Fig. 3 |. Expression of a gene from a genomic locus in proximity to nuclear speckles leads to increased mRNA splicing.
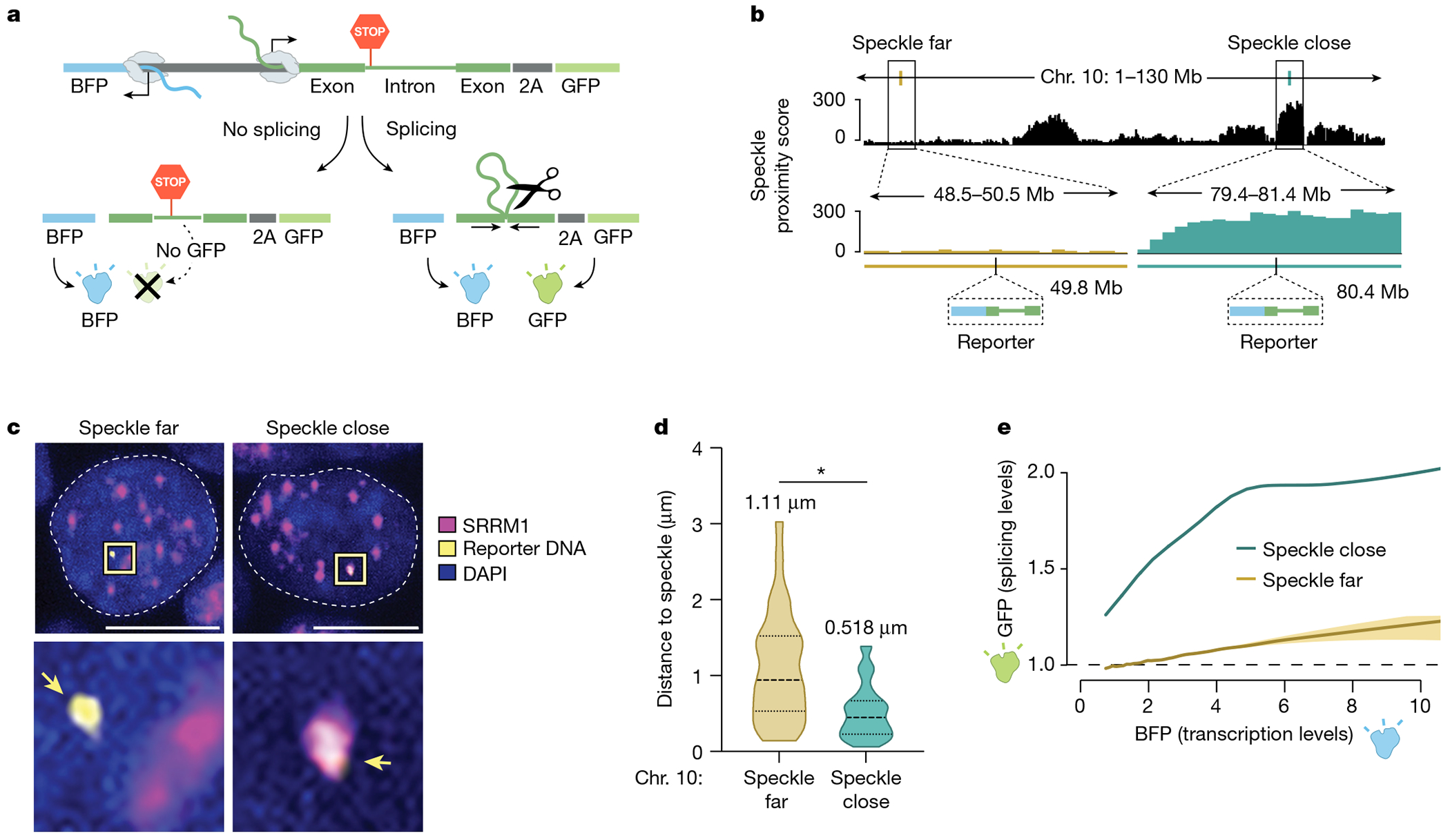
a, Schematic of the bidirectional reporter assay using a fluorescence-based readout. The splicing reporter contains an exon–intron–exon minigene fused in-frame to a GFP that is translated when spliced but not when unspliced. The spliced GFP reporter is linked to a bidirectionally transcribed BFP reporter that is expressed and translated regardless of whether it is spliced. b, SPRITE speckle proximity score (100-kb bin) for the two genomic regions on mouse chromosome 10 where the reporter was integrated. c, Representative images and zoom-in images of SRRM1 immunofluorescence combined with DNA FISH for cells containing the two integrated reporters. Scale bar, 10 μm. n = 85 cells over two biological replicates. d, Violin plots of the distance of DNA FISH spots of speckle-far and speckle-close integrated loci to the nearest nuclear speckle (immunofluorescence of SRRM1) across multiple cells (n = 28 speckle-far and n = 57 speckle-close). Mean distance is displayed above each distribution. *P < 0.0001. e, GFP expression (fluorescence intensity) as a function of BFP transcription levels for speckle-close and speckle-far integrated loci. We estimated the variation of these measurements using a bootstrap procedure from ten random bootstraps generated from these data (Methods). n = 744,019 cells analysed for speckle-close and n = 158,971 cells analysed for speckle-far. P value from two-sided t-test. Illustrations in a and b created by Inna-Marie Strazhnik, Caltech.
