Abstract
The fusion peptide of the avian sarcoma/leukosis virus (ASLV) envelope protein (Env) is internal, near the N terminus of its transmembrane (TM) subunit. As for most internal viral fusion peptides, there is a proline near the center of this sequence. Robson-Garnier structure predictions of the ASLV fusion peptide and immediate surrounding sequences indicate a region of order (β-sheet), a tight reverse turn containing the proline, and a second region of order (α-helix). Similar motifs (order, turn or loop, order) are predicted for other internal fusion peptides. In this study, we made and analyzed 12 Env proteins with substitutions for the central proline of the fusion peptide. Env proteins were expressed in 293T cells and in murine leukemia virus pseudotyped virions. We found the following. (i) All mutant Envs form trimers, but when the bulky hydrophobic residues phenylalanine or leucine are substituted for proline, trimerization is weakened. (ii) Surprisingly, the proline is required for maximal processing of the Env precursor into its surface and TM subunits; the amount of processing correlates linearly with the propensity of the substituted residue to be found in a reverse turn. (iii) Nonetheless, proteolytically processed forms of all Envs are preferentially incorporated into pseudotyped virions. (iv) All Envs bind receptor with affinity greater than or equal to wild-type affinity. (v) Residues that support high infectivity cluster with proline at intermediate hydrophobicity. Infectivity is not supported by mutant Envs in which charged residues are substituted for proline, nor is it supported by the trimerization-defective phenylalanine and leucine mutants. Our findings suggest that the central proline in the ASLV fusion peptide is important for the formation of the native (metastable) Env structure as well as for membrane interactions that lead to fusion.
All enveloped viruses enter cells by fusion of their membranes with a target cell membrane. Specialized viral glycoproteins mediate this process. For each virus, the fusion-mediating glycoprotein contains a sequence, termed the fusion peptide, which interacts with and destabilizes the target membrane. Candidate fusion peptides have been identified as hydrophobic sequences, approximately 16 to 26 residues in length, that are conserved within a virus family and that can be modeled (although they need not exist or function) as an α-helix with a strongly hydrophobic face (48). In many cases the identity of the fusion peptide has been confirmed by mutational analysis and/or by cross-linking with photoreactive lipids (14, 27). Although many studies have probed the structure of synthetic fusion peptides in model membranes (7, 8, 12, 13, 26, 37, 39, 42, 44), the structure of a fusion peptide during an actual virus-cell fusion event remains uncertain (12, 13, 20).
Avian sarcoma/leukosis virus subtype A (ASLV-A) is a model retrovirus. The single viral glycoprotein of ASLV-A, EnvA, and its single target cell receptor appear to be sufficient to initiate membrane fusion (11, 24, 29). EnvA is composed of two subunits, gp85 (also known as the surface [SU] subunit) and gp37 (the transmembrane [TM] subunit) that are joined by a disulfide bond (31). gp85 provides the receptor binding function, while gp37 contains the TM domain and an internal, hydrophobic fusion peptide (30, 31).
Like most retroviruses, ASLV-A fuses at neutral pH, apparently at the cell surface, and receptor binding appears to initiate the process (11, 29). This is in contrast to the more extensively characterized influenza virus, which fuses in response to low pH in endosomes. Nonetheless, the fusion mechanism of the ASLV EnvA is thought to share steps in common with that of the influenza virus hemagglutinin (HA). When the influenza virus HA is exposed to low pH, its fusion peptide is exposed and binds to the target membrane. These events are accompanied by major conformational changes in the coiled-coil core of the HA trimer (5, 6). We have recently proposed a model for how these conformational changes drive the membrane fusion event (28, 49; http://www.people.Virginia.EDU/∼jag6n/whitelab.html). Like many (but not all) viral fusion proteins, EnvA is predicted to use a coiled-coil mechanism (6, 45). Therefore, we propose that, once triggered, steps leading to EnvA mediated fusion will be similar to those used by HA.
For many viral fusion proteins (e.g., the influenza virus HA and the human immunodeficiency virus Env), the fusion peptide resides at the N terminus of the TM subunit, generated during processing of the fusion protein precursor (28). For others, such as the Ebola virus glycoprotein (GP) and the flavivirus glycoprotein, the fusion peptide is internal, within the polypeptide chain (28). We have recently confirmed that the fusion peptide of EnvA is an apolar sequence found 20 residues from the N terminus of gp37 (30). While N-terminal fusion peptides have been extensively studied (13), little is known about the structure or function of internal fusion peptides.
A common feature of internal viral fusion peptides is a proline at or near their centers (48). Little has yet been done to delineate the roles of these prolines. We therefore generated a series of mutant EnvAs with different residues substituted for this proline (P29 of the TM subunit) and studied the effects of these mutations on EnvA proteolytic processing, delivery to the cell surface, incorporation into murine leukemia virus (MLV) pseudotyped viruses, binding to receptor, and infectivity. Our data suggest that the central proline is important for processing the precursor protein, pr95, into its subunits, gp85 and gp37, as well as for virus-cell fusion.
MATERIALS AND METHODS
Structure prediction.
Sequences for proteins containing putative or confirmed internal fusion peptides were obtained as follows. Our ASLV-A Env cDNA was sequenced in its entirety (S. E. Delos, unpublished results). All other sequences were obtained from GenBank: Ebola virus GP, U31033; Semliki Forest virus E1 protein (SFV-E1), X74425; vesicular stomatitis virus G protein (VSV-G), M35207; tick-borne encephalitis virus envelope protein (TBE-E), A02208; mouse fertilin α, U22056; macaque fertilin α, X79808-9. Each sequence was subjected to structure prediction using the Robson-Garnier algorithm in the Genetics Computer Group program package (21).
Mutagenesis.
