Abstract
The protein kinase inhibitor H7 blocks influenza virus replication, inhibits production of the matrix protein (M1), and leads to a retention of the viral ribonucleoproteins (vRNPs) in the nucleus at late times of infection (K. Martin and A. Helenius, Cell 67:117–130, 1991). We show here that production of assembled vRNPs occurs normally in H7-treated cells, and we have used H7 as a biochemical tool to trap vRNPs in the nucleus. When H7 was removed from the cells, vRNP export was specifically induced in a CHO cell line stably expressing recombinant M1. Similarly, fusion of cells expressing recombinant M1 from a Semliki Forest virus vector allowed nuclear export of vRNPs. However, export was not rescued when H7 was present in the cells, implying an additional role for phosphorylation in this process. The viral NS2 protein was undetectable in these systems. We conclude that influenza virus M1 is required to induce vRNP nuclear export but that cellular phosphorylation is an additional factor.
Influenza virus has a segmented genome consisting of eight single-stranded, negative-sense RNAs packaged into helical viral ribonucleoprotein (vRNP) complexes (for a review, see reference 19). The nucleoprotein (NP) is the most abundant structural component of the vRNPs. In the virus particle, the vRNPs are connected to each other and with the viral envelope by the matrix protein, M1. The viral NS2 protein is associated with M1-containing vRNPs (37). The vRNPs display bidirectional traffic through the nuclear envelope (33). During virus entry, rapid import of incoming vRNPs occurs from the cytosol to the nucleoplasm through the nuclear pore complexes (22). Replication and transcription of viral RNAs then commence inside the nucleus (see reference 19 for a review). Viral mRNAs are exported to the cytosol and used for translation of nonstructural and structural viral proteins. Many of these proteins are then imported to the nucleus to support continued viral replication and assembly of progeny vRNPs. Replication of viral RNA appears to occur in proximity to the nuclear matrix (3, 20)—the insoluble “skeleton” of the nucleus.
Export of vRNPs to the cytosol occurs late in infection. There is evidence that export requires the synthesis of M1 (21), one of several late viral proteins. Why M1 is essential for export of vRNPs from the nucleus to the cytosol is not clear. It may escort the vRNPs from the nucleus and through the nuclear pores, or it may be needed to release the bound vRNPs from the nuclear matrix (38). Another possibility is that, by associating with vRNPs in the cytosol, M1 may prevent their reimport into the nucleus (32).
In this study, we have examined the requirement of M1 for nuclear export of vRNPs. We made use of a protein kinase inhibitor, H7 (11), which blocks the synthesis of M1 and other late virus proteins such as hemagglutinin (15, 18). In H7-treated cells, synthesis of the early viral proteins, including NP, occurs and viral transcription is not significantly affected (18). The inhibitory effect of H7 is thought to occur at the level of viral mRNA export from the nucleus; while mRNAs for the early viral proteins are apparently exported normally, the mRNAs for the late viral proteins are selectively retained (30). Another effect of H7 is that vRNPs appear to be retained in the nucleus (21).
We show here that in the presence of H7, vRNPs are assembled in the nucleus. Following removal of H7, vRNPs can be induced for nuclear export by expression of recombinant M1. NS2 was undetectable in these assays and was apparently unnecessary for export in this system. These data show the essential role of M1 in vRNP nuclear export and indicate a role for cellular phosphorylation in this process.
MATERIALS AND METHODS
Cells and virus. (i) Cell culture.
CHO and HeLa cells were passaged twice weekly and maintained in the alpha modification of Eagle's medium (αMEM) containing 7% fetal calf serum (FCS), 100 U of penicillin per ml, and 10 μg of streptomycin per ml. L929 cells were also passaged twice weekly and grown in αMEM containing 10% FCS–penicillin–streptomycin.
(ii) Virus infections.
Influenza A virus (strain WSN) (34) was used for all experiments. Stocks of virus were prepared from the supernatant of Madin-Darby bovine kidney (MDBK) cells, and plaque titers were determined on MDBK or MDCK cells. For virus infection, stocks were diluted in RPMI 1680 medium, containing 0.2% bovine serum albumin, buffered to pH 6.8 with HEPES (RPMI-bovine serum albumin), and virus (2 to 5 PFU/ml) was adsorbed for 60 min at 37°C. Cells were then washed, and cells were maintained in αMEM containing 2% FCS. For radiolabeling studies, cells were incubated with 20 μCi of Tran35S-label (Amersham) per ml for 30 min.
