Abstract
Recombinant adeno-associated virus type 2 (AAV) is a common vector used in human gene therapy protocols. We characterized the humoral immune response to AAV and observed that 80% of normal human subjects have anti-AAV antibodies and that 18% have neutralizing antibodies. To analyze the effect of neutralizing antibodies on AAV readministration, we attempted to deliver recombinant AAV expressing human factor IX (AAV-hFIX) intraportally into the livers of mice which had been preexposed to AAV and shown to harbor a neutralizing antibody response. While all naive control mice expressed hFIX following administration of AAV-hFIX, none of the mice with preexisting immunity expressed hFIX, even after transient immunosuppression at the time of the second administration with anti-CD4 or anti-CD40L antibodies. This suggests that preexisting immunity to AAV, as measured by a neutralizing antibody response, may limit AAV-mediated gene delivery. Using human sera in an enzyme-linked immunosorbent assay for AAV and a capsid peptide scan library to block antibody binding, we mapped seven regions of the AAV capsid containing immunogenic epitopes. Using pools of these peptides to inhibit the binding of neutralizing antibodies, we have identified a subset of six peptides which potentially reconstitute a single neutralizing epitope. This information may allow the design of reverse genetic approaches to circumvent the preexisting immunity that can be encountered in some individuals.
Recombinant adeno-associated virus type 2 (AAV) vectors represent a promising gene delivery system because of their nonpathogenicity, ability to stably transduce both dividing and nondividing cells including cells from lung (5), liver (21, 22), brain (13), and muscle (8, 9, 23), and genome-integrating capability which results in long-term protein expression (16, 22). AAV-mediated gene delivery can be potentially obstructed by a host's immune response to its component proteins. In the case of recombinant AAV vectors, the primary target of the immune response is the capsid of the vector particle since these vectors do not encode any viral proteins. Several groups have shown that the failure of AAV readministration to generate further transduction events correlated with the presence of virus-neutralizing antibodies generated in response to a previous exposure to the virus. Manning et al. demonstrated that transient depletion of helper T cells during the initial exposure to AAV with anti-CD4 antibodies allowed successful readministration of AAV vectors to skeletal muscle (14). Similarly, immunosuppression during the initial exposure with anti-CD40L antibodies (which block T-cell activation of B cells) or CTLA4Ig (which inhibits T-cell activation by interfering with CD28-B7 interactions) facilitated transgene expression in mouse lung (6) and also allowed readministration of adenovirus to the mouse liver (10).
The liver is a potential target for gene therapy including treatment for hemophilia (21, 22). Since this treatment will likely require delivery to individuals with established preexisting immunity to AAV (1) or repeat vector delivery, and because conclusions regarding vector delivery cannot be extrapolated from tissue to tissue, we examined the effect of preexisting immunity on the delivery of AAV to the liver. In addition, we transiently immunosuppressed the mice concomitantly with readministration of the “therapeutic” AAV, a protocol which closely reflects the reality of a clinical situation in which patients already have immunity, rather than during the primary exposure as reported by others.
To delineate further the specificity of the AAV neutralizing antibody response in humans, we used serum samples and a capsid peptide scan (pepscan) in blocking enzyme-linked immunosorbent assays (ELISAs) to map linear antibody epitopes on AAV. Using pools of immunogenic peptides identified in the linear scan, we then identified six peptides that block the effect of neutralizing sera and a neutralizing mouse monoclonal antibody. This information may allow genetic manipulation to circumvent the host immune response for successful AAV vector delivery to patients with preexisting immunity. The immunogenic epitopes described here also corroborate previous genetic and structural data and identify exposed capsid regions potentially involved in the binding of AAV to cellular receptors.
MATERIALS AND METHODS
Construction and production of AAV vectors.
AAV vectors expressing green fluorescent protein (GFP) (11), β-galactosidase (LacZ) (15), and human factor IX (hFIX) were constructed and generated as described previously (22). Titers were determined by dot blot analysis.
Assessment of AAV readministration in mice.
Eight-week-old C57BL/6 were purchased from Taconic (Germantown, N.Y.). Mice were immunized with 5 × 1010 particles of AAV-LacZ intravenously and monitored weekly for neutralizing antibodies, using serum obtained by retro-orbital bleeding. Readministration of AAV-hFIX (5 × 1010 particles) was done intraportally in a volume of 100 μl that was infused over 30 s (22). Serum was collected retro-orbitally every 2 weeks and analyzed for hFIX expression as described below.