A pCB6 vector containing cDNA for wild-type ASLV EnvA was used for rapid high-level expression in 293T cells as described previously (30, 35, 46). The mutations P29A, P29D, P29N, P29Q, P29E, P29R, P29S, P29T, P29F, and P29L were engineered at residue P29 of the gp37 subunit of EnvA as follows. The sequence between the unique EcoRI and AflII sites within the EnvA gene was amplified by PCR. A common primer for the 5′ end of the fragment, (5′-GGCAAGGAATTCCC-3′), encompassing the EcoRI site, was used. To generate random mutants, two bases within the codon for P29 were substituted with all four possible nucleotides. Hence, the reverse primer, encompassing the AflII site (underlined) and the mutant codon (X denotes randomly substituted bases), was 5′-GTCTCTCAATTTCTCTTAAGGCTTGCGCAGCTGCTACCCCCXXGGCTAAG-3′. The PCR fragments were digested with EcoRI and AflII and substituted for the identical fragment of the parent cDNA, and the mutant residue was identified by sequence analysis. Two additional mutants, P29G and P29V, were prepared using a QuickChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. The primers for P29G were 5′-GCATCTATCCTAGCCGGAGGGGTAGCAGCTGCG-3′ (forward) and 5′-CGCAGCTGCTACCCCTCCGGCTAGGATAGATGC-3′ (reverse). Those for P29V were 5′-GCATCTATCCTAGCCGTAGGGGTAGCAGCTGCG-3′ (forward) and 5′-CGCAGCTGCTACCCCTACGGGTAGGATAGATGC-3′ (reverse). The sequence of the entire EcoRI-AflII fragment was confirmed after substitution into the wild-type cDNA in the pCB6 vector.
Reagents for expression and detection of EnvA.
293T and PG950 cells have been described elsewhere (24). PG950 cells express the ASLV-A receptor, Tva (23). Cells were maintained in Dulbecco's modified Eagle's medium containing 10% supplemented calf serum (HyClone, Ogden, Utah) and 500 mg of Geneticin per liter, supplemented with 1× glucose, 1× pyruvate, and 1× penicillin-streptomycin (Gibco-BRL). pCB6 plasmids encoding the envelope protein of ASLV subtype C (EnvC) and the EnvA cleavage-negative mutant Acl have been described elsewhere (24). The anti-MLV Gag, anti-Ngp37, and anti-EnvA tail antibodies have been described elsewhere (30). s47, a functional recombinant fragment of the ASLV-A receptor, was a gift from R. Peters and D. Agard, University of California at San Francisco.
Transfection, induction, and harvesting of cells.
293T cells were transfected with 10 μg of plasmid DNA by the calcium phosphate method (30). At 28 h posttransfection (hpt), cells were treated with 10 mM sodium butyrate to induce Env expression. Cells were harvested at 48 hpt, lysed with a lysis buffer (1% NP-40, 10 mM HEPES [pH 7.3], 130 mM NaCl) containing freshly added protease inhibitors (5 μM phenylmethylsulfonyl fluoride, 5 μg of pepstatin A, 10 μg of leupeptin, 20 μg of aprotinin, 50 μg of antipain, 2 mM benzamidine, 50 μg of soybean trypsin inhibitor, and 2.5 μM iodoacetamide), and pelleted at 4°C for 10 min at high speed in a microcentrifuge to remove insoluble cell debris. Supernatants were transferred to clean tubes, and fresh aliquots of protease inhibitors were added.
Biotinylation of s47.
One hundred microliters of s47 (0.2 to 0.5 mg/ml) was incubated on ice with 100 μl of N-hydroxysuccinimide-LC-biotin (2 mg/ml; Sigma) in phosphate-buffered saline (PBS) for 2 to 4 h, quenched with 50 μl of 1 M glycine, and stored at 4°C. Biotinylated s47 is stable at 4°C for at least 1 to 2 months (Delos, unpublished results).
Sucrose gradients and immunoblots.
Cell lysates (200 μl) were layered onto 12-ml 10 to 30% continuous sucrose gradients prepared in octylglucoside buffer (40 mM n-octylglucoside [Boehringer Mannheim] in 20 mM HEPES [pH 7.3]–130 mM NaCl) and centrifuged at 275,000 × g for 17 h at 4°C in an SW41 rotor. Fractions of 0.5 ml were collected from the top, precipitated with CHCl3-methanol, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing reducing agent, resolved on SDS-gels, transferred to nitrocellulose, and probed with the anti-Ngp37 antibody.
Quantitation of cell surface expression.
The amounts of wild-type and mutant EnvA proteins at the cell surface were determined as follows. Approximately 2 × 105 transiently transfected 293T cells were incubated on ice with the anti-Ngp37 antibody in a total volume of 100 μl of PBS–0.02% azide–2% FCS (fetal calf serum), washed twice with PBS–0.02% azide, and incubated with fluoresceinated goat anti-rabbit antibody in PBS-azide-FCS as described previously (30). Cells were then washed twice, fixed in PBS containing 3% paraformaldehyde, and analyzed by fluorescence-activated cell sorting (FACS) at the University of Virginia Core Facility, using a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). Values from five independent experiments were averaged.
s47 coimmunoprecipitation.
To assay for s47 binding to EnvA, 100-μl aliquots of cell lysates were mixed with 2 μl of biotinylated s47 and incubated at 4°C for 30 min. The mixture was then precleared with preimmune serum bound to protein A-agarose beads and immunoprecipitated with the affinity-purified anti-EnvA tail antibody bound to protein A-agarose beads. Samples were resolved by SDS-PAGE (15% acrylamide), transferred to nitrocellulose, and probed with horseradish peroxidase-conjugated streptavidin.
Quantitation of s47 binding to EnvA mutants.
To characterize the binding between s47 and EnvA, approximately 2 × 105 transiently transfected 293T cells expressing wild-type EnvA were incubated for 30 min on ice with biotinylated s47 in concentrations from 0.1 nM to 1.0 μM in a total volume of 100 μl in PBS–0.02% azide–2% FCS, washed twice with PBS–0.02% azide, and incubated for 30 min on ice with avidin-Oregon Green 488 (Molecular Probes) in PBS–0.02% azide–2% FCS. Cells were then washed twice, fixed in PBS containing 3% paraformaldehyde, and subjected to FACS analysis as described above. KD represents the concentration of s47 at which the fluorescence was half of the maximum value: fluorescence50 = [(fluorescencemax − fluorescencemin)/2] + fluorescencemin. Binding curves were also obtained for selected mutant EnvAs. The remaining mutant EnvAs were examined at saturating concentrations of s47 (100 nM), and their binding values were normalized to those obtained for wild-type EnvA.
Radiolabeling, endoglycosidase F treatment, and quantitation of EnvA processing.