Drug treatment.
H7 was obtained from Alexis Biochemicals. We found that the required concentration of H7 was highly dependent on the particular batch of drug obtained, and in order to reproduce the results of Martin and Helenius (21), H7 concentrations between 25 and 75 μM were needed. H7 was added at the time of infection (0 h). Cycloheximide (Sigma) was used at a concentration of 0.5 mM.
Expression of M1.
A CHO cell line, called CHO-M1, was developed that stably expressed M1 under the control of a cytomegalovirus (CMV) promoter. The M1 gene from influenza A/WSN virus was obtained from Peter Palese (pBSM1). The M1 fragment was excised with XbaI and was subcloned into the XbaI site of the expression vector pCB6 (13) to generate the plasmid pCB6-M1. The pCB6-M1 plasmid was transfected into CHO cells using calcium phosphate precipitation (35), and M1 was expressed from the CMV promoter (17). Cells stably expressing M1 were isolated by G418 selection, maintained in CHO medium containing 400 μg of G418 (Gibco) per ml, and passaged twice weekly. Cell clones expressing M1 were selected by immunofluorescence after overnight induction of expression with 10 mM sodium butyrate (9). Quantitation by Western blot analysis and densitometry showed that the amount of M1 expressed after a 12-h induction was approximately 2% of that produced during normal influenza virus infection (data not shown).
M1 was also expressed using the previously described recombinant Semliki Forest virus (SFV) that carries the M1 gene (2). By 8 h postinfection (p.i.) with SFV-M1, the amount of M1 was approximately 60% of that synthesized during an 8-h influenza virus infection.
SDS-PAGE and Western blotting.
For analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), cell lysates were homogenized by brief sonication, or by 10 strokes through a 25-gauge needle, and centrifuged at 8,000 × g in a microcentrifuge. Samples were analyzed by Western blotting, using the monoclonal antibody H16 L10 4R5 (American Type Culture Collection), or using the polyclonal anti-influenza virus serum (IBO; 1:1,000 dilution for 2 h at room temperature). As secondary antibodies, we used goat anti-mouse– or goat anti-rabbit–horseradish peroxidase or goat anti-mouse–alkaline phosphatase (1:5,000 dilution for 30 min at room temperature). Blots were developed using chemiluminescent detection (ECL; Amersham) or standard alkaline phosphate reagents. Alternatively, samples were immunoprecipitated with a polyclonal anti-NS2 antibody (kindly provided by Peter Palese) and analyzed by SDS-PAGE followed by autoradiography (34).
Indirect immunofluorescence microscopy.
Immunofluorescence microscopy was carried out essentially as described previously (34). Briefly, cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, quenched with 50 mM NH4Cl–PBS, and permeabilized for 5 min with 0.1% Triton X-100–PBS. After being blocked in 10% goat serum, cells were incubated with primary and secondary antibodies for 30 min each and mounted in Moviol. Influenza virus NP was detected using a monoclonal antibody (H16 L10 4R5; American Type Culture Collection). M1 was detected using the polyclonal serum 7648 (34). NS2 was detected using a polyclonal antibody kindly supplied by Peter Palese (Mt. Sinai School of Medicine). As secondary antibodies, we used Oregon Green-labeled goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG (Molecular Probes) and Texas Red-labeled goat anti-rabbit and goat anti-mouse IgG (Molecular Probes). Hoechst 33258 (Molecular Probes) was used at a concentration of 1 μg/ml. Cells were viewed using a Zeiss Axioplan 2 fluorescence microscope fitted with a 40× or 63× objective lens, and images were captured using TMAX 400 film (Kodak). Negatives were scanned and digitally processed using Adobe Photoshop software.
Isolation and characterization of vRNPs from cell nuclei.
