Abstract
Previously, we and others have demonstrated that the process of reverse transcription of human immunodeficiency virus type 1 (HIV-1) is disturbed in nondividing macrophages and quiescent T lymphocytes. Here we analyzed which phase of the cell cycle in macrophages is crucial for early steps in the HIV-1 replication cycle. HIV-1 Ba-L-inoculated macrophages arrested early in the G1 phase by n-butyrate contained incomplete products of reverse transcription. In gamma-irradiated macrophages, reverse transcription was successfully completed but proviral integration could not be detected. In these cells, nuclear import was disturbed as reflected by the absence of two-long-terminal-repeat circles. In macrophages arrested late in G1 phase by aphidicolin or 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB), reverse transcription was unaffected. Proviral integration occurred efficiently in DRB-treated macrophages, whereas integrated proviral DNA could not be detected after aphidicolin treatment. Arrest at G2 phase of the cell cycle by nocodazole did not affect reverse transcription or proviral integration. Treatment of macrophages with hydroxyurea (HU), which reduces the intracellular deoxynucleoside triphosphate (dNTP) pool by blocking the de novo synthesis of dNTP, resulted in a dose-dependent inhibition of HIV-1 reverse transcription. This could partially be restored by the addition of nucleoside precursors. Addition of nucleoside precursors enhanced both reverse transcription and cell proliferation. However, the disturbed reverse transcription observed in the nonproliferating and n-butyrate-treated macrophages could not be restored by addition of nucleoside precursors. Similar to observations in quiescent T lymphocytes, incomplete proviral DNA species were arrested in the cytoplasm of the macrophages. Our results indicate that also in primary macrophages the intracellular nucleotide pools and other cellular factors that coincide with late G1 phase of the cell cycle may contribute to efficient reverse transcription and nuclear localization.
Oncoretroviruses depend on cycling cells for their replication. Passage through G1/S phase of the cell cycle is essential for efficient reverse transcription (6, 16, 26, 28, 29, 46). The same has been demonstrated for human immunodeficiency virus type 1 (HIV-1). In nondividing cells the process of reverse transcription is disturbed, resulting in incomplete proviral DNA species both in quiescent T cells (50, 51) and in nondividing primary macrophages (32, 33, 43). Early events in cellular activation and cell proliferation have been demonstrated to regulate reverse transcription (31, 34, 43).
For oncoretroviruses, it has also been demonstrated that nuclear transport of the large preintegration complex occurs only during mitosis, when the nuclear membrane is permeabilized (28, 35, 41, 46). In that respect, HIV-1 was described to be different. HIV-1 nuclear transport did not require nuclear membrane permeabilization as seen during mitosis, and its replication was therefore considered to be cell cycle independent. Indeed, several studies have suggested that the presence of nuclear localization signals (NLS) in Vpr, integrase, and the matrix protein of Gag supports active nuclear transport of the preintegration complex in an ATP-dependent process (4, 5, 13, 18–21, 27, 40, 48, 49). However, we and others could not confirm the NLS function of the matrix protein of gag (14, 15, 33).
Here we studied the phase of the cell cycle that supports early steps of the HIV-1 replication cycle in primary macrophages. In addition, we studied the effect of changes in deoxynucleoside triphosphate (dNTP) levels on the efficiency of HIV-1 reverse transcription.
MATERIALS AND METHODS
Isolation and culture of primary macrophages.
Monocytes were obtained from peripheral blood mononuclear cells (PBMC) of HIV-1-seronegative blood donors by centrifugal elutriation as described previously (11). Monocytes were cultured at a cell concentration of 106/ml in endotoxin-free Iscove's modified Dulbecco's medium supplemented with 10% pooled human serum, penicillin (100 U/ml), and streptomycin (100 mg/ml) and maintained at 37°C in a humidified atmosphere supplemented with 5% CO2. To obtain monocyte-derived macrophages (MDM), cells were allowed to adhere to plastic and cultured for 6 days to allow differentiation.
Inhibition of the cell cycle in primary macrophages.
MDM were cultured in the presence of cell cycle inhibitors or agents affecting the intracellular nucleotide pools starting 24 h before inoculation.
Nocodazole (0.5 to 100 μg/ml) and 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB; 0.5 to 100 μg/ml) were from Biomol Research Laboratories Inc. Aphidicolin (0.5 to 5 μg/ml) was from Merck. Hydroxyurea (HU; 0.1 to 5 mM) was from Calbiochem. n-Butyrate (1 to 10 mM) and the nucleoside precursors (dN) deoxyadenosine, deoxycytidine, deoxyguanidine, and thymidine (10 to 100 μM) were from Sigma.