293T cells were transfected with cDNA encoding wild-type or mutant Env as described above. At 26 hpt, cells were washed with PBS and incubated in Cys- and Met-deficient medium for 2 h. Cells were then labeled overnight at 37°C with 100 μCi of Tran35S-label (Amersham) per plate in Cys- and Met-deficient medium containing 10 mM sodium butyrate. Cells were harvested, lysed, and immunoprecipitated with the anti-EnvA tail antibody as described above. Immunoprecipitated proteins were eluted from the immunobeads by boiling for 3 min in N-glycosidase F buffer (1% n-octylglucoside–0.2% SDS–40 mM Tris-HCl [pH 8.0]–5 mM EDTA to which 1% β-mercaptoethanol was added immediately before use). The elution was repeated, and the two eluants were pooled. Samples were then treated with 1 U of N-glycosidaseF (Boehringer Mannheim) for 2 h on ice. Samples were resolved by SDS-PAGE, and the dried gels were analyzed using a Molecular Dynamics PhosphorImager and the ImageQuant program (Molecular Dynamics). Ratios of the SU to pr95 bands were obtained and normalized to the ratio for wild-type (P) EnvA.
EnvA incorporation into pseudotyped virions and infectivity assay.
EnvA-bearing MLV pseudotyped virions encoding β-galactosidase were prepared by a three-plasmid transfection method and concentrated from infected culture supernatants as described elsewhere (30). The assay measures a single-round of infection because only the β-galactosidase cDNA contains the appropriate signal for packaging by MLV capsids. Expression of β-galactosidase is used as a measure of infection. To measure EnvA incorporation, pseudotyped virions were resuspended in SDS sample buffer; equal amounts were resolved by SDS-PAGE on two parallel gels, transferred to nitrocellulose, and probed with either an antibody against MLV Gag (which recognizes the capsid protein present in virions) or the anti-Ngp37 antibody. Blots were developed by enhanced chemiluminescence and scanned on a Molecular Dynamics densitometer, and the MLV capsid and EnvA bands were quantified using ImageQuant. For each experiment, the ratio of gp37 to MLV capsid was determined and normalized to the wild-type ratio. Care was taken to ensure that band intensities were within the linear range.
Virus from aliquots of the same culture supernatants was titered for infectivity on PG950 cells as previously described (30). At 48 h postinfection, PG950 cells were fixed, stained for β-galactosidase, and counted to determine the number of infected (blue) cells.
RESULTS
Predicted structure of the ASLV Env and other internal fusion peptides.
The fusion-mediating subunit (TM; also referred to as gp37) of the ASLV-A glycoprotein, EnvA, contains an internal fusion peptide conserved among all ASLV subtypes (30). A common feature among many internal fusion peptides, including that of ASLV Env, is a proline at or near the center of the sequence (48). The ASLV Env fusion peptide was originally proposed after examining the EnvA sequence for a hydrophobic sequence, conserved among ASLV Envs, that could be modeled as an α-helix with a strongly hydrophobic face (30). (This analysis does not require that the fusion peptide exist as an α-helix either before or during fusion.) To determine possible structures for the ASLV Env and other internal viral fusion peptides, we modeled several using the Robson-Garnier algorithm (21). We also modeled the candidate fusion peptide of fertilin α (ADAM 1), a protein postulated to be involved in sperm-egg fusion (3, 4).
In Fig. 1, we present the modeling. We refer to each of these sequences as the fusion peptide region. We show the prediction for the ASLV Env fusion peptide region, the subject of this study. We present the analysis for the fusion peptide region of Ebola virus GP because the structure of its fusion-mediating subunit has been predicted to be highly similar to that of ASLV Env (19), because it has been subjected to mutational analysis (32), and because its interaction with phosphatidylinositol-containing liposomes has been studied (44). We also analyzed the fusion peptide regions of VSV-G and SFV-E1 because they have been characterized by mutational analysis (18, 33, 50, 55), and, for VSV-G, by photo-cross-linking to target membrane lipids (14). We analyzed the fusion peptide region of TBE-E because the structure of the TBE-E ectodomain has been solved (43) and because the identity of the fusion peptide has been confirmed by mutational analysis (F. Heinz, personal communication). As seen in Fig. 1, each of these internal viral fusion peptide regions can be modeled as a segment of order (α-helix or β-sheet), a central segment of turn or random coil, and finally a second segment of order, suggesting that these peptides may adopt a bent or turn structure either before, during, or after fusion. The prediction of an order-turn-order motif is consistent with the structure of the fusion peptide region of the TBE-E glycoprotein in its native, likely metastable state (43), with the exception that the structure conforms to a β-strand–turn–β-strand motif instead of the predicted β-strand–turn–α-helix motif. The candidate internal fusion peptide regions from a variety of species of fertilin α also conform to the order-turn-order motif (Fig. 1; C. Rea, S. E. Delos, and J. M. White, unpublished results).
FIG. 1.
Predicted structures for internal fusion peptide regions. Protein sequences were subjected to structure prediction using the Robson-Garnier algorithm (21) in the Genetics Computer Group program package as described in Materials and Methods. b, β-sheet; ., random coil; h, α-helix; t, reverse turn. The segments of order (α-helix or β-sheet) are indicated by gray boxes; segments of random coil or reverse turn are indicated by white boxes; prolines are starred. The crystal structure of TBE-E reveals extension of the ordered sequences within the fusion peptide region into the predicted turn segment (43). The residues that actually form the turn are underlined. Fusion peptides, as described in the literature, are in boldface (ASLV EnvA [30], Ebola virus G2 [Ebola-GP] [19, 32, 44], VSV-G [14, 50, 55], SVF-E1 [33], TBE-E [43], mouse fertilin α [51]; macaque fertilin α [40].)
The central segments of the ASLV-A Env and the mouse fertilin α (candidate) fusion peptide regions contain four residues and would therefore be expected to form a tight turn. The longer central turn segments of the Ebola virus GP, VSV-G, SFV-E1, TBE-E, and macaque fertilin α fusion peptides might form more complex turn structures as is seen for the TBE-E fusion peptide (43). We refer to the loop or turn region within internal fusion peptide regions as the “turn” to distinguish this subregion from the overall predicted “loop” composed of the entire order-turn-order motif of the fusion peptide region.
Expression, trimerization, and processing of mutant EnvAs.
