Abstract
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch transactivate genes by interacting with the transcription factor RBP-Jκ. The viral protein EBNA2 may hence be regarded as a functional equivalent of an activated Notch receptor. Until now, nothing has been known about the physiological role of Notch signaling in B cells. Here we investigated whether activated Notch can induce the same phenotypic changes as EBNA2 in Burkitt's lymphoma cells. An estrogen receptor fusion protein of the intracellular part of mouse Notch 1 (mNotch1-IC), mimicking in the presence of estrogen a constitutively active Notch receptor, was stably transfected into the Burkitt's lymphoma cell lines BL41-P3HR1 and HH514. Northern blot analysis revealed that the LMP2A gene is induced by Notch-IC in the presence of estrogen, whereas increased expression of LMP1 could be detected only if cycloheximide was simultaneously added. Concerning the cellular genes regulated by EBNA2, Notch-IC was able to upregulate CD21 but not CD23 expression. Immunoglobulin μ (Igμ) expression, which is downregulated by EBNA2, was also negatively regulated by Notch-IC. Similarly to EBNA2, Notch-IC was able to repress c-myc expression, which is under the control of the immunoglobulin heavy-chain locus in Burkitt's lymphoma cells with a t(8;14) translocation. The data show that Notch-IC is able to participate in gene regulation in B cells.
Epstein-Barr virus (EBV), a human herpesvirus, is associated with several human malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, T-cell lymphoma, gastric carcinoma, Hodgkin lymphoma, and immunoblastic lymphoma in immunocompromised individuals. The virus has the ability to immortalize primary B cells in vitro. In these immortalized lymphoblastoid cell lines, only a few viral genes are expressed, coding for six nuclear proteins (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNA-LP) and three latent membrane proteins (LMP1, LMP2A, and LMP2B) (for a review, see reference 35). Five of the nuclear antigens (EBNA1, -2, -3A, and -3C seem to be absolutely required for B-cell immortalization (10, 23, 32, 68), whereas EBNA-LP and LMP2A seem to affect the efficiency of the process (6, 23, 47).
EBNA2 plays a pivotal role in B-cell immortalization, since a natural occurring EBV mutant, the P3HR1 strain, carrying a deletion of the EBNA2 gene, has lost the ability to transform primary B cells. Reintroduction of the EBNA2 gene in the viral genome by homologous recombination or complementation can restore the immortalizing capacity of the virus (23). EBNA2 contributes to B-cell immortalization most likely by its ability to act as transcriptional modulator of cellular and viral gene expression. It activates the transcription of the B-cell activation markers CD21 and CD23 (9, 11, 70) and the tyrosine kinase c-Fgr (37) and downregulates the expression of the immunoglobulin heavy-chain locus (Igμ) (30). In addition, EBNA2 transactivates the viral promoters of the three latent membrane proteins LMP1, LMP2A, and LMP2B and the Cp promoter, which regulates the transcription of the EBNA genes (17, 64, 75, 76). EBNA2 does not bind to DNA directly (45, 77) but is recruited to EBNA2-responsive elements by interacting with the transcriptional factors RBP-Jκ (21, 25, 69, 78) and PU.1/Spi-1 (30a, 42). RBP-Jκ binding sites are present in all EBNA2-regulated promoters known so far, whereas binding of PU.1 could be identified only within the LMP1 promoter. Binding of RBP-Jκ is essential but not sufficient to confer EBNA2 responsiveness to the EBNA2-regulated promoters (49). RBP-Jκ was originally purified and characterized by Matsunami et al. (48) and Hamaguchi et al. (22). The protein is highly conserved in evolution from nematodes to humans. In Drosophila, the homologous gene is known as suppressor of hairless (SuH). RBP-Jκ/SuH acts downstream of the receptor Notch. Activation of the Notch receptor by binding of its ligand Delta, Jagged, or Serrate (in Drosophila) leads to proteolytic cleavage of the receptor at the inner side of the membrane (59). The intracellular domain (Notch-IC) is then translocated to the nucleus, where it activates genes by interacting with RBP-Jκ (34, 39, 43, 62). EBNA2 may thus be regarded as a functional homolog of the activated Notch protein. In Drosophila, the role of Notch signaling in the development of the nervous system has been extensively studied. Notch signaling plays an important role in cell fate decisions. It prevents cells from adopting the neural fate and directs them toward the epidermal fate (24). Notch is expressed throughout Drosophila development and influences cell fate decision not only in the nervous system but also in many other tissues (reviewed in references 2 and 36). Notch signals influence developmental processes also in vertebrates. Four homologs, Notch1 (Tan1), Notch2, Notch3, and Notch4 (int3) have been cloned in mice and humans (16, 41, 57, 72, 73). Notch1 plays an essential role during embryogenesis, since Notch1-/- mice show growth retardation at day 9.5 and die before day 11.5 of gestation with widespread cell death (65). Additionally, Notch signaling is implicated in myogenesis and neurogenesis (13a, 38, 55). Notch is also believed to play a role in cell type determination at multiple steps of hematopoietic differentiation. Notch1 is expressed in human bone marrow hematopoietic precursors and may be involved in the renewal and differentiation of these cells (51). Notch1 and Notch2 were reported to inhibit granulocytic differentiation (4, 52). In T cells, Notch1 affects the choice between CD4 and CD8 cells and between alpha/beta versus gamma/delta T-cell lineages (58, 71). Recently it was shown that Notch signalling is critically involved in the maturation of thymocytes (13, 56a). Until now, nothing has been known about Notch signaling in B cells.
EBNA2 is involved in activation processes of B cells by upregulating the B-cell activation markers CD21 and CD23. Concomitantly with the upregulation of CD21 and CD23, expression of Igμ is downregulated. Since both EBNA2 and activated Notch interact with the transcription factor RBP-Jκ, leading to gene activation, it was of interest to see whether an activated Notch receptor can induce the same phenotypic changes in B cells as EBNA2. Therefore, we stably transfected an expression vector coding for a Notch-IC–estrogen receptor (ER) fusion protein in EBV-positive but EBNA2-negative Burkitt's lymphoma cell lines and examined whether the EBNA2-regulated cellular (CD21, CD23, and Igμ) and viral (LMP1 and LMP2A) genes are also regulated by Notch-IC. We show here that Notch1-IC is active in B cells and participates in gene regulation in a manner very similar but not identical to that of EBNA2.