For transient immunosuppression by anti-CD4 antibody, mice were injected with 100 μg of rat anti-mouse CD4 (clone GK1.5; Pharmingen, San Diego, Calif.) by intraperitoneal injection at days −3, 0, and +3 relative to the second injection of AAV. For anti-CD40L treatment, mice received 100 μg of antibody (clone MR1; Pharmingen) by intraperitoneal injection at days −3, 0, +3, and +6 relative to the secondary injection.
Detection of serum hFIX by ELISA.
Microtiter plates were coated (100 μl/well) with a solution containing monoclonal anti-hFIX (2 mg/ml; Boehringer Mannheim, Indianapolis, Ind.) diluted in 89 mM boric acid–90 mM NaCl (pH 8.3) (BBS). Plates were incubated overnight at 4°C and then washed five times with BBS containing 0.025% (vol/vol) Tween 20 (BBST). Plates were blocked for 2 h at room temperature with 1% (wt/vol) nonfat milk (100 μl/well) in BBS (BBM) and washed twice with BBST. hFIX standard was diluted in BBS containing 20% of mice serum in order to maintain the same conditions for test serum and standards. hFIX and test samples diluted in BBM (1:5) were used at serial concentrations of 50 μl/well. After incubation for 2 h at room temperature, plates were washed five times, 50 μl of a 1:100 dilution of horseradish peroxidase-conjugated goat anti-hFIX antibody (Affinity Biologicals Inc.) was added, and the mixture was incubated for 1.5 h at room temperature. Plates were then washed five times with BBST and twice with BBS. Color was developed for 25 min at room temperature with 50 μl of a buffer containing p-nitrophenyl phosphate in 34 mM citric acid–67 mM dibasic sodium phosphate–0.1% (vol/vol) hydrogen peroxide (pH 5.0; 1 mg/ml). Color development was stopped with 2 M sulfuric acid (50 μl/well), and the optical density was measured at 490 nm.
Detection of anti-AAV antibodies by ELISA.
Ninety-six-well MaxiSorp flat surface Nunc-Immuno plates were coated with 5 × 107 particles of AAV in 100 μl of 0.1 M carbonate (pH 9.6) per well, incubated overnight at 4°C, and washed twice with washing buffer from an AMPAK amplification kit (DAKO, Carpenteria, Calif.). After blocking with 3% bovine serum albumin (BSA) in washing buffer for 2 h at room temperature, the plates were washed once and incubated for 1 h at room temperature with donor serum at 1:100 dilution in washing buffer–1% BSA in a total volume 100 μl/well. Next the plates were washed five times, and alkaline phosphatase-conjugated mouse anti-human antibodies (Zymed, San Francisco, Calif.) were added at 1:800 dilution in washing buffer–1% BSA (100 μl/well). The plates were incubated for 1 h at room temperature and washed with washing buffer four times. For color development and further amplification of the signal, an AMPAK amplification kit was used. Absorbance was measured at 490 nm.
Detection of neutralizing anti-AAV antibodies.
293 cells were seeded in a 24-well plate at a density of 105 cells/well in 1 ml of Iscove's modified Dulbecco medium media (JRH). The cells were allowed to adhere for 2 h at 37°C. The medium was then removed by aspiration before 6 × 106 particles of adenovirus dl309 (4) were added in a final volume of 200 μl per well. The cells were further incubated at 37°C for 1 h and then washed twice in the same medium before the following mix was added. AAV-GFP (1 μl = 5 × 108 total particles or 9 × 106 transducing units) was incubated with serum sample diluted in phosphate-buffered saline (PBS) for 2 h at 4°C in a total volume of 25 μl. The final dilution of the test serum was 1:100 or 1:1,000. This mix was added to the washed cells in a final volume of 200 μl and incubated for 1 h at 37°C; 400 μl of medium was then added to each well, and cells were incubated overnight. Cells were collected, washed in PBS-BSA (1%), and analyzed by fluorescence-activated cell sorting. Percent inhibition was calculated with a no-antibody control sample as a reference. Another control was anti-AAV guinea pig sera that showed maximal inhibition.
