Abstract
To improve embryo transfer success and increase the chances of live birth in assisted reproductive methods, there is a growing demand for the use of pre-implantation genetic testing (PGT). However, the invasive approaches used in PGT have led to in vitrofertilization failure and abortions, increasing anxiety levels for parents. To address this, non-invasive PGT methods have been introduced, such as the detection of DNA in blastocoel fluid of blastocysts and spent culture media (SCM). These methods have proven to be minimally invasive and effective in detecting aneuploidy in the chromosomes of human embryos. This review aims to explore the different approaches to pre-implantation diagnosis, including invasive and non-invasive methods, with a particular focus on non-invasive PGT (niPGT). The search strategy involved gathering data from scientific databases such as PubMed, Google Scholar, and Science Direct using relevant keywords. The search was conducted until January 2023. In total, 22 studies have successfully reported the detection and amplification of cell-free DNA in the embryonic SCM. It is important to note that niPGT has some limitations, which include differences in indicators such as cell-free DNA amplification rate, concordance, level of maternal DNA contamination, sensitivity, and specificity between SCM samples and biopsied cells. Therefore, more extensive and detailed research is needed to fully understand niPGT's potential for clinical applications.
Keywords: Spent culture media, Non-invasive pre-implantation genetic testing, Biopsy methods, Cell-free embryonic DNA.
1. Introduction
Pre-implantation genetic testing (PGT) gives us information about the genetic health of the embryo and has garnered more attention over the past 2 centuries. The development of safe and accurate diagnostic methods is the predominant key to achieving a healthy birth (1, 2). A wide range of new methods have been used in this field, which will be explained in the following sections. They can be used as assisted reproductive methods to help patients with infertility and repeated miscarriages to have a healthy baby. The prevalence of infertility varies in different countries and is estimated at approximately 9% and the rate of clinical abortions is estimated to be about 10–15% of all diagnosed pregnancies. Also, one of the causes of miscarriage and infertility is the increase and decrease in the number of chromosomes, which is called aneuploidy (3).
Therefore, PGT is one of the diagnostic techniques in the field of assisted reproductive technologies that can help increase the pregnancy rate in these patients. This study aims to introduce PGT, its types, and applications, focusing on non-invasive techniques that have received special attention recently. The data collection process in scientific databases such as PubMed, Google Scholar, and Science Direct using selected keywords started in February 2022 and continued until January 2023. The keywords included the following: invasive and non-invasive PGT, pre-implantation genetic diagnosis, PGT of aneuploidy (PGT-A), blastocyst biopsy methods, cell-free DNA (cfDNA), spent culture media/medium, and assisted reproduction. We found 1128 articles and analyzed 22 original articles evaluating the niPGT method, published in English until January 2023 (Figure 1). Animal studies, case reports, and review articles were excluded from the results.
Figure 1.
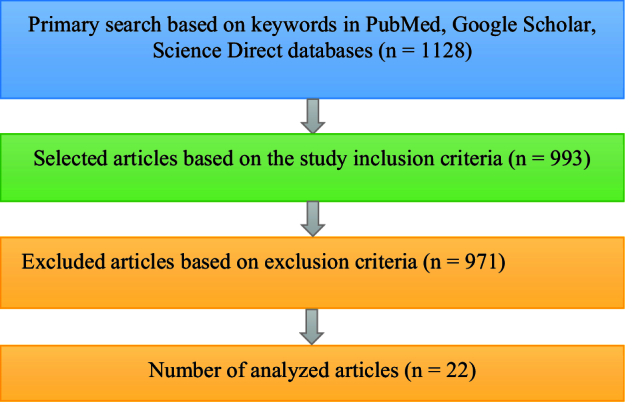
Flowchart of the methodology used in our study.
2. Overview of PGT and its approaches
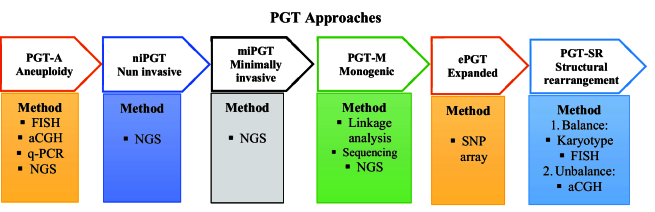
The successful rate of in vitro fertilization (IVF) depends on multiple factors including the genetic complement of the embryo. Pre-implantation genetic screening (PGS) and genetic evaluation of the embryo with the use of the fluorescence in situ hybridization technique have been applied in IVF, since 1995 (4). Across these years, PGS have utilized several cytogenetics molecular techniques like comparative genomic hybridization, array-comparative genomic hybridization, single-nucleotide polymorphism array, quantitative-polymerase chain reaction (q-PCR), and next-generation sequencing. Although, it has some drawbacks it can improve pregnancy rates by the election of euploid embryos for transfer (5, 6). PGT (PGD) refers to the detection of known mutations or chromosomal rearrangements in embryos when one or both parents are carriers (7). Hence, it is clinically important to prevent the transmission of inherited disorders in the family to the future offspring (8). Since 2017, PGS and PGD were re-termed as PGT (9), and it is widely used in IVF centers around the world to select euploid embryos (Figure 2) (10).
PGT-A is a technique that is used to screen the numerical chromosome abnormalities in the embryo. It is used instead of PGS, comprehensive chromosome screening, and aneuploidy screening to identify euploid chromosomes during IVF (11). PGT for single gene/monogenic disorders (PGT-M), formerly known as PGD, is an early genetic diagnosis test for a single defective gene that can be inherited from a parent in a carrier family or formed during germ-line development (12–14). A balanced structural rearrangement is a type of chromosomal structural variant involving translocations, insertions, Robertsonian, and inversions of chromosomes without cytogenetically apparent gain or loss. Although the carriers are not phenotypically affected, it can impact meiotic segregation leading to unbalanced gametes and an increased risk of miscarriage. PGT for structural rearrangement can improve the reproductive product in affected couples and reduce the time to achieve a successful live birth (15–17). Expanded PGT combines polygenic risk score algorithms with the newest molecular biology techniques statutes (18–20).