MATERIALS AND METHODS
Cell lines and culture conditions.
The cell line BL41-P3HR1 was obtained after infection of the EBV-negative Burkitt's lymphoma cell line BL41 with the P3HR1 strain of EBV (9). HH514 is a single-cell clone of P3HR1 (56). The cell line BL41-P3HR1-5E was obtained after stable transfection of BL41-P3HR1 cells with a plasmid encoding an ER-EBNA2 fusion protein (33).
The cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml). For induction of Notch-IC and EBNA2, β-estradiol was added to the cell culture medium at a final concentration of 1 μM. All cell lines were incubated at 37°C in an atmosphere of 5% CO2 and diluted 1:3 with fresh medium twice a week.
Plasmid constructs.
The mouse (m) Notch1-IC–ER expression plasmid was generated by ligation of a KpnI/ApaII fragment of the pSG5 mNotch1-IC expression vector (28) with a KpnI/BamHI fragment of the c-Myc-ER expression plasmid HH275-3 (26). The ApaII and BamHI sites were refilled with Klenow polymerase.
The LMP1 promoter-luciferase construct LMPluc0 has been described by Laux et al. (42), the LMP2A promoter luciferase construct pTP1(-804) has been described by Zimber-Strobl et al. (77), and the RBP-Jκ multimer luciferase construct Ga 981-16 has been described by Strobl et al. (60).
Stable transfection.
BL41-P3HR1 and HH514 cells were transfected by electroporation using a Bio-Rad gene pulser at 960 μF and 250 V. After transfection, the cells were seeded in 96-well flat-bottom plates and selected in RPMI medium supplemented with hygromycin B (150 μg/ml), antioxidant mix (7, 18), and HEPES buffer (1%).
Transient transfection.
Electroporation was carried out as described previously (77), with slight modifications. Cells were electroporated in RPMI 1640 medium without fetal calf serum at room temperature; 500 μl of fetal calf serum was added to the cells immediately after electroporation. Cells were resuspended in 10 ml of prewarmed RPMI 1640 containing 10% fetal calf serum and standard supplements.
Luciferase assays.
Cells were harvested and lysed as described previously (49). Ten microliters of each probe was mixed with 150 μl of test buffer (25 mM glycylglycine [pH 7.8], 5 mM ATP, 15 mM MgSO4) on a 96-well plate. After addition of 100 μl of 11 mM luciferin in 0.5 M Tris-HCl (pH 7.8) to each reaction, the bioluminescence in relative light units (RLU) was measured with a Micro Lumat LB 96 P (Berthold, Wildbach, Germany). The RLU values were subsequently standardized by the protein concentrations of the corresponding cell lysates.
Protein immunoblots.
For Western blot analysis, cellular extracts were prepared by sonification in H8 lysis buffer (20 mM Tris [pH 7.0], 2 mM EGTA, 2 mM EDTA, 6 mM β-mercaptoethanol, 50 mM NaF, 100 mM NaCl, 1% sodium dodecyl sulfate [SDS]). Protein concentrations were determined, and equal amounts of protein separated on an SDS–10% polyacrylamide gel. Proteins were transferred onto nitrocellulose filters (Amersham Hybond ECL), and the filters were incubated with an antibody raised against the ER binding domain (Dianova, Hamburg, Germany). Immunoreactive proteins were detected by peroxidase-coupled secondary antibodies and enhanced chemoluminescence (ECL system; Amersham).
Northern blot analysis.
Northern blot analysis was performed as described previously (30). As radioactive probes, the following DNA fragments were used: (i) for LMP1, a 1.5-kb XhoI/Xba fragment of the LMP1 gene; (ii) for LMP2A, 2.0 kb of cDNA (76); (iii) for CD21, 1.6 kb of cDNA (74); (iv) for c-Myc, a 1.3-kb EcoRI/HindIII fragment of the c-myc cDNA (1); (v) for Igμ, a 1.2-kb EcoRI-EcoRI fragment of the constant region (15).
Signal intensities were determined by high-resolution scanning of the autoradiographs and subsequent quantification with the software TINA (Fujix BAS 1000 system; Fuji). Signal intensities of one blot were standardized by dividing all determined data by the one with the lowest measurable intensity. Lanes marked with a minus displayed no detectable signal (background level).
FACS (fluorescence-activated cell sorting) analysis.
Cells (106) were incubated with an excess of unlabeled mouse monoclonal antibodies recognizing CD21, CD23, and surface IgM (sIgM; Dianova). Fluorescein isothiocyanate-conjugated goat anti-mouse F(ab′)2 fragments (DAKO, Hamburg, Germany) were used as secondary antibody for staining of positive cells. Dead cells were identified and excluded from the analysis after propidium iodide staining (0.1 μg/ml). Samples were analyzed with a FACScan (Becton Dickinson).
RESULTS
Stable transfection of a conditional mNotch1-IC gene in Burkitt's lymphoma cells.