To arrest MDM in G1 phase of the cell cycle, MDM were gamma irradiated with 3,000 rads.
Virus.
For cell-free infection of MDM, the macrophage-tropic HIV-1 variant Ba-L (25) was used. Infectious titers of the virus stock were quantified by determination of the 50% tissue culture infectious dose (TCID50). Before inoculation, the virus stocks were DNase (200 ng/ml; RQ1; Promega Corp., Madison, Wis.) treated for 45 min in medium supplemented with 6 mM MgCl2 and then filtered through a 0.22-μm-pore-size filter to distinguish newly synthesized proviral DNA. As a control for the efficacy of DNase treatment, MDM were incubated with 3′-azido-3′-deoxythymidine (AZT; 2.5 to 25 μM) starting 24 h prior to inoculation. For cell-free infection of MDM, an inoculum of 500 TCID50s/106 cells was used and 48 h after inoculation the MDM were washed with phosphate-buffered saline and harvested for DNA isolation.
Analysis of cell proliferation.
To allow separation of proliferating and nonproliferating MDM, cells were incubated with bromodeoxyuridine (BrdU; 20 mM; Sigma) for 48 h during inoculation. Cells were harvested and subsequently fixed with paraformaldehyde (2%, 10 min, 0°C) and ethanol (70%, 30 min, 0°C). DNA was denaturated with HCl (4 N, 30 min, 0°C), and incorporated BrdU was visualized by staining with a fluorescein isothiocyanate-labeled monoclonal antibody specific for BrdU (Becton Dickinson) as described previously (43). The BrdU-negative and BrdU-positive cell fractions were separated with a fluorescence-activated cell sorter (FACS).
DNA isolation.
DNA from cell fractions obtained after BrdU staining and FACS sorting was isolated with a QIAamp blood kit (Qiagen). Total DNA was extracted from MDM by lysis of 106 cells in buffer L6 (2) and subsequently precipitated with isopropanol and washed with 70% ethanol, after which the DNA was dissolved in 100 μl of water.
For the extraction of cytoplasmic and nuclear fractions, MDM were lysed in ice-cold lysis buffer containing 0.1 M NaCl, 10 mM Tris-HCl (pH 7.9), 0.5% Nonidet P-40, and 1.5 mM MgCl2. Cells were kept on ice for 10 min, and subsequently the cytoplasmic fraction and the nuclear fraction were separated by centrifugation (10 min at 2,700 × g). DNA was isolated from the cytoplasmic and nuclear fractions as described above.
HIV-1 DNA standards were prepared by serial dilutions of in vitro-, HIV-1-infected phytohemagglutinin (PHA)-stimulated PBMC in carrier DNA.
PCR analysis.
For all PCR primer sets, the MgCl2 concentration and thermocycling were optimized. A two-step nested PCR amplifying a conserved 125-bp sequence of the pol region was used to detect proviral pol DNA in DNA samples obtained from the FACS-sorted cell fraction. The HIV-1 pol region was amplified in the presence of 3 mM MgCl2; primer pair pol-D and pol-F were used in the first step and primer pair pol-E and pol-B were used in the second step (3).
To monitor the process of reverse transcription, a PCR assay amplifying the R/U5 fragment, a conserved pol fragment, and the R/PBS region, representing, respectively, an early, intermediate, and late product in reverse transcription, were used. The HIV-1 R/U5 region was amplified in the presence of 2 mM MgCl2 with primers M667 and AA55 (50). To amplify a conserved sequence of the HIV-1 pol region in the presence of 3 mM MgCl2, primer pair pol-D and pol-F was used (3). The HIV-1 R/PBS region was amplified in the presence of 3 mM MgCl2 with primers M667 and M661 (50). As a control for the general efficiency of PCR amplification of the DNAs, all DNAs were subjected to PCR analysis in the presence of 3 mM MgCl2 with primer set PC03 and PC04, amplifying part of the human β-globin gene (42). For amplification of regions in R/U5, pol, R/PBS, and the β-globin gene, the following PCR cycles were used: 1 cycle of 5 min at 95°C and 30 cycles of 1 min at 95°C, 1 min 30 s at 50°C, and 1 min 30 s at 72°C, followed by an extra 5-min extension at 72°C and subsequent cooling to 4°C.