Epitope mapping of anti-AAV antibodies.
A set of 91 overlapping peptides (15-mers) spanning the entire 735-amino-acid AAV-VP1 capsid protein sequence (GenBank accession no. AF043303) were synthesized by using the pin synthesis strategy (Chiron Mimotopes, Clayton, Australia). These peptide sequences overlap by five amino acids, thus generating all possible 10-mers of VP1. Two control peptides were also synthesized to verify purity and assess yield. Peptides were resuspended in PBS at a concentration of 5 mg/ml and stored at −20°C.
ELISA was performed in the presence of 1 μl (corresponding to a final concentration of approximately 20 μM) of individual peptides or 10 μl of peptide pools which were present at the antibody incubation stage. Similarly, 1 μl of each peptide was added to the 25 μl antibody–AAV-GFP mix in the neutralizing assay to assess the ability to block the binding of neutralizing antibodies to AAV-GFP.
RESULTS
Frequency of neutralizing antibodies in humans.
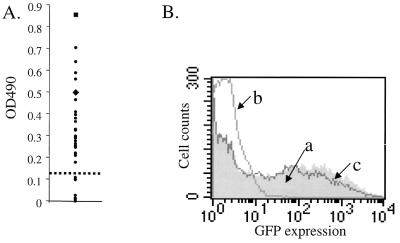
We obtained 50 human serum samples (from the Stanford University Hospital, Palo Alto, Calif.) to characterize the presence and specificity of anti-AAV antibodies in the normal population. ELISAs using these sera to probe AAV particles showed that 40 of 50 samples (80%) were positive for anti-AAV antibodies (Fig. 1A). In AAV neutralization assays, 18% of the Stanford samples were positive for the presence of neutralizing antibodies (Fig. 1B). In a population of Sardinians, we found that neutralizing antibodies were present at a frequency of 52% (data not shown), similar to a previously reported rate (1). Recently Xiao et al. reported a frequency of 27% in 77 individuals tested (30). This points to a significant population-to-population variability in the anti-AAV immune response.
FIG. 1.
Characterization of the anti-AAV immune response in humans. (A) Representative ELISA using 30 (of the total of 50 tested) serum samples to probe AAV particles. ●, test samples. Controls, including an anti-AAV monoclonal antibody (■) and anti-AAV serum raised in guinea pigs (⧫), verified the specificity of detection in this assay. The dashed line represents the background cutoff. (B) Representative neutralization assay analysis. AAV-GFP was used to infect 293 cells in the presence of human serum. Plot a (shaded), normal GFP expression in the absence of serum; plot b, example of a neutralizing serum sample that blocks AAV uptake and prevents GFP expression; plot c, example of a serum that does not inhibit AAV-GFP uptake into the cells.
Effect of a preexisting anti-AAV immune response on administration of AAV vectors.
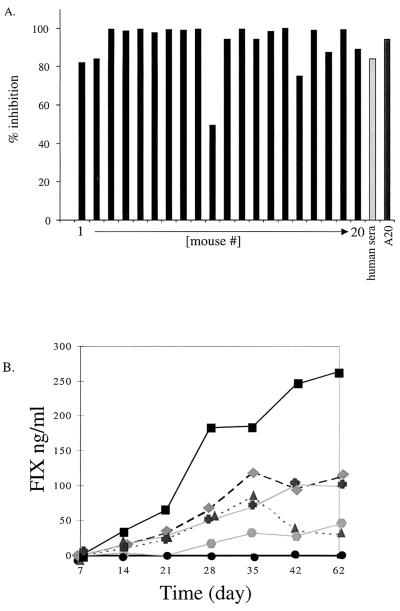
To assess the effect of neutralizing antibodies on readministration of AAV vectors, we immunized 20 C57BL/6 mice intravenously with an AAV vector expressing the Escherichia coli lacZ transgene. Neutralizing antibodies that developed with the kinetics of a normal immune response over the course of 2 weeks were detected in all mice. There was minor variation in the titer, but at 42 days postexposure each mouse elicited neutralizing antibodies at levels comparable to those found in human sera (Fig. 2A). A secondary therapeutic dose of AAV-hFIX vector (5 × 1010 viral particles) was delivered by an intraportal route to all 20 preexposed mice and 5 naive mice. Five of these preexposed mice were also treated with anti-CD40L antibodies and five mice were treated with anti-CD4 antibodies at the time of the second administration. The conditions for immunosuppression were identical to those previously shown to block B-cell responses successfully (14).