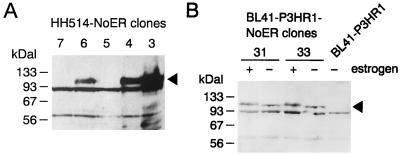
To study whether Notch signaling can induce expression of EBNA2-regulated genes, we have stably introduced a cDNA coding for mNotch1-IC in established B-cell lymphoma cell lines. Several studies have demonstrated that truncated Notch molecules harboring only the intracytoplasmic domain behave as constitutive active forms of Notch (20, 44, 61). Therefore, expression of Notch-IC should result in effects comparable to those induced after activation of Notch by binding of its ligand Delta or Jagged. To be able to analyze the same cell clone in the absence and presence of activated Notch, we fused the carboxy terminus of Notch1-IC with the ER hormone binding domain, rendering protein function dependent on the presence of estrogen. The mNotch1-IC–ER fusion under transcriptional control of the simian virus 40 early promoter/enhancer was cloned onto an episomal oriP vector encoding the gene for hygromycin phosphotransferase (63). The P3HR1 virus-infected Burkitt's lymphoma cell lines BL41-P3HR1 and P3HR1 (subclone HH514) were transfected with the mNotch1-IC–ER expression plasmid and selected by hygromycin. Single-cell clones were analyzed for mNotch1-IC–ER expression by immunostaining of Western blots with a monoclonal antibody that recognizes an epitope within the ER binding domain. Nineteen BL41-P3HR1 and 18 HH514 single-cell clones were analyzed; of these, three and seven clones, respectively, expressed the Notch1-IC–ER protein. In Fig. 1, Notch1-IC–ER expression is shown in three positive (cl3, cl4, and cl6) and two negative HH514 (cl5 and cl7) clones in the presence of estrogen and in two positive BL41-P3HR1 clones (cl31 and cl33) in the presence and absence of estrogen. The antibody detected a protein with the expected molecular mass of 110 kDa. The protein was absent in the untransfected cell line BL41-P3HR1. Addition of estrogen for 48 h resulted in a shift in the electrophoretic mobility, most likely due to posttranslational modification.
FIG. 1.
Expression of a Notch1-IC–ER fusion protein in BL41-P3HR1 and HH514 cells. Extracts from hygromycin-resistant single-cell clones derived after transfection of HH514 (A) and BL41-P3HR1 (B) cells with a plasmid encoding an mNotch1-IC–ER fusion protein were separated by gel electrophoresis, Western blotted, and analyzed by immunostaining with a monoclonal antibody specific for an epitope within the hormone binding domain of the ER. Extracts were prepared from HH514-NoER cl3 to cl7 cultivated in the presence of estrogen and from BL41-P3HR1-NoER cl31 and cl33 grown in both the absence and presence of estrogen. An extract from the untransfected BL41-P3HR1 cell line was included as a control. The arrow head marks the position of the band specific for the mNotch1-IC–ER fusion protein.
The stably introduced mNotch1-IC–ER transactivates a multimerized RBP-Jκ site dependent on the presence of estrogen.
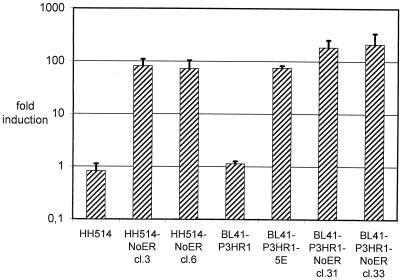
We have shown previously that in transient transfection assays a promoter reporter gene construct carrying a multimerized RBP-Jκ binding site can be transactivated by mNotch1-IC (60). To analyze whether the stably introduced mNotch1-IC–ER is functionally active, we transiently transfected the promoter reporter gene construct containing the multimerized RBP-Jκ binding site into the mNotch1-IC–ER-expressing BL41-P3HR1-NoER (cl31 and cl33) and HH514-NoER (cl3 and cl6) clones and the corresponding parental cell lines BL41-P3HR1 and HH514 and measured luciferase activities in the presence and absence of estrogen. In parallel, BL41-P3HR1-5E cells expressing ER-EBNA2, were transfected with the same promoter reporter gene construct as a positive control. As shown in Fig. 2, the promoter carrying the multimerized RBP-Jκ binding sites could be transactivated after activation of the mNotch1-IC–ER by addition of estrogen. The activation of the promoter reporter gene construct was dependent on the presence of the RBP-Jκ binding site (data not shown). Addition of estrogen to the parental cell lines HH514 and BL41-P3HR1 did not influence promoter activity. Activation of the ER-EBNA2 fusion protein resulted in an induction comparable with the mNotch1-IC–ER protein. This indicates that the mNotch1-IC–ER protein is functionally active in Burkitt's lymphoma cells and can be regulated by estrogen.
FIG. 2.
Activation of a promoter reporter construct containing a multimerized RBP-Jκ binding site by an mNotch1-IC–ER fusion protein in the presence of estrogen. Cells from the parental cell lines HH514 and BL41-P3HR1 and from two HH514-NoER and two BL41-P3HR1-NoER clones, which have been shown to express the mNotch1-IC–ER fusion protein (Fig. 1), were transiently transfected with a promoter reporter construct containing a multimerized RBP-Jκ site. As a positive control, a single-cell clone derived from BL41-P3HR1 expressing an ER-EBNA2 fusion protein (BL41-P3HR1-5E) was included in the experiment. After transfection, each cell suspension was divided in two parts and cultivated with and without estrogen for 2 days. Subsequently luciferase activities (RLU) were determined. The bars represent arithmetic mean values of the promoter activation by estrogen (RLU with estrogen/RLU without estrogen) from at least three (two for parental cell lines) independent experiments. Positive standard deviations are indicated by lines.
mNotch1-IC activates the LMP1 promoter in a transient transfection assay but induces endogenous LMP1 expression only marginally.
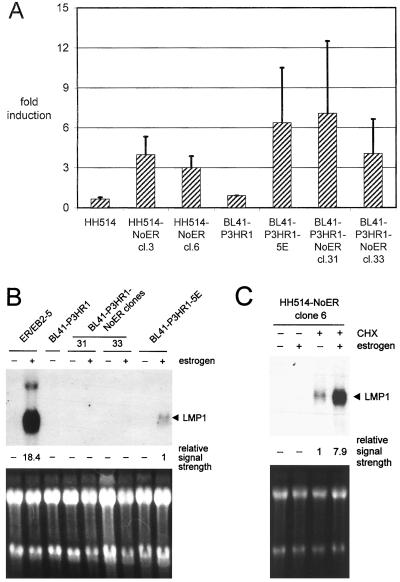
To analyze whether the stably introduced mNotch1-IC–ER is also able to regulate EBNA2-regulated viral promoters, an LMP1 promoter reporter gene construct (LMPLuc0) was transfected in BL41-P3HR1-NoER cl31 and cl33 and HH514-NoER cl3 and cl6. As a negative control, an LMP1 promoter reporter gene construct without the EBNA2-responsive region (LMPLuc9) was transfected (data not shown). To exclude a direct influence of estrogen on LMP1 promoter activity, the parental cell lines HH514 and BL41-P3HR1 were transfected in parallel. The stably transfected mNotch1-IC–ER protein could activate the LMP1 promoter about three- to sevenfold after addition of estrogen provided that the EBNA2-responsive region was present (Fig. 3A). In comparison, activation of the ER-EBNA2 fusion protein (cell clone BL41-P3HR1-5E) led to an about sixfold increase of the luciferase activity. Addition of estrogen to the parental cell lines had no effect on LMP1 promoter activity. These data suggest that mNotch1-IC is able to induce the LMP1 promoter.