To specifically detect integrated proviral DNA, a nested PCR with primers specific for ubiquitous repeats found in the human genome and HIV-1 was used. This Alu HIV-1 PCR was performed in the presence of 1.5 mM MgCl2 with primer pair Alu 278 (45) and p24-3I (3) in the first step and primer pair Alu 278 and M661 in the second step. For amplification, the following PCR cycles were used: 1 cycle of 5 min at 94°C, 3 min at 61°C, and 5 min at 72°C; 35 cycles of 30 s at 94°C, 1 min at 61°C, and 5 min at 72°C; and an extra 15-min extension at 72°C and subsequent cooling to 4°C. Alu HIV-1 PCR products were detected by PCR with M667 and AA55 to increase sensitivity.
To visualize positive PCR amplifications, PCR products were separated on 1% agarose gels, blotted onto GeneScreen membranes, and hybridized with [α-32P]dATP-, end-labeled oligonucleotide pol-C (3) for fragments amplified with pol-D-pol-F and pol-E-pol-B; LTR-B (12) for fragments amplified with M667-AA55, M667-M661, and nested HIV-1 Alu PCR; and RS06 (42) for fragments amplified with PC03-PC04. Dependent on the specific activity of the probes, autoradiography was performed for 1 to 24 h at −70°C with intensifying screens.
RESULTS
Effect of cell cycle arrest on HIV-1 reverse transcription and proviral integration.
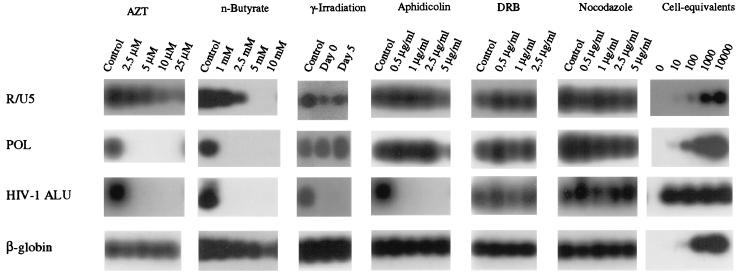
To determine which stage of the cell cycle is essential for early steps in the replication cycle, MDM were cultured in the presence of cell cycle inhibitors starting 24 h before inoculation. Alternatively, MDM were gamma irradiated directly after isolation or 24 h prior to inoculation. Total DNA was isolated 48 h after inoculation. Subsequently, the processes of reverse transcription and proviral integration were analyzed by PCR. PCRs amplifying the R/U5 region of the long terminal repeat (LTR) or a part of the pol gene were used to detect, respectively, early and intermediate products of reverse transcription, and a nested HIV-1 Alu PCR was used to detect integrated proviral DNA. To distinguish between newly synthesized proviral DNA and proviral DNA present in the inoculum, increasing concentrations of AZT (2.5 to 25 μM) were added to the MDM cultures 24 h before inoculation. R/U5 products were present in the AZT-treated cultures, whereas no pol products could be detected, thus confirming the detection of only newly synthesized proviral DNA in the untreated culture (Fig. 1).
FIG. 1.
Analysis of reverse transcription and proviral integration in growth-arrested MDM. MDM were treated with the following cell cycle inhibitors starting 24 h before inoculation: n-butyrate (1 to 10 mM), aphidicolin (0.5 to 5 μg/ml), DRB (0.5 to 2.5 μg/ml), and nocodazole (0.5 to 5 μg/ml). Alternatively, cells were subjected to gamma irradiation (3,000 rads) performed either directly after isolation (day 0) or 24 h before inoculation (day 5). MDM were inoculated at day 6 after isolation with 500 TCID50s of DNase-treated HIV-1 Ba-L. As a control for the efficacy of the DNase treatment, zidovudine (2.5 to 25 μM)-treated MDM were inoculated with Ba-L and analyzed for the presence of proviral DNA. DNA was extracted 48 h after inoculation and was subjected to PCR analysis. To specifically detect integrated proviral DNA, a nested HIV-1 Alu PCR with primers specific for ubiquitous repeats found in the human genome and HIV-1 was used. As a control for the general efficiency of PCR amplification of the DNAs, all DNAs were subjected to PCR analysis amplifying part of the human β-globin gene. The results are representative of four independent experiments. Serial dilutions of in vitro-, HIV-1-infected PHA-stimulated PBMC in carrier DNA were used as DNA standards.
First, we analyzed the effect of n-butyrate, which arrests early in the G1 phase of the cell cycle (8), on early events in the virus replication cycle. When MDM were treated with increasing concentrations of n-butyrate (1 to 10 mM), a dose-dependent inhibition of reverse transcription, as demonstrated by decreasing amounts of R/U5 proviral DNA, could be observed. pol proviral DNA could not be demonstrated in n-butyrate-treated cells (Fig. 1). When autoradiograms were overexposed, pol proviral DNA could be detected in MDM treated with 1 mM n-butyrate.