Minimally invasive PGT uses cell-free embryonic DNA in spent culture medium (SCM) combined with blastocoel fluid (BF) to increase the amount of assayable cell-free embryonic nuclear DNA. Due to the aforementioned requirements, a new approach has recently been presented that is an alternative to TE biopsy and has attracted a lot of attention (21). This method, called non-invasive PGT (niPGT), examines the cfDNA released by an embryo into its adjacent culture medium (22). The clinical application of niPGT is based on the elucidation of cfDNA origin and the representation degree of the whole embryo (23). However, the cfDNA collection requires less skill and makes a lower risk to embryos because trophectoderm (TE) biopsy could affect embryo health and its potential implantation rate. Different studies have reported various concordance rates between TE and SCM samples. Thus, the accuracy, specificity, and sensitivity of niPGT must be confirmed in the larger clinical trials (11), and further investigations are needed to find out the accurate mechanisms underlying the release of embryonic DNA also the whole clinical potential of niPGT remains unknown and needs more discovery (24).
Figure 2.
Overview of different PGT technical approaches. PGT: Pre-implantation genetic testing, PGT-A: PGT of aneuploidy, FISH: Fluorescence in situ hybridization, aCGH: Array comparative genomic hybridization, q-PCR: Quantitative-polymerase chain reaction, NGS: Next-generation sequencing, SNP: Single nucleotide polymorphism, PGD: Pre-implantation genetic, niPGT: Non-invasive pre-implantation genetic testing, miPGT: Minimally invasive pre-implantation genetic testing, PGT-M: PGT for monogenic disorders, ePGT: Expanded pre-implantation genetic testing, PGT-SR: Pre-implantation genetic testing for structural rearrangement.
3. Types of biopsy methods for PGT
It is fair to say that PGT methods based on biopsies of polar bodies, blastomeres, or TE cells have been somewhat successful (25). Although isolation of the first polar body (PB) for PGT purposes is often considered less invasive than other methods (26). The most common form of biopsy in PGT comprised removing one or 2 blastomeres during the cleavage stage, which was frequently done on day 3 when the embryo typically had 6–10 cells. However, this strategy has several drawbacks including the presence of chromosomal mosaicism and the phenomenon of allele dropout (27). Also, PGT-A based on TE biopsy is currently the most widely used genetic test identifying de novo aneuploidy in embryos in clinical IVF.
The removal of 5–10 TE cells in the blastocyst biopsy usually does not affect the inner cell mass (ICM) cells; therefore, it is safer than other methods and is gradually replacing them (28). There are 3 main challenges of PGT associated with TE biopsy samples: 1) laborious and time-consuming biopsy procedure, 2) invasiveness, 3) subject to sampling bias -TE biopsy may not accurately represent the ICM and the rest of the TE. The initial report about the discovery of suitable DNA for amplification and genetic testing in the BF has aroused great curiosity among scientists. This discovery has opened up the possibility of a new era of minimally invasive PGT (29).
4. The necessity of using non-invasive methods in PGT
Although embryo biopsy is presently the method of choice over the world, it has significant drawbacks as well. First, an embryo biopsy can only be done during a particular stage of the embryo's growth. Because blastomere removal impacts developing embryos, day 3 (D3) cleaved embryos with fewer than 6 blastomeres are not candidates for biopsy. Similarly, to the quantity of biopsied cells has a detrimental impact on the implantation ability of blastocysts with inadequate TE quality. Secondly, while performing an embryo biopsy, only a small portion of the entire embryo is removed, which increases the risk of genetic misdiagnosis (false positives or negatives) in cases of embryo mosaicism. Third, invasive operations reduce an embryo's ability to reproduce. Embryo growth is less than ideal when a biopsy is performed at the cleavage stage. Because TE biopsy needs in vitro culture up until the blastocyst stage, there are worries related to its safety. Fourth, time- and money-consuming intrusive procedures are required. These difficult approaches call for advanced technical abilities, ongoing training, and suitable tools (such as laser equipment) (19, 24, 29, 30).
5. Origin, importance, and analytical workflow of cfDNA in SCM
Recently, studies have shown that DNA released by the embryo into the culture medium can be used for genetic testing as a noninvasive method (31). There are different views on the question regarding what the origin of cfDNA is. In general, it is believed that processes such as apoptosis, cell lysis, cell debris, or other mechanisms during embryonic development in embryo culture may cause the DNA of cells to break and form cfDNA fragments (32). Investigations revealed 2 types of DNA fragments: one between 160–220 bp and the other between 300–400 bp (33). Another study has reported apoptosis in ICM and TE between euploid cells and aneuploid cells in mouse models, and in aneuploid embryos, the percentage of apoptotic cells in ICM has been observed to be higher than in TE. Therefore, cfDNA is mainly formed through apoptosis and originates mainly from the ICM. However, more detailed and extensive studies on the origin of this type of DNA are still needed (34).