FIG. 3.
The stably expressed mNotch1-IC–ER fusion protein induces the LMP1 promoter in an estrogen-dependent manner. (A) Cells from two HH514-NoER clones, two BL41-P3HR1-NoER clones, and the parental cell lines were transiently transfected with an LMP1 promoter-luciferase construct. As a positive control, a single-cell clone derived from BL41-P3HR1 cells expressing an ER-EBNA2 fusion protein (BL41-P3HR1-5E) was included in the experiment. After transfection, cells were cultivated in the presence or absence of estrogen. Luciferase activities were determined as RLU 2 days after transfection. The bars represent arithmetic mean values of the promoter activation by estrogen (RLU with estrogen/RLU without estrogen) from at least three (two for parental cell lines) independent experiments. Positive standard deviations are indicated by lines. (B) The endogenous LMP1 mRNA expression was analyzed by Northern blotting. The cell lines indicated were cultivated in parallel with and without estrogen. RNAs were separated, blotted, and hybridized with a probe specific for LMP1. (C) The LMP1 mRNA expression of one single HH514-NoER clone (clone 6) was analyzed by Northern blotting in the presence and absence of estrogen and cycloheximide (CHX) as indicated. (B and C) The intensities of the LMP1 signals were quantified and standardized (the weakest measurable signal was set to 1) and are listed below the blots. As a control for RNA quantity and integrity, the ethidium bromide-stained RNA gels are shown.
We next examined whether the endogenous LMP1 gene is induced by Notch-IC. To this end, we performed Northern blot analysis for LMP1 RNA in the presence and absence of estrogen. As shown in Fig. 3B, mNotch1-IC–ER was not able to induce LMP1 RNA in BL41-P3HR1 cl31 and cl33. The LMP1 transcript could, however, be weakly visualized after activation of the ER-EBNA2 protein. LMP1 transcription was significantly lower in the ER-EBNA2-transfected Burkitt lymphoma cell line BL41-P3HR1-5E than in the lymphoblastoid cell line ER/EB2-5, which also harbors an ER-EBNA2 fusion protein. This is in accordance with our previous results which showed that LMP1 in BL41-P3HR1 cells could not be induced by EBNA2 to a level comparable to that in EBV-immortalized lymphoblastoid cell lines (11). LMP1 has been reported to be expressed in P3HR1 cells even in the absence of EBNA2 at a very low level (11a). Therefore, we next examined whether mNotch1-IC is able to induce LMP1 transcription in the Notch-ER-transfected P3HR1 subclone HH514. RNA was prepared from cells cultivated without or with estrogen (for activation of mNotch1-IC) and without or with cycloheximide (for RNA stabilization). Without cycloheximide treatment, LMP1 RNA was detectable only after long exposure and at similar amounts in untreated and estrogen-treated cells (data not shown). After cycloheximide treatment, a significant increase in the amount of LMP1 RNA became visible after activation of the mNotch1-IC–ER fusion protein (Fig. 3C).
mNotch1-IC induces the LMP2A promoter and upregulates expression of the endogenous LMP2A gene.
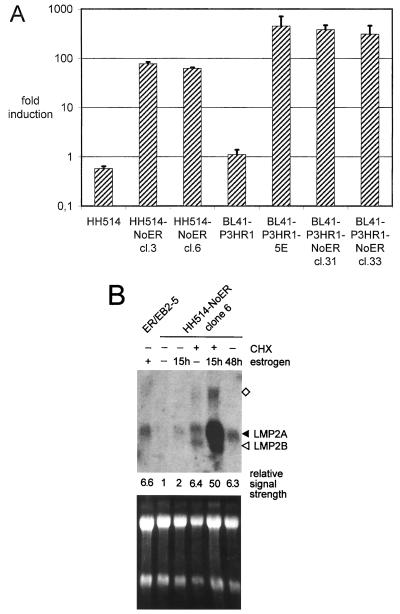
EBNA2 induces not only LMP1 but also LMP2A transcription. Therefore, we examined whether Notch-IC can activate the viral LMP2A gene. First we tested whether the LMP2A promoter can be transactivated by Notch1-IC–ER. The LMP2A promoter luciferase construct TP1luc/-804 was transiently transfected into the parental cell lines HH514 and BL41-P3HR1, into HH514-NoER cl3 and cl6 and BL41-P3HR1-NoER cl31 and cl33, and into the ER-EBNA2 fusion protein-expressing cell clone BL41-P3HR1-5E. Luciferase activity was determined in the presence and absence of estrogen (Fig. 4A). In all Notch1-IC–ER- and ER-EBNA2-expressing cell clones tested, the LMP2A promoter could be efficiently transactivated by addition of estrogen, whereas in the parental cell lines no induction was seen.
FIG. 4.