Previously, we demonstrated that HIV-1 is unable to replicate in primary macrophages arrested at G1 phase of the cell cycle by gamma irradiation (43). Here, we determined the level at which HIV-1 replication is disturbed in gamma-irradiated primary macrophages. Gamma irradiation of MDM with 3,000 rads was performed at day 0 (directly after cell isolation) or at day 5, which was 24 h before inoculation. In gamma-irradiated MDM, normal signals were obtained for proviral DNA representing either the R/U5 region of the LTR or a pol fragment. However, no signal was obtained with the HIV-1 Alu PCR, indicating the absence of integrated proviral DNA (Fig. 1).
Next, we analyzed the effect of cell cycle inhibitors which block late in the G1 phase. Aphidicolin specifically inhibits DNA polymerases α and δ, whereas DRB blocks RNA polymerase II. Normal reverse transcription as demonstrated by the presence of R/U5 and pol proviral DNA could be detected in MDM treated with increasing concentrations of aphidicolin (0.5 to 5 μg/ml) or DRB (0.5 to 2.5 μg/ml). However, a positive signal for the HIV-1 Alu PCR representing integrated proviral DNA was observed in the DRB-treated MDM but not in the aphidicolin-treated MDM (Fig. 1).
Finally, MDM were treated with increasing concentrations of nocodazole (0.5 to 5 μg/ml), which inhibits microtubule depolymerization and consequently arrests cells in the G2 phase of the cell cycle. Even in the presence of 5 μg of nocodazole per ml, the amount of R/U5 and pol proviral DNA was comparable to that in the control MDM, indicating normal reverse transcription. Furthermore, a positive signal in the HIV-1 Alu PCR pointed to efficient proviral integration these MDM (Fig. 1).
Effect of cellular dNTP synthesis on reverse transcription.
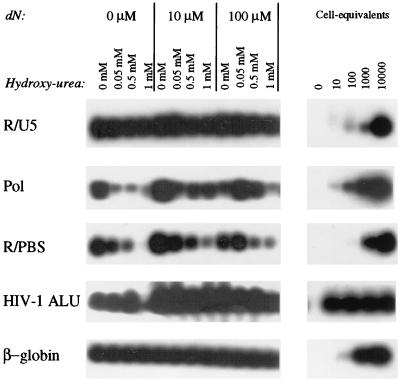
HIV-1 proviral DNA synthesis depends on intracellular dNTP pools. Here, we studied the effect of changes in the intracellular dNTP pools on reverse transcription. HU decreases the intracellular dNTP pool by blocking de novo dNTP synthesis through inhibition of the enzyme ribonucleotide reductase. First we analyzed the effect of HU treatment of MDM on reverse transcription. In MDM treated before inoculation with increasing concentrations of HU (0.05 to 1 mM), normal levels of R/U5 proviral DNA could be observed. However, the levels of intermediate and late products of reverse transcription as demonstrated by the amounts of pol and R/PBS proviral DNA were decreased (Fig. 2). Although the process of reverse transcription was diminished in MDM treated with 0.05 and 0.5 mM HU, proviral integration could still be observed using the nested HIV-1 Alu PCR (Fig. 2). In MDM treated with 3 to 5 mM HU, only R/U5 proviral DNA could be detected, indicative of initiation of reverse transcription (data not shown).
FIG. 2.
Analysis of reverse transcription and proviral integration in HU-treated arrested MDM in the presence or absence of dN. MDM were treated with HU (0.05 to 1 mM) in combination with dN (10 to 100 μM) starting 24 h before inoculation. For further details on the PCR analysis, see the legend to Fig. 1. An additional PCR was performed to detect the R/PBS region. The results are representative of four independent experiments.
HU blocks de novo dNTP synthesis but also stimulates dNTP synthesis through the salvage pathway by activating thymidine kinase and deoxycytodine kinase (22–24). Here we analyzed whether addition of extracellular dN could restore HU-induced inhibition of reverse transcription. MDM were treated with increasing concentrations of HU in the presence of additional dN (10 to 100 μM) starting 24 h before inoculation. PCR analysis revealed that addition of low concentrations of dN during inoculation increased the levels of pol and R/PBS proviral DNA, which is indicative of enhanced reverse transcription (Fig. 2). Addition of extracellular dN to HU-treated MDM could partially restore the process of reverse transcription, as demonstrated by an enhanced signal for pol and R/PBS proviral DNA and subsequent proviral integration (Fig. 2).
Effect of dN on HIV-1 reverse transcription in nonproliferating macrophages.