Subconfluent monolayers of CHO cells were washed with PBS and scraped into PBS containing 0.1% Triton X-100, and cells were lysed by 20 passes through a 25-gauge needle. A chromatin extract (5, 28) was prepared by treatment of the cell lysate with 600 U of EcoRI (Gibco-BRL) per ml for 30 min at 37°C, followed by addition of NaCl to a final concentration of 1 M. EcoRI was used in chromatin extraction in place of DNase I, as described by Deppert and Schirmbeck (5). A postnuclear supernatant was then prepared by two centrifugation steps at 750 × g for 15 min, and vRNPs were isolated essentially as described previously (14). Briefly, supernatants were loaded onto a step glycerol gradient (1 ml of 70%, 0.75 ml of 50%, 0.375 ml of 40% and 1.8 ml of 33% [wt/vol] glycerol). The gradient was centrifuged at 45,000 rpm for 4 h at 4°C in a SW50.1 rotor. Fractions (300 μl) were collected by top aspiration, and vRNPs were concentrated from the fractions by centrifugation at 100,000 × g for 30 min at 4°C in a TLA100 rotor. vRNP pellets were resuspended and analyzed by SDS-PAGE and Western blotting.
RESULTS
Isolation of vRNPs from cell nuclei.
Assembled influenza virus vRNPs migrate to a defined sedimentation coefficient on a glycerol gradient and can be separated from monomeric NP molecules (14, 16). Previous analyses of influenza virus vRNPs have used detergent-treated virions or cytosolic lysates (2, 14) and have not attempted to analyze vRNPs in the nuclei of cells at late times of infection. In order to examine nuclear vRNPs, we first needed to achieve quantitative extraction of the vRNPs from nuclei.
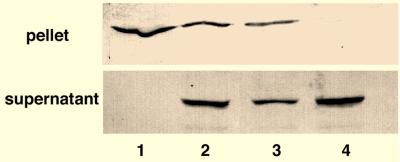
CHO cells were infected with influenza virus and first homogenized in PBS. Lysates were centrifuged at 5,000 × g for 10 min, and a pellet and a supernatant fraction were isolated. When the supernatant and pellet were analyzed by Western blotting, NP was found in the insoluble (nuclear) fraction (Fig. 1, lane 1). To attempt to quantitatively extract NP from the nuclear material, we lysed cells with the nonionic detergent Triton X-100 (0.1% [vol/vol]). In this case, NP also remained in the nuclear fraction, with the remainder of the NP being found in the soluble (cytoplasmic) fraction (Fig. 1, lane 2). Cells were next extracted with a zwitterionic detergent, 0.1% (wt/vol) CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), which is known to extract bound nuclear material (6). NP was again retained in the nuclear fraction after CHAPS treatment (data not shown). To attempt to release the tightly bound vRNPs from the nucleus, we performed a high-salt chromatin extraction procedure (5, 28). In this case, we achieved complete extraction of vRNPs, and all the NP detectable by Western blotting was found in the soluble fraction (Fig. 1, lane 4). Overall, these experiments show that influenza virus vRNPs are tightly associated with the nuclear matrix and must be treated with high salt to extract cellular chromatin in order to be released.
FIG. 1.
Extraction of vRNPs from infected cell nuclei. The figure shows SDS-PAGE analysis of influenza virus-infected cells treated with PBS (lane 1), PBS–0.1% Triton X-100 (lane 2), Triton X-100–EcoRI (lane 3), or Triton X-100–EcoRI–1 M NaCl (lane 4). Cells were infected with influenza virus for 8 h, lysed, and separated into an insoluble (pellet) and a soluble (supernatant) fraction. vRNPs were detected by Western blotting using anti-NP antibodies.
Effect of H7 on vRNP and viral protein synthesis.
H7 has previously been reported to prevent M1 synthesis, while under the same drug treatment, NP is synthesized in the cytoplasm and retained in the nucleus (21). We wished to confirm that authentic vRNPs are still formed in the presence of H7. To analyze the production of vRNP complexes in H7-treated cells, we infected cells with influenza virus either in the presence or in the absence of H7. Total vRNPs were extracted from the soluble and chromatin fractions of cells and analyzed by glycerol gradient centrifugation (2, 14). vRNPs were visible by Western blotting with anti-NP antibodies and sedimented in their expected positions in the glycerol gradient, peaking in fractions 12 and 13 (Fig. 2). The peak vRNP fractions were the same whether cells were treated with H7 or remained untreated. No soluble monomeric NP (which would run in load fractions 1 to 3) was detectable by Western blot analysis in the presence or absence of H7. As the gradient fractions containing total cellular vRNP are the same as those previously shown to contain authentic vRNP structures from virions (14, 16), we conclude that vRNP structures are still formed in H7-treated cells.
FIG. 2.