The stably expressed mNotch1-IC–ER fusion protein induces LMP2A transcription in an estrogen-dependent manner. (A) Cells from two HH514-NoER clones, two BL41-P3HR1-NoER clones, and the parental cell lines were transiently transfected with an LMP2A promoter-luciferase construct. A single-cell clone derived from BL41-P3HR1 expressing an ER-EBNA2 fusion protein (BL41-P3HR1-5E) was included as a positive control. After transfection, each cell suspension was divided in two parts and cultivated with and without estrogen for 2 days. Subsequently luciferase activities (RLU) were determined. The bars represent arithmetic mean values of the promoter activation by estrogen (RLU with estrogen/RLU without estrogen) from at least three (two for controls) independent experiments. Positive standard deviations are indicated by lines. (B) Endogenous LMP2 mRNA expression was analyzed by Northern blotting. ER/EB2-5 cells were cultivated in the presence (+) or absence (−) of estrogen for 4 days. HH514-NoER cells were routinely kept without estrogen. The mNotch1-IC–ER fusion protein was induced by addition of estrogen for 15 or 48 h. Where indicated, cells were grown in the presence of cycloheximide (CHX) for 16 h. RNAs were prepared, separated on a 1% agarose gel, blotted, and hybridized with a probe specific for LMP2. Bands specific for LMP2A and LMP2B are marked by arrowheads. The band marked by the square might represent an antisense transcript. The intensities of the LMP2A signals were quantified and standardized (the weakest measurable signal was set to 1) and are listed below the blot. As a control for RNA quantity and integrity, the ethidium bromide-stained RNA gel is shown at the bottom.
To examine whether transcription of the endogenous viral LMP2A gene is induced by Notch1-IC, RNA was prepared from HH514-NoER cl6 cells, untreated and treated with estrogen for 15 and 48 h. To stabilize LMP2A transcripts, RNA of cells treated with cycloheximide was prepared in parallel. Cell clones of BL41-P3HR1 could not be included in this analysis because the viral genome is integrated into the cellular genome through the terminal repeats and therefore a functional transcription unit is not created in this cell line (29, 76). Analysis of the HH514-NoER clones (Fig. 4B) showed that addition of estrogen led to low but significant induction of LMP2A transcription. Levels of LMP2A RNA were higher after 48 h than after 15 h of estrogen treatment. As described before (76), cycloheximide treatment stabilized the LMP2A RNA and thus facilitated the interpretation of the induction experiment.
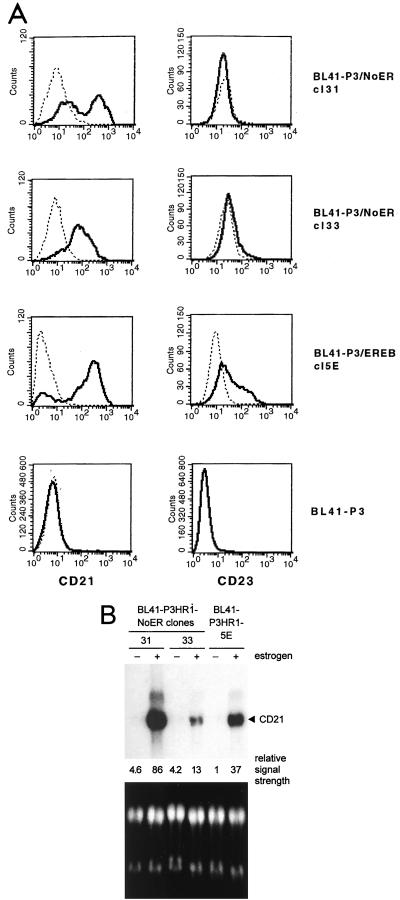
mNotch1-IC upregulates expression of CD21 but not CD23 on the cell surface and transcriptionally regulates CD21.
To see whether mNotch1-IC can induce the same phenotypic changes as EBNA2, we analyzed CD21 and CD23 expression in the mNotch1-IC–ER-expressing BL41-P3HR1-NoER cl31 and cl33 by flow cytometric (FACS) analysis before and after addition of estrogen. BL41-P3HR1-5E cells expressing the ER-EBNA2 fusion protein and BL41-P3HR1 cells were used as positive and negative controls, respectively. Activation of mNotch1-IC as well as EBNA2 revealed a significant increase in the expression of CD21 on the cell surface. However, in contrast to EBNA2, mNotch1-IC was not able to activate expression of CD23 (Fig. 5A).
FIG. 5.
Notch1-IC upregulates CD21 but not CD23 in stably transfected BL cells. (A) CD21 and CD23 expression was analyzed in the cell lines indicated by flow cytometry before (dotted line) and after (thick line) treatment with estrogen for 48 h. (B) Endogenous CD21 mRNA expression was analyzed by Northern blotting. RNAs were prepared from the cells indicated which had been cultivated in the absence (−) or presence (+) (48 h) of estrogen. RNAs were separated, blotted, and hybridized with a probe specific for CD21. The intensities of the CD21 signals were quantified and standardized (the weakest measurable signal was set to 1) and are listed below the blot. As a control for RNA quantity and integrity, the ethidium bromide-stained RNA gel is shown at the bottom.
To see whether CD21 induction by mNotch1-IC is regulated at the RNA or the protein level, Northern blot analysis was performed with RNAs of the mNotch1-IC–ER- and EBNA2-ER-expressing BL41-P3HR1 cell clones grown in the presence and absence of estrogen. As shown in Fig. 5B, mNotch1-IC clearly induced an increase in the CD21 RNA levels. The results were in good agreement with those observed by FACS analysis. Cl 31, with higher mean fluorescence after addition of estrogen, revealed also a higher amount of CD21 RNA compared to cl33. These results indicate that mNotch1-IC induces expression of CD21 but not of CD23.
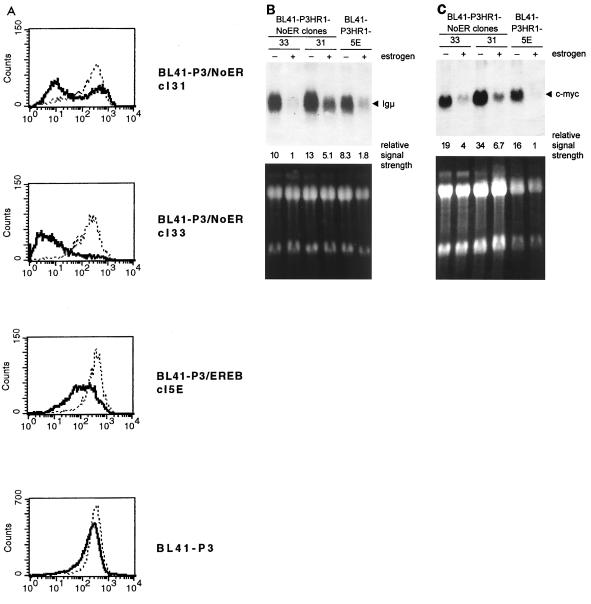
mNotch1-IC downregulates Igμ and c-myc in Burkitt's lymphoma cells.