FIG. 2.
AAV vector delivery into mice with preexisting immunity. (A) Analyses of neutralization antibodies from murine sera collected 42 days after AAV-LacZ administration. Neutralization assays were performed in parallel with serum from each mouse, a positive human serum sample (Ser3), and an AAV-neutralizing monoclonal antibody (A20) at a dilution of 1:500. (B) Serum hFIX levels were measured for 62 days after introduction of AAV-hFIX intraportally into 20 preexposed and 5 naive mice. All 20 mice pretreated with AAV-LacZ (●) showed no hFIX expression over the entire 62 days of follow-up testing. The five naive mice (■,  ,
,  ,
,  , and
, and  ) expressed levels varying from 50 to 250 ng of hFIX per ml.
) expressed levels varying from 50 to 250 ng of hFIX per ml.
The five naive mice that received AAV-hFIX all expressed hFIX for the entire 62-day period of evaluation, while none of the mice with neutralizing immunity, including those that were transiently immunosuppressed, expressed hFIX over the same period (Fig. 2B). Since AAV-mediated transgene expression is typically observed 2 to 3 weeks after vector introduction (16), it is unlikely that the neutralizing immunity simply causes a further lag in gene expression. Thus, a preexisting neutralizing response clearly prevents the readministration of AAV vectors to the liver, and this inhibition of gene transfer cannot be overcome by transient immunosuppression.
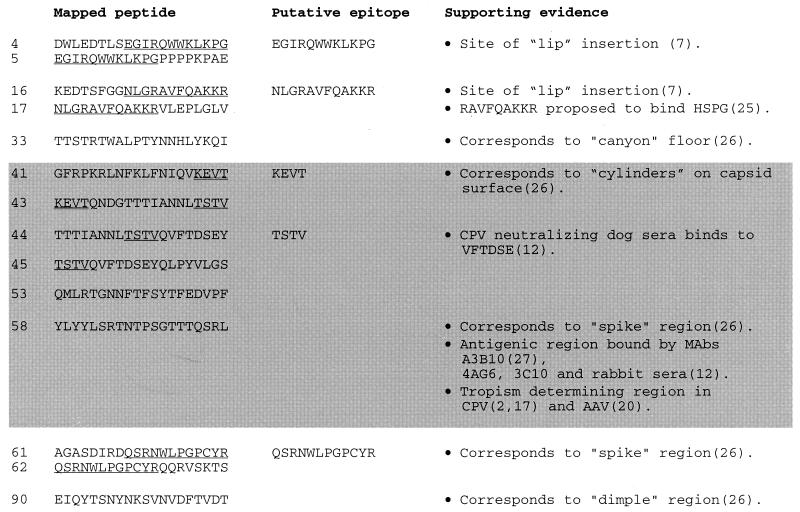
Mapping of anti-AAV antibody epitopes.
We attempted to further characterize the specificity of neutralizing antibodies by identifying their cognate epitopes. We used the positive human serum samples in an ELISA with a capsid pepscan library. Antibodies can bind to epitopes composed of amino acid residues from separated portions of the linear amino acid sequence that are spatially juxtaposed in a folded protein (conformational epitopes) or to adjacent residues on the amino acid sequence of a protein (linear epitopes). Peptides that could block antibody binding in an ELISA identify linear antibody epitopes.