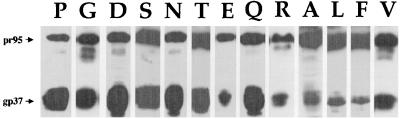
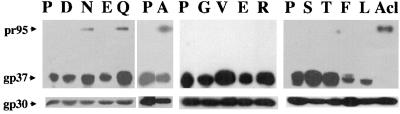
We predict that the central prolines seen in many, but not all, internal fusion peptides (e.g., five out of the seven shown in Fig. 1) are important for fusion. As a first test of this hypothesis, we generated a series of mutants in which we substituted 12 different residues for the P at the center of the EnvA fusion peptide. We first examined the expression of each mutant EnvA by immunoblot analysis of cell lysates using an antibody against the N terminus of gp37, anti-Ngp37 (30). This antibody recognizes both the precursor, pr95, and the processed TM subunit, gp37. As can be seen in Fig. 2, all mutants were expressed to high levels. Each mutant was processed, although the amount of processing appeared to vary from mutant to mutant (see below).
FIG. 2.
Expression of mutant EnvAs. 293T cells were transfected with pCB6-EnvA DNA, induced, harvested, and lysed as described in Materials and Methods. Samples were resolved by SDS-PAGE and processed for Western blot analysis with the anti-Ngp37 antibody. Both the uncleaved pr95 (upper band) and cleaved gp37 (lower band) are indicated.
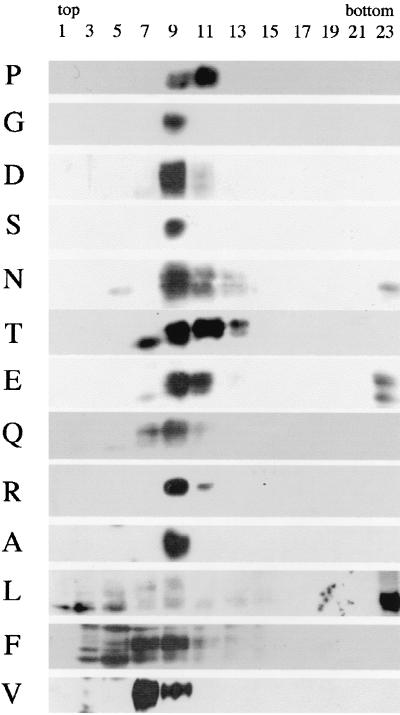
Mature EnvA is a trimer of SU and TM subunits that are generated from the precursor, pr95, by proteolytic processing. Since trimerization is required for EnvA to exit the endoplasmic reticulum (15), and since proteolytic processing of pr95 is required to produce an Env that can mediate fusion and infection (24), we next examined whether mutant EnvAs form trimers and quantitated their extent of processing. As shown in Fig. 3, wild-type EnvA (P), which forms proteolytically processed trimers (25), sedimented in fractions 9 to 11 near the center of a sucrose gradient. All mutant EnvAs also formed proteolytically processed trimers, as evidenced by positive blotting material in fractions 9 to 11. However, for EnvAs with the bulky hydrophobic residues L and F in place of P, a significant amount of protein was found in lighter fractions, suggesting that for these mutant Envs, the trimer interface was weakened. A significant amount of degraded protein (lower bands on the blot) was also observed for F and L. Some aggregated material was seen for N, E, and L. In these experiments, no attempt was made to normalize the amount of material placed on the gradient, nor were the ECL exposures identical. Thus, no conclusions regarding the relative expression levels of the mutants can be drawn from these data.
FIG. 3.
Trimerization of mutant EnvAs. Cell lysates were prepared as described for Fig. 2 and subjected to sucrose density centrifugation as described in Materials and Methods. Fractions of 500 μl were collected and processed for Western blot analysis with the anti-Ngp37 antibody. The gp37 band is shown.
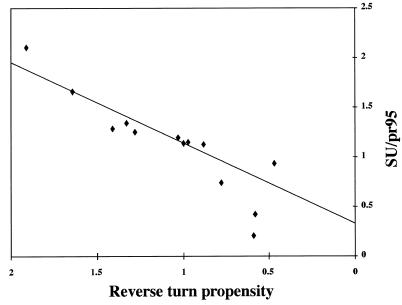
As mentioned above, the amount of processing appeared to differ among the mutant EnvAs. Because processing is a pivotal event for the function of EnvA, we quantitated the amount of processing for each mutant as described in Materials and Methods. We then analyzed the processing data as a function of the relative hydrophobicity, side chain volume, and preferred structure (α-helix, β-sheet, or reverse turn) of the mutant residue (10). Of the parameters examined, the extent of processing correlated only with the probability that the mutant residue be found in a reverse turn (Table 1). A plot of the extent of processing as a function of the reverse turn probability of the mutant residue is shown in Fig. 4. Surprisingly, the ability of a given mutant EnvA to be processed into SU (gp85) and TM (gp37) subunits correlated linearly with the reverse turn probability. The two data points that fell below the line were for F and L, the mutants that are defective in trimerization (Fig. 3). The fact that wild-type EnvA (P) is processed better than any of the mutants suggests that a major reason for the conservation of proline at residue 29 of the TM subunit is for optimal processing of the precursor, pr95, into SU and TM subunits, an event that is required to enable EnvA to mediate infectivity (24). We expect that this processing event is a rate-limiting step in the ASLV life cycle. However, in the MLV pseudotyping system used below, high levels of protein are expressed and processed ASLV Envs are preferentially incorporated. Hence, with the MLV pseudotyping system, the processing defects are overcome and subsequent EnvA-mediated events can be examined.
TABLE 1.
Reverse turn propensity and processing of proline mutants
| Mutant | Reverse turn propensitya | [SU]/[pr95] (mean ± SE)b |
|---|---|---|
| P | 1.91 | 2.10 ± 0.23 |
| G | 1.64 | 1.66 ± 0.03 |
| D | 1.41 | 1.28 ± 0.05 |
| S | 1.33 | 1.34 ± 0.12 |
| N | 1.28 | 1.25 ± 0.12 |
| T | 1.03 | 1.19 ± 0.01 |
| E | 1.00 | 1.13 ± 0.08 |
| Q | 0.97 | 1.15 ± 0.24 |
| R | 0.88 | 1.12 ± 0.04 |
| A | 0.78 | 0.74 ± 0.02 |
| L | 0.59 | 0.21 ± 0.02 |
| F | 0.58 | 0.42 ± 0.05 |
| V | 0.47 | 0.93 ± 0.23 |
From reference 10.
Determined as described in Materials and Methods.
FIG. 4.