We have shown previously that EBNA2 is able to downregulate Igμ (30). To see whether mNotch1-IC can also negatively regulate Igμ expression, we determined the amount of IgM on the cell surface of BL41-P3HR1 cells transfected with mNotch1-IC–ER (cl31 and cl33) in the presence and absence of estrogen by FACS analysis. BL41-P3HR1-5E cells expressing the ER-EBNA2 fusion protein and BL41-P3HR1 cells were used as positive and negative controls, respectively. As shown in Fig. 6A, mNotch1-IC was also able to downregulate IgM on the cell surface significantly.
FIG. 6.
In Burkitt's lymphoma cells, both Igμ and c-myc are transcriptionally downregulated by the estrogen-activated mNotch1-IC–ER fusion protein. (A) sIgM expression was analyzed in the cell lines indicated by flow cytometry before (dotted line) and after (thick line) treatment with estrogen for 48 h. Endogenous Igμ (B) and c-myc (C) mRNA expression was analyzed by Northern blotting. Cell lines were cultivated without (−) or with (+) estrogen for 48 h. RNAs were separated, blotted, and hybridized with probes specific for the Igμ or c-myc. The intensities of the Igμ and c-myc signals were quantified and standardized (the weakest measurable signal was set to 1) and are listed below the blots. As controls for RNA quantity and integrity, the ethidium bromide-stained RNA gels are shown at the bottom. In the c-myc blot, RNA quantity of the BL41-P3HR1-5E control (− and + estrogen) was reduced ∼2-fold compared to the other cell lines.
In Burkitt's lymphoma cells carrying a t(8;14) translocation, the proto-oncogene c-myc is activated by juxtaposition to Igμ. We have shown previously that in Burkitt's lymphoma cells with t(8;14) translocations EBNA2 downregulates c-myc expression concomitantly with Igμ, presumably by interacting with the heavy-chain immunoglobulin enhancers (30). It was thus obvious to ask whether mNotch1-IC downregulates Igμ as well as c-myc expression in Burkitt's lymphoma cells. Northern blot analysis was performed with RNAs of BL41-P3HR1-NoER cl31 and cl33 cells. RNA of the ER-EBNA2-transfected cell clone BL41-P3HR1-5E was analyzed as a positive control. The filters were hybridized with radioactive probes specific for Igμ and c-myc (Fig. 6B and C). Igμ RNA expression clearly correlated with the FACS data. RNA levels were significantly lower in the presence than in the absence of estrogen, indicating that Notch1-IC as well as EBNA2 downregulated Igμ at the transcriptional level. Like the Igμ levels, c-myc RNA levels decreased by addition of estrogen in the NoER and in the ER-EBNA2-expressing cell clones, indicating that in Burkitt's lymphoma cells with the c-myc gene juxtaposed to the IgH locus by a t(8;14) translocation, c-myc expression is negatively coregulated with Igμ by Notch1-IC as well as EBNA2.
DISCUSSION
Both EBNA2 and the activated form of the Notch receptor (Notch-IC) interact with the transcription factor RBP-Jκ, leading to gene activation. Therefore, the viral protein EBNA2 can be regarded as a functional equivalent of an activated Notch receptor. EBNA2 is absolutely necessary for initiation and maintenance of B-cell immortalization by EBV. In Burkitt's lymphoma cells, EBNA2 can induce characteristic phenotypic changes like upregulation of CD21 and CD23 and the downregulation of IgM. Additionally, expression of the viral membrane proteins LMP1 and LMP2A is dependent on the presence of EBNA2 in immortalized B cells. If in fact EBNA2 mimics an activated Notch receptor, it may be postulated that Notch-IC can induce phenotypic changes in B cells similar to those induced by EBNA2. To test this hypothesis, we transfected mNotch-IC–ER) in the EBNA2-negative P3HR1-EBV-infected Burkitt's lymphoma cell lines BL41-P3HR1 and HH514. A Notch-IC–ER fusion protein was used to rule out artifacts due to clonal selection, allowing analysis of the same cell clone in the presence and absence of a functional Notch-IC protein. This paper deals with the effect of activated Notch on viral as well as cellular genes in B cells.
First, we studied the expression of the viral LMP1 and LMP2A genes. In the absence of EBNA2, the LMP1 promoter is inactive in BL41-P3HR1 cells and has very low basal activity in HH514 cells. In both cell lines, Notch-IC was not able to induce LMP1 transcription to a detectable level. This finding was in variance to transient transfection assays in which the LMP1 promoter was clearly induced by activated Notch (27). The inability of Notch1-IC to induce LMP1 expression after stable transfection is reminiscent to the situation in EBNA2-transfected P3HR1 virus carrying BL cell lines (11). Although it is well established that the LMP1 promoter is positively regulated by EBNA2 in B cells, we have been unable to demonstrate induction of the endogenous LMP1 gene after stable transfection of the EBNA2 gene into BL41-P3HR1 cells (11) presumably because the LMP1 promoter is rendered unresponsive to EBNA2 by methylation. To amplify the effect of Notch-IC–ER, we studied induction of the endogenous LMP1 gene in stably transfected HH514 cells after addition of estrogen and cycloheximide and found that LMP1 induction by Notch-IC–ER can in fact be visualized under these conditions. Most likely, induction of LMP1 by Notch1-IC is very weak and can be visualized only if degradation of LMP1 RNA is inhibited by cycloheximide.
