promises to uncover exciting new variation in nuclear division. For example, the fungus U. maydis does not exhibit the same NE shedding mechanism in meiosis as is observed in mitosis [67].
Even in well-studied model organisms such as budding yeast (S. cerevisiae), fission yeast (S. pombe), and fruit flies (D. melanogaster), new forms of nuclear division have only recently been identified [18,55–57]. Investigation of multiple life stages of the same organism has the potential to reveal more instances of nuclear division-type switching. Meiosis represents a particularly exciting area of study, due to its ancient origin in the last eukaryotic common ancestor and its vital role in resetting lifespan [68]. Studying nuclear division in different cell types or environmental contexts for the same organism promises to illuminate the selective pressures that drive the diverse dynamics of nuclear division across evolutionary timescales.
Acknowledgments
We would like to thank Ben Styler, Cyrus Ruediger, Tina Sing, Jessica Leslie, Alena Bishop, Tianyao Xiao, Naohiro Kuwayama, and the rest of the Brar and Ünal labs for their thoughtful feedback.
Funding Statement
This work was supported by funds from NIH [R01AG071801] and Astera Institute to EÜ; a National Science Foundation Graduate Research Fellowship [DGE2146752, DGE1752814] and a National Institutes of Health Traineeship [T32 GM007232] to MEW and GAK is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation [DRG-2500-2].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
MEW reviewed the literature and wrote the first and subsequent drafts of the manuscript. GAK and EÜ reviewed the drafts, provided input and critique for the revisions and helped with the assembly of the final manuscript. EÜ provided supervision and acquired funding.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- [1].Lin DH, Hoelz A.. The structure of the nuclear pore complex (an update). Annu Revi Biochem. 2019;88(June):725–12. doi: 10.1146/annurev-biochem-062917-011901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Frey S, Görlich D. Asaturated FG-Repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130(3):512–523. doi: 10.1016/j.cell.2007.06.024 [DOI] [PubMed] [Google Scholar]
- [3].Popken P, Ghavami A, Onck PR, et al. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol Biol Cell. 2015;26(7):1386–1394. doi: 10.1091/mbc.E14-07-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Timney BL, Raveh B, Mironska R, et al. Simple rules for passive diffusion through the nuclear pore complex. J Cell Bio. 2016;215(1):57–76. doi: 10.1083/jcb.201601004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wing CE, Yee Joyce Fung H, Min Chook Y. Karyopherin-mediated nucleocytoplasmic transport. Nat Rev Mol Cell Biol. 2022;23(5):307–328. doi: 10.1038/s41580-021-00446-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu J, Hetzer MW. Nuclear pore complex maintenance and implications for age-related diseases. Trends Cell Biol. 2022;32(3):216–227. doi: 10.1016/j.tcb.2021.10.001 [DOI] [PubMed] [Google Scholar]
- [7].Martins F, Sousa J, Pereira CD, et al. Nuclear envelope dysfunction and its contribution to the aging process. Aging Cell. 2020;19(5):e13143. doi: 10.1111/acel.13143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dey G, Baum B. Nuclear envelope remodelling during mitosis. Curr Opinion Cell Biol. 2021;70(June):67–74. doi: 10.1016/j.ceb.2020.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Bio. 2007;179(4):593–600. doi: 10.1083/jcb.200708054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ungricht R, Kutay U. Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol. 2017;18(4):229–245. doi: 10.1038/nrm.2016.153 [DOI] [PubMed] [Google Scholar]