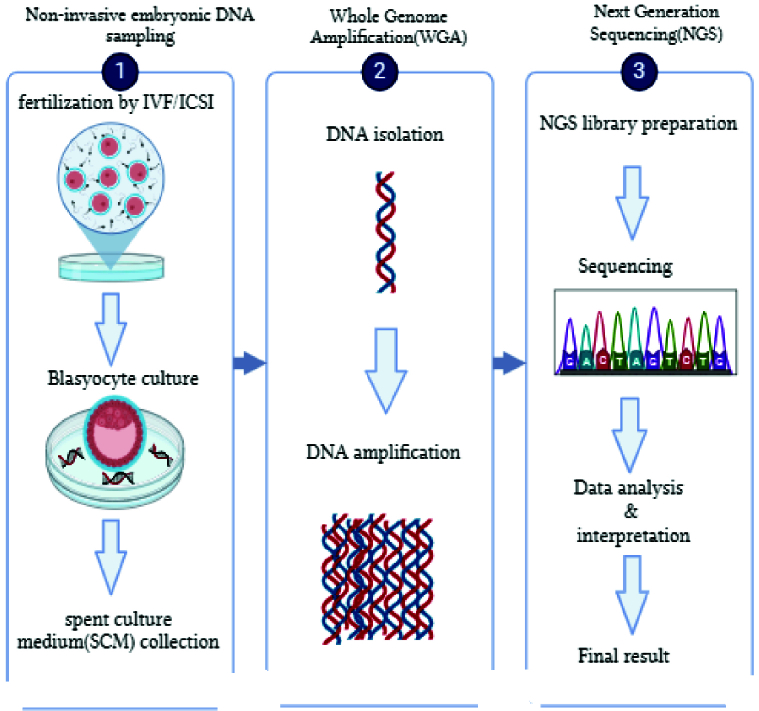
The recent finding of cfDNA in biological fluids has unlocked new perspectives for advancing non-invasive examinations within reproductive medicine. Indeed, researchers have identified cfDNA in the BF and used culture medium of embryos undergoing IVF procedures (18). Developing research has revealed the existence of cfDNA in various bodily fluids, blastocyst fluid, and the medium utilized to culture in vitro embryos (Figure 3). These findings have opened new ways for incorporating non-invasive methodologies into assisted reproductive technology. The utilization of cfDNA, which can be detected by the embryonic developmental culture material known as SCM, appears to be the most promising option for non-invasive PGT. A multitude of research groups have identified cfDNA and are currently undergoing assessment as a method for evaluating the chromosomal status of in vitro cultured embryos. A recent evaluation of 15 published studies has demonstrated that the detection of DNA in a SCM is a safe and efficacious approach for determining the chromosomal status of developing embryos. Nevertheless, the diverse methodologies employed in distinct studies have the potential to compromise the validity of the results, making it implausible to directly correlate the findings (18, 35–37).
Figure 3.
The workflow for analysis of embryonic cfDNA in blastocyst culture medium. ICSI: Intracytoplasmic sperm injection, IVF: In vitro fertilization. (The image was designed using BioRender.com database).
6. Current studies on niPGT and their summary
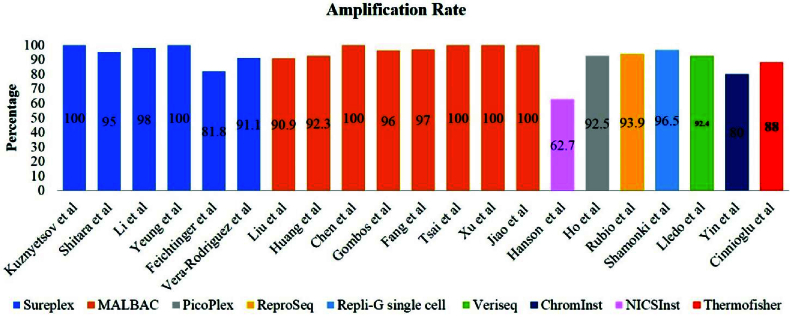
PGF for aneuploidy abnormalities is an increasingly used approach in helping couples with fertility-related problems. At present, the main drawback of the PGT-A method is its invasiveness, because it can lead to irreparable risks to the embryo (38). Therefore, there is an increasing need to develop non-invasive or minimally invasive approaches such as niPGT-A and minimally invasive PGT-A for clinical practice. Moreover, the successful amplification of the TSPY1 gene on the Y chromosome in the culture medium to determine fetal sex led to the development of non-invasive pre-implantation approaches to the management of sex-linked diseases (39). A subsequent report demonstrated successful detection of the alpha thalassemia gene using quantitative-polymerase chain reaction. Interestingly, their finding showed that the detection rate obtained from SCM was higher than that in the embryo biopsy (88.6% and 82.1%, respectively) (40). Other studies used next-generation sequencing-based techniques to investigate cfDNA presented in the culture medium (Figure 4) (41, 42). Another research team achieved 100% successful amplification rate by using the noninvasive multiple annealing and looping-based amplification cycle (MALBAC)-NGS protocol on 42 SCM samples between days 3 and 5 (22).
Also, 100% successful DNA isolation rate was reported for 47 samples analyzed through a combined protocol of blastocyst culture medium and BF. After studying 166 SCM samples, a DNA amplification failure rate of 37.3% was observed. They concluded that the current form of niPGT-A makes its clinical application practically impossible (43). However, it seems that one of the weaknesses of their study was the low sample size and the use of a special amplification method called noninvasive chromosome screening inst library preparation kit Genomics. In a recent multicenter study with a high sample size, the amplification rate in embryonic culture medium samples was evaluated around 97.4% (1267/1301) (44, 45). Consequently, one of the reasons for the apparent difference between the successful rates of DNA amplification in these studies may be due to differences in the study design.
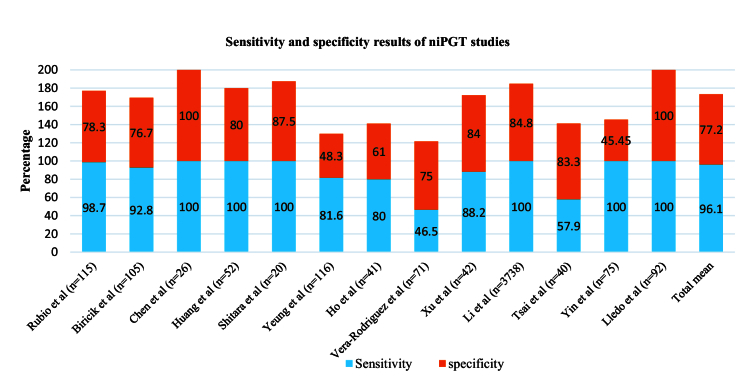
Also, in studies related to niPGT, different levels of sensitivity and specificity have been reported (Figure 5). The highest level of sensitivity, even up to 100%, was observed in some studies, and the highest rate of specificity has been reported in the study of other researches (8, 46). One of the reasons for the high sensitivity and specificity of these studies can be related to the type of whole genome amplification (WGA) method used for DNA amplification and culture time; most of these studies have chosen MALBAC or SurePlex methods and D5/D6 culture time (47). Additionally, to obtain the concordance rate, it is necessary to compare niPGT results with embryo biopsy results. Studies have used different biopsy methods for this comparison, including PB biopsy, TE biopsy (16, 31, 48, 49), BF (50, 51), and whole embryo (28). For the PB biopsy, a lower concordance rate was obtained (27%), while for the BF 87.5% rate was observed. Also, when TE biopsy was used, the concordance rate was between 3.5–93.8% and these differences were due to other reasons, such as the type of cfDNA amplification method, the WGA method used, the type of culture medium and its volume, the level of maternal contamination, etc. can happen, which will be discussed further (52).