Gradient isolation of vRNPs from H7-treated and untreated cells. Cells were infected with influenza virus for 8 h, in the presence or absence of H7. Cells were lysed in PBS–Triton X-100–EcoRI–1 M NaCl and separated into an insoluble (pellet) and a soluble (supernatant) fraction. Soluble material was loaded onto a discontinuous glycerol gradient and centrifuged to isolate vRNPs. Gradient fractions were analyzed by SDS-PAGE, and vRNPs were detected by Western blotting using anti-NP antibodies. Fractions 1 to 3 and 9 to 16 are shown.
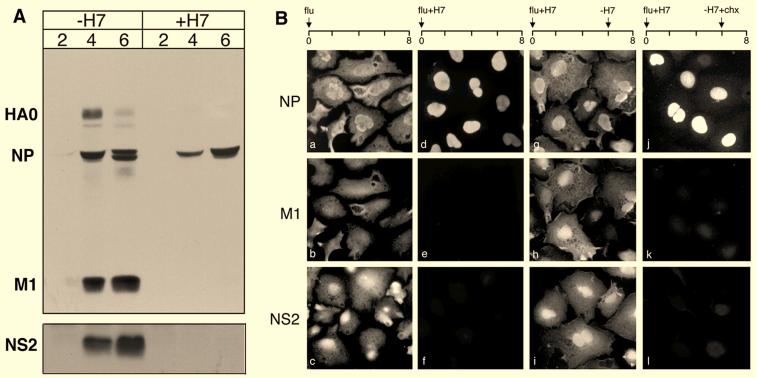
To analyze the effects of H7 on vRNP trafficking, cells were infected with virus, and at 2, 4, and 6 h p.i., cells were analyzed by SDS-PAGE. Cells were also infected in parallel and analyzed by immunofluorescence microscopy at 8 h p.i. As shown previously by Martin and Helenius (21), our Western blotting analysis (Fig. 3A) showed that NP was synthesized in H7-treated cells, although at lower levels than in untreated cells. Also, as previously reported (15, 21), synthesis of late proteins such as M1 and hemagglutinin was inhibited and was undetectable by Western blotting (Fig. 3A). NS2, another late protein, was efficiently immunoprecipitated from infected radiolabeled, control cell lysates, but no synthesis was detectable after H7 treatment (Fig. 3A). These findings were confirmed by immunofluorescence microscopy (Fig. 3B). Therefore, in the presence of H7 the late viral proteins (M1 and NS2) were not detected at 8 h p.i. (Fig. 3B, panels e and f). The early protein (NP) was strongly labeled and, as previously reported (21), confined to the nucleus (Fig. 3B, panel d).
FIG. 3.
Effect of H7 on viral protein synthesis. (A) SDS-PAGE analysis of influenza virus-infected cells in the presence (+H7) or absence (−H7) of H7 at 2, 4, and 6 h p.i.. HA0, NP, and M1 were detected by Western blotting using the anti-influenza virus antibody IBO. NS2 was detected by immunoprecipitation with anti-NS2 sera. (B) Immunofluorescence microscopy of influenza virus-infected cells using anti-NP, anti-M1, and anti-NS2 antibodies. The panels showing anti-NP and anti-M1 staining represent the same field, whereas panels showing anti-NS2 were a separate sample processed in parallel. Cells were analyzed at 8 h p.i. and were infected in the absence of H7 (a to c), in the presence of H7 (+H7) (d to f), with H7 washed out (−H7) at 6 h p.i. (g to i), and with H7 washed out at 6 h and cycloheximide (−H7+chx) added (j to l).
To determine whether the effects of H7 were reversible, influenza virus-infected cells were treated with H7 and transferred at 6 h p.i. to medium without H7. Cells were then analyzed by immunofluorescence microscopy at 8 h p.i. Under these conditions, both M1 and NS2 were detected (Fig. 3B, panels h and i), showing that late virus proteins were synthesized after H7 washout. In addition, immunofluorescence microscopy showed that NP now had a predominantly cytoplasmic location, indicating that the nuclear export of vRNPs had occurred when H7 was removed (Fig. 3B, panel g).
To confirm that the late viral proteins were indeed needed for vRNP nuclear export, we repeated the H7 washout experiment, but immediately after removal of the kinase inhibitor, cycloheximide was added to prevent further protein synthesis. Under these conditions, the synthesis of the late viral proteins M1 and NS2 was not observed (Fig. 3B, panels k and l), and the vRNPs remained confined to the nucleus (Fig. 3B, panel j). These data were consistent with the finding that synthesis of one or more of the late proteins is needed for vRNP export from the nucleus (21).