- [11].Sazer S, Lynch M, Needleman D. Deciphering the evolutionary history of open and closed mitosis. Curr Biol. 2014;24(22):R1099–1103. doi: 10.1016/j.cub.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boettcher B, Barral Y. The cell biology of open and closed mitosis. Nucleus. 2013;4(3):160–165. doi: 10.4161/nucl.24676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].De Souza Colin PC, Osmani SA. Mitosis, not just open or closed. Eukaryot Cell. 2007;6(9):1521–1527. doi: 10.1128/EC.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hauke D, McAinsh AD. Exotic Mitotic Mechanisms. Open Biol. 2012;2(12):120140. doi: 10.1098/rsob.120140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Heath IB. Variant mitoses in lower eukaryotes: Indicators of the evolution of mitosis. Int Rev Cytol. 1980;64:1–80. doi: 10.1016/s0074-7696(08)60235-1 [DOI] [PubMed] [Google Scholar]
- [16].Makarova M, Oliferenko S. Mixing and matching nuclear envelope remodeling and spindle assembly strategies in the evolution of mitosis. Curr Opinion Cell Biol. 2016;41(August):43–50. doi: 10.1016/j.ceb.2016.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].King GA, Goodman JS, Schick JG, et al. Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast.” edited by Noboru mizushima, Vivek Malhotra, Noboru mizushima, tokuko haraguchi, and c Patrick lusk. Elife. 2019a;8(August):e47156. doi: 10.7554/eLife.47156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Duan T, Cupp R, Geyer PK. Drosophila female germline stem cells undergo mitosis without nuclear breakdown. Curr Biol. 2021;31(7):1450–1462.e3. doi: 10.1016/j.cub.2021.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Roubinet C, White IJ, Baum B. Asymmetric nuclear division in neural stem cells generates sibling nuclei that differ in size, envelope composition, and chromatin organization. Curr Biol. 2021;31(18):3973–3983.e4. doi: 10.1016/j.cub.2021.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stafstrom JP, Staehelin LA. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the drosophila embryo. Eur J Cell Biol. 1984;34(1):179–189. [PubMed] [Google Scholar]
- [21].Ferrandiz N, Downie L, Starling GP, et al. Endomembranes promote chromosome missegregation by ensheathing misaligned chromosomes. J Cell Bio. 2022;221(6):e202203021. doi: 10.1083/jcb.202203021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lang C, Grava S, van den Hoorn T, et al. Mobility, microtubule nucleation and structure of microtubule-organizing centers in multinucleated hyphae of ashbya gossypii. Mol Biol Cell. 2010;21(1):18–28. doi: 10.1091/mbc.e09-01-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fujita I, Nishihara Y, Tanaka M, et al. Telomere-nuclear envelope dissociation promoted by rap1 phosphorylation ensures faithful chromosome segregation. Curr Biol. 2012;22(20):1932–1937. doi: 10.1016/j.cub.2012.08.019 [DOI] [PubMed] [Google Scholar]
- [24].Pieper GH, Sprenger S, Teis D, et al. ESCRT-III/Vps4 controls heterochromatin-nuclear envelope attachments. Dev Cell. 2020;53(1):27–41.e6. doi: 10.1016/j.devcel.2020.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ptak C, Saik NO, Premashankar A, et al. Phosphorylation-dependent mitotic SUMOylation drives nuclear envelope–chromatin Interactions. J Cell Bio. 2021;220(12):e202103036. doi: 10.1083/jcb.202103036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yam C, Gu Y, Oliferenko S. Partitioning and remodeling of the Schizosaccharomyces Japonicus mitotic nucleus require chromosome tethers. Curr Biol. 2013;23(22):2303–2310. doi: 10.1016/j.cub.2013.09.057 [DOI] [PubMed] [Google Scholar]
- [27].Castagnetti S, Oliferenko S, Nurse P. Fission yeast cells undergo nuclear division in the absence of spindle microtubules. PLOS Biol. 2010;8(10):e1000512. doi: 10.1371/journal.pbio.1000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yam C, He Y, Zhang D, et al. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr Biol. 2011;21(15):1314–1319. doi: 10.1016/j.cub.2011.06.052 [DOI] [PubMed] [Google Scholar]
- [29].Zhang D, Oliferenko S. Remodeling the nuclear membrane during closed mitosis. Curr Opinion Cell Biol. 2013;25(1):142–148. doi: 10.1016/j.ceb.2012.09.001 [DOI] [PubMed] [Google Scholar]