Figure 4.
The rate of successful amplification of niPGT studies by various WGA methods. MALBAC: Multiple annealing and looping-based amplification cycle, NICS: Noninvasive implantation capability screening.
Figure 5.
Sensitivity and specificity of results in niPGT studies.
7. Tips and tricks in niPGT using SCM
Our analysis of various studies led to the identification of important tips and tricks in niPGT using SCM, which are summarized below:
Types of embryo culture medium systems
Embryo culture systems are commercially divided into 2 types: single-step and 2-step culture media. 2-step or sequential culture medium, takes into account the needs of the embryo in its different stages of growth and development, its essential nutrients are provided in the culture medium. In single-step culture medium, the nutritional requirements of the embryo were mixed and added to the culture medium in one step. The embryos then chose the nutrients based on their needs. The type of culture medium is important in 2 aspects: 1) Degradation of cfDNA in culture medium decreases the DNA quality, as a result, the amount of time spent on cfDNA degradation can be diminished by changing the culture medium. 2) By changing the culture medium, the amount of maternal DNA contamination produced by cumulus cells can be reduced. In our investigations, it was found that most of the studies used a 2-step system to analyze the SCM of the embryos (Table I). In some studies, however, no significant difference was observed, by comparing both types of culture medium system regarding their effect on the accuracy of niPGT-A results (44).
Table 1.
Summary of studies evaluating the amplification and analysis outcomes of cell-free DNA in the SCM
|
| |||||||||||
| Author, yr (Ref) | Number of samples | Amplification rate | Concordance rate | Discordance rate | Culture volume (µL) | Culture medium (single/2-stage) | Fertilization method | Manipulation of embryo | Culture time | Amplification method | WGA method |
| Kuznyetsov et al. , 2020 (29) | 47 | 100 (47/47) | 97.8 (45/47) | 2.2 (2/47) | 25 | 2 steps | ICSI | Cryopreservation on D4 | D4/D5/D6 | SurePlex (Illumina) | NGS |
| Chen et al. , 2022 (53) | 26 | 100 (26/26) | 80.8 (21/26) | 19.2 (5/26) | 10 | 2 steps | ICSI | Cryopreservation | D5/D6/D7 | MALBAC (Yikon) | NGS |
| Gombos et al. , 2021 (35) | 753 | 95–96 | - - | 20–40 | 2 steps | ICSI | Assisted hatching on D3 | D3 | MALBAC (Yikon) | NGS | |
| Huang et al. , 2022 (51) | 166 | 62.7 (104/166) | 59.6 (62/104) | 40.4 (42/104) | 30 | 2 steps | IVF and ICSI | Cryopreservation D3 | D5/D6/D7 | NICSInst Genomics (Yikon) | NGS |
| Shitara et al. , 2021 (31) | 20 | 95 (19/20) | 56.3 (9/16) | 43.7 (7/16) | - 2 steps | IVF | Cryopreservation on D5/D6 | D5/D6 | SurePlex (Illumina) | NGS | |
| Brouillet et al. , 2020 (54) | 3738 | 98 (3662/3738) | 87.2 | 12.8 | 30 | - ICSI | Cryopreservation on D3 | D3-D5/D6 | SurePlex (BlueGnome) | NGS | |
| Yeung et al. , 2019 (52) | 168 | 100 (26/26) | 87.8 (21/26) | 19.2 (5/26) | 10 | 2 steps | ICSI | Assisted hatching on D3 | D3-D5/D6 | SurePlex (Illumina) | NGS |
| Tsai et al. , 2022 (55) | 40 | 100 (40/40) | 67.7 (21/31) | 32.3 (10/31) | 30 | Single step | IVF/ICSI | Cryopreservation | D3-D5/D6 | MALBAC (Yikon) | NGS |
| Alteri et al. , 2023 (28) | 57 | 96.5 (55/57) | 33.3 (2/6) | - 15 | 2 steps | ICSI | Assisted hatching on D3 | D3-D5/D6 | Repli-G single-cell kit (Qiagen) | aCGH | |
| Xu et al. , 2016 (56) | 42 | 100 (42/42) | 85.7 (36/42) | 14.3 (6/42) | 30 | 2 steps | ICSI | Cryopreservation on D3 | D3-D5 | MALBAC (Yikon) | NGS |
| Jiao et al. , 2019 (30) | 62 | 100 (62/62) | 76.19 (16/21) | - 12 | - ICSI | Freeze-thaw. Laser collapse | D5/D6 | MALBAC (Yikon) | NGS | ||
| Feichtinger et al. , 2017 (57) | 22 | 81.8 (55/57) | 72.2 (13/18) | 27.8 (5/18) | 25 | Single step | ICSI | Cryopreservation on D3 | D1-D5/D6 | SurePlex (Illumina) | aCGH |
| Vera-Rodriguez et al. , 2017 (58) | 56 | 91.1 (51/56) | 33.3 (17/51) | - 20 | 2 steps | ICSI | Assisted hatching on D3 | D3-D5 | SurePlex + ReproSeq (Illumina) | NGS | |
| Rubio et al. , 2020 (44) | 1301 | 97.5 (79/80) | 78.2 (866/1108) | 137 false positive, 92 false negative | 10 | A single step or 2 steps | IVF/ICSI | Cryopreservation + cryopreservation on D3 | D6/D7 | - NGS | |
| Liu et al. , 2020 (59) | 88 | 90.9 (80/88) | 64.52 | 35.48 | 30 | Single step | ICSI | Cryopreservation | D1-D5 | MALBAC (Yikon) | NGS |
| Ho et al. , 2018 (60) | 41 | 92.5 (37/40) | 65 (26/40) | 8 false positive, 3 false negative | 25 | Single step | ICSI | Cryopreservation + assisted hatching on D3 | D3-D5 | PicoPlex (Rubicon) | NGS |
| Rubio et al. , 2019 (47) | 115 | 93.9 (108/115) | Overall: 7/78 (108/85) D5: 63 (27/17) D6/7: 84 (81/68) | 8 false positive, 1 false negative | 10 | Single step | IVF | - D4-D5/D6/D7 | ReproSeq (Life Technologies) | NGS | |
| Xu et al. , 2016 (50) | 170 | 97 | - - | 20–25 | 2 steps | ICSI | Cryopreservation | D3-D5/D6 | MALBAC (Yikon) | NGS | |
| Yin et al. , 2021 (48) | 75 | 80 (60/75) | 89.83 (53/59) | 10.17 (6/59) | 25 | 2 steps | IVF | Cryopreservation | D5 | ChromInst (Yikon) | NGS |
| Lledo et al. , 2018 (37) | 92 | 92.4 (85/92) | 74.6 (62/83) | 25.4 (21/83) | 20 | 2 steps | IVF | Assisted hatching on D3 | D5-D6 | SurePlex | NGS |