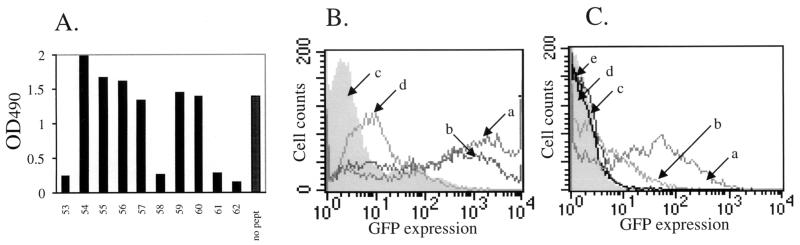
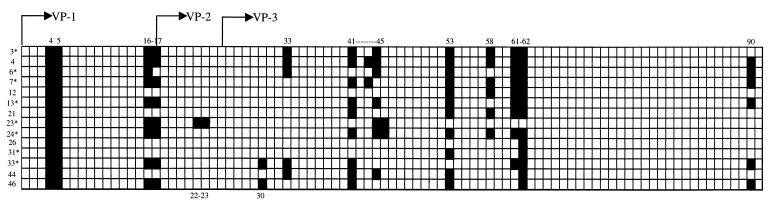
The AAV capsid is composed of three related proteins, VP1, VP2, and VP3 (in order of decreasing size), present at a ratio of 1:1:10, respectively, and derived from a single gene by alternative splicing and alternative start codon usage (19). Since VP2 and VP3 are subfragments of VP1, we generated a 91-peptide pepscan of AAV capsid protein VP1, composed of 15-mers overlapping by 5 amino acids and thus containing all possible 10-mers of the 735-amino-acid sequence. We initially used nine pools of 10 to 11 peptides to screen 14 positive human serum samples for blocking of antibody binding in an ELISA. Since most pools showed some blocking activity, we then used individual peptides. A representative analysis is shown in Fig. 3A. Seven regions of immunogenic sequence were clearly identified with the majority of serum samples (Fig. 4 and 5). Some peptides were able to block antibody binding of all serum samples (e.g., peptides 4 and 5), some could do so in the majority of sera (e.g., peptides 16, 17, and 61), and some could do so in only a few serum samples (e.g., peptide 33). Several tandem peptide pairs or triplets blocked binding presumably due to a shared, overlapping epitope sequence. We did not identify a unique linear epitope profile that correlated to the presence of neutralizing antibodies in the sera, suggesting the presence of multiple neutralizing antibodies or that anti-AAV neutralizing antibodies recognize a conformational epitope. This conformation epitope cannot be identified in a linear scan but can be potentially reconstituted by using a mixture of linear epitopes which potentially represent the separated portions of amino acid sequence that are juxtaposed in a folded capsid. We therefore used pools of linear epitopes to block the neutralizing activity of positive human serum.
FIG. 3.
Mapping of anti-AAV antibody epitopes by ELISA and neutralization assay blocking analyses. (A) Representative analysis of peptide blocking of antibody binding in ELISA using Ser3 and peptides 53 to 62. Positive blocking was considered to cause an inhibition of at least 50% relative to a no-peptide control (no pept). (B) Pools of peptides block neutralizing antibodies. AAV antibody neutralizing assays were performed under the following conditions: plot a, AAV-GFP only; plot b, AAV-GFP with 14-peptide pool; plot c (shaded), AAV-GFP plus neutralizing Ser24; plot d, AAV-GFP plus neutralizing Ser24 in the presence of 14-peptide pool. (C) Plot a, AAV-GFP; plot b, AAV-GFP plus neutralizing Ser24 in the presence of peptide pool 2; plot c, AAV-GFP with neutralizing Ser24; plot d, (shaded), AAV-GFP plus neutralizing Ser24 in the presence of peptide pool 1; plot e, AAV-GFP plus neutralizing Ser24 in the presence of negative peptide pool.
FIG. 4.
Summary of antibody epitope mapping. Each box represents a 15-amino-acid peptide sequence from AAV VP1 starting at MAADGY and ending with LTRNL. A total of 91 peptides overlapping by five amino acids were used. The VP2 sequence begins with TAPGK (amino acid 149, peptide 17), and the VP3 sequence begins with MATGS (amino acid 203, peptide 25). Blackened boxes represent a blocking of antibody binding by this peptide in ELISA. Blocking peptide numbers are shown for reference above and below the grid. Serum sample designations are shown for reference to the left of the grid. Asterisks mark those sera that were positive for neutralizing antibodies.
FIG. 5.
Sequences of immunogenic peptides identified by peptide blocking ELISA experiments. Overlapping sequences from two positive peptides are underlined and shown as putative epitopes, and overlapping sequences from three juxtaposed peptides are double underlined. The shaded area corresponds to the conformational epitope sequences.
Identification of neutralizing antibody epitopes.