Processing of mutant EnvAs into SU (gp85) and TM (gp37) subunits. Cells expressing wild-type and mutant EnvAs were radiolabeled with Tran35S-label, lysed, subjected to immunoprecipitation using the anti-EnvA tail antibody, and treated with N-glycosidase F. Proteins were resolved by SDS-PAGE, visualized using a PhosphorImager, and quantified using ImageQuant. The data were subjected to linear regression analysis and fit best to the linear equation y = 0.81× + 0.33 with r2 = 0.84. The data (triangles) represent the ratio of gp85 to unprocessed pr95. The solid line represents the linear least squares fit of the data. The two points significantly below the line are for the poorly trimerized F and L mutants.
Incorporation of mutant EnvAs into pseudotyped virions.
We use single-cycle infectivity of MLV pseudotype virions as a measure of the ability of mutant EnvAs to mediate virus-cell fusion (30, 35, 46). We first assessed the relative ability of each mutant EnvA to be incorporated into MLV pseudotyped virions as described in Materials and Methods. As shown in Fig. 5, all mutant EnvAs were incorporated into pseudotyped virions. Importantly, with the exception of the genetically engineered cleavage site mutant, Acl (24), the proteolytically processed form of each EnvA was selectively incorporated (compare the density of pr95 to that of gp37 in Fig. 5 with that in Fig. 2). Furthermore, in this overexpression system, most mutant EnvAs were incorporated into the pseudotyped virions to similar levels as wild-type EnvA (Fig. 5 and Table 2). Even for the F and L mutants, those impaired in trimerization and processing, a significant amount of processed EnvA was incorporated into pseudotyped virions. For reasons that are not apparent, the Q mutant was consistently incorporated to a greater extent than the others; the Q mutant does not exhibit increased expression at the cell surface (Table 2). Because of the incorporation of approximately wild-type levels of processed forms of all mutant EnvAs, we can use single-cycle infection of MLV-pseudotyped virions bearing these mutant EnvAs to score their relative ability to support infectivity.
FIG. 5.
Incorporation of mutant EnvAs into MLV-pseudotyped virions. EnvA-MLV pseudotyped virions were prepared and concentrated as described in Materials and Methods. Virions were diluted into SDS sample buffer, resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the anti-Ngp37 antibody. Parallel blots were probed with an antibody recognizing MLV capsid (gp30).
TABLE 2.
Cell surface expression, virion incorporation, and receptor binding of proline mutantsa
| Mutant | Relative cell surface expression | Relative virion incorporation | Relative s47 saturation binding | KD for S47 binding (nM) |
|---|---|---|---|---|
| R | 0.7 ± 0.1 | 0.7 ± 0.3 | 0.9 ± 0.1 | ND |
| D | 0.9 ± 0.1 | 1.2 ± 0.1 | 1.2 ± 0.4 | ND |
| E | 0.9 ± 0.1 | 0.9 ± 0.2 | 1.0 ± 0.2 | ND |
| N | 0.8 ± 0.0 | 1.4 ± 0.3 | 0.9 ± 0.3 | ND |
| Q | 1.0 ± 0.1 | 2.1 ± 0.3 | 0.9 ± 0.1 | ND |
| S | 0.9 ± 0.1 | 1.0 | 1.0 ± 0.5 | 14 |
| T | 0.8 ± 0.1 | 1.0 ± 0.3 | 0.8 ± 0.1 | 5 |
| P | 1.0 | 1.0 | 1.0 | 17 |
| G | 1.1 ± 0.1 | 0.8 ± 0.1 | 1.2 ± 0.0 | 10 |
| A | 0.7 ± 0.1 | 0.8 ± 0.2 | 0.5 ± 0.0 | 7 |
| V | 1.4 ± 0.1 | 1.1 ± 0.1 | 0.7 ± 0.0 | 16 |
| L | 0.8 ± 0.0 | 0.4 | 0.9 ± 0.1 | ND |
| F | 0.7 ± 0.0 | 0.6 | 0.2 ± 0.1 | 2 |
| Acl | 1.3 ± 0.2 | 1.0 ± 0.3 | ND | ND |
| HA | ND | ND | ND | ND |
Data were obtained and normalized to that for wild-type EnvA (P) as described in Materials and Methods. They are presented in order of increasing residue hydrophobicity. (Hydrophobicity tables from different sources rank residues differently, both in order and by extent of hydrophobic nature. We present our data in an order of increasing hydrophobicity representing the consensus ordering from the tables in reference 16, 10, 34, and 17. When plotted versus hydrophobicity for each of the four individual hydrophobicity scales, a similar correlation between high infectivity and intermediate hydrophobicity was obtained [data not shown].) All values are reported as fractions of the P values ± standard error of the mean. Because values were normalized to wild type for each experiment, the error in P is embedded in the values for the other errors. Where no errors are given for relative virion incorporation (other than P), a single blot was quantitated. Otherwise, results were averaged from two to five independent experiments. ND, not determined.
Binding to s47.
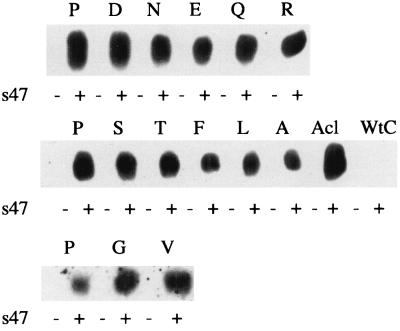
To determine whether mutation of the central P of the fusion peptide of gp37 affected the ability of the gp85 subunit to bind receptor, we assessed the ability of each mutant to bind s47, a soluble, recombinant form of Tva (the cellular receptor for ASLV-A) (29, 54). We recently showed that s47 is able to bind EnvA and trigger fusion-activating conformational changes in EnvA (29). We used two assays to quantitate s47 binding to the EnvA: coimmunoprecipitation of s47 with EnvA and, more rigorously, a FACS-based binding assay. We first showed that s47 could be immunoprecipitated with each mutant EnvA. Lysates from cells expressing wild-type or mutant EnvA proteins were incubated with biotinylated s47 and immunoprecipitated with the anti-EnvA tail antibody. SDS-gels of the immunoprecipitates were then probed with horseradish peroxidase-conjugated streptavidin. As seen in Fig. 6, biotinylated s47 was coimmunoprecipitated with wild-type EnvA, with the cleavage-deficient mutant Acl, and with each of the P29X mutants, suggesting that mutations at P29 do not impair receptor binding.
FIG. 6.