| Huang et al. , 2022 (51) | 52 | 92.3 (48/52) | 93.8 (45/48) | 6.2 (3/48) | 25 | Single step | ICSI | Assisted hatching on D3 | D4-D5/D6/D7 | Modified MALBAC (Yikon) | NGS |
| Cinnioglu et al. , 2023 (61) | 50 | 88 (44/50) | 78.9 (30/38) | 21 (8/38) | 30 | - - | - D5-D6 | Thermofisher | NGS | ||
| SCM: Spent culture medium, WGA: Whole-genome amplification, ICSI: Intracytoplasmic sperm injection, D: Days, NGS: Next-generation sequence, IVF: In vitro fertilization, aCGH: Array comparative genomic hybridization, MALBAC: Multiple annealing and looping-based amplification cycle, NICSInst: Noninvasive chromosome screening inst library preparation kit | |||||||||||
The volume of embryo culture medium
In general, studies on the amount of SCM collected to amplify DNA present in culture media have reported a wide range. This factor is important, because it potentially has a direct effect on the proportion of extra-embryonic DNA collected and changes the initial concentration of the template. In the niPGT method, determining the optimal volume for embryo culture and subsequent analysis is challenging and problematic. These challenges include:
1. Inhibitory effect of culture medium components on DNA amplification during WGA and PCR.
2. Design of the most commercial WGA protocols with very small volumes (usually 10 µL) for use in SCM. Using more volume leads to increased costs. To overcome these challenges, some authors have suggested embryo culture in smaller volumes (47). This reduction in culture volume requires careful and detailed examinations because it is vital to ensure normal growth and survival and no risk to the embryo.
Timing of SCM collection
The timing of SCM collection during embryo culture is also considered one of the crucial factors that may lead to different results of the analysis of niPGT. In a study by Lane et al., they observed that by comparing the time of collecting the medium, a higher accuracy for SCM samples D4–D5 compared to D3–D5 was obtained (62). Also, another study reported SCM consistency and decreased levels of maternal contamination of D4–D6/7 significantly higher than D4–D5. In figure 6, the results for concordance rate, false positive and negative rate, and maternal DNA contamination are shown for different sampling methods (63). Sampling method 1, which was performed by collecting SCM on D4–D6/7, showed the highest concordance rate (84%), the lowest false positive rate, and maternal contamination (8.5% and 5%, respectively).
Figure 6.
Comparison of different sampling methods and their results in a study by Huang et al. (51).
Methods of cfDNA amplification
There are several types of WGA techniques used to amplify cfDNA in SCM, including multiple annealing and looping amplification cycles (MALBAC) and sureplex/picoplex, repli-G single cell, multiple displacement amplification, and veriseq. As shown in figure 4, several niPGT-A studies have used the MALBAC method to amplify cfDNA in SCM with a successful rate of over 90%, and using the PicoPlex/SurePlex method, a successful amplification rate of over 80% has been reported. Therefore, using the aforementioned methods for cfDNA amplification in SCM is more common and practical. Also, the high application of the MALBAC method for cfDNA analysis can be due to its high genome coverage and very low allele deletion ratio (64).
Limitations of performing niPGT
Recent developments in the field of cfDNA discovery in BF and SCM have given rise to a greater interest in the niPGT field to make a less invasive medical diagnosis of embryo health status, but it has some limitations. One of the biggest challenges of niPGT is a lower quantity and lesser quality of the cell-free genetic material, and its unspecified origin (65). But some researchers believe that despite the fact that embryonic and maternal DNA differences are still ineffectively characterized, this problem does not make a threat to diagnostic validity of niPGT for monogenic and X-linked disease. In these cases, the only limitation might be the incomplete representation of the whole embryonic genome by cfDNA (21). Hence, low abundance and poor integrity, make technical challenges for genetic analysis as in this time it is not clear which method is more accurate to analyze the extra-embryonic DNA and to get a precise clinical diagnosis, the origin of extra-embryonic DNA should be definite and the factors that create discordance with results must be discovered.
8. Conclusion
In this study, we investigated the PGT approach and its types, especially the non-invasive methods, and explained the factors affecting the rate of amplification, concordance, sensitivity, specificity, and current limitations in the field of using cfDNA in PGT. Much attention has been paid to non-invasive PGT due to its cost-effectiveness, no damage to the fetus, and no need for biopsy. However, important challenges in its clinical application include differences in results obtained from niPGT compared to results obtained from biopsy samples, maternal DNA contamination, fetal mosaicism, and the relatively low abundance of DNA present in SCM. Therefore, it seems necessary to conduct more research to evaluate the potential of using PGT in clinical practice.
Data availability
Not applicable.