M1 is the only late viral protein needed to promote vRNP export.
We have previously shown that M1 promotes vRNP export (21). But it is currently unknown whether M1 is sufficient for this process. To address this issue, we used H7 as a biochemical tool to suppress late viral protein synthesis in influenza virus-infected cells and block vRNP export. We then expressed recombinant M1 in these cells and monitored nuclear export. CHO-M1 or CHO wild-type (wt) cells were first infected with influenza virus and treated with sodium butyrate to induce M1 synthesis in the CHO-M1 cells. H7 was added at the time of infection to suppress the expression of viral M1 and other late proteins. H7 had no effect on the level of M1 synthesized under the CMV promoter, showing that the effects of H7 on mRNA export are limited to mRNAs produced by influenza virus infection. H7 was removed from the infected CHO-M1 and CHO wt cells and replaced with cycloheximide to prevent further protein synthesis.
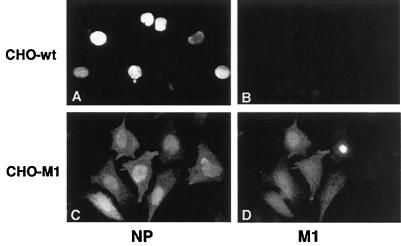
When the infected cells were examined by immunofluorescence microscopy, vRNPs remained confined to the nucleus in influenza virus-infected CHO wt cells (Fig. 4A and B). However, in CHO-M1 cells, the vRNPs were present in the cytoplasm (Fig. 4C and D). Since M1 was the only late virus protein present in these cells, we concluded that the expressed M1 could rescue influenza virus vRNP export and transport.
FIG. 4.
Stable expression of M1 promotes vRNP export. wt CHO (A and B) and CHO-M1 (C and D) cells were infected with influenza virus in the presence of H7. At 6 h p.i., H7 was washed out by transferring cells to medium lacking H7 but containing cycloheximide. Cells were analyzed at 8 h p.i. by immunofluorescence microscopy with a monoclonal anti-NP antibody (A and C) or a polyclonal anti-M1 antibody (B and D). The presynthesized M1 in CHO-M1 cells was sufficient to include vRNP export.
M1 and the regulation of vRNP export in heterokaryons.
To further characterize the role of M1, we used heterokaryons obtained by fusion of influenza virus-infected and noninfected cells. As we have previously shown (32), one advantage of this technique is that one can monitor vRNP shuttling into and out of nuclei. We used heterokaryons to determine whether the lack of vRNP accumulation in the cytosol of H7-treated cells was due to inhibition of vRNP export or to induction of rapid reimport after export. The distinction was important because we have shown that one of M1's functions is to prevent import of exported vRNPs (32).
HeLa cells were infected with influenza virus and treated with H7 to accumulate nuclear vRNPs and to suppress late viral protein synthesis. L929 cells, used as fusion partners, either were uninfected or were infected with SFV-M1. The two cell types were fused at 6 h post-influenza virus infection using polyethylene glycol (32). Immediately after fusion, H7 was withdrawn and cycloheximide was added. The cells were analyzed by immunofluorescence microscopy after 1 additional h (Fig. 5).
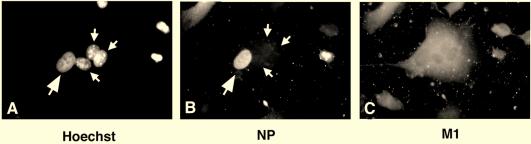
FIG. 5.
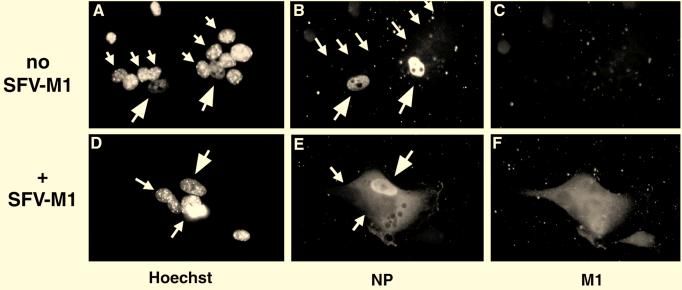
M1 provided by fusion of SFV-M1-infected cells promotes vRNP export. A cell fusion approach was used to study the effect of H7 and M1 expression on the nuclear export of vRNPs. HeLa cells (large arrows) were infected with influenza virus in the presence of H7, and L929 cells (small arrows) were fused at 6 h p.i. L929 cells were either uninfected (A to C; no SFV-M1) or infected with SFV-M1 (D to F; +SFV-M1). After fusion, cells were transferred to medium containing cycloheximide. Cells were analyzed by fluorescence microscopy using the DNA stain Hoechst 33258 (A and D), with anti-NP antibodies (B and E), and with anti-M1 antibodies (C and F). In the absence of M1, no shuttling of vRNPs occurred, but vRNP export occurred rapidly upon fusion of M1-expressing cells.