A pool of 14 peptides (peptides 4, 5, 16, 17, 33, 61, 62, 41, 43, 44, 45, 53, 58, and 90) that blocked antibody binding in the ELISA was initially used to block activity of neutralizing sera. This pool inhibited the neutralizing effect of seven different neutralizing positive sera (Ser3, Ser6, Ser7, Ser13, Ser23, Ser24, and Ser31) to the same extent (Fig. 3B). Interestingly, the peptides alone (without serum) were able to reduce AAV uptake, suggesting that this series of peptides contain mimetic sequences involved in the binding of AAV to its cognate receptor. Since neutralizing antibodies generally work by blocking the binding of virus to cell surface receptors, this supports the identification of these peptides as neutralizing epitope sequences. This pool was then divided into two pools of seven peptides. Pool 1 contained peptides 4, 5, 16, 17, 33, 61, and 62, and pool 2 contained peptides 41, 43, 44, 45, 53, 58, and 90, thus maintaining juxtaposed peptides that likely contain a single epitope within the same pool. As shown in Fig. 3C, pool 1 had no effect on neutralizing activity (of Ser24), whereas pool 2 partially reversed the neutralizing effect. A control negative pool of seven peptides (peptides 7, 8, 9, 10, 11, 12, and 85) showed no inhibition. Removal of peptide 90 from pool 2 had no effect on this inhibition, implying that the core neutralizing pool of peptides is composed of peptides 41, 43, 44, 45, 53, and 58. This same pattern was observed with five serum samples (Ser3, Ser6, Ser7, Ser23, and Ser24) and also with a neutralizing anti-AAV mouse monoclonal antibody, A20 (28, 29). The blocking of a monoclonal antibody suggests that the six peptide sequences identified here reconstitute a single conformation epitope. As shown in Fig. 5, an overlap analysis and the expendability of peptide 42 points to sequences KEVT and TSTV as key residues within this conformational epitope.
DISCUSSION
AAV-reactive antibodies are present in a large portion of our population, and a significant percentage of individuals have neutralizing antibodies against AAV. Our work shows that this neutralizing response is sufficient to prevent AAV-mediated gene delivery to the liver and that this immune barrier cannot be overcome by transient immunosuppression. Anti-CD4 and anti-CD40L antibodies have been shown to inhibit T-cell activation and B-cell activation through T-cell helper function, respectively. Therefore, these treatments would be expected to inhibit the initiation and expansion of a primary immune response and facilitate viral delivery of transgenes as previously reported (6, 10, 14). These reagents should be less effective during a secondary immune response in which a high level of neutralizing antibodies already exists, as is the case in humans and the experimental mice described here. In addition, these reagents do not specifically target viral immunity but lead to a potentially deleterious general immunosuppression. A more effective and specific strategy would be to prevent antibody binding to the viral particle. This can be achieved by using the epitope sequence information reported here.
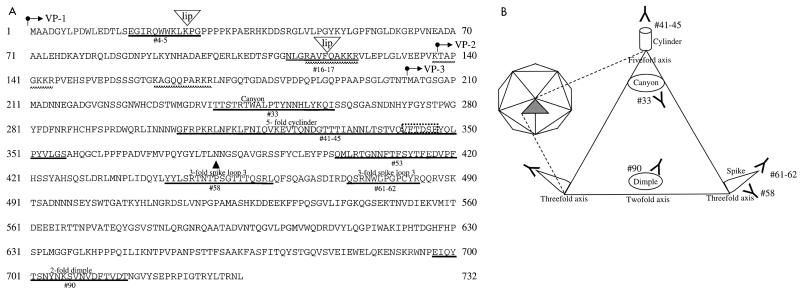
The immunogenic epitopes identified here would be expected to be on exposed surfaces of the AAV capsid since neutralizing antibodies generally bind to the virus surface to prevent virus binding to cellular receptors and viral uptake into the cell. The alignment of canine parvovirus (CPV) VP2 with the AAV sequence (beginning at amino acid 176) and superimposition on the CPV structure (3) allows the structural location of the antigenic sites identified in this study to be extrapolated (Fig. 6A). As summarized in Fig. 5 and 6, several of the B-cell epitopes identified correspond to exposed regions of AAV. The three-dimensional atomic structure of CPV has been determined (26). The virus is a T=1 icosahedral structure (depicted in Fig. 6B) composed of 60 subunits of VP1, VP2, and VP3 and is characterized by several exposed structural regions that are referred to with previously reported nomenclature (3, 26). A cylinder structure protrudes from each fivefold axis and is encircled by a canyon. Each threefold axis also has a protruding spike formed by four loops, and each twofold axis contains a depression termed a dimple. Epitope 33 lies in the canyon, and epitope 41-45 is located on the cylinder structure. Epitopes 58 and 61-62 are found on the spike region, and epitope 90 is located at the twofold dimple. In addition, neutralizing epitope 58 has been shown to bind monoclonal antibodies (12, 27) and rabbit sera (12). Furthermore, this region contains critical residues that have been shown to determine the tropism of CPV (2, 17), and different AAV subtypes (20).