Author contributions
SM Kalantar, F Montazeri, and N Karami designed the study and conducted the paper.
N Karami, F Iravani and F Montazeri had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of presented data.
F Iravani, S Bakhshandeh Bavarsad, SM Hoseini and S Asadollahi: Drafting of the manuscript.
SM Hoseini, S Asadollahi, and S Bakhshandeh Bavarsad reviewed the article and made the necessary corrections as requested by the referees.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- Cheng W-L, Hsiao Ch-H, Tseng H-W, Lee T-P. Noninvasive prenatal diagnosis. Taiwan J Obstet Gynecol. 2015;54:343–349. doi: 10.1016/j.tjog.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Nguyen MTM, Knoppers BM. Milunsky A, Milunsky JM. Genetic disorders and the fetus: Diagnosis, prevention, and treatment. 8. US: Wiley-Blackwell Publisher; Prenatal and preimplantation diagnosis: International policy perspectives; 2015 pp. [Google Scholar]
- Fleddermann L, Hashmi SS, Stevens B, Murphy L, Rodriguez-Buritica D, Friel LA, et al. Current genetic counseling practice in the United States following positive non-invasive prenatal testing for sex chromosome abnormalities. J Genet Couns. 2019;28:802–811. doi: 10.1002/jgc4.1122. [DOI] [PubMed] [Google Scholar]
- Dahdouh EM, Balayla J, García-Velasco JA. Comprehensive chromosome screening improves embryo selection: A meta-analysis. Fertil Steril. 2015;104:1503–1512. doi: 10.1016/j.fertnstert.2015.08.038. [DOI] [PubMed] [Google Scholar]
- El Hachem H, Crepaux V, May-Panloup P, Descamps P, Legendre G, Bouet P-E. Recurrent pregnancy loss: Current perspectives. Int J Womens Health. 2017;9:331–345. doi: 10.2147/IJWH.S100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutzler AG. Theory and practice of preimplantation genetic screening (PGS) Eur J Med Genet. 2019;62:103670. doi: 10.1016/j.ejmg.2019.103670. [DOI] [PubMed] [Google Scholar]
- Sullivan-Pyke Ch, Dokras A. Preimplantation genetic screening and preimplantation genetic diagnosis. Obstet Gynecol Clin. 2018;45:113–125. doi: 10.1016/j.ogc.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Chen H-F, Chen S-U, Ma G-C, Hsieh S-T, Tsai H-D, Yang Y-S, et al. Preimplantation genetic diagnosis and screening: Current status and future challenges. J Formos Med Assoc. 2018;117:94–100. doi: 10.1016/j.jfma.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Munné S. Status of preimplantation genetic testing and embryo selection. Reprod Biomed Online. 2018;37:393–396. doi: 10.1016/j.rbmo.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Greco E, Litwicka K, Minasi MG, Cursio E, Greco PF, Barillari P. Preimplantation genetic testing: Where we are today. Int J Mol Sci. 2020;21:4381. doi: 10.3390/ijms21124381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesahat F, Montazeri F, Hoseini SM. Preimplantation genetic testing in assisted reproduction technology. J Gynecol Obstet Hum Reprod. 2020;49:101723. doi: 10.1016/j.jogoh.2020.101723. [DOI] [PubMed] [Google Scholar]
- Chada AR, Crawford S, Hipp HS, Kawwass JF. Trends and outcomes for preimplantation genetic testing for monogenic disorders in the United States, 2014-2018. Fertil Steril. 2022;118:1190–1193. doi: 10.1016/j.fertnstert.2022.08.854. [DOI] [PubMed] [Google Scholar]
- Parikh FR, Athalye AS, Kulkarni DK, Sanap RR, Dhumal SB, Warang DJ, et al. Evolution and utility of preimplantation genetic testing for monogenic disorders in assisted reproduction: A narrative review. J Hum Reprod Sci. 2021;14:329–339. doi: 10.4103/jhrs.jhrs_148_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycke M, Berckmoes V. Preimplantation genetic testing for monogenic disorders. Genes (Basel) 2020;11:871. doi: 10.3390/genes11080871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu-Brull E, Rodrigo L, Peinado V, Mercader A, Campos-Galindo I, Bronet F, et al. Interchromosomal effect in carriers of translocations and inversions assessed by preimplantation genetic testing for structural rearrangements (PGT-SR) J Assist Reprod Genet. 2019;36:2547–2555. doi: 10.1007/s10815-019-01593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TF, Chen MJ, Lee MS, Huang CC, Ho ST, Cheng EH, et al. Comparison of clinical outcome between day 5 and day 6 single blastocyst transfers in cycles undergoing preimplantation genetic testing for aneuploidy. Taiwan J Obstet Gynecol. 2023;62:429–433. doi: 10.1016/j.tjog.2023.03.005. [DOI] [PubMed] [Google Scholar]
- Dong Z, Wang H, Chen H, Jiang H, Yuan J, Yang Z, et al. Identification of balanced chromosomal rearrangements previously unknown among participants in the 1000 genomes project: Implications for interpretation of structural variation in genomes and the future of clinical cytogenetics. Genet Med. 2018;20:697–707. doi: 10.1038/gim.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakourou G, Mamas T, Vrettou C, Traeger-Synodinos J. An update on non-invasive approaches for genetic testing of the preimplantation embryo. Curr Genomics. 2022;23:337–352. doi: 10.2174/1389202923666220927111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Hao Y, Chen D, Ji D, Zhu W, Zhu X, et al. Non-invasive preimplantation genetic testing for putative mosaic blastocysts: A pilot study. Hum Reprod. 2021;36:2020–2034. doi: 10.1093/humrep/deab080. [DOI] [PubMed] [Google Scholar]
- Treff NR, Zimmerman R, Bechor E, Hsu J, Rana B, Jensen J, et al. Validation of concurrent preimplantation genetic testing for polygenic and monogenic disorders, structural rearrangements, and whole and segmental chromosome aneuploidy with a single universal platform. Eur J Med Genet. 2019;62:103647. doi: 10.1016/j.ejmg.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Farra C, Choucair F, Awwad J. Non-invasive pre-implantation genetic testing of human embryos: An emerging concept. Hum Reprod. 2018;33:2162–2167. doi: 10.1093/humrep/dey314. [DOI] [PubMed] [Google Scholar]