In contrast to LMP1, endogenous LMP2A was clearly induced by Notch-IC. The increase of LMP2A RNA was low after 15 h and higher after activation of Notch-IC for 48 h. Cycloheximide treatment dramatically increased induction of LMP2A RNA, indicating that Notch-IC acts directly on the LMP2A promoter and that the short-lived LMP2A RNA is stabilized by cycloheximide. The observation that Notch-IC can induce LMP2A transcription in B cells may be relevant for EBV latency in vivo as well as for the role of EBV in the development of Hodgkin lymphomas. It has been shown that EBV persists in memory B cells (3) and that stimulation of the B-cell receptor is able to break latency and induce the lytic cycle of the virus (66). Furthermore, it has been shown that LMP2A expression can block induction of the lytic cycle after cross-linking of the immunoglobulin receptor (50). LMP2A appears to be expressed in germinal center cells (D. A. Thorley-Lawson et al., personal communication) and may be the only viral gene expressed in latently infected B cells in vivo (53, 55a). LMP2A, potentially induced by activated Notch, may block induction of the lytic cycle by antigen and drive EBV-infected cells into the memory compartment or prevent them from leaving the memory compartment by viral reactivation. Additionally, LMP2A is expressed in EBV-positive Hodgkin lymphoma cells (12). Hodgkin lymphoma is a tumor of the centrocytic stage of B-cell differentiation. The majority of Hodgkin Reed-Sternberg (HRS) cells have lost the ability to code for B-cell receptor (BCR) complexes due to stop codons within the variable regions of immunoglobulins (5, 31). Normally, receptorless B lymphocytes are rapidly eliminated by apoptosis (40). LMP2A expression, presumably through its ITAM signaling motifs (46), was recently shown to inhibit the elimination of receptorless cells (8). In both HRS and memory B cells, LMP2 is expressed independently of EBNA2 by mechanisms currently not understood. Notch signaling may be the underlying mechanism leading to induction of LMP2A in latently infected normal B cells or HRS cells.
In addition to the viral genes, we have studied the modulation of known EBNA2-regulated cellular genes through Notch-IC. We could show that Notch-IC upregulates CD21 expression and downregulates sIgM as has been shown for EBNA2. However, in contrast to EBNA2, Notch-IC was not able to upregulate CD23 expression. The observation that Notch-IC regulates sIgM and CD21 expression raises the question of whether Notch may play a physiological role during B-cell activation. Signals from the BCR and CD21 are involved in the early activation of naive B cells and in selection processes in the germinal center. The human complement receptor 2 (CD21) is complexed with CD19 and CD81 and acts as a BCR coreceptor enhancing signals from the BCR (67). Simultaneous ligation of CD19-CD21 and membrane immunoglobulin in vitro lowers the threshold required for signaling through the BCR (14, 54). Therefore, engagement of CD21 on B cells seems to be important if suboptimal concentrations of antigen are present or if the affinity of antibody is low. In the germinal center, signals from the CD21 receptor seem to contribute to the survival of B cells (19). Activation of Notch during B-cell development might physiologically modulate B cell receptor-CD21 signaling.
We have shown previously that in Burkitt's lymphoma cells carrying a t(8;14) translocation, the c-myc gene is downregulated by EBNA2, most likely by elements regulating immunoglobulin heavy-chain expression. Here we have shown that Notch-IC can also downregulate Igμ and c-myc gene expression in the Burkitt's lymphoma cell line BL41-P3HR1. We have been unable, however, to identify the cis-acting elements involved in this regulation in transient transfection experiments using the IgH intron as well as 3′ enhancer. We have also not found RBP-Jκ binding sites in the IgH enhancers which might mediate this effect.
In summary, we have shown that Notch1-IC shares many of the properties of EBNA2 and induces a similar although not identical set of target genes in B cells as EBNA2. The next question is whether Notch-IC can substitute for EBNA2 in B-cell proliferation by EBV. Experiments are under way to address this important question.
ACKNOWLEDGMENTS
We thank T. Henkel for providing the pSG5-mNotch1IC expression plasmid.
This work was supported by Die Deutsche Forschungsgemeinschaft (Forschergruppe Multiproteinkomplexe and Leibniz-Programm), the European Union (Molekulare Pathogenese menschlicher Tumorvirusinfektionen), and Fonds der Chemischen Industrie.
REFERENCES
- 1.Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 2.Artavanis Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 3.Babcock G J, Decker L L, Volk M, Thorley-Lawson D A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 4.Bigas A, Martin D I, Milner L A. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol. 1998;18:2324–2333. doi: 10.1128/mcb.18.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braeuninger A, Kuppers R, Strickler J G, Wacker H H, Rajewsky K, Hansmann M L. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci USA. 1997;94:9337–9342. doi: 10.1073/pnas.94.17.9337. . (Erratum, 94:14211.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brielmeier M, Mautner J, Laux G, Hammerschmidt W. The latent membrane protein 2 gene of Epstein-Barr virus is important for efficient B cell immortalization. J Gen Virol. 1996;77:2807–2818. doi: 10.1099/0022-1317-77-11-2807. [DOI] [PubMed] [Google Scholar]
- 7.Brielmeier M, Bechet J M, Falk M H, Pawlita M, Polack A, Bornkamm G W. Improving stable transfection efficiency: antioxidants dramatically improve the outgrowth of clones under dominant marker selection. Nucleic Acids Res. 1998;26:2082–2085. doi: 10.1093/nar/26.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 9.Calender A, Billaud M, Aubry J P, Banchereau J, Vuillaume M, Lenoir G M. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc Natl Acad Sci USA. 1987;84:8060–8064. doi: 10.1073/pnas.84.22.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordier M, Calender A, Billaud M, Zimber U, Rousselet G, Pavlish O, Banchereau J, Tursz T, Bornkamm G W, Lenoir G M. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Cordier B M, Calender A, Vuillaume M, Bornkamm G W, Lenoir G M. Expression of the Epstein-Barr virus (EBV) latent membrane protein is tightly regulated, independently of EB nuclear antigen 2 and of EBV integration or copy number. Virus Res. 1993;27:55–69. doi: 10.1016/0168-1702(93)90112-z. [DOI] [PubMed] [Google Scholar]
- 12.Deacon E M, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson A B, Young L S. Epstein-Barr virus and Hodgkin's disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deftos M L, He Y W, Ojala E W, Bevan M J. Correlating notch signaling with thymocyte maturation. Immunity. 1998;9:777–786. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, Mak T W, Rossant J, Conlon R A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 14.Dempsey P W, Fearon D T. Complement: instructing the acquired immune system through the CD21/CD19 complex. Res Immunol. 1996;147:71–5. doi: 10.1016/0923-2494(96)87176-8. [DOI] [PubMed] [Google Scholar]