In HU-treated MDM, the addition of extracellular dN resulted in enhanced reverse transcription. This suggests that the probably low dNTP levels in the nonproliferating MDM could also explain the low efficiency of reverse transcription. We therefore analyzed whether addition of dN could overcome the block in reverse transcription in the nonproliferating subfraction of MDM (32, 33, 43).
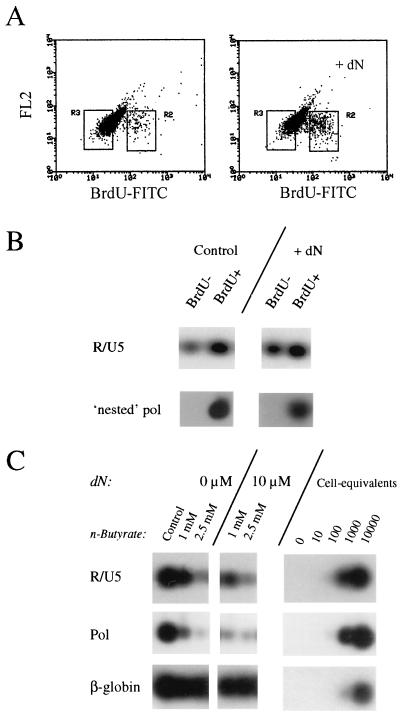
Five-day-cultured MDM were supplemented with 10 μM dN and exposed to a DNase-treated Ba-L inoculum of 1,000 TCID50s/106 MDM. To discriminate between proliferating and nonproliferating macrophages during inoculation, thymidine in the dN mixture was replaced by BrdU. Forty-eight hours after inoculation, cells were harvested and BrdU incorporation was visualized by a BrdU-specific monoclonal antibody and subsequent FACS analysis. When MDM were cultured in medium alone, 3.3% of the cells had traversed S phase. BrdU staining of MDM cultured in the presence of 10 μM dN revealed an increase of proliferating cells up to 10.6% (Fig. 3A).
FIG. 3.
Analysis of reverse transcription in nonproliferating MDM and n-butyrate-treated MDM in the presence of dN. (A) MDM proliferation was analyzed by BrdU incorporation in the presence or absence of dN. (B) MDM were sorted into BrdU-negative (gate R3) and BrdU-positive (gate R2) populations, and these cell fractions were analyzed for the presence of HIV-1 proviral DNA. (C) MDM were treated with n-butyrate (1 and 2.5 mM) in the presence or absence of dN (10 μM) starting 24 h before inoculation. Proviral DNA synthesis was analyzed by R/U5 and pol PCR. Serial dilutions of in vitro-, HIV-1-infected PHA-stimulated PBMC in carrier DNA were used as DNA standards.
To analyze whether addition of dN could support HIV-1 reverse transcription also in nonproliferating cells, BrdU-negative and BrdU-positive cells were separated by FACS sorting (Fig. 3A, gates R3 and R2, respectively). BrdU-negative and BrdU-positive cell fractions were mixed with HIV-1-negative PBMC to improve DNA isolation, and DNA cell equivalents of 4 × 104 of the BrdU-negative and 4 × 103 of the BrdU-positive cell fractions obtained from the control and the dN-treated macrophages were used for PCR analysis in the presence of proviral DNA. The presence of proviral DNA corresponding to the R/U5 region could be demonstrated in the BrdU-negative as well as the BrdU-positive cell fractions, whereas pol proviral DNA was observed only in the BrdU-positive populations of both the control and the dN-treated cultures (Fig. 3B). This indicates that in dN-treated MDM efficient reverse transcription is still restricted to the proliferating subpopulation. Next, we analyzed whether the incomplete reverse transcription observed in n-butyrate-treated MDM was a consequence of low intracellular dNTP pools. Macrophages were treated with n-butyrate (1 and 2.5 mM) in the presence or absence of dN (10 μM) starting 24 h before inoculation. PCR analysis for the presence of R/U5 and pol proviral DNA confirmed that the process of reverse transcription was inhibited in n-butyrate-treated MDM and that it could not be restored by addition of dN (Fig. 3C).
Incomplete proviral DNA species are arrested in the cytoplasm of macrophages.