FIG. 6.
Structural locations of the immunogenic regions of AAV. (A) Amino acid sequences of the overlapping VP1, VP2, and VP3 proteins that form the AAV capsid. The arrows indicate the start point of the protein sequences of VP1, VP2, and VP3. Identified epitopes are underlined in bold and marked with the corresponding peptide designation; “lip” denotes the insertion site of four amino acids that result in lip mutants (7). The basic regions proposed to interact with heparan sulfate proteoglycan are marked with a checkered line. The structural regions extrapolated from the CPV structure are marked above the corresponding sequence. ▴, key residues involved in determining tropism of CPV; dashed box, the VFTDSE sequence recognized by CPV-neutralizing dog serum. (B) Schematic representation of parvovirus structure (adapted from reference 12) that shows the approximate structural locations of the epitopes identified in this study. The icosohedral structure (left) is composed of 60 icosohedral units (shaded triangle) formed by VP1, VP2, and VP3. The expanded triangle represents one icosohedral unit.
AAV mutants that produce 0.01 to 1% of the normal virus yield have been described (7). These low-infectious-particle-yield (lip) mutants were generated by random insertion of 8- or 9-bp sequences, which results in an in-frame addition of four amino acids. Two of the three lip mutations map to and disrupt epitopes identified here, suggesting that these regions form surface-exposed domains that are critical for virus binding and uptake. Furthermore, one of several regions of basic amino acid motifs that have been identified and proposed to interact with the glucosaminoglycan component of the heparan sulfate proteoglycan receptor (25) of AAV forms part of epitope 16-17 (Fig. 6A). Our results lend support to the lip mapping data and the proposed receptor binding motifs, and they point to the importance of these identified regions for AAV infection.
The epitopes identified in this study reveal exposed capsid structures that assimilate previous genetic and structural analyses, shed light on AAV structure, suggesting a high structural conservation between AAV and CPV, and suggest contact points of AAV with its recently identified receptors (18, 24, 25). Furthermore, they provide a starting point for genetic modification of the capsid to circumvent the humoral response and to improve the design of future AAV vectors.
ACKNOWLEDGMENTS
We thank Tammy Langer, Sandra Powell, Vishalini Vimal, and Peggy Winters for technical assistance.