Coimmunoprecipitation of s47 with mutant EnvAs. Lysates of EnvA-expressing cells were prepared as described for Fig. 2 and incubated with biotinylated s47 for 30 min at 4°C. The mixtures were then immunoprecipitated with the anti-EnvA tail antibody (anti-EnvC tail antibody for EnvC, the Env protein of ASLV, Prague C, used as a negative control), resolved by SDS-PAGE, transferred to nitrocellulose, and probed with horseradish peroxidase-conjugated streptavidin.
To examine the receptor binding ability of each mutant EnvA in more detail, we used a FACS-based assay similar to that described by Young and coworkers (56). Binding of different amounts of s47 to cells expressing wild-type EnvA was measured as described in Materials and Methods. From the average of three experiments, we calculated a KD of 17 ± 2 nM for wild-type EnvA. The binding of s47 to cells expressing each mutant EnvA was then measured at saturating concentrations of s47. The results are presented in Table 2. For most mutant EnvAs, s47 binding was close to wild-type binding and variations correlated with differences in surface expression. However, mutants A, V, and F appeared to bind s47 less well than wild-type EnvA. To further assess the ability of these mutants to bind s47, binding curves were obtained. In each case, although less s47 was bound at saturating concentrations, the measured affinity was greater than or equal to wild-type affinity (Table 2). Thus, mutations at P29 of the TM subunit of EnvA do not impair receptor binding.
Infectivity of mutant EnvAs.
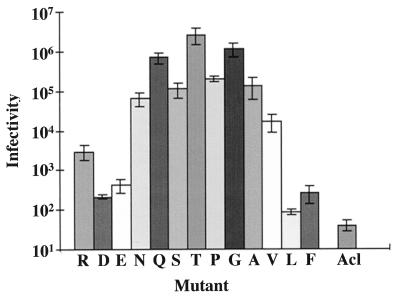
MLV-pseudotyped virions, prepared as described above, were titered for infectivity on PG950 cells, cells that stably express Tva, the EnvA receptor. The results are presented in Fig. 7. The infectivity data correlated best with the hydrophobicity of the substituted residue; no correlation was seen if the data were plotted as a function of preferred structure (reverse turn, α-helix, or β-sheet) or with the side chain volume of the substituted residue (not shown). As seen in Fig. 7, residues of intermediate hydrophobicity (Q, S, T, P, G, and A) appeared to function best. Both residues of high (e.g., V) and low (e.g., R) hydrophobicity were impaired by 1 to 2 logs in their ability to support infection. The acidic residues, D and E, showed even greater defects, being impaired by 3 logs. The data approximate a bell-shaped curve centered about T. The unexpectedly high level of infection supported by Q is likely the consequence of its heightened incorporation into virions (see above and Table 2). The low levels of infection seen for F and L may be a consequence of both their lower level of incorporation into virions and their relative hydrophobicity. We observed a similar correlation between the hydrophobicity of the substituted residue and the ability of the corresponding EnvA to support cell-cell fusion (Delos and White, unpublished results).
FIG. 7.
Infectivity of mutant EnvAs. EnvA-MLV pseudotyped virions encoding β-galactosidase were prepared as for Fig. 4. Serial dilutions of pseudotyped viral supernatants were added to Tva-expressing NIH 3T3(PG950) cells, incubated at 37°C for 48 h, and assayed for β-galactosidase as described in Materials and Methods. Data from at least three independent experiments were averaged and are presented as a function of increasing hydrophobicity of the mutant residue. Ranking of relative hydrophobicity is described in the footnote to Table 2. Error bars represent standard error of the mean.
DISCUSSION
A common feature of many internal fusion peptides is a proline at or near their centers (48). The work reported here represents the first comprehensive study of the roles of a central proline within an internal fusion peptide. We generated 12 mutations at P29 within the fusion peptide of the TM subunit of the ASLV-A envelope protein, EnvA, and examined their effects on EnvA proteolytic processing, trimerization, cell surface expression, receptor binding, virion incorporation, and infectivity. We found the following. With the exception of the bulky hydrophobic residues F and L, all mutant EnvAs were proteolytically processed, trimerized, and efficiently expressed at the cell surface. Processing correlated linearly with the reverse turn probability of the mutant residue. P, the wild-type residue, has the highest reverse turn probability, and wild-type EnvA was the best processed. All mutant EnvAs were incorporated into MLV pseudotyped virions and bound the ASLV-A receptor, Tva, with high affinity. For all mutants, the processed form of EnvA was preferentially incorporated. Infectivity of the pseudotyped particles approximated a bell-shaped pattern as a function of residue hydrophobicity; the optimal residues for infectivity are, like proline, of moderate hydrophobicity. Our findings have implications for the roles of the central proline of the internal fusion peptide of ASLV EnvA in setting up the metastable state of the fusion protein and in target bilayer interactions.
The central proline is required for optimal processing of EnvA.
The proteolytic processing of the ASLV EnvA into its SU and TM subunits is critical to viral infectivity (24). Since the degree of processing of the mutant EnvAs correlated linearly with the propensity of the substituted residue to be found in a reverse turn (Fig. 4), our findings suggest that a compelling selective force for the choice of P at the center of the ASLV fusion peptide is to position the proteolytic cleavage site, 29 residues upstream, for optimal accessibility to the processing protease.
Most viral fusion proteins require a proteolytic processing event to render them fusion competent. For HA, it has been shown that this processing event has two related consequences: (i) it allows conversion of the lowest energy state of HA0 to a metastable state that can then be converted to a lower-energy fusion-active state, and (ii) it causes the fusion peptide to be hidden in the trimer interface until HA is exposed to its fusion trigger. Processing of pr95 into SU and TM may set up a metastable state in EnvA and may position the fusion peptide so that it can be liberated in response to a trigger. As for HA (9), after processing, the fusion peptide may occupy a hydrophobic pocket within the trimer interface and may be maintained in this conformation by interactions between gp85 and gp37 that clamp EnvA in a metastable state. If this is the case, it may explain why substitution of P with the bulky hydrophobic residues, F and L, appears to weaken the trimer interface.
For reasons that are not yet apparent, processed EnvA was preferentially incorporated into MLV pseudotyped particles (Fig. 2 and 5). Even for mutants where little EnvA was processed (e.g., F and L), little unprocessed material was found in pseudotyped virions (Fig. 5). In the case of a normal ASLV infection, we predict that processing will be a rate-limiting step in the production of infectious particles. However, because in our transient transfection system the amount of expressed EnvA is very high, a significant amount of processed EnvA reaches the cell surface and is incorporated into pseudotype virions (Fig. 2, Fig. 5, and Table 2).