In heterokaryons formed between infected HeLa cells and uninfected L929 cells, M1 was not detectable (Fig. 5C). The vRNPs remained in the influenza virus-infected HeLa cell nuclei (Fig. 5B). Since no NP staining was seen in the L929-derived nucleus, no nucleocytoplasmic shuttling of vRNPs was taking place. However, in heterokaryons formed between influenza virus-infected HeLa cells and SFV-M1-infected L929 cells, M1 was detectable throughout the heterokaryon (Fig. 5F). The vRNPs were visible in the cytoplasm (Fig. 5E). These results indicated that M1 is needed for export of vRNPs from the nucleus and not needed merely to prevent reimport of exported, shuttling vRNPs.
As the kinase inhibitor H7 has been shown to affect localization of NP when expressed transiently in cells, we wished to address the possibility that cellular phosphorylation is needed for vRNP nuclear trafficking, in addition to a requirement for M1. We therefore repeated the experiments above, but did not wash out H7, prior to cycloheximide addition and M1 expression. We found that even though high levels of M1 were produced and localized throughout the heterokaryon (Fig. 6C), vRNPs were confined to the nucleus as long as H7 was present in the cells (Fig. 6B). These experiments show that M1 cannot induce vRNP nuclear export while cellular phosphorylation is inhibited by H7.
FIG. 6.
M1 cannot promote vRNP nuclear export in the presence of H7. HeLa cells (large arrows) were infected with influenza virus in the presence of H7, and L929 cells (small arrows) were fused at 6 h p.i. L929 cells were infected with SFV-M1. After fusion, cells were transferred to medium containing cycloheximide, in the presence of H7. Cells were analyzed by fluorescence microscopy using the DNA stain Hoechst 33258 (A), with anti-NP antibodies (B), and with anti-M1 antibodies (C). In the presence of H7, no export of vRNPs occurred, despite high levels of M1.
DISCUSSION
These experiments confirmed that the influenza virus M1 protein is fundamental to a late event in the virus life cycle—the transfer of vRNPs from the nucleus to the cytosol. Other late proteins were evidently not essential for this process in this system. In addition, the results showed that M1 was directly required for the export of vRNPs to the cytosol and not only to prevent reuptake of the vRNPs from the cytoplasm to the nucleus. We also found a role for cellular phosphorylation in addition to a requirement for M1.
We found that vRNPs were tightly associated with the nuclear matrix and were extracted from nuclei only by a high-salt extraction procedure designed to release cellular chromatin. These results are in close agreement with the findings of Bukrinskaya et al. (3), who found that input vRNPs were associated with chromatin 1 h after infection. Although there are some shape changes (particularly circularization) in the vRNPs which are associated with high-salt treatment, the particles remain essentially intact (10, 16) and appear to sediment normally on glycerol gradients.
A tight association with the nuclear matrix may suggest that nuclear export of vRNPs is in fact a two-stage process. The first event would be the release of the vRNPs from cellular chromatin, which may rely on specific viral factors or nuclear retention motifs on the vRNPs. The phenomenon of nuclear retention has received comparatively little attention, compared to the processes of nuclear import and export—although nuclear retention signals have been identified, which are distinct from nuclear import signals or nuclear export signals (NESs) (23). It is interesting to note that the original nuclear transport sequence on influenza virus NP was originally proposed to act as a retention or accumulation sequence, rather than an import sequence per se (4). However, the factors responsible for tight affinity of vRNPs for the nuclear matrix remain poorly understood—although M1-mediated displacement of vRNPs from the nuclear matrix has been proposed as a trigger for export (38).