REFERENCES
- 1.Blacklow N R, Hoggan M D, Rowe W P. Serologic evidence for human infection with adenovirus-associated viruses. J Natl Cancer Inst. 1968;40:319–327. [PubMed] [Google Scholar]
- 2.Chang S F, Sgro J Y, Parrish C R. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J Virol. 1992;66:6858–6867. doi: 10.1128/jvi.66.12.6858-6867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman M S, Rossmann M G. Structure, sequence, and function correlations among parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbert C L, Standaert T A, Aitken M L, Alexander I E, Russell D W, Miller A D. Transduction by adeno-associated virus vectors in the rabbit airway: efficiency, persistence, and readministration. J Virol. 1997;71:5932–5941. doi: 10.1128/jvi.71.8.5932-5941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halbert C L, Standaert T A, Wilson C B, Miller A D. Successful readministration of adeno-associated virus vectors to the mouse lung requires transient immunosuppression during the initial exposure. J Virol. 1998;72:9795–9805. doi: 10.1128/jvi.72.12.9795-9805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzog R W, Hagstrom J N, Kung S H, Tai S J, Wilson J M, Fisher K J, High K A. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herzog R W, Yang E Y, Couto L B, Hagstrom J N, Elwell D, Fields P A, Burton M, Bellinger D A, Read M S, Brinkhous K M, Podsakoff G M, Nichols T C, Kurtzman G J, High K A. Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector. Nat Med. 1999;5:56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 10.Kay M A, Meuse L, Gown A M, Linsley P, Hollenbaugh D, Aruffo A, Ochs H D, Wilson C B. Transient immunomodulation with anti-CD40 ligand antibody and CTLA4Ig enhances persistence and secondary adenovirus-mediated gene transfer into mouse liver. Proc Natl Acad Sci USA. 1997;94:4686–4691. doi: 10.1073/pnas.94.9.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein R L, Meyer E M, Peel A L, Zolotukhin S, Meyers C, Muzyczka N, King M A. Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp Neurol. 1998;150:183–194. doi: 10.1006/exnr.1997.6736. [DOI] [PubMed] [Google Scholar]
- 12.Langeveld J P, Casal J I, Vela C, Dalsgaard K, Smale S H, Puijk W C, Meloen R H. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J Virol. 1993;67:765–772. doi: 10.1128/jvi.67.2.765-772.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel R J, Spratt S K, Snyder R O, Leff S E. Midbrain injection of recombinant adeno-associated virus encoding rat glial cell line-derived neurotrophic factor protects nigral neurons in a progressive 6-hydroxydopamine-induced degeneration model of Parkinson's disease in rats. Proc Natl Acad Sci USA. 1997;94:14083–14088. doi: 10.1073/pnas.94.25.14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning W C, Zhou S, Bland M P, Escobedo J A, Dwarki V. Transient immunosuppression allows transgene expression following readministration of adeno-associated viral vectors. Hum Gene Ther. 1998;9:477–485. doi: 10.1089/hum.1998.9.4-477. [DOI] [PubMed] [Google Scholar]
- 15.McCown T J, Xiao X, Li J, Breese G R, Samulski R J. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 1996;713:99–107. doi: 10.1016/0006-8993(95)01488-8. [DOI] [PubMed] [Google Scholar]
- 16.Miao C H, Snyder R O, Schowalter D B, Patijn G A, Donahue B, Winther B, Kay M A. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. [DOI] [PubMed] [Google Scholar]
- 17.Parker J S, Parrish C R. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol. 1997;71:9214–9222. doi: 10.1128/jvi.71.12.9214-9222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 19.Rose J A, Maizel J V, Jr, Inman J K, Shatkin A J. Structural proteins of adenovirus-associated viruses. J Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutledge E A, Halbert C L, Russell D W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder R O, Miao C, Meuse L, Tubb J, Donahue B A, Lin H F, Stafford D W, Patel S, Thompson A R, Nichols T, Read M S, Bellinger D A, Brinkhous K M, Kay M A. Correction of hemophilia B in canine and murine models using recombinant adeno-associated viral vectors. Nat Med. 1999;5:64–70. doi: 10.1038/4751. [DOI] [PubMed] [Google Scholar]
- 22.Snyder R O, Miao C H, Patijn G A, Spratt S K, Danos O, Nagy D, Gown A M, Winther B, Meuse L, Cohen L K, Thompson A R, Kay M A. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- 23.Snyder R O, Spratt S K, Lagarde C, Bohl D, Kaspar B, Sloan B, Cohen L K, Danos O. Efficient and stable adeno-associated virus-mediated transduction in the skeletal muscle of adult immunocompetent mice. Hum Gene Ther. 1997;8:1891–1900. doi: 10.1089/hum.1997.8.16-1891. [DOI] [PubMed] [Google Scholar]
- 24.Summerford C, Bartlett J S, Samulski R J. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 25.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsao J, Chapman M S, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T J, Rossmann M G, Compans R W, et al. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 27.Wikoff W R, Wang G, Parrish C R, Cheng R H, Strassheim M L, Baker T S, Rossmann M G. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure. 1994;2:595–607. doi: 10.1016/s0969-2126(00)00062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt J A. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wistuba A, Weger S, Kern A, Kleinschmidt J A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao W, Chirmule N, Berta S C, McCullough B, Gao G, Wilson J M. Gene therapy vectors based on adeno-associated virus type 1. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]