- Xu J, Fang R, Chen L, Chen D, Xiao J-P, Yang W, et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci. 2016;113:11907–11912. doi: 10.1073/pnas.1613294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeriy K, Svetlana M, Ran A, Rina A, Gelareh M, Zenon I, et al. [Google Scholar]
- Leaver M, Wells D. [Google Scholar]
- De Rycke M, Goossens V, Kokkali G, Meijer-Hoogeveen M, Coonen E, Moutou C. ESHRE PGD consortium data collection XIV-XV: Cycles from January 2011 to December 2012 with pregnancy follow-up to October 2013. Hum Reprod. 2017;32:1974–1994. doi: 10.1093/humrep/dex265. [DOI] [PubMed] [Google Scholar]
- Montag M, Van der Ven K, Rösing B, Van der Ven H. [Google Scholar]
- Piyamongkol W, Bermúdez MG, Harper JC, Wells D. Detailed investigation of factors influencing amplification efficiency and allele drop‐out in single cell PCR: Implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003;9:411–420. doi: 10.1093/molehr/gag051. [DOI] [PubMed] [Google Scholar]
- Alteri A, Cermisoni GC, Pozzoni M, Gaeta G, Cavoretto PI, Vigano P. Obstetric, neonatal, and child health outcomes following embryo biopsy for preimplantation genetic testing. Hum Reprod Update. 2023;29:291–306. doi: 10.1093/humupd/dmad001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznyetsov V, Madjunkova S, Abramov R, Antes R, Ibarrientos Z, Motamedi G, et al. Minimally invasive cell-free human embryo aneuploidy testing (miPGT-A) utilizing combined spent embryo culture medium and blastocoel fluid-towards development of a clinical assay. Sci Rep. 2020;10:7244. doi: 10.1038/s41598-020-64335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Shi B, Sagnelli M, Yang D, Yao Y, Li W, et al. Minimally invasive preimplantation genetic testing using blastocyst culture medium. Hum Reprod. 2019;34:1369–1379. doi: 10.1093/humrep/dez075. [DOI] [PubMed] [Google Scholar]
- Shitara A, Takahashi K, Goto M, Takahashi H, Iwasawa T, Onodera Y, et al. Cell-free DNA in spent culture medium effectively reflects the chromosomal status of embryos following culturing beyond implantation compared to trophectoderm biopsy. PLoS One. 2021;16:e0246438. doi: 10.1371/journal.pone.0246438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond ER, Shelling AN, Cree LM. Nuclear and mitochondrial DNA in blastocoele fluid and embryo culture medium: Evidence and potential clinical use. Hum Reprod. 2016;31:1653–1661. doi: 10.1093/humrep/dew132. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li N, Wang L, Sun H, Ma M, Wang H, et al. Molecular analysis of DNA in blastocoele fluid using next-generation sequencing. J Assist Reprod Genet. 2016;33:637–645. doi: 10.1007/s10815-016-0667-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton H, Graham SJ, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7:11165. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos K, Galik B, Kalacs KI, Godony K, Varnagy A, Alpar D, et al. NGS-based application for routine non-invasive pre-implantation genetic assessment in IVF. Int J Mol Sci. 2021;22:2443. doi: 10.3390/ijms22052443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer HA. Preimplantation genetic testing for aneuploidy (PGT-A): The biology, the technology and the clinical outcomes. Aust N Z J Obstet Gynaecol. 2019;59:317–324. doi: 10.1111/ajo.12960. [DOI] [PubMed] [Google Scholar]
- Lledo B, Ortiz JA, Morales R, Garcia-Hernandez E, Ten J, Bernabeu A, et al. Comprehensive mitochondrial DNA analysis and IVF outcome. Hum Reprod Open. 2018;2018:hoy023. doi: 10.1093/hropen/hoy023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WY, von Versen-Höynck F, Kapphahn KI, Fleischmann RR, Zhao Q, Baker VL. Maternal and neonatal outcomes associated with trophectoderm biopsy. Fertil Steril. 2019;112:283–290. doi: 10.1016/j.fertnstert.2019.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Aït-Ahmed O, El Messaoudi S, Thierry AR, Hamamah S. Non-invasive pre-implantation genetic diagnosis of X-linked disorders. Med Hypotheses. 2014;83:506–508. doi: 10.1016/j.mehy.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Wu H, Ding C, Shen X, Wang J, Li R, Cai B, et al. Medium-based noninvasive preimplantation genetic diagnosis for human α-thalassemias-SEA. Medicine (Baltimore) 2015;94:e669. doi: 10.1097/MD.0000000000000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, Yuan Z, Hu H, Zhang W, Wang G, Wang X. Genome-wide discovery of circulating cell-free DNA methylation biomarkers for colorectal cancer detection. Clin Epigenetics. 2023;15:119. doi: 10.1186/s13148-023-01518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamonki MI, Jin H, Haimowitz Z, Liu L. Proof of concept: preimplantation genetic screening without embryo biopsy through analysis of cell-free DNA in spent embryo culture media. Fertil Steril. 2016;106:1312–1318. doi: 10.1016/j.fertnstert.2016.07.1112. [DOI] [PubMed] [Google Scholar]
- Hanson BM, Tao X, Hong KH, Comito CE, Pangasnan R, Seli E, et al. Noninvasive preimplantation genetic testing for aneuploidy exhibits high rates of deoxyribonucleic acid amplification failure and poor correlation with results obtained using trophectoderm biopsy. Fertil Steril. 2021;115:1461–1470. doi: 10.1016/j.fertnstert.2021.01.028. [DOI] [PubMed] [Google Scholar]