The second stage of influenza virus nuclear export would be the translocation of the vRNPs across the nuclear pore. By analogy to the export of cellular proteins and RNPs, this is likely to rely on NESs and a cellular recognition system (24). Recently, Palese and colleagues suggested that NS2 (nuclear export protein, or NEP) is responsible for vRNP nuclear export via its interaction with M1 (27). NS2 was shown to contain a functional, leucine-based NES and to be able to replace the effector domain of human immunodeficiency virus (HIV) Rev, a protein that allows full-length transcripts of the HIV genome to be exported from the nucleus (31). NS2 was also shown to interact with a family of nucleoporin-related molecules termed RIP or Rab (1, 8). These are involved in nuclear export of molecules containing Rev-like, leucine-based NESs (26, 29). Finally, it was shown that, when anti-NS2 antibodies were injected into cells, vRNPs failed to exit from the nucleus (27). In our experiments, we found that vRNP nuclear export could occur in the absence of detectable NS2. However, it remains formally possible that very low levels of NS2 may be present in our cells, which are undetectable by the assays used, or that NS2 plays a nonessential role for which cellular factors can substitute. Thus, the role of NS2 remains unclear, especially in light of results from other investigators who have isolated mutant influenza viruses which contain little or no NS2 but are fully replication competent (36). It is possible that instead of acting directly in vRNP export, NS2 may play a role in nuclear export of the late viral mRNAs. NS2 could, for example, assist the export of the unspliced M1 message in a manner analogous to that of HIV type 1 Rev, which is needed for nuclear export of the unspliced HIV type 1 RNA (1, 7). It is currently unknown whether M1 interacts with any nuclear export receptors. Given the increasing complexity of nuclear transport pathways and the ability of viruses to make use of these cellular mechanisms, it is quite possible that vRNPs may have more than one route of nuclear exit.
In the experiments described above, the effects of H7 on viral M1 expression were circumvented by supplying M1 from an alternative expression system. By modifying the protocol somewhat, we made an observation that added an interesting facet to our understanding of vRNP export. We found that the expressed M1 could promote vRNP transport only if the H7 was washed out and replaced with cycloheximide. If H7 was left in the culture and cycloheximide was added, no export occurred even though M1 was present. One explanation for this effect is that to be transport competent, components of the vRNP must be phosphorylated. NP is an extensively phosphorylated protein that undergoes changes in phosphorylation pattern depending on the infectious cycle (15). It is known that, when expressed alone, its nuclear localization is affected by kinase inhibitors (25; G. Whittaker, unpublished results). Our results indicate that assembled vRNPs also show phosphorylation-regulated nuclear localization. As H7 is a broad-acting kinase inhibitor, it is possible that it has multiple targets in influenza virus-infected cells. The available data suggest two distinct roles for H7; first, a role in late mRNA nuclear export and regulation of late protein synthesis, and second, a more direct role in vRNP nuclear export.
It is becoming clear that posttranslational modification, such as phosphorylation, may regulate several events in nuclear import and export (see reference 12 for a review). Our studies suggest that a combination of M1 and phosphorylation may be used to regulate nuclear export of the vRNPs during infection by influenza virus. Further analysis of influenza virus nuclear export in defined assays will allow this question to be addressed and elucidate the specific requirements for this crucial step in the virus life cycle.
ACKNOWLEDGMENTS
We thank Ruth Collins for critical reading of the manuscript and Melanie Ebersold and Amy Glaser for experimental help. We also thank Peter Palese for providing antibodies and plasmids.
M.B. was supported by fellowships from the Life and Health Insurance Medical Research Training Fund and the National Institutes of Health (MSTP training grant GM 07205). The support of the NIH (grant AI 18599 to A.H.) and the USDA Animal Health and Disease Research Program (to G.R.W.) is also acknowledged.