Although many internal fusion peptides have a proline near their centers, only a subset may need to fold with a tight reverse turn about the proline. EnvA has an unusually short (four-residue) predicted turn region in its fusion peptide. The predicted turn region for most internal fusion peptides is seven or more residues, a length compatible with more flexible structures capable of accommodating a broader range of residues. For example, mutations at either of the two prolines within the internal fusion peptide of the Ebola virus G2 protein do not appear to affect processing (32).
Roles of the central proline in target bilayer interactions.
The ability of each mutant Env to support infection of receptor-bearing cells by EnvA-MLV pseudotyped virions correlated best with the hydrophobicity of the mutant residue. Residues of intermediate hydrophobicity (S, T, P, G, and A) supported high levels of infection. The decrease in infectivity seen with residues of high hydrophilicity (R, D, and E) or high hydrophobicity (V, F, and L) could not be attributed to impaired receptor binding (Table 2 and data not shown). We measured an apparent KD for the EnvA-s47 interaction of 17 ± 2 nM. This affinity is lower than those reported using other Tva or EnvA reagents (2, 56). Future work is necessary to determine whether these differences stem from experimental differences or whether our data suggest that additional residues, not present in s47, contribute to the binding affinity between EnvA and Tva.
We surmise that our infectivity data reflect the relative ability of mutant EnvAs to support virus-cell fusion. This supposition is supported by the similarity between the ability of each mutant EnvA to support infectivity (Fig. 7) and its ability to support cell-cell fusion. Although the signal-to-noise ratio for the cell-cell fusion assay is low, mutant EnvAs with S, T, G, or A in place of P29 support cell-cell fusion, whereas those with substitutions of residues of high (V, L, and F) or low (R, D, E, and N) hydrophobicity exhibit poor cell-cell fusion (Delos and White, unpublished results). These observations parallel recent findings on the initial residue of the (N-terminal) fusion peptide of the influenza virus HA. For all type A influenza viruses, this residue is G, a residue of intermediate hydrophobicity. Substitution with residues of higher (V) or lower (E) hydrophobicity abolish fusion (13, 22, 41, 47). It thus may be that a residue of intermediate hydrophobicity is needed for initial interactions of fusion peptides with target cell membranes. We propose that P29 is at the apex of a fusion peptide loop which, by analogy with several viral fusion proteins (1, 45) and based on the predicted similarities between the Ebola virus G2 and ASLV EnvA TM subunit, would sit on top of a triple coiled-coil structure. In this configuration, P29 could be in an ideal position to make an initial interaction with the target membrane.
Other internal fusion peptides.
Ebola virus G2 is predicted to be similar in structure to the TM subunit of ASLV. The Ebola virus G1 and ASLV SU subunits are, however, quite different. Not unexpectedly, the activation requirements for these two proteins appear to be different: the Ebola virus GP requires low pH, whereas the ASLV protein requires association with its receptor, Tva (24, 29, 52). The ASLV Env requires proteolytic processing to be infectious; Ebola virus GP may not (24, 53). Thus, comparison of these two proteins may be limited to functions that reside in TM and G2. Mutational analysis of the G2 fusion peptide has recently been reported; in this study, residues throughout the fusion peptide were mutated to either R or A, and the infectivity of the mutant G proteins was studied using VSVΔG pseudotyped virus (32). The G2 fusion peptide has two prolines, at residues 533 and 537. For each of these prolines, the P-to-A mutation slightly and the P-to-R mutation severely impaired infectivity. These results are consistent with our findings for EnvA.
The candidate internal fusion peptides of SFV-E1, VSV-G, and TBE-E have been studied by mutational analysis (18, 33, 50; Heinz, personal communication). The most extensive studies have been on VSV-G. The region encompassing the candidate fusion peptide of VSV-G conforms well to the order-turn-order motif (Fig. 1). Mutation of P127, within the predicted turn segment of the candidate fusion peptide region (Fig. 1, the most C-terminal proline of the VSV-G fusion peptide), to D significantly decreased fusion (50) and abolished infectivity (18). These results are also consistent with our results for EnvA. The SFV-E1 fusion peptide has been extensively mutagenized (33). Unfortunately, the only mutant with a substitution for the proline (to D) could not be expressed at the cell surface, and so no information regarding fusion could be obtained (33).
Membrane interactions of synthetic peptides representing the candidate internal fusion peptides of Ebola virus G2 and guinea pig fertilin α have been reported (36, 38, 44). In each case, the peptides preferentially interacted with acidic phospholipid bilayers. For fertilin α, a peptide in which the central PP doublet was replaced with AA was less fusogenic and more lytic (38). These results suggest that the central prolines are important for the fusogenic properties of the synthetic fertilin α candidate internal fusion peptide.
Concluding remarks.
By systematically mutating the proline at residue 29 of the fusion peptide of the TM subunit of ASLV EnvA, we have defined two roles for this central proline. First, our data suggest that the proline, by its strong propensity to be found in a reverse turn, and thereby likely affecting the structure and packing of the fusion peptide, is the optimal residue at this position for the critical proteolytic processing event which converts pr95 to its metastable state composed of SU and TM subunits. Second, our data suggest that the proline is one of the optimal residues at this position to support virus-cell fusion; the other residues that support infection (S, T, G, and A) are all of intermediate hydrophobicity. Our data are consistent with a model in which the fusion peptide region adopts an order-turn-order configuration in the precursor state (Fig. 1). The fusion peptide region may retain the order-turn-order conformation as it approaches the target membrane, with the proline providing an initial point of membrane interaction. The fusion peptide may adopt other structures during later steps of the fusion process.
ACKNOWLEDGMENTS
We thank Lukas Tamm for critical reading of the manuscript.
The work of J.M.G. was performed while J.M.G. and J.M.W. were at The University of California, San Francisco. This work was supported by NIH grant AI22470 to J.M.W.