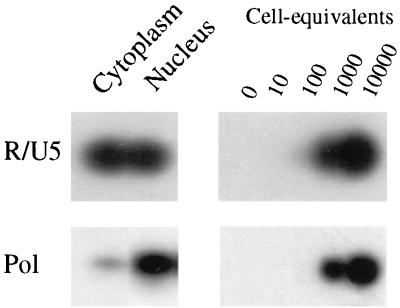
Previously it has been demonstrated that in quiescent T lymphocytes, incomplete proviral DNA species were arrested in the cytoplasm (50, 51). Here we studied whether this was also the case in primary macrophages. DNA was extracted from cytoplasmic and nuclear fractions obtained from MDM 48 h after inoculation and subsequently analyzed for the presence of proviral DNA. Early products of reverse transcription as demonstrated by R/U5 PCR were equally distributed among the cytoplasmic and nuclear fractions, whereas pol proviral DNA was predominantly present in the nuclear fraction of the macrophages (Fig. 4). Equal amounts of R/U5 and pol proviral DNA products could be demonstrated in the nuclear fraction. This may imply that elongated reverse transcription products are immediately transported to the nucleus and that the incomplete proviral DNA species are arrested in the cytoplasm.
FIG. 4.
Analysis of proviral DNA in nuclear and cytoplasmic fractions of MDM. Cell fractions were isolated from MDM 48 h after inoculation. DNA extracted from these fractions was analyzed for the presence of HIV-1 proviral DNA. Serial dilutions of in vitro-, HIV-1-infected PHA-stimulated PBMC in carrier DNA were used as DNA standards.
DISCUSSION
We previously have demonstrated that a small subpopulation of primary macrophages is able to proliferate and that only this subpopulation supports reverse transcription upon inoculation with HIV-1 (32, 43). Here we analyzed which phase of the cell cycle is essential for efficient support of reverse transcription in primary macrophages. When MDM were arrested early in G1 phase of the cell cycle by n-butyrate, the process of reverse transcription was inhibited in a dose-dependent manner. Gamma-irradiated MDM arrested in G1 phase of the cell cycle (30) efficiently supported reverse transcription, but proviral integration could not be observed, which is in agreement with the absence of virus replication as demonstrated previously (43). Efficient virus replication has been demonstrated in gamma-irradiated T-cell lines (5, 36). However, in contrast to the G1 arrest of primary cells, gamma irradiation arrests cell lines in G2 phase of the cell cycle, thus indicating that only later stages of cell cycle do provide cellular conditions for nuclear transport and proviral integration. We were unable to show accumulation of two-LTR circles in gamma-irradiated macrophages (data not shown), which may suggest that not proviral integration but already the process of nuclear transport is disturbed. When MDM were arrested late in G1 phase of the cell cycle by aphidicolin, reverse transcription was unaffected but proviral integration was disturbed. During proviral integration, integrase joins the viral DNA and the host cell DNA, after which the gaps in the host cell DNA are filled in by the host DNA repair mechanism (17, 47). Aphidicolin specifically inhibits DNA polymerases α and δ, which are involved in the host cell DNA repair mechanism (7, 10). Thus, by interfering with the DNA repair mechanism, aphidicolin probably prevents proviral integration. Previously, we demonstrated that aphidicolin treatment during inoculation did not interfere with HIV-1 replication (33, 43). In these studies, aphidicolin was removed 48 h after inoculation, which indicates that the DNA repair mechanism can be restored upon removal of aphidicolin and that proviral integration can subsequently be completed.
Arrest of MDM late in G1 phase or G2 phase of the cell cycle by DRB or nocodazole had no effect on reverse transcription or proviral integration.
Previous studies have demonstrated that the presence of a functional Vpr and of NLS in the MA protein of gag and integrase supports nuclear transport of the preintegration complex in the absence of cell proliferation (4, 5, 13, 18–21, 27, 40, 48, 49). Despite the use of a wild-type virus variant, we observed cytoplasmic arrest of full-length proviral DNA in gamma-irradiated macrophages, whereas efficient proviral integration and thus nuclear transport were observed when MDM were arrested late in G1 phase of the cell cycle. This indicates that not only reverse transcription but also nuclear transport relies on cellular conditions coinciding with late G1 phase of the cell cycle. In agreement, nuclear transport of wild-type HIV-1 is also observed in growth-arrested but activated T cells and not in quiescent T lymphocytes (5). Apparently, the ATP levels required for the active nuclear transport of the preintegration complex are not present in resting macrophages and T lymphocytes.
HIV-1 entirely depends on the intracellular dNTP pool for DNA synthesis, and the observed low dNTP pools in quiescent cells (22–24) might explain the disturbed reverse transcription in nondividing cells. Treatment with HU, which blocks de novo dNTP synthesis, has been shown to inhibit HIV-1 replication in acutely infected PBMC and primary macrophages (37). Here we demonstrated that in agreement with the observation in PBMC (37), HU treatment of MDM interferes with reverse transcription. The impaired HIV-1 proviral DNA synthesis could partially be restored by addition of dN, which are phosphorylated by nucleoside-specific kinases as part of the salvage dNTP synthesis. In agreement with a previous study by O'Brien et al. (39), the addition of dN alone also resulted in enhanced reverse transcription in MDM, which coincided with the enhancement of cell proliferation. Addition of extracellular dN was unable to support reverse transcription in nonproliferating macrophages and in macrophages arrested in G1 phase of the cell cycle by n-butyrate. This indicates that cellular factors other than nucleotide pools are required for HIV-1 reverse transcription in primary macrophages. Furthermore, we demonstrated that similar to previous observations in quiescent T lymphocytes (50, 51), incomplete proviral DNA species were arrested in the cytoplasm of the macrophages.