- Rubio C, Navarro-Sanchez L, Garcia-Pascual CM, Ocali O, Cimadomo D, Venier W, et al. Multicenter prospective study of concordance between embryonic cell-free DNA and trophectoderm biopsies from 1301 human blastocysts. Am J Obstet Gynecol. 2020;223:751. doi: 10.1016/j.ajog.2020.04.035. [DOI] [PubMed] [Google Scholar]
- Farra C, Choucair F, Awwad J. Corrigendum. Non-invasive pre-implantation genetic testing of human embryos: an emerging concept Hum Reprod. 2019;34:590. doi: 10.1093/humrep/dey385. [DOI] [PubMed] [Google Scholar]
- Biricik A, Cotroneo E, Minasi MG, Greco PF, Bono S, Surdo M, et al. Cross-validation of next-generation sequencing technologies for diagnosis of chromosomal mosaicism and segmental aneuploidies in preimplantation embryos model. Life (Basel) 2021;11:340. doi: 10.3390/life11040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C, Rienzi L, Navarro-Sanchez L, Cimadomo D, Garcia-Pascual CM, Albricci L, et al. Embryonic cell-free DNA versus trophectoderm biopsy for aneuploidy testing: Concordance rate and clinical implications. Fertil Steril. 2019;112:510–519. doi: 10.1016/j.fertnstert.2019.04.038. [DOI] [PubMed] [Google Scholar]
- Yin B, Zhang H, Xie J, Wei Y, Zhang C, Meng L. Validation of preimplantation genetic tests for aneuploidy (PGT-A) with DNA from spent culture media (SCM): Concordance assessment and implication. Reprod Biol Endocrinol. 2021;19:41. doi: 10.1186/s12958-021-00714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Shi W, Li Y, Tong J, Xue X, Zhao Z, et al. SCM is potential resource for non-invasive preimplantation genetic testing based on human embryos single-cell sequencing. Gene. 2023;882:147647. doi: 10.1016/j.gene.2023.147647. [DOI] [PubMed] [Google Scholar]
- Xu J, Fang R, Chen L, Chen D, Xiao JP, Yang W, et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci U S A. 2016;113:11907–11912. doi: 10.1073/pnas.1613294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rong L, Zeng L, Hu L, Shi J, Cai L, et al. Embryo selection through non-invasive preimplantation genetic testing with cell-free DNA in spent culture media: A protocol for a multicentre, double-blind, randomised controlled trial. BMJ Open. 2022;12:e057254. doi: 10.1136/bmjopen-2021-057254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung QSY, Zhang YX, Chung JPW, Lui WT, Kwok YKY, Gui B, et al. A prospective study of non-invasive preimplantation genetic testing for aneuploidies (NiPGT-A) using next-generation sequencing (NGS) on spent culture media (SCM) J Assist Reprod Genet. 2019;36:1609–1621. doi: 10.1007/s10815-019-01517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xu Y, Ding C, Wang Y, Fu Y, Cai B, et al. The inconsistency between two major aneuploidy-screening platforms-single-nucleotide polymorphism array and next-generation sequencing-in the detection of embryo mosaicism. BMC Genomics. 2022;23:62. doi: 10.1186/s12864-022-08294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillet S, Martinez G, Coutton C, Hamamah S. Is cell-free DNA in spent embryo culture medium an alternative to embryo biopsy for preimplantation genetic testing? A systematic review. Reprod Biomed Online. 2020;40:779–796. doi: 10.1016/j.rbmo.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Tsai NC, Chang YC, Su YR, Lin YC, Weng PL, Cheng YH, et al. Validation of non-invasive preimplantation genetic screening using a routine IVF laboratory workflow. Biomedicines. 2022;10:1386. doi: 10.3390/biomedicines10061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Pan W. Binomial mixture model based association testing to account for genetic heterogeneity for GWAS. Genet Epidemiol. 2016;40:202–209. doi: 10.1002/gepi.21954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feichtinger M, Vaccari E, Carli L, Wallner E, Madel U, Figl K, et al. Non-invasive preimplantation genetic screening using array comparative genomic hybridization on spent culture media: A proof-of-concept pilot study. Reprod Biomed Online. 2017;34:583–589. doi: 10.1016/j.rbmo.2017.03.015. [DOI] [PubMed] [Google Scholar]
- Vera-Rodriguez M, Rubio C. Assessing the true incidence of mosaicism in preimplantation embryos. Fertil Steril. 2017;107:1107–1112. doi: 10.1016/j.fertnstert.2017.03.019. [DOI] [PubMed] [Google Scholar]
- Liu YL, Yu TN, Wang PH, Tzeng CR, Chen CH, Chen CH. Could PGT-A pick up true abnormalities that have clinical relevance? Retrospective analysis of 1043 embryos. Taiwan J Obstet Gynecol. 2020;59:496–501. doi: 10.1016/j.tjog.2020.05.005. [DOI] [PubMed] [Google Scholar]
- Ho JR, Arrach N, Rhodes-Long K, Ahmady A, Ingles S, Chung K, et al. Pushing the limits of detection: Investigation of cell-free DNA for aneuploidy screening in embryos. Fertil Steril. 2018;110:467–475. doi: 10.1016/j.fertnstert.2018.03.036. [DOI] [PubMed] [Google Scholar]
- Cinnioglu C, Glessner H, Jordan A, Bunshaft S. A systematic review of noninvasive preimplantation genetic testing for aneuploidy. Fertil Steril. 2023;120:235–239. doi: 10.1016/j.fertnstert.2023.06.013. [DOI] [PubMed] [Google Scholar]
- Lane M, Zander-Fox D, Hamilton H, Jasper M, Hodgson B, Fraser M, et al. [Google Scholar]
- Huang J, Yao Y, Jia J, Zhu X, Ma J, Wang J, et al. Chromosome screening of human preimplantation embryos by using spent culture medium: Sample collection and chromosomal ploidy analysis. J Vis Exp. 2021;175:e62619. doi: 10.3791/62619. [DOI] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomic M, Vrtacnik Bokal E, Stimpfel M. Non-invasive preimplantation genetic testing for aneuploidy and the mystery of genetic material: A review article. Int J Mol Sci. 2022;23:3568. doi: 10.3390/ijms23073568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.