- [30].Makarova M, Gu Y, Chen J-S, et al. Temporal regulation of lipin activity diverged to account for differences in mitotic programs. Curr Biol. 2016;26(2):237–243. doi: 10.1016/j.cub.2015.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shah H, Olivetta M, Bhickta C, et al. Life cycle-coupled evolution of mitosis in close relatives of animals. 2023. bioRxiv. doi: 10.1101/2023.05.10.540163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sagolla MS, Dawson SC, Mancuso JJ, et al. Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite giardia intestinalis. J Cell Sci. 2006;119(23):4889–4900. doi: 10.1242/jcs.03276 [DOI] [PubMed] [Google Scholar]
- [33].Straube A, Weber I, Steinberg G. A novel mechanism of nuclear envelope break-down in a fungus: Nuclear migration strips off the envelope. Embo J. 2005;24(9):1674–1685. doi: 10.1038/sj.emboj.7600644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cuylen S, Blaukopf C, Politi AZ, et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 2016;535(7611):308–312. doi: 10.1038/nature18610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cuylen-Haering S, Petrovic M, Hernandez-Armendariz A, et al. Chromosome clustering by ki-67 excludes cytoplasm during nuclear assembly. Nature. 2020;587(7833):285–290. doi: 10.1038/s41586-020-2672-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Margalit A, Segura-Totten M, Gruenbaum Y, et al. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Nat Acad Sci. 2005;102(9):3290–3295. doi: 10.1073/pnas.0408364102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Samwer M, Schneider MWG, Hoefler R, et al. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell. 2017;170(5):956–972.e23. doi: 10.1016/j.cell.2017.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Olmos Y, Hodgson L, Mantell J, et al. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522(7555):236–239. doi: 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vietri M, Schink KO, Campsteijn C, et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522(7555):231–235. doi: 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- [40].Leadbeater BSC. The Choanoflagellates: evolution, Biology and Ecology. Cambridge: Cambridge University Press; 2015. doi: 10.1017/CBO9781139051125 [DOI] [Google Scholar]
- [41].Lee KK, Gruenbaum Y, Spann P, et al. C. Elegans nuclear envelope proteins emerin, man1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11(9):3089–3099. doi: 10.1091/mbc.11.9.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, schizosaccharomyces pombe. J Cell Bio. 1993;120(1):141–151. doi: 10.1083/jcb.120.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].West RR, Vaisberg EV, Ding R, et al. cut11 +: A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in schizosaccharomyces pombe. Mol Biol Cell. 1998;9(10):2839–2855. doi: 10.1091/mbc.9.10.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fernández-Álvarez A, Bez C, O’Toole ET, et al. Mitotic nuclear envelope breakdown and spindle nucleation are controlled by interphase contacts between centromeres and the nuclear envelope. Dev Cell. 2016;39(5):544–559. doi: 10.1016/j.devcel.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ader NR, Chen L, Surovtsev IV, et al. An ESCRT grommet cooperates with a diffusion barrier to maintain nuclear integrity. Nat Cell Biol. 2023;25(10):1465–1477. doi: 10.1038/s41556-023-01235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dey G, Culley S, Curran S, et al. Closed mitosis requires local disassembly of the nuclear envelope. Nature. 2020;585(7823):119–123. doi: 10.1038/s41586-020-2648-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].De Souza Colin PC, Osmani AH, Hashmi SB, et al. Partial nuclear pore complex disassembly during closed mitosis in aspergillus nidulans. Current Biology: CB. 2004;14(22):1973–1984. doi: 10.1016/j.cub.2004.10.050 [DOI] [PubMed] [Google Scholar]