Transduction of nondividing cells using HIV-1-based retroviral vectors has been demonstrated to have a very low efficiency, due to inefficient reverse transcription (38). The limiting dNTP pools in nondividing cells can be bypassed by in vitro induction of intravirion reverse transcription (52–54), which has been demonstrated to enhance gene delivery by HIV-1-based retroviral vectors in neural cells (1). However, quiescent T lymphocytes inoculated with virus in which endogenous reverse transcription was induced still require stimulation to support virus replication (9), which is in agreement with our present observations that cellular activation is also essential for a post-reverse-transcription step.
Recently, it has been demonstrated that expression of nuclear factor of activated T lymphocytes (NFAT) supports efficient reverse transcription in quiescent T lymphocytes (31). NFAT expression is an early event in T-lymphocyte activation which initiates a cascade of events leading to suitable cellular conditions for reverse transcription and proviral integration without the induction cell proliferation (31). NFAT expression has also been demonstrated in primary macrophages (44). Whether NFAT expression in primary macrophages also creates appropriate cellular conditions for reverse transcription and nuclear transport remains to be established. The identification of the actual cellular cofactors involved in reverse transcription and nuclear transport will be of great importance for the use of HIV-1-based retroviral vectors for gene delivery in nondividing cells of different origins.
REFERENCES
- 1.Blömer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van der Noordaa J. A rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1991;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruisten S M, Koppelman M H G M, Van der Poel C L, Huisman J G. Enhanced detection of HIV-1 sequences using PCR and a liquid hybridization technique. Vox Sang. 1991;61:24–29. doi: 10.1111/j.1423-0410.1991.tb00922.x. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen I S Y, Temin H M. Establishment of infection by spleen necrosis virus: inhibition in stationary cells and the role of secondary infection. J Virol. 1982;41:183–191. doi: 10.1128/jvi.41.1.183-191.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciarrocchi G, Jose J G, Linn S. Further characterization of a cell-free system for measuring replicative and repair DNA synthesis with cultured human fibroblasts and evidence for the involvement of DNA polymerase alpha in DNA repair. Nucleic Acids Res. 1979;7:1205–1219. doi: 10.1093/nar/7.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darzynkiewicz Z, Traganos F, Xue S B, Melamed M R. Effect of n-butyrate on cell cycle progression and in situ chromatin structure of L1210 cells. Exp Cell Res. 1981;136:279–293. doi: 10.1016/0014-4827(81)90006-9. [DOI] [PubMed] [Google Scholar]
- 9.Dornadula G, Zhang H, Shetty S, Pomerantz R J. HIV-1 virions produced from replicating peripheral blood lymphocytes are more infectious than those from nonproliferating macrophages due to higher levels of intravirion reverse transcripts: implications for pathogenesis and transmission. Virology. 1999;253:10–16. doi: 10.1006/viro.1998.9465. [DOI] [PubMed] [Google Scholar]
- 10.Dresler S L, Gowans B J, Robinson-Hill R M, Hunting D J. Involvement of DNA polymerase delta in DNA repair synthesis in human fibroblasts at late times after ultraviolet irradiation. Biochemistry. 1988;27:6379–6383. doi: 10.1021/bi00417a028. [DOI] [PubMed] [Google Scholar]
- 11.Figdor C G, Bont W S, Touw I, De Roos J, Roosnek E E, De Vries J. Isolation of functionally different human monocytes by counter-flow centrifugation elutriation. Blood. 1982;60:46–54. [PubMed] [Google Scholar]
- 12.Fouchier R A M, Brouwer M, Kootstra N A, Huisman J G, Schuitemaker H. HIV-1 macrophage-tropism is determined at multiple steps of the viral replication cycle. J Clin Investig. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Albright A V, Gonzalez-Scarano F, Malim M H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6039. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed E O, Englund G, Martin M. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritsch E F, Temin H M. Inhibition of viral DNA synthesis in stationary chicken embryo fibroblasts infected with avian retroviruses. J Virol. 1977;24:461–469. doi: 10.1128/jvi.24.2.461-469.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 18.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of non-dividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 21.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 22.Gao W-Y, Agbaria R, Driscoll J S, Mitsuya H. Divergent anti-human immunodeficiency virus activity and anabolic phosphorylation of 2′,3′-dideoxynucleoside analogs in resting and activated human cells. J Biol Chem. 1994;269:12633–12638. [PubMed] [Google Scholar]