REFERENCES
- 1.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 2.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinskaya A G, Vorkunova G K, Vorkunova N K. Cytoplasmic and nuclear input virus RNPs in influenza virus-infected cells. J Gen Virol. 1979;45:557–567. doi: 10.1099/0022-1317-45-3-557. [DOI] [PubMed] [Google Scholar]
- 4.Davey J, Dimmock N J, Colman A. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell. 1985;40:667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- 5.Deppert W, Schirmbeck R. The nuclear matrix and virus function. Int Rev Cytol. 1995;162A:485–537. doi: 10.1016/s0074-7696(08)61237-1. [DOI] [PubMed] [Google Scholar]
- 6.Efthymiadis A, Briggs L J, Jans D A. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J Biol Chem. 1998;273:1623–1628. doi: 10.1074/jbc.273.3.1623. [DOI] [PubMed] [Google Scholar]
- 7.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 8.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 9.Gorman C M, Howard B H. Expression of recombinant plasmids in mammalian cells is enhanced by sodium butyrate. Nucleic Acids Res. 1983;11:7631–7648. doi: 10.1093/nar/11.21.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heggeness M H, Smith P R, Ulmanen I, Krug R M, Choppin P W. Studies on the helical nucleocapsid of influenza virus. Virology. 1982;118:466–470. doi: 10.1016/0042-6822(82)90367-1. [DOI] [PubMed] [Google Scholar]
- 11.Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolonesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 12.Hood J K, Silver P A. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 13.Hunziker W, Harter C, Matter K, Mellman I. Basolateral sorting in MDCK cells requires a distinct cytoplasmic domain determinant. Cell. 1991;66:907–920. doi: 10.1016/0092-8674(91)90437-4. [DOI] [PubMed] [Google Scholar]
- 14.Kemler I, Whittaker G, Helenius A. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology. 1994;202:1028–1033. doi: 10.1006/viro.1994.1432. [DOI] [PubMed] [Google Scholar]
- 15.Kistner O, Muller K, Scholtissek C. Differential phosphorylation of the nucleoprotein of influenza A viruses. J Gen Virol. 1989;70:2421–2431. doi: 10.1099/0022-1317-70-9-2421. [DOI] [PubMed] [Google Scholar]
- 16.Klumpp K, Ruigrok R W H, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kretzschmar E, Bui M, Rose J K. Membrane association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology. 1996;220:37–45. doi: 10.1006/viro.1996.0283. [DOI] [PubMed] [Google Scholar]
- 18.Kurokawa M, Ochiai H, Nakajima K, Niwayama S. Inhibitory effect of protein kinase C inhibitor on the replication of influenza type A virus. J Gen Virol. 1990;71:2149–2155. doi: 10.1099/0022-1317-71-9-2149. [DOI] [PubMed] [Google Scholar]
- 19.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–87. [Google Scholar]
- 20.López-Turiso J A, Martínez C, Tanaka T, Ortín J. The synthesis of influenza virus negative-strand RNA takes place in insoluble complexes present in the nuclear matrix fraction. Virus Res. 1990;16:325–336. doi: 10.1016/0168-1702(90)90056-h. [DOI] [PubMed] [Google Scholar]
- 21.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 22.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakielny S, Fischer U, Michael W M, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 25.Neumann G, Castrucci M R, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neville M, Stutz F, Lee L, Davis L, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 27.O'Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staufenbiel M, Deppert W. Preparation of nuclear matrices from cultured cells: subfractionation of nuclei in situ. J Cell Biol. 1984;98:1886–1894. doi: 10.1083/jcb.98.5.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 30.Vogel U, Kunerl M, Scholtissek C. Influenza A virus late mRNAs are specifically retained in the nucleus in the presence of a methyltransferase or a protein kinase inhibitor. Virology. 1994;198:227–233. doi: 10.1006/viro.1994.1025. [DOI] [PubMed] [Google Scholar]
- 31.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 32.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittaker G, Bui M, Helenius A. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 1996;6:67–71. doi: 10.1016/0962-8924(96)81017-8. [DOI] [PubMed] [Google Scholar]
- 34.Whittaker G, Kemler I, Helenius A. Hyperphosphorylation of mutant influenza virus matrix (M1) protein causes its retention in the nucleus. J Virol. 1995;69:439–445. doi: 10.1128/jvi.69.1.439-445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. DNA-mediated transfer of adenine phosphoribosyl-transferase locus into mammalian cells. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolstenholme A J, Barrett T, Nichol S T, Mahy B W J. Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins. J Virol. 1980;35:1–7. doi: 10.1128/jvi.35.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda J, Nakada S, Kato A, Toyoda T, Ishihama A. Molecular assembly of influenza: association of the NS2 protein with virion matrix. Virology. 1993;196:249–255. doi: 10.1006/viro.1993.1473. [DOI] [PubMed] [Google Scholar]
- 38.Zhirnov O P, Klenk H-D. Histones as a target for influenza virus matrix protein M1. Virology. 1997;235:302–310. doi: 10.1006/viro.1997.8700. [DOI] [PubMed] [Google Scholar]