- [48].Suresh S, Markossian S, Osmani AH, et al. Mitotic nuclear pore complex segregation involves nup2 in aspergillus nidulans. J Cell Bio. 2017;216(9):2813–2826. doi: 10.1083/jcb.201610019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mitic K, Meyer I, Gräf R, et al. Temporal changes in nuclear envelope permeability during semi-closed mitosis in dictyostelium amoebae. Cells. 2023;12(10):1380. doi: 10.3390/cells12101380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Theisen U, Straube A, Steinberg G, et al. Dynamic rearrangement of nucleoporins during fungal “open” mitosis. Mol Biol Cell. 2008;19(3):1230–1240. doi: 10.1091/mbc.E07-02-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lénárt P, Rabut G, Daigle N, et al. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Bio. 2003;160(7):1055–1068. doi: 10.1083/jcb.200211076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Winey M, Yarar D, Giddings TH, et al. Nuclear pore complex number and distribution throughout the saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8(11):2119–2132. doi: 10.1091/mbc.8.11.2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shelton SN, Smith SE, Unruh JR, et al. A distinct inner nuclear membrane proteome in saccharomyces cerevisiae gametes. G3: Genes | Genomes | Genetics. 2021;11(12):jkab345. doi: 10.1093/g3journal/jkab345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Solnica-Krezel L, Burland TG, Dove WF. Variable pathways for developmental changes of mitosis and cytokinesis in physarum polycephalum. J Cell Bio. 1991;113(3):591–604. doi: 10.1083/jcb.113.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Arai K, Sato M, Tanaka K, et al. Nuclear compartmentalization is abolished during fission yeast meiosis. Curr Biol. 2010;20(21):1913–1918. doi: 10.1016/j.cub.2010.09.004 [DOI] [PubMed] [Google Scholar]
- [56].Asakawa H, Kojidani T, Mori C, et al. Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr Biol. 2010;20(21):1919–1925. doi: 10.1016/j.cub.2010.09.070 [DOI] [PubMed] [Google Scholar]
- [57].King GA, Wettstein R, Varberg JM, et al. Meiotic nuclear pore complex remodeling provides key insights into nuclear basket organization. J Cell Bio. 2022;222(2):e202204039. doi: 10.1083/jcb.202204039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sipiczki M. Where does fission yeast sit on the tree of life? Genome Bio. 2000;1(2):reviews1011.1–reviews1011.4. doi: 10.1186/gb-2000-1-2-reviews1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Flor-Parra I, Belén Iglesias-Romero A, Salas-Pino S, et al. Importin α and vNEBD control meiotic spindle disassembly in fission yeast. Cell Rep. 2018;23(4):933–941. doi: 10.1016/j.celrep.2018.03.073 [DOI] [PubMed] [Google Scholar]
- [60].Ünal E, Kinde B, Amon A. Gametogenesis eliminates age-induced cellular damage and resets lifespan in yeast. Science. 2011;332(6037):1554–1557. doi: 10.1126/science.1204349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chu DB, Gromova T, Newman TAC, et al. The nucleoporin nup2 contains a meiotic-autonomous region that promotes the dynamic chromosome events of meiosis. Genetics. 2017;206(3):1319–1337. doi: 10.1534/genetics.116.194555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Komachi K, Burgess SM. The Nup2 meiotic-autonomous region relieves inhibition of nup60 to promote progression of meiosis and sporulation in Saccharomyces Cerevisiae. Genetics. 2022;221(1):iyac045. doi: 10.1093/genetics/iyac045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Iwamoto M, Mori C, Kojidani T, et al. Two distinct repeat sequences of nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr Biol. 2009;19(10):843–847. doi: 10.1016/j.cub.2009.03.055 [DOI] [PubMed] [Google Scholar]
- [64].Iwamoto M, Osakada H, Mori C, et al. Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate tetrahymena. J Cell Sci. 2017;130(10):1822–1834. doi: 10.1242/jcs.199398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Patterson DJ. The diversity of eukaryotes. Am Natur. 1999;154(S4):S96–124. doi: 10.1086/303287 [DOI] [PubMed] [Google Scholar]
- [66].Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].O’Donnell KL, McLaughlin DJ. Ultrastructure of meiosis in Ustilago maydis. Mycologia. 1984;76(3):468–485. doi: 10.1080/00275514.1984.12023868 [DOI] [Google Scholar]
- [68].Speijer D, Lukeš J, Eliáš M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc Natl Acad Sci USA. 2015;112(29):8827–8834. doi: 10.1073/pnas.1501725112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