- 23.Gao W-Y, Cara A, Gallo R C, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao W-Y, Shirasaka T, Johns D G, Broder S, Mitsuya H. Differential phosphorylation of azidothymidine, dideoxycytidine, and dideoxyinosine in resting and activated peripheral blood mononuclear cells. J Clin Investig. 1993;91:2326–2333. doi: 10.1172/JCI116463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gartner S, Markovits P, Markovits D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 26.Harel J, Rassart E, Jolicoeur P. Cell cycle dependence of synthesis of unintegrated viral DNA in mouse cells newly infected with murine leukemia virus. Virology. 1981;110:202–207. doi: 10.1016/0042-6822(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 27.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu T W, Taylor J M. Effect of aphidicolin on avian sarcoma virus replication. J Virol. 1982;44:493–498. doi: 10.1128/jvi.44.2.493-498.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphries E H, Temin H M. Requirement for cell division for initiation of transcription of Rous sarcoma virus RNA. J Virol. 1974;14:531–546. doi: 10.1128/jvi.14.3.531-546.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Kacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 31.Kinoshita S, Chen B K, Kaneshima H, Nolan G P. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 32.Kootstra N A, Schuitemaker H. Proliferation dependent replication in primary macrophages of macrophage-tropic HIV-1 variants. AIDS Res Hum Retrovir. 1998;14:339–345. doi: 10.1089/aid.1998.14.339. [DOI] [PubMed] [Google Scholar]
- 33.Kootstra N A, Schuitemaker H. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of GAG and Vpr is comparable to wild type HIV-1 in primary macrophages. Virology. 1999;253:170–180. doi: 10.1006/viro.1998.9482. [DOI] [PubMed] [Google Scholar]
- 34.Korin Y D, Zack J A. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Simm M, Potash M J, Volsky D J. Human immunodeficiency virus type 1 DNA synthesis, integration, and efficient viral replication in growth-arrested T cells. J Virol. 1993;67:3969–3977. doi: 10.1128/jvi.67.7.3969-3977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lori F, Malykh A, Cara A, Sun D, Weinstein J N, Lisziewicz J, Gallo R C. Hydroxyurea as an inhibitor of human immunodeficiency virus type 1 replication. Science. 1994;266:801–805. doi: 10.1126/science.7973634. [DOI] [PubMed] [Google Scholar]
- 38.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien W A, Namazi A, Kalhor H, Mao S H, Zack J A, Chen I S Y. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roe T Y, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 43.Schuitemaker H, Kootstra N A, Fouchier R A M, Hooibrink B, Miedema F. Productive HIV-1 infection of macrophages restricted to the cell fraction with proliferative capacity. EMBO J. 1994;13:5929–5936. doi: 10.1002/j.1460-2075.1994.tb06938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw K T Y, Ho A M, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan P G. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc Natl Acad Sci USA. 1995;92:11205–11209. doi: 10.1073/pnas.92.24.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonza S, Maerz A, Deacon N, Meanger J, Mills J, Crowe S. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J Virol. 1996;70:3863–3869. doi: 10.1128/jvi.70.6.3863-3869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varmus H E, Padgett T, Heasley S, Simon G, Bishop J M. Cellular functions are required for synthesis and integration of avian sarcoma virus-specific DNA. Cell. 1977;11:307–319. doi: 10.1016/0092-8674(77)90047-2. [DOI] [PubMed] [Google Scholar]
- 47.Vink C, Groenink M, Elgersma Y, Fouchier R A M, Tersmette M, Plasterk R H A. Analysis of the junctions between human immunodeficiency virus type 1 proviral DNA and human DNA. J Virol. 1990;64:5626–5627. doi: 10.1128/jvi.64.11.5626-5627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 51.Zack J A, Haislip A M, Krogstad P, Chen I S Y. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Dornadula G, Wu Y, Havlir D, Richman D D, Pomerantz R J. Kinetic analysis of intravirion reverse transcription in the blood plasma of human immunodeficiency virus type 1-infected individuals: direct assessment of resistance to reverse transcriptase inhibitors in vivo. J Virol. 1996;70:628–634. doi: 10.1128/jvi.70.1.628-634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, Zhang Y, Spicer T P, Abbott L Z, Abbott M, Poiesz B J. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retrovir. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]