- 15.Eick D, Piechaczyk M, Henglein B, Blanchard J M, Traub B, Kofler E, Wiest S, Lenoir G M, Bornkamm G W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985;4:3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 17.Fahraeus R, Rymo L, Rhim J S, Klein G. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature. 1990;345:447–449. doi: 10.1038/345447a0. [DOI] [PubMed] [Google Scholar]
- 18.Falk M H, Meier T, Issels R D, Brielmeier M, Scheffer B, Bornkamm G W. Apoptosis in Burkitt lymphoma cells is prevented by promotion of cysteine uptake. Int J Cancer. 1998;75:620–625. doi: 10.1002/(sici)1097-0215(19980209)75:4<620::aid-ijc21>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 19.Fischer M B, Goerg S, Shen L, Prodeus A P, Goodnow C C, Kelsoe G, Carroll M C. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science. 1998;280:582–585. doi: 10.1126/science.280.5363.582. [DOI] [PubMed] [Google Scholar]
- 20.Fortini M E, Artavanis-Tsakonas S. Notch: neurogenesis is only part of the picture. Cell. 1993;75:1245–1247. doi: 10.1016/0092-8674(93)90611-s. [DOI] [PubMed] [Google Scholar]
- 21.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamaguchi Y, Matsunami N, Yamamoto Y, Honjo T. Purification and characterization of a protein that binds to the recombination signal sequence of the immunoglobulin J kappa segment. Nucleic Acids Res. 1989;17:9015–9026. doi: 10.1093/nar/17.22.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 24.Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- 25.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 26.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 27.Höfelmayr H, Strobl L J, Stein C, Laux G, Marschall G, Bornkamm G W, Zimber-Strobl U. Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J Virol. 1999;73:2770–2780. doi: 10.1128/jvi.73.4.2770-2780.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurley E A, Agger S, McNeil J A, Lawrence J B, Calendar A, Lenoir G, Thorley-Lawson D A. When Epstein-Barr virus persistently infects B-cell lines, it frequently integrates. J Virol. 1991;65:1245–1254. doi: 10.1128/jvi.65.3.1245-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jochner N, Eick D, Zimber-Strobl U, Pawlita M, Bornkamm G W, Kempkes B. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin mu gene: implications for the expression of the translocated c-myc gene in Burkitt's lymphoma cells. EMBO J. 1996;15:375–382. [PMC free article] [PubMed] [Google Scholar]
- 30a.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanzler H, Kuppers R, Hansmann M L, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kempkes B, Zimber-Strobl U, Eissner G, Pawlita M, Falk M, Hammerschmidt W, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 (EBNA2)-oestrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J Gen Virol. 1996;77:227–237. doi: 10.1099/0022-1317-77-2-227. [DOI] [PubMed] [Google Scholar]
- 34.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 36.Kimble J, Simpson P. The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997;13:333–361. doi: 10.1146/annurev.cellbio.13.1.333. [DOI] [PubMed] [Google Scholar]
- 37.Knutson J C. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J Virol. 1990;64:2530–2536. doi: 10.1128/jvi.64.6.2530-2536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 39.Kopan R, Schroeter E H, Weintraub H, Nye J S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam K P, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 41.Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 42.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- 44.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 45.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longnecker R, Kieff E. A second Epstein-Barr virus membrane protein (LMP2) is expressed in latent infection and colocalizes with LMP1. J Virol. 1990;64:2319–2326. doi: 10.1128/jvi.64.5.2319-2326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsunami N, Hamaguchi Y, Yamamoto Y, Kuze K, Kangawa K, Matsuo H, Kawaichi M, Honjo T. A protein binding to the J kappa recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature. 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- 49.Meitinger C, Strobl L J, Marschall G, Bornkamm G W, Zimber-Strobl U. Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element. J Virol. 1994;68:7497–7506. doi: 10.1128/jvi.68.11.7497-7506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller C L, Lee J H, Kieff E, Longnecker R. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc Natl Acad Sci USA. 1994;91:772–776. doi: 10.1073/pnas.91.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milner L A, Kopan R, Martin D I, Bernstein I D. A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. Blood. 1994;83:2057–2062. [PubMed] [Google Scholar]
- 52.Milner L A, Bigas A, Kopan R, Brashem Stein C, Bernstein I D, Martin D I. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;93:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mongini P K, Vilensky M A, Highet P F, Inman J K. The affinity threshold for human B cell activation via the antigen receptor complex is reduced upon co-ligation of the antigen receptor with CD21 (CR2) J Immunol. 1997;159:3782–3791. [PubMed] [Google Scholar]
- 55.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 55a.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabson M, Gradoville L, Heston L, Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982;44:834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald H R, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 57.Robbins J, Blondel B J, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 59.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 60.Strobl L J, Höfelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber-Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 61.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 62.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 63.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sung N S, Kenney S, Gutsch D, Pagano J S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991;65:2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 66.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 67.Tedder T F, Engel P. CD20: a regulator of cell-cycle progression of B lymphocytes. Immunol Today. 1994;15:450–454. doi: 10.1016/0167-5699(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 68.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang F, Gregory C D, Rowe M, Rickinson A B, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci USA. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes B J, Cado D, Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 72.Weinmaster G, Roberts V J, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- 73.Weinmaster G, Roberts V J, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 74.Weis J J, Fearon D T, Klickstein L B, Wong W W, Richards S A, de Bruyn Kops A, Smith J A, Weis J H. Identification of a partial cDNA clone for the C3d/Epstein-Barr virus receptor of human B lymphocytes: homology with the receptor for fragments C3b and C4b of the third and fourth components of complement. Proc Natl Acad Sci USA. 1986;83:5639–5643. doi: 10.1073/pnas.83.15.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woisetschlaeger M, Jin X W, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci USA. 1991;88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zimber-Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]