REFERENCES
- 1.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 2.Balliet J W, Berson J, D'Cruz C M, Huang J, Crane J, Gilbert J M, Bates P. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1999;73:3054–3061. doi: 10.1128/jvi.73.4.3054-3061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigler D, Chen M, Waters S, White J M. A model for sperm-egg binding and fusion based on ADAMs and integrins. Trends Cell Biol. 1997;7:220–225. doi: 10.1016/S0962-8924(97)01058-1. [DOI] [PubMed] [Google Scholar]
- 4.Blobel C P, Wolfsberg T G, Turck C W, Myles D G, Primakoff P, White J M. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang D, Cheng S, Trivedi V D. Biophysical characterization of the structure of the amino-terminal region of gp41 of HIV-1. J Biol Chem. 1999;274:5299–5309. doi: 10.1074/jbc.274.9.5299. [DOI] [PubMed] [Google Scholar]
- 8.Chang D K, Cheng S F, Chien W J. The amino-terminal fusion domain peptide of human immunodeficiency virus type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as a helix with a conserved glycine at the micelle-water interface. J Virol. 1997;71:6593–6602. doi: 10.1128/jvi.71.9.6593-6602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Lee K H, Steinhauer D A, Stevens D J, Skehel J J, Wiley D C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 10.Creighton T E. Proteins: structural and molecular principles. W. H. New York, N.Y: Freeman and Co.; 1984. [Google Scholar]
- 11.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies S M, Kelly S M, Price N C, Bradshaw J P. Structural plasticity of the feline leukaemia virus fusion peptide: a circular dichroism study. FEBS Lett. 1998;425:415–418. doi: 10.1016/s0014-5793(98)00274-9. [DOI] [PubMed] [Google Scholar]
- 13.Durell S R, Martin I, Ruysschaert J M, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 14.Durrer P, Gaudin Y, Ruigrok R W H, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J Biol Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 15.Einfeld D A, Hunter E. Mutational analysis of the oligomer assembly domain in the transmembrane subunit of the Rous sarcoma virus glycoprotein. J Virol. 1997;71:2383–2389. doi: 10.1128/jvi.71.3.2383-2389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberg D, McLachlan A D. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 17.Engleman D M, Steitz T A, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 18.Fredericksen B L, Whitt M A. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallaher W R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 20.Gallaher W R, Segrest J P, Hunter E. Are fusion peptides really “sided” insertional helices? Cell. 1992;70:531–532. doi: 10.1016/0092-8674(92)90423-a. [DOI] [PubMed] [Google Scholar]
- 21.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 22.Gething M-J, Doms R W, York D, White J M. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert J M, Bates P, Varmus H E, White J M. The receptor for the subgroup A avian leukosis sarcoma virus binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;67:6889–6892. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert J M, Hernandez L D, Chernov-Rogan T, White J M. Generation of a water soluble oligomeric ectodomain of the Rous sarcoma virus envelope glycoprotein. J Virol. 1993;67:6889–6892. doi: 10.1128/jvi.67.11.6889-6892.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray C, Tamm L K. pH-induced conformational changes of membrane-bound influenza hemagglutinin and its effect on target lipid bilayers. Protein Sci. 1998;7:2359–2373. doi: 10.1002/pro.5560071113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harter C, Bächi T, Semenza G, Brunner J. Hydrophobic photolabeling identifies BHA2 as the subunit mediating the interaction of bromelain-solubilized influenza virus hemagglutinin with liposomes at low pH. Biochemistry. 1988;27:1856–1864. doi: 10.1021/bi00406a010. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez L D, Peters R R, Delos S E, Young Y A T, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez L D, White J M. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72:3259–3267. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter E, Hill E, Hardwick M, Bhown A, Schwartz D E, Tizard R. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J Virol. 1983;46:920–936. doi: 10.1128/jvi.46.3.920-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito H, Watanabe S, Sanchez A, Whitt M A, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 34.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 35.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muga A, Neugebauer W, Hirama T, Surewicz W K. Membrane interactive and conformational properties of the putative fusion peptide of PH-30, a protein active in sperm-egg fusion. Biochemistry. 1994;33:4444–4448. doi: 10.1021/bi00181a002. [DOI] [PubMed] [Google Scholar]
- 37.Nieva J L, Nir S, Muga A, Goni F M, Wilschut J. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: different structural requirements for fusion and leakage. Biochemistry. 1994;33:3201–3209. doi: 10.1021/bi00177a009. [DOI] [PubMed] [Google Scholar]
- 38.Niidome T, Kimura M, Chiba T, Ohmori N, Mihara H, Aoyagi H. Membrane interaction of synthetic peptides related to the putative fusogenic region of PH-30α, a protein in sperm-egg fusion. J Pep Res. 1997;49:563–569. doi: 10.1111/j.1399-3011.1997.tb01164.x. [DOI] [PubMed] [Google Scholar]
- 39.Pereira F B, Goni F M, Muga A, Nieva J L. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: dose and sequence effects. Biophys J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry A C F, Gichuhi P M, Jones R, Hall L. Cloning and analysis of monkey fertilin reveals novel α subunit isoforms. Biochem J. 1995;307:843–850. doi: 10.1042/bj3070843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiao H, Armstrong T, Melikyan G B, Cohen F S, White J M. A specific point mutation at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rafalski M, Lear J D, DeGrado W F. Phospholipid interactions of synthetic fusion peptides representing the N-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 43.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz-Arguello M B, Goni F M, Pereira F B, Nieva J L. Phosphatidylinositol-dependent membrane fusion induced by a putative fusogenic sequence of Ebola virus. J Virol. 1998;72:1775–1781. doi: 10.1128/jvi.72.3.1775-1781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 46.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C. Studies of membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 49.White J M. Membrane fusion: the influenza paradigm. Cold Spring Harbor Symp Quant Biol. 1995;60:581–588. doi: 10.1101/sqb.1995.060.01.062. [DOI] [PubMed] [Google Scholar]
- 50.Whitt M A, Zagouras P, Crise B, Rose J K. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J Virol. 1990;64:4907–4913. doi: 10.1128/jvi.64.10.4907-4913.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfsberg T G, Straight P D, Gerena R L, Huovila A-P J, Primakoff P, Myles D G, White J M. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with adisintegrin and metalloprotease domain. Dev Biol. 1995;169:378–383. doi: 10.1006/dbio.1995.1152. [DOI] [PubMed] [Google Scholar]
- 52.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wool-Lewis R J, Bates P. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young J A, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L, Ghosh H P. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J Virol. 1994;68:2186–2193. doi: 10.1128/jvi.68.4.2186-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zingler K, Young J